ABSTRACT
The syphilis spirochete Treponema pallidum is an important human pathogen but a highly enigmatic bacterium that cannot be cultivated in vitro. T. pallidum lacks many biosynthetic pathways and therefore has evolved the capability to exploit host-derived metabolites via its periplasmic lipoprotein repertoire. We recently reported a flavin-trafficking protein in T. pallidum (Ftp_Tp; TP0796) as the first bacterial metal-dependent flavin adenine dinucleotide (FAD) pyrophosphatase that hydrolyzes FAD into AMP and flavin mononucleotide (FMN) in the spirochete’s periplasm. However, orthologs of Ftp_Tp from other bacteria appear to lack this hydrolytic activity; rather, they bind and flavinylate subunits of a cytoplasmic membrane redox system (Nqr/Rnf). To further explore this dichotomy, biochemical analyses, protein crystallography, and structure-based mutagenesis were used to show that a single amino acid change (N55Y) in Ftp_Tp converts it from an Mg2+-dependent FAD pyrophosphatase to an FAD-binding protein. We also demonstrated that Ftp_Tp has a second enzymatic activity (Mg2+-FMN transferase); it flavinylates protein(s) covalently with FMN on a threonine side chain of an appropriate sequence motif using FAD as the substrate. Moreover, mutation of a metal-binding residue (D284A) eliminates Ftp_Tp’s dual activities, thereby underscoring the role of Mg2+ in the enzyme-catalyzed reactions. The posttranslational flavinylation activity that can target a periplasmic lipoprotein (TP0171) has not previously been described. The observed activities reveal the catalytic flexibility of a treponemal protein to perform multiple functions. Together, these findings imply mechanisms by which a dynamic pool of flavin cofactor is maintained and how flavoproteins are generated by Ftp_Tp locally in the T. pallidum periplasm.
IMPORTANCE
Treponema pallidum, the syphilis spirochete, exploits its periplasmic lipoproteins for a number of essential physiologic processes. One of these, flavin-trafficking protein (Ftp), not only exploits its catalytic center to mediate posttranslational flavinylation of proteins (to create flavoproteins) but also likely maintains the periplasmic flavin pool via its unique ability to hydrolyze FAD. This functional diversity within a single lipoprotein is quite remarkable and reflects the enzymatic versatility of the treponemal lipoproteins, as well as molecular parsimony in an organism with a limited genome. Ftp-mediated protein flavinylation in the periplasm also likely is a key aspect of a predicted flavin-dependent Rnf-based redox homeostasis system at the cytoplasmic membrane of T. pallidum. In addition to its importance in T. pallidum physiology, Ftp homologs exist in other bacteria, thereby expanding our understanding of the bacterial periplasm as a metabolically active subcellular compartment for flavoprotein biogenesis as well as flavin homeostasis.
INTRODUCTION
Treponema pallidum, the causative agent of syphilis, cannot be cultivated continuously in vitro (1). Although T. pallidum is responsible for one of the most prevalent sexually transmitted infections worldwide (2, 3), it is among the most poorly understood of all human bacterial pathogens. The relatively small size of the T. pallidum genome (ca. 1 Mb) accounts for the fact that the spirochete lacks many of the genes encoding biosynthetic pathways (4). The organism thus is dependent on an extracellular supply of glucose, purines, amino acids, fatty acids, and many other cofactors and vitamins. The many enigmatic features of the T. pallidum outer envelope (5–7) raise key questions regarding how T. pallidum obtains these essential nutrients from its obligate human host (8–11).
Flavin is an essential cofactor required for metabolic processes within all living organisms (12, 13). Riboflavin is a direct precursor of the cofactors flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD); it is synthesized by plants and many microorganisms but not by mammals (13–15). Bacterial spirochetes, such as T. pallidum, lack the riboflavin biosynthesis pathway. We recently described in T. pallidum an ATP-binding cassette (ABC) type of riboflavin uptake system (RfuABCD) that ostensibly is essential for the organism’s survival within its human host (11). We also recently identified a previously unknown Mg2+-dependent FAD pyrophosphatase (TP0796, or Ftp_Tp) which processes FAD into AMP and FMN in the periplasm (16, 17). These findings, along with our recent description of putative treponemal flavoproteins and a presumptive flavin-based redox system in T. pallidum (16), underscore the potential importance of flavins in the physiology and metabolism of T. pallidum. We consider this aspect of T. pallidum’s parasitic strategy as its “flavin-centric” lifestyle.
FAD pyrophosphatase (EC 3.6.1.18) catalyzes the hydrolysis of FAD, forming AMP and FMN. To date, the Ftp (TP0796) of T. pallidum is the first bacterial FAD pyrophosphatase shown to have a strict requirement for Mg2+ for its catalytic activity (16). Other Ftp homologs (formerly known as ApbE proteins) are present in the genomes of numerous bacteria (16–18) and in lower eukaryotes, such as Trypanosoma spp. (agents of sleeping sickness and Chagas disease) and Leishmania spp. (agent of leishmaniasis), but the eukaryotic homologs appear to be fused with a multidomain fumarate reductase (19, 20). Previous studies have shown that some of the Ftp family proteins bind FAD (16, 18) and that the Ftp protein from Vibrio harveyi transfers the FMN portion of FAD to a subunit of the integral inner membrane Nqr redox pump (17). The crystal structure of Ftp from T. pallidum displays a highly conserved Ftp fold and an active site/FAD-binding site of all known Ftp-like proteins (16).
In this study, we focused on elucidating the role of the active center of Ftp_Tp in Mg2+-dependent FAD hydrolysis and in potential FMN transferase (EC 2.7.1.180) activities (e.g., flavinylation of flavoproteins). Using both biochemical and structural properties of Ftp_Tp variants, we identified the critical residues required for both enzymatic activities by Ftp_Tp. Ftp_Tp is unique in that it appears not only to use its bimetal catalytic center for maintaining a periplasmic flavin pool via its FAD hydrolytic activity but also to modulate posttranslational flavinylation (covalent attachment of an FMN moiety on a threonine residue of a protein). Given the wide distribution of Ftp orthologs in bacteria, these results have broad implications for bacterial physiology, and they underscore the potential importance of the bacterial periplasm for flavin homeostasis and flavin utilization.
RESULTS AND DISCUSSION
Reconstitution of the T. pallidum flavinylation reaction by Ftp_Tp in Escherichia coli.
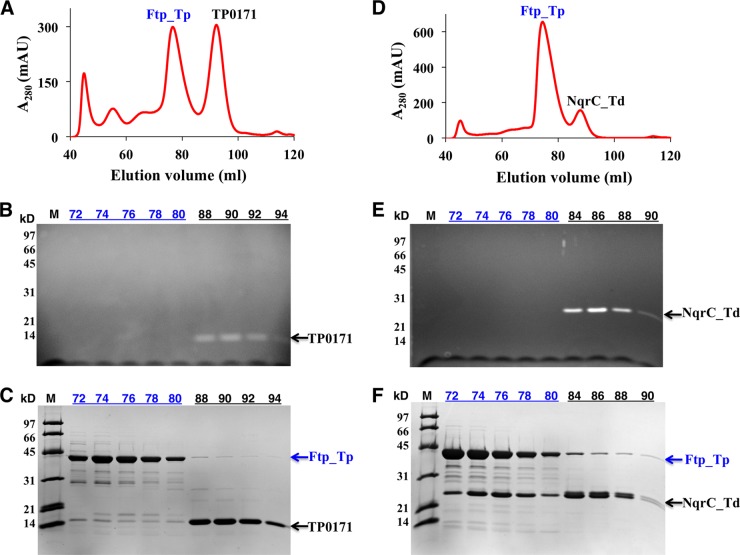
Recently, it has been reported that the Ftp proteins of Vibrio harveyi (Ftp_Vh) and Klebsiella pneumoniae (Ftp_Kp) can flavinylate NqrC subunits of their respective Nqr redox systems via the covalent attachment of FMN to the threonine side chain of an appropriate sequence motif (17). Bioinformatics revealed only two putative FMN-binding proteins (TP0171, a periplasmic lipoprotein, and TP0572, a putative integral membrane protein) in the treponemal genome that contain potential flavinylation sequence motifs (Fig. 1). Treponema denticola, an oral spirochete, appears to encode a putative Nqr-type redox pump and a flavinylation sequence motif in its NqrC ortholog (NqrC_Td) (Fig. 1). Therefore, TP0171 and NqrC_Td were employed to study Ftp_Tp’s putative flavinylation activity. Whereas recombinant NqrC_Td and TP0171 purified as nonyellow apoproteins from Escherichia coli (data not shown), coexpression of either the TP0171-Ftp_Tp or NqrC_Td-Ftp_Tp pair in E. coli resulted in yellow proteins after Ni2+ affinity purification. Further purification of the protein mixtures by gel filtration chromatography resulted in two peaks, one a colorless sample and the other yellow. SDS-PAGE analyses confirmed that the flavin was covalently attached to both TP0171 and NqrC_Td (Fig. 2), confirming the role of Ftp_Tp in protein flavinylation. As expected from the high sequence identity of their Ftp proteins, Ftp_Tp of T. pallidum can flavinylate NqrC_Td of T. denticola, though with somewhat reduced efficiency, as observed in the mass spectra showing a proportion of unflavinylated NqrC_Td (Fig. 3B). However, the flavinylation reaction likely proceeds via weak interactions between Ftp_Tp and the flavinylated proteins (NqrC_Td or TP0171), because stable elution complexes (i.e., single peaks) were not observed upon gel filtration chromatography (Fig. 2A and D).
FIG 1 .

Partial primary sequence alignment of putative flavinylated bacterial proteins. Shown in red is the modified threonine residue, and in green are the conserved residues of the flavinylation motif. Aligned are the Ftp-mediated posttranslational flavinylation sites of various bacterial proteins from Nqr and Rnf complexes (17) (current study), plus two T. pallidum proteins that were not previously identified as potential flavinylation substrates.
FIG 2 .
Purification and SDS-PAGE characterization of coexpressed (Ftp_Tp-TP0171 and Ftp_Tp-NqrC_Td) recombinant proteins isolated from E. coli. The gel filtration chromatographic profiles (A and D), UV illumination of peak fractions separated by SDS-PAGE (B and E), and Coomassie blue stain of gel after UV illumination (C and F) of coexpressed Ftp_Tp-TP0171 (A, B, C) and Ftp_Tp-NqrC_Td (D, E, F) pairs are shown. The number in each lane represents the elution volume (in milliliters) examined.
FIG 3 .
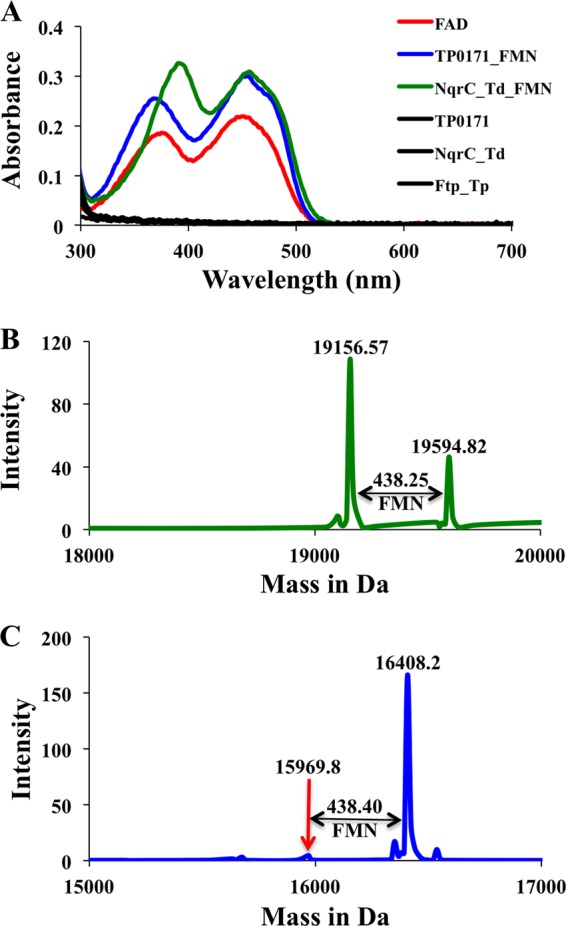
Characterization and identification of the covalently bound flavin. (A) The UV-visible absorbance spectra of Ftp_Tp-mediated flavinylated protein TP0171 and NqrC_Td as isolated from E. coli. The UV-visible absorbance spectra of free FAD and singly expressed Ftp_Tp, TP0171, and NqrC_Td are shown for comparison. Q-TOF mass spectra of flavinylated proteins obtained from NqrC_Td (B) and TP0171 (C). The mass differences correspond to covalently linked FMN.
The UV-visible absorbance spectra of flavinylated NqrC_Td and TP0171 showed pronounced dual-absorbance maxima (~370 and 450 nm) and shoulders around 470 nm, indicative of bound flavins (Fig. 3). The yellow color associated with the proteins flavinylated by Ftp_Tp cannot distinguish between bound riboflavin, FMN, and FAD. Therefore, to identify the flavin covalently bound to NqrC_Td and TP0171, the yellow proteins were subjected to quantitative time of flight mass spectrometry (Q-TOF MS) analyses. As shown in Fig. 3, mass differences between the flavinylated and nonflavinylated proteins confirmed that the covalently bound flavin was FMN. Taken together, our results reveal the function of Ftp_Tp as a periplasmic flavinylation/lipoprotein modification enzyme.
Evidence for Mg2+-dependent phosphoester-threonyl-FMN posttranslational modification of proteins by Ftp_Tp.
Recombinant NqrC_Td and TP0171 purified as nonyellow apoprotein samples; therefore, they were used to study the in vitro flavin transferase activity of Ftp_Tp. As shown in Fig. 4, recombinant wild-type Ftp_Tp was able to flavinylate TP0171 in an Mg2+-dependent manner in the presence of FAD (lanes 4 and 5), suggesting that this type of posttranslational flavinylation reaction is indeed a protein-dependent FMN transferase activity rather than an autocatalytic one. EDTA strongly inhibited the flavin transferase activity (lane 7); thus, the activity of Ftp_Tp is Mg2+ dependent. As expected, AMP, which is the reaction product of Ftp_Tp’s FAD pyrophosphatase activity (16), also inhibits the FMN transferase activity (lane 6). In addition, Ftp_Tp can flavinylate NqrC_Td (lane 11), a substrate from the closely related organism T. denticola, although not as efficiently as it can modify T. pallidum substrates. Remarkably, Ftp_Td, unlike Ftp_Tp, failed to flavinylate the TP0171 lipoprotein (lane 12), highlighting the species specificity of Ftp activity for lipoprotein modification.
FIG 4 .
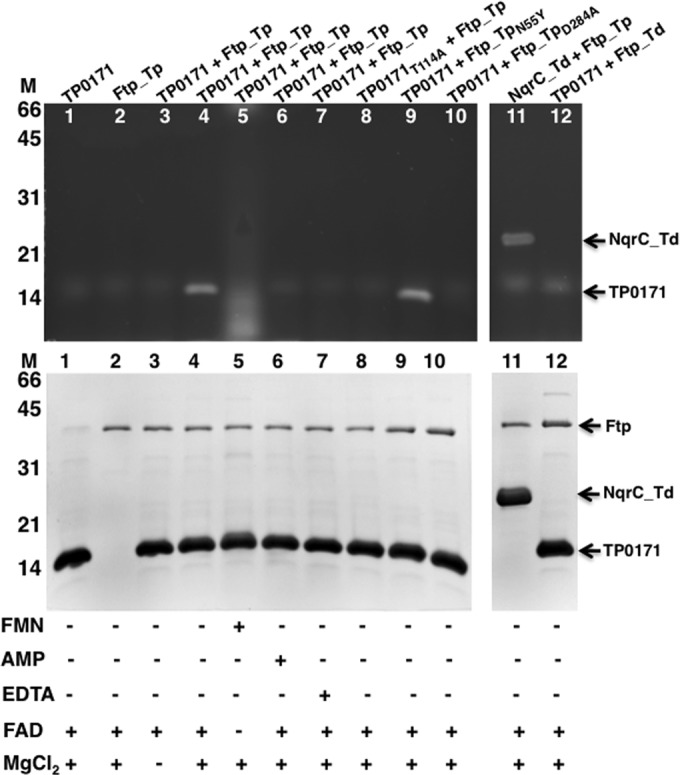
SDS-PAGE characterization of flavinylation reactions followed by UV illumination and Coomassie blue staining. UV illumination of unstained gel is shown at the top, and the Coomassie-stained gel is shown below. Protein molecular markers are on the left side. Ftp_Tp (wild type and mutants) reacted with TP0171 (wild type and mutants) and NqrC_Td under various indicated conditions. The nonspecific diffuse bands (observed in all lanes, including the control reactions) are in vitro artifacts of the flavinylation reactions.
In the flavinylation reaction, the side-chain hydroxyl of a conserved threonine in the appropriate protein substrate could serve as the catalytic nucleophile that attacks the diphosphate to cleave the FAD and transfer the FMN as a phosphoester-threonyl-FMN. To investigate this hypothesis, a variant of TP0171 (TP0171T114A) was generated by site-directed mutagenesis. Similar to the wild type, the variant expressed and purified as a soluble protein. However, it failed to become flavinylated when incubated with Ftp_Tp in the presence of FAD and MgCl2 (Fig. 4, lane 8).
These results show that the threonine residue of the flavinylation motif is critical for FMN attachment and further support earlier studies of protein flavinylation in other bacterial species (17, 21). Ftp_Tp’s posttranslational flavinylation reaction involves a protein-protein interaction, and we have previously demonstrated in in vivo cross-linked T. pallidum that there are many such interactions involving Ftp_Tp (16). These findings likely are of broad importance to bacterial periplasmic flavin homeostasis, because Ftp orthologs are widespread in bacteria. Although the precise physiological role of flavinylated TP0171 is not known, it may serve as a periplasmic redox protein because of its NqrC-type FMN-binding motif.
Structural and biochemical analyses of Ftp_Tp that affect FAD pyrophosphatase and FMN transferase activities.
We have previously shown that Ftp_Tp hydrolyzes FAD into FMN and AMP in an Mg2+-dependent manner (16). Further, crystallographic investigations of Ftp_Tp revealed the disposition of the active site in atomic detail (16). Although Ftp’s FMN transferase activity is metal dependent, there is a paucity of information regarding the mechanism of protein flavinylation by Ftp-like proteins. To identify the residues of Ftp_Tp critical for its FAD pyrophosphatase and FMN transferase catalytic activities, we pursued structure-guided mutagenesis. Based on the substrate, product, and inhibitor-bound structures (16), 9 amino acid mutations (N55Y, K165A and E, S240A, E244A, R245A, H256A, D284A, and T288A) of Ftp_Tp were generated. Note that the residue numbering reflects the assignment of a lipid-modified cysteine residue as 1 in the recombinant proteins (16). We measured the extent of FMN formation in single-turnover reactions that were catalyzed by both wild-type Ftp_Tp and its variants. As shown in Fig. 5, the wild-type enzyme generated ~0.7 FMN per Ftp_Tp in the single-turnover reactions; nonstoichiometry of this activity is likely due to the nonrelease of the AMP product in a percentage of the protein, as heterologously expressed in E. coli (16). From the observed catalytic activities/turnover rates and the information derived from the Ftp_Tp structures (16), residues can be classified as four types: (i) metal binding, mutation of the residues (D284A and T288A) in the first coordination sphere of the 2 Mg2+ sites abolished FAD pyrophosphatase activity; (ii) substrate binding, mutation of the isoalloxazine ring-binding residue N55Y also abolished FAD pyrophosphatase activity; (iii) critical catalytic, mutation of residues that may activate a water molecule for nucleophilic attack (S240A, E244A) or neutralize the charge on the leaving group during attack (R245A, H256A) displayed reduced activity; and (iv) auxiliary catalytic, mutation of K165 enhanced the FAD pyrophosphatase activity.
FIG 5 .
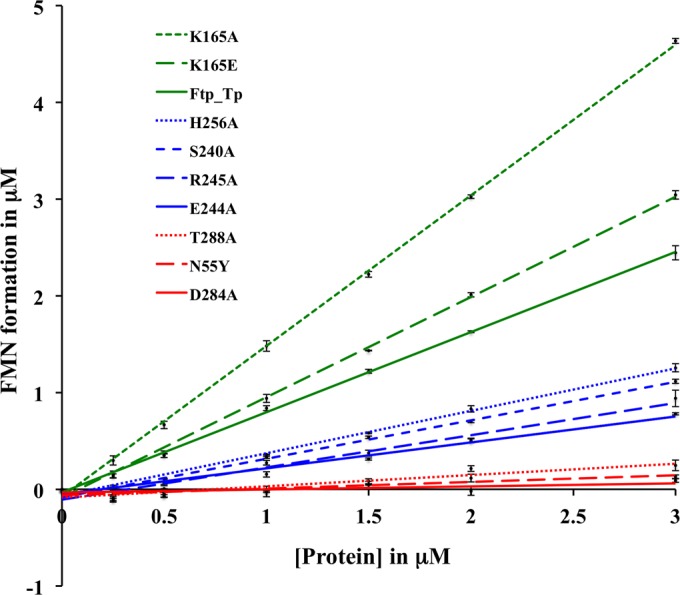
Stoichiometry of FMN formation by wild-type T. pallidum Ftp (Ftp_Tp) and its variants (indicated in color) as a function of protein concentration in the single-turnover reactions. Points are plotted as the means from two samples at each protein concentration and represent FMN concentrations derived from the change in fluorescence experiments using an FMN standard curve. Standard deviations of the data points, <0.09 µM. Note that the FAD turnover by wild-type Ftp_Tp was published previously (16).
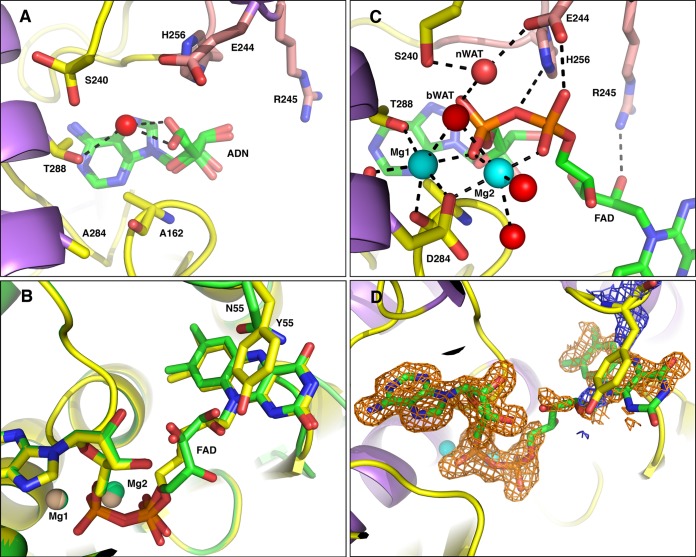
The elimination of activity in metal-binding residue mutants is not surprising. It is likely that the primary functions of the metal ion in site 1 are to maintain protein structural integrity and to neutralize some of the negative charge and properly position the diphosphate moiety of FAD for catalysis. In addition, no FMN transferase activity was observed with the Ftp_TpD284A mutant (Fig. 4, lane 10), suggesting a role for Mg2+ in metal site 1 in the covalent modification reaction. Given the lack of catalytic turnover, we attempted to obtain a product-bound crystal structure of the Ftp_TpD284A mutant by incubating the protein with Mg2+ and AMP; however, upon inspection of the electron density map, we determined that the structure obtained was in fact a complex with adenosine (ADN) (presumably scavenged by the protein as an impurity in our AMP stocks), and no metal ions were located in the active site (Fig. 6A). The adenosine contaminant, rather than AMP, bound preferentially to the enzyme due to the disruption of metal site 1 and the subsequent lack of charge neutralization. Thus, we believe that the differences in the coordination of the FAD diphosphate moiety bound to Ftp_Tp and to Ftp_Se can be attributed to Ftp_Se’s lack of coordinated metal ions to neutralize the negative charge (16, 18).
FIG 6 .
Active-site geometries of Ftp_Tp mutant complexes. The carbon atoms of protein side chains are yellow, the carbon atoms from residues of the β-hairpin insert are salmon, the nucleotide atoms are green, Mg2+ ions are cyan spheres, water is red spheres, nitrogens are blue, and oxygens are red. Black dotted lines represent metal first-coordination-sphere contacts and important hydrogen bonding interactions. The candidate nucleophilic water is labeled nWAT, and the bridging water is labeled bWAT. For clarity, some protein residues have been selectively removed from the images. (A) Ftp_TpD284A complexed with adenosine (ADN), which is modeled in two conformations. (B) Ftp_TpN55Y complexed with Mg2+ and FAD shown with yellow carbon atoms, aligned to the Ftp_Tp wild-type Mg2+-FAD complex shown with green carbon atoms and salmon Mg2+ ions. (C) Ftp_TpN55Y complex with Mg2+ and FAD. (D) Omit electron density around FAD ligand. Shown in orange mesh is the |mFo − DFc| electron density calculated after omitting the ligand from the model, contoured at the 2σ level and superimposed on the FAD of the Ftp_TpN55Y complex structure. This map was calculated by omitting the FAD from the model and conducting three rounds of maximum-likelihood positional and B-factor refinement. Shown in blue mesh is the |2mFo − DFc| electron density from the same calculation, contoured at the 0.8 σ level and superimposed on the Y55 residue of the Ftp_TpN55Y complex structure.
Mutation of the isoalloxazine ring-binding residue (N55Y) resulted in complete loss of FAD hydrolyase activity, yet Ftp_TpN55Y was able to flavinylate TP0171 in an Mg2+-dependent manner (Fig. 4, lane 9). A crystallographic structure of Ftp_Se revealed that the analogous residue (Y78) forms a pi-stacking interaction with the isoalloxazine ring of bound FAD, and the authors hypothesize that this interaction is required for FAD binding (18). To test this hypothesis, we obtained a crystal structure of Ftp_TpN55Y with bound Mg2+ and FAD and found that it agreed most closely with the wild-type Ftp_Tp Mg2+-FAD complex (Fig. 6B), with some minor differences in the ribityl conformation. Coordination of metal ions, waters, and active-site residues near the site of pyrophosphate hydrolysis in the Ftp_TpN55Y structure is almost indistinguishable from that observed for wild-type Ftp_Tp (Fig. 6C; see also Table S1 in the supplemental material). A large degree of mobility is suggested for the ribityl portion of the FAD and the Y55 side chain, as the electron densities for these regions are extremely weak to nonexistent (Fig. 6D), and thus it is not possible to state with certainty whether the pi stacking observed in the Ftp_Se structure occurs in our complex. The inhibition of spontaneous FAD hydrolysis in Ftp proteins that contain a bulky aromatic residue near the isoalloxazine ring is likely due to steric hindrance and not specifically to the pi stacking observed only in the metal-free Ftp_Se structure.
A single amino acid mutation (Ftp_TpN55Y) leads to a switch from an Mg2+-dependent FAD pyrophosphatase to an FAD-binding Ftp that can still flavinylate its protein substrate (Fig. 4, lane 9, and 6B). Thus, we have identified a single amino acid in Ftp_Tp that modulates the differences in Ftp’s activities; one subset of residues at this position favors an Ftp that simply binds FAD (for subsequent FMN transfer to a suitable protein substrate), whereas another subset confers on Ftp its ability to have both hydrolytic and FMN transfer activities.
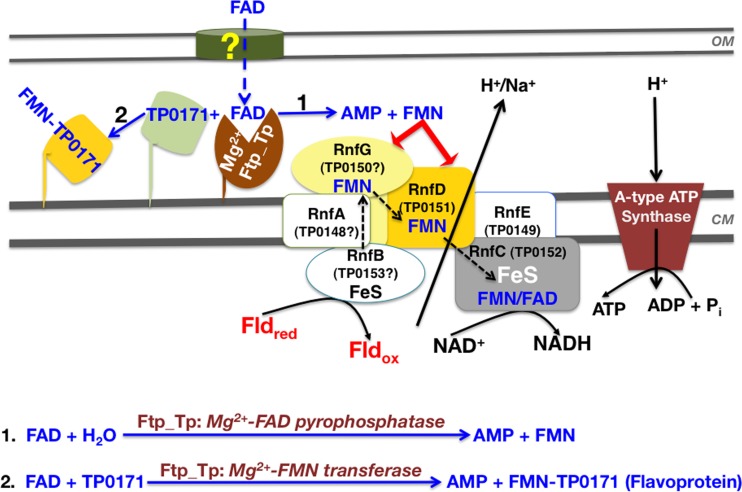
Ftp_Tp was initially characterized as an Mg2+-dependent FAD pyrophosphatase (16, 18). Our current study now demonstrates that Ftp_Tp also plays an important role in protein flavinylation (Fig. 2, 3, and 4). However, it is important to point out that these two diverse reactions appear not to be coupled. That is, concerning Ftp_Tp’s pyrophosphatase activity, FAD is cleaved to form FMN and AMP (Fig. 7, pathway 1); this reaction likely contributes to maintaining T. pallidum’s periplasmic flavin pool. For protein flavinylation, Ftp_Tp also utilizes FAD as a substrate, but to transfer an FMN moiety to an acceptor protein (e.g., TP0171; Fig. 7, pathway 2). However, the two diverse functions are both dependent on the same bimetal center of Ftp_Tp, as evidenced by our structural and biochemical analyses. Because the Ftp-like proteins are widespread in bacteria and found in all Rnf- and/or Nqr redox-containing organisms, the FMN transferase activity likely is responsible for generating redox-active flavoproteins. However, none of T. pallidum’s predicted (TP0151 [also known as RnfD_Tp] and TP0152 [also known as RnfC_Tp]) or as-yet-unidentified Rnf components (see Fig. 7) contain a readily identifiable NqrC/RnfG-type FMN-binding motif (Fig. 1). This suggests that T. pallidum may bind FMN covalently via a noncanonical mechanism or noncovalently after its generation by Ftp_Tp’s FAD pyrophosphatase activity (Fig. 7). An intriguing question is why has a single amino acid change led to an additional FAD hydrolytic activity by treponemal Ftp? Organisms with reduced genomes, such as T. pallidum, often evolve proteins that participate in multiple functions to compensate for the loss of other important proteins/pathways (22).
FIG 7 .
Proposed model of periplasmic flavoprotein biogenesis and flavin homeostasis in the T. pallidum periplasm. The model is predicated on Ftp_Tp’s dual function in posttranslational protein flavinylation described herein and flavin homeostasis modulated by Ftp_Tp’s Mg2+-FAD pyrophosphatase activity (16). The yellow question mark indicates the unknown import mechanism of host-derived FAD across the outer membrane. FAD in the T. pallidum periplasm is either hydrolyzed into AMP and FMN to maintain the flavin pool (scheme 1) and/or utilized by Ftp_Tp’s Mg2+-FMN transferase activity to generate flavoproteins (e.g., TP0171) (scheme 2). The product inhibition of Ftp_Tp’s FAD pyrophosphatase activity (16) likely maintains the FAD pool for FMN transferase activity. The scheme also proposes a hypothetical energy conservation pathway that likely couples a noncanonical flavin-based Rnf redox pump to T. pallidum’s A-type ATP synthase (see text for explanation). Note that the function of the redox protein(s) depends on periplasmic flavin and the dual activities of Ftp_Tp. Assignments of the unidentified Rnf subunits (designated by “?”) ostensibly encoding the T. pallidum Rnf complex (the tp0153-tp0148 operon) are based tentatively on an rnfBCDGEA type of system (24) and the predicted transmembrane helices of TP0148, TP0149, and TP0150. Also, TP0149 has ~30% identity to Pseudomonas brassicacearum RnfE (according to the KEGG gene function identification tool). OM, outer membrane; CM, cytoplasmic membrane; ox, oxidized; red, reduced.
Functional significance of Ftp and posttranslational protein flavinylation.
Although most bacteria can synthesize riboflavin, pathogenic spirochetes like T. pallidum almost assuredly rely on an essential ABC-type RfuABCD system for the exploitation of host-derived riboflavin (11). Riboflavin serves as the precursor for the flavin prosthetic group (FMN and FAD), and their redox-active isoalloxazine ring system is widely used by living organisms for a diversity of fundamental oxidation reduction processes (13). In addition to their role as redox catalysts, flavins are also found in some nonredox enzymes, such as hydrolases, transferases, isomerases, and lyases (13, 16). Although almost 90% of flavin enzymes contain noncovalently bound flavins (13), there is a small group of enzymes where the flavin ring is covalently linked to an amino acid residue, such as Cys, Tyr, Thr, or His (23). Most covalent flavin attachment is thought to be posttranslational and autocatalytic; only recently, an Ftp-mediated attachment of FMN to a threonine residue found in components of redox-driven ion pumps (RnfG and NqrC) was discovered (13, 17).
The mechanism of flavin homeostasis in the bacterial periplasm remained largely unclear until our recent discovery of an FAD-hydrolyzing enzyme (16). In the bacterial cytosol, FAD and FMN are synthesized from riboflavin (vitamin B2) via the bifunctional FAD synthase. Recently, two flavin- and quinone-based redox-driven Na+ pumps (Nqr and Rnf) have been discovered that are believed to be of central importance to the bioenergetics of many pathogenic bacteria, and often they are the only ion-motive electron transport chain in these organisms (24). Flavinylation of the electron transfer subunits of these systems requires periplasmic flavin trafficking. Whereas the FMN transferase activities of FAD binding by Ftp proteins have been investigated, the FAD pyrophosphatase activities of the FAD-hydrolyzing Ftp proteins have not been analyzed. We herein now have shown that Ftp_Tp not only plays a pivotal role in flavinylation of a periplasmic soluble flavoprotein (TP0171) but also can flavinylate the NqrC subunit of a T. denticola quinone-based Nqr redox pump (NqrC_Td). Thus, the dual activities of FAD hydrolysis and flavinylation by treponemal Ftp prompt a model by which a dynamic pool of flavin cofactor is maintained and flavoproteins are generated locally in the periplasm (Fig. 7). Although how flavin enters the treponemal periplasm is yet to be unveiled, T. pallidum likely exploits host-derived FAD to balance and maintain its flavin pools in the periplasm via Ftp_Tp. A salient question emanating from our studies is how the identification of a flavinylation pathway can be reconciled in the context of its physiological impact on T. pallidum. In other bacteria, flavin-based ion motive forces (Na+/H+-Rnf/Nqr) are essential for ATP synthesis, rotation of the flagellar motor, and accumulation of nutrients that are taken up by symporters (25). Two components (TP0151 and TP0152, encoded as RnfD_Tp and RnfC_Tp, respectively) of a putative Na+/H+-translocating Rnf redox pump have been predicted in T. pallidum (16, 25). In addition, TP0149 may be an RnfE ortholog because it has ~30% identity to the RnfE subunit of Pseudomonas brassicacearum (KEGG gene function identification tool). Of note, although the initial T. pallidum genome sequence annotated its ATPase as a V-type ATPase (4) that translocates H+ at the expense of ATP, the recently updated databases (NCBI protein and UniProtKB) now indicate it to be a V-type ATP synthase that produces ATP from ADP in the presence of a proton gradient. In addition, more recently Mayer and Müller have proposed from a phylogenetic analysis that ATPase genes from bacteria previously annotated as V-type ATPases are actually A-type ATP synthases that synthesize ATP at the expense of an electrochemical ion gradient (26). As such, from this point on, the treponemal ATPase should be classified and named as an A-type ATP synthase. The mechanism of potential coupling of the ion pump/gradient to an ATP synthase for energy generation in T. pallidum remains uncertain (Fig. 7). However, reverse transcription (RT)-PCR analyses have shown that RnfC_Tp, RnfD_Tp, and putative RnfE_Tp are cotranscribed within a set of genes (tp0147 to tp0153) (data not shown) that are conserved in all treponemal subspecies (data included in the KEGG gene cluster) and thus are likely to constitute a noncanonical Rnf redox/ion pump (Rnf_Tp) (Fig. 7) that either lacks a defined flavinylation motif or contains one or more cryptic flavinylation sites. Alternatively, a noncanonical Rnf system may carry a noncovalently bound FMN cofactor. Although we have reported that the Ftp_Tp-type protein exists in other bacterial species (16), it is not known to what extent a noncanonical-type Rnf might be found in other bacteria. In this regard, it is particularly noteworthy that a new type of archael Rnf complex was recently described that lacks a flavinylation motif in its RnfD subunit (27).
A plausible extension of our results herein is to assume that T. pallidum’s acquisition of host-derived flavin and its flavin homeostasis are separate processes, with the periplasmic pool maintained by Ftp_Tp’s dual Mg2+-dependent FAD pyrophosphatase/FMN transferase activities (16) and the cytosolic flavin pools maintained via the RfuABCD system (11). We propose that these two pathways are central to overall flavin homeostasis and bioenergetics in treponemes. In this proposed “flavin-centric” lifestyle, a putative flavin-based redox system would generate an electrochemical gradient, which could drive subsequent ATP synthesis by an A-type ATP synthase (Fig. 7) under energy limitation (24, 26). It has long been held that T. pallidum’s only ATP-generating system likely is glycolysis (28), fueled by the virtually limitless supply of glucose available in the human body. However, it has always been perplexing as to how the two net ATPs generated per molecule of glucose via glycolysis can satisfy the entire energy needs of the pathogen; this, along with the presumed absence of a TCA cycle (4), leads to an alternative hypothesis that ATP also may be generated in this quinone-free bacterium via its putative flavin-based energy conservation pathway (i.e., flavin-based Rnf redox system coupled to its A-type ATP synthase). This notion is underscored by the idea that energy-limited treponemes probably cannot afford such a large molecular machinery simply to burn/hydrolyze ATP, as opposed to utilizing it as an energy conservation pathway in the presence of a coupling redox/ion pump. A periplasmic Ftp_Tp for protein flavinylation, a flavin-based membrane redox pump (Rnf_Tp), and an A-type ATP synthase, all required for this overall proposed energy conservation pathway (Fig. 7), warrant further future investigation for their importance to T. pallidum physiology and metabolism. Ultimately, clarification of many aspects of our proposed model (Fig. 7) will likely rely on studies performed in other related, but heterologous, bacteria, such as T. denticola, which is genetically manipulable.
It is becoming increasingly clear that Ftp protein-mediated flavin homeostasis and posttranslational flavinylation likely play a wider role in the periplasm than previously appreciated. Moreover, Ftp likely is essential in that it provides the requisite flavins to both flavoproteins and the flavin-based redox pump, which also is consistent with the fact that the phosphoester-threonyl-FMN posttranslational modification is found only in bacteria. As such, this study expands our comprehension of the role of the bacterial periplasm as a metabolically active subcellular compartment, not only for flavoprotein biogenesis, but also for overall membrane redox bioenergetics. Finally, it is possible that the catalytic core of Ftp may prove useful as a new platform for structure-based drug discovery of broad-spectrum antimicrobials that kill bacterial pathogens without harming the human host.
MATERIALS AND METHODS
Reagents.
Unless otherwise noted, chemicals were either purchased from Sigma-Aldrich or Hampton Research. All oligonucleotide primers employed in this study were synthesized at Integrated DNA Technologies (Coralville, IA).
Bioinformatics.
Motif and gene cluster search tools available at GenomeNet (http://www.genome.jp/) were used for FMN-binding motif identification and gene cluster analyses, respectively (29). NCBI’s CD (conserved domain) analysis was also performed to identify the FMN-binding domain (30).
Protein preparation.
Recombinant TP0796 (Ftp_Tp) protein preparation was as previously described (16). The Ftp ortholog from Treponema denticola (TDE2614, referred to as Ftp_Td) was employed in this study. In addition, the putative flavoproteins TP0171 (also known as TP15) from T. pallidum and TDE0836 (also known as NqrC) from T. denticola (NqrC_Td) were used. Recombinant plasmids for ftp_Td (encoding residues 28 to 372), tp0171 (encoding residues 19 to 143), and nqrC_Td (encoding residues 29 to 190) were generated using the polymerase incomplete primer extension (PIPE) cloning method (31). Genes encoding truncated versions of the proteins (without their predicted N-terminal transmembrane helices in the case of the NqrC homolog or signal peptides, including the N-terminal acylated Cys residue in the cases of Ftp homolog and TP0171 lipoproteins) from their respective genomic DNA were amplified by PCR using pfuTurbo DNA polymerase (Agilent Technologies) and primers encoding the predicted 5′ and 3′ termini of the genes (PIPE-inserts). The expression vector pSpeedET (DNASU, Arizona), which encodes an N-terminal TEV-protease-cleavable expression and purification hexahistidine tag (MGSDKIHHHHHHENLYFQG), was PCR amplified with PIPE-vector primers. PIPE-inserts for the respective gene insert and PIPE-vector were individually mixed to anneal the amplified DNA fragments together. E. coli HK100 competent cells were transformed with the mixtures (PIPE-vector and -insert) and selected for kanamycin resistance on LB agar plates. Cloning junctions/fragments were verified by DNA sequencing. Protein expression was performed in LB medium with l-arabinose as the inducer. The procedures for expression and purification of the recombinant proteins were essentially as previously described (11, 16).
Site-directed mutagenesis.
Mutations were introduced into the plasmids carrying wild-type sequences using a QuikChange site-directed mutagenesis kit (Agilent Technologies). All mutants/variants were confirmed by DNA sequencing. Mutant proteins were expressed and purified as described for the wild-type proteins (16).
Protein concentration determination and UV-visible absorption spectroscopy.
Protein concentrations were determined in buffer A (20 mM HEPES [pH 7.5], 0.1 M NaCl, 2 mM β-octylglucoside) from their deduced extinction coefficients using the ProtParam utility of Expasy (32). UV-visible absorption spectra of FAD and flavinylated proteins in buffer A were recorded over the scan range of 300 to 700 nm using a NanoDrop 2000c (Thermo Scientific).
FAD pyrophosphatase assay.
FAD pyrophosphatase activity was assayed by measuring the production of FMN formation, as described previously (16). Briefly, the standard 200-µl reaction mixture containing 1 µM enzyme/protein, 5 mM MgCl2, and 10 µM FAD in buffer A was allowed to incubate for 5 min at 37°C before the change in fluorescence intensity was measured (16).
Covalent flavinylation in E. coli.
In E. coli, flavinylation was performed by coexpressing either the Ftp_Tp-TP0171 or Ftp_Tp-TDE0836 (also known as NqrC_Td) plasmid pair in HK100 cells (DNASU, Arizona). For this experiment, ftp_Tp was recloned in an ampicillin resistance vector (pProEx HTb; Invitrogen), and the cotransformants were selected both for kanamycin and ampicillin. Expression cultures were grown at 37°C in LB medium supplemented with both kanamycin (40 µg/ml) and ampicillin (100 µg/ml). At an optical density at 600 nm (OD600) of ~0.5, the temperature was dropped to 16°C and expression was induced by adding both isopropyl-β-d-thiogalactopyranoside (IPTG; 0.6 mM) and l-arabinose (0.2%) followed by overnight incubation (16°C). Cell pellets were harvested for protein isolation. The yellow protein sample was a mixture of flavinylated and unflavinylated protein and was purified by an Ni2+ affinity column, and the flavinylated protein was separated from the unflavinylated protein by gel filtration chromatography (11, 16). Flavin bound covalently either to TP0171 or NqrC_Td was visualized under UV light (see below).
Flavinylation assay.
Purified proteins in buffer A were incubated with the indicated concentrations of exogenous FAD and MgCl2 in a 100-µl reaction volume for 1 h at 30°C. Approximately 100 µM NqrC protein homolog was incubated in buffer A containing ~20 µM Ftp, 5 mM MgCl2, and 1 mM FAD. Reactions were stopped by adding an equal volume of 2× SDS-PAGE sample buffer, and the mixtures were boiled for 5 min. An approximately 20-µl sample of boiled reaction mixture was separated on a 12.5% SDS PAGE gel and visualized by UV illumination with a Gel Logic 200 imaging system (Kodak) before Coomassie blue staining. Sometimes, boiled reaction mixtures were kept frozen until use.
Mass spectrometry.
Covalently bound flavin to Ftp was identified by mass spectrometry (16).
Crystallization and data collection.
The crystallization and data collection of mutant crystals of Ftp_Tp were performed as described previously (16). All the mutants and ternary complex crystals were routinely obtained in 2 to 3 days by crystallizing Ftp in the presence of 5 mM MgCl2 and/or 1 mM AMP/FAD using 0.1 M morpholineethanesulfonic acid (MES) (pH 6.5) and 0.7 M Na-acetate as the precipitant, the conditions identical to the wild-type protein (16).
Synchrotron X-ray diffraction data were collected at sector 19 (Structural Biology Center) of the Advanced Photon Source and were indexed, integrated, and scaled using the HKL-3000 program package (33). Data collection statistics are provided in Table S2 in the supplemental material.
Phase determination and structure refinement.
Phases for the Ftp_Tp mutant structures were obtained via isomorphous replacement using the apo structures (16) (PDB identifier 4IFU). Manual model rebuilding was performed in the Coot program (34), and refinement was performed in the Phenix program (35). Model refinement statistics are provided in Table S2 in the supplemental material.
Protein Data Bank accession numbers.
The coordinates and structure factors for the Ftp_Tp (D284A)-ADN complex (4XDR) and the Ftp_Tp (N55Y)-FAD complex (4XDT) have been deposited in the Protein Data Bank.
SUPPLEMENTAL MATERIAL
Active-site geometric parameters for T. pallidum FtpN55Y structure.
Data collection, phasing, and refinement statistics for T. pallidum Ftp structures.
ACKNOWLEDGMENTS
This research was supported by an NIH grant (AI056305) to M.V.N. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center, at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Footnotes
Citation Deka RK, Brautigam CA, Liu WZ, Tomchick DR, Norgard MV. 2015. Evidence for posttranslational protein flavinylation in the syphilis spirochete Treponema pallidum: structural and biochemical insights from the catalytic core of a periplasmic flavin-trafficking protein. mBio 6(3):e00519-15. doi:10.1128/mBio.00519-15.
REFERENCES
- 1.Norris SJ. 1993. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Treponema pallidum Polypeptide Research Group. Microbiol Rev 57:750–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerbase AC, Rowley JT, Heymann DH, Berkley SF, Piot P. 1998. Global prevalence and incidence estimates of selected curable STDs. Sex Transm Infect 74(Suppl 1):S12–S16. [PubMed] [Google Scholar]
- 3.Cohen MS, Hawkes S, Mabey D. 2006. Syphilis returns to China … with a vengeance. Sex Transm Dis 33:724–725. doi: 10.1097/01.olq.0000245917.47692.b7. [DOI] [PubMed] [Google Scholar]
- 4.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 5.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD. 2009. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol 191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol 403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radolf JD. 1995. Treponema pallidum and the quest for outer membrane proteins. Mol Microbiol 16:1067–1073. doi: 10.1111/j.1365-2958.1995.tb02332.x. [DOI] [PubMed] [Google Scholar]
- 8.Deka RK, Brautigam CA, Yang XF, Blevins JS, Machius M, Tomchick DR, Norgard MV. 2006. The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum. J Biol Chem 281:8072–8081. doi: 10.1074/jbc.M511405200. [DOI] [PubMed] [Google Scholar]
- 9.Deka RK, Brautigam CA, Goldberg M, Schuck P, Tomchick DR, Norgard MV. 2012. Structural, bioinformatic, and in vivo analyses of two Treponema pallidum lipoproteins reveal a unique TRAP transporter. J Mol Biol 416:678–696. doi: 10.1016/j.jmb.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brautigam CA, Deka RK, Schuck P, Tomchick DR, Norgard MV. 2012. Structural and thermodynamic characterization of the interaction between two periplasmic Treponema pallidum lipoproteins that are components of a TPR-protein-associated TRAP transporter (TPAT). J Mol Biol 420:70–86. doi: 10.1016/j.jmb.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deka RK, Brautigam CA, Biddy BA, Liu WZ, Norgard MV. 2013. Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. mBio 4:00615-12. doi: 10.1128/mBio.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer M, Bacher A. 2005. Biosynthesis of flavocoenzymes. Nat Prod Rep 22:324–350. doi: 10.1039/b210142b. [DOI] [PubMed] [Google Scholar]
- 13.Macheroux P, Kappes B, Ealick SE. 2011. Flavogenomics—a genomic and structural view of flavin-dependent proteins. FEBS J 278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 14.Abbas CA, Sibirny AA. 2011. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol Mol Biol Rev 75:321–360. doi: 10.1128/MMBR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deka RK, Brautigam CA, Liu WZ, Tomchick DR, Norgard MV. 2013. The TP0796 lipoprotein of Treponema pallidum is a bimetal-dependent FAD pyrophosphatase with a potential role in flavin homeostasis. J Biol Chem 288:11106–11121. doi: 10.1074/jbc.M113.449975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertsova YV, Fadeeva MS, Kostyrko VA, Serebryakova MV, Baykov AA, Bogachev AV. 2013. Alternative pyrimidine biosynthesis protein ApbE is a flavin transferase catalyzing covalent attachment of FMN to a threonine residue in bacterial flavoproteins. J Biol Chem 288:14276–14286. doi: 10.1074/jbc.M113.455402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyd JM, Endrizzi JA, Hamilton TL, Christopherson MR, Mulder DW, Downs DM, Peters JW. 2011. FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins. J Bacteriol 193:887–895. doi: 10.1128/JB.00730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besteiro S, Biran M, Biteau N, Coustou V, Baltz T, Canioni P, Bringaud F. 2002. Succinate secreted by Trypanosoma brucei is produced by a novel and unique glycosomal enzyme, NADH-dependent fumarate reductase. J Biol Chem 277:38001–38012. doi: 10.1074/jbc.M201759200. [DOI] [PubMed] [Google Scholar]
- 20.Coustou V, Besteiro S, Rivière L, Biran M, Biteau N, Franconi JM, Boshart M, Baltz T, Bringaud F. 2005. A mitochondrial NADH-dependent fumarate reductase involved in the production of succinate excreted by procyclic Trypanosoma brucei. J Biol Chem 280:16559–16570. doi: 10.1074/jbc.M500343200. [DOI] [PubMed] [Google Scholar]
- 21.Backiel J, Juárez O, Zagorevski DV, Wang Z, Nilges MJ, Barquera B. 2008. Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae. Biochemistry 47:11273–11284. doi: 10.1021/bi800920j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelkar YD, Ochman H. 2013. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics 193:303–307. doi: 10.1534/genetics.112.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heuts DP, Scrutton NS, McIntire WS, Fraaije MW. 2009. What’s in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J 276:3405–3427. doi: 10.1111/j.1742-4658.2009.07053.x. [DOI] [PubMed] [Google Scholar]
- 24.Biegel E, Schmidt S, González JM, Müller V. 2011. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci 68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häse CC, Fedorova ND, Galperin MY, Dibrov PA. 2001. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev 65:353–370. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer F, Müller V. 2014. Adaptations of anaerobic archaea to life under extreme energy limitation. FEMS Microbiol Rev 38:449–472. doi: 10.1111/1574-6976.12043. [DOI] [PubMed] [Google Scholar]
- 27.Welte C, Deppenmeier U. 2014. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta 1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Norris SJ, Cox DL, Weinstock GM. 2001. Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J Mol Microbiol Biotechnol 3:37–62. [PubMed] [Google Scholar]
- 29.Tanabe M, Kanehisa M. 2012. Using the KEGG database resource. Curr Protoc Bioinformatics Chapter 1:Unit 1.12. doi: 10.1002/0471250953.bi0112s38. [DOI] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klock HE, Koesema EJ, Knuth MW, Lesley SA. 2008. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 32.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the Expasy server, p 571–607. In Walker JM (ed), The proteomics protocols handbook. Humana Press, New York, NY. [Google Scholar]
- 33.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Active-site geometric parameters for T. pallidum FtpN55Y structure.
Data collection, phasing, and refinement statistics for T. pallidum Ftp structures.