ABSTRACT
Swarming bacteria are challenged by the need to invade hostile environments. Swarms of the flagellated bacterium Paenibacillus vortex can collectively transport other microorganisms. Here we show that P. vortex can invade toxic environments by carrying antibiotic-degrading bacteria; this transport is mediated by a specialized, phenotypic subpopulation utilizing a process not dependent on cargo motility. Swarms of beta-lactam antibiotic (BLA)-sensitive P. vortex used beta-lactamase-producing, resistant, cargo bacteria to detoxify BLAs in their path. In the presence of BLAs, both transporter and cargo bacteria gained from this temporary cooperation; there was a positive correlation between BLA resistance and dispersal. P. vortex transported only the most beneficial antibiotic-resistant cargo (including environmental and clinical isolates) in a sustained way. P. vortex displayed a bet-hedging strategy that promoted the colonization of nontoxic niches by P. vortex alone; when detoxifying cargo bacteria were not needed, they were lost. This work has relevance for the dispersal of antibiotic-resistant microorganisms and for strategies for asymmetric cooperation with agricultural and medical implications.
IMPORTANCE
Antibiotic resistance is a major health threat. We show a novel mechanism for the local spread of antibiotic resistance. This involves interactions between different bacteria: one species provides an enzyme that detoxifies the antibiotic (a sessile cargo bacterium carrying a resistance gene), while the other (Paenibacillus vortex) moves itself and transports the cargo. P. vortex used a bet-hedging strategy, colonizing new environments alone when the cargo added no benefit, but cooperating when the cargo was needed. This work is of interest in an evolutionary context and sheds light on fundamental questions, such as how environmental antibiotic resistance may lead to clinical resistance and also microbial social organization, as well as the costs, benefits, and risks of dispersal in the environment.
INTRODUCTION
To thrive in heterogeneous and competitive environments, microorganisms employ both cooperative and competitive strategies. Swarming bacteria collectively migrate over surfaces driven by their flagella (1). Swarming offers a competitive advantage in invading some habitats (2, 3); however, the costs and benefits remain largely uncertain. Swarming is studied in monoculture, but more than one swarming species can associate (coswarming), with mutual benefits (4, 5). There are also instances where a motile eukaryote (e.g., Acanthamoeba spp. [6] and slugs of Dictyostelium spp. [7]) transports bacteria. Examples where bacteria provide the driving force to move sessile cargo microorganisms are rare. However, swarms of the bacterium Paenibacillus vortex (8, 9) can transport conidia of the filamentous fungus Aspergillus fumigatus (10) over long distances (tens of centimeters). The conidia gain by moving to a more favorable location, while the bacteria use germinating mycelia to cross air gaps. The idea that a bacterial swarm could be considered a moving ecosystem, incorporating sessile cargo microorganisms with costs and benefits, motivated this work. We approached this problem with respect to the contribution of antibiotic resistance by the cargo bacterium, particularly the spread of beta-lactamase (BL) genes encoding enzymes that degrade beta-lactam antibiotics (BLAs) (11).
The spread of antibiotic resistance in bacteria is of widespread concern. The BLAs include major classes of front-line therapeutics (12). Resistance to BLAs is reaching a crisis point, for example with Gram-negative pathogens having extended-spectrum beta-lactamases (ESBLs) and carbapenem resistance. Interactions between different lineages and species of bacteria are relevant to the prevalence of antibiotic resistance within a population. Marine Vibrionaceae show population-level interactions mediated by antibiotic production and resistance (13), suggesting that populations defined by a common microhabitat and an antibiotic-producing/resistance network may represent socially cohesive units. Complex relationships, including reciprocity of both antagonism and cooperation, have been found between Streptomyces strains isolated from the same soil granule (14). The idea that soil bacteria represent a reservoir of resistance genes available for exchange with clinical pathogens is supported by the discovery of identical antibiotic resistance cassettes, including ones active against BLAs, in soil bacteria and human pathogens (15). Taken together, these observations suggest that networks of microbial interactions within the environment are important in the emergence and spread of antibiotic resistance.
Bacteria on surfaces may be in physiological states that confer tolerance to antimicrobials, notably within a biofilm, but also when swarming (1, 16). P. vortex shows a limited phenotypic resistance to a diverse range of antibiotics when swarming (17). Multiple strategies exist within isogenic populations of bacteria to temporarily protect themselves from antibiotics (16, 17, 18), including ones linked to swarming. For example, swarming Bacillus subtilis undergoes spatial and population shifts between motile and nonmotile cells, with the net effect that both colony expansion and growth are less vulnerable to antibiotics (19). Salmonella enterica serovar Typhimurium has decreased permeability to some antibiotics when swarming (20). Further, phenotypic tolerance of antibiotics by P. vortex is due to a highly motile but poorly reproducing subpopulation, the explorer morphotype (17). Despite a degree of antibiotic tolerance, swarming P. vortex cannot thrive (i.e., disperse and expand their population) in the presence of high levels of BLAs.
Here, we show that swarming P. vortex can transport a different species of bacteria (cargo). When the cargo expresses a BL, transport opens up new niches for P. vortex. In the presence of a BLA on an agar plate, both transporter and cargo species gain from moving as a consortium in an asymmetric but mutually beneficial relationship: the former drives dispersal and the latter detoxifies. Additionally, P. vortex employs a bet-hedging strategy; when this bacterium invades new territory, it does so first with swarming cells without cargo bacteria and subsequently with swarming groups containing cargo. Therefore, we characterize the relationship as a mutualism of convenience, in which P. vortex uses the cargo bacteria when needed.
RESULTS
Transport of a BL-expressing cargo by P. vortex facilitates the dispersal and growth of both species in the presence of high levels of the BLA ampicillin.
When inoculated on a 14-cm-diameter Mueller-Hinton (MH) agar plate and incubated at 37°C under aerobic conditions, P. vortex was capable of swarming continuously and reaching the edge of the plate within 12 h. When P. vortex was inoculated on a plate containing ampicillin (Amp; 40 to 200 µg ml−1), limited swarming was observed (a few millimeters) without growth. When an Escherichia coli strain (VL001; see Table S1 in the supplemental material) expressing a BL was inoculated on an MH plate containing Amp, a static colony formed. In other words, in the presence of Amp neither species could use the full resources of the agar plate: the E. coli strain could not spread and P. vortex was antibiotic sensitive. However, when inoculated together (in an equal ratio of cells) on an MH agar plate containing 200 µg ml−1 Amp, the two species could colonize the plate within 72 h (Fig. 1a).
FIG 1 .
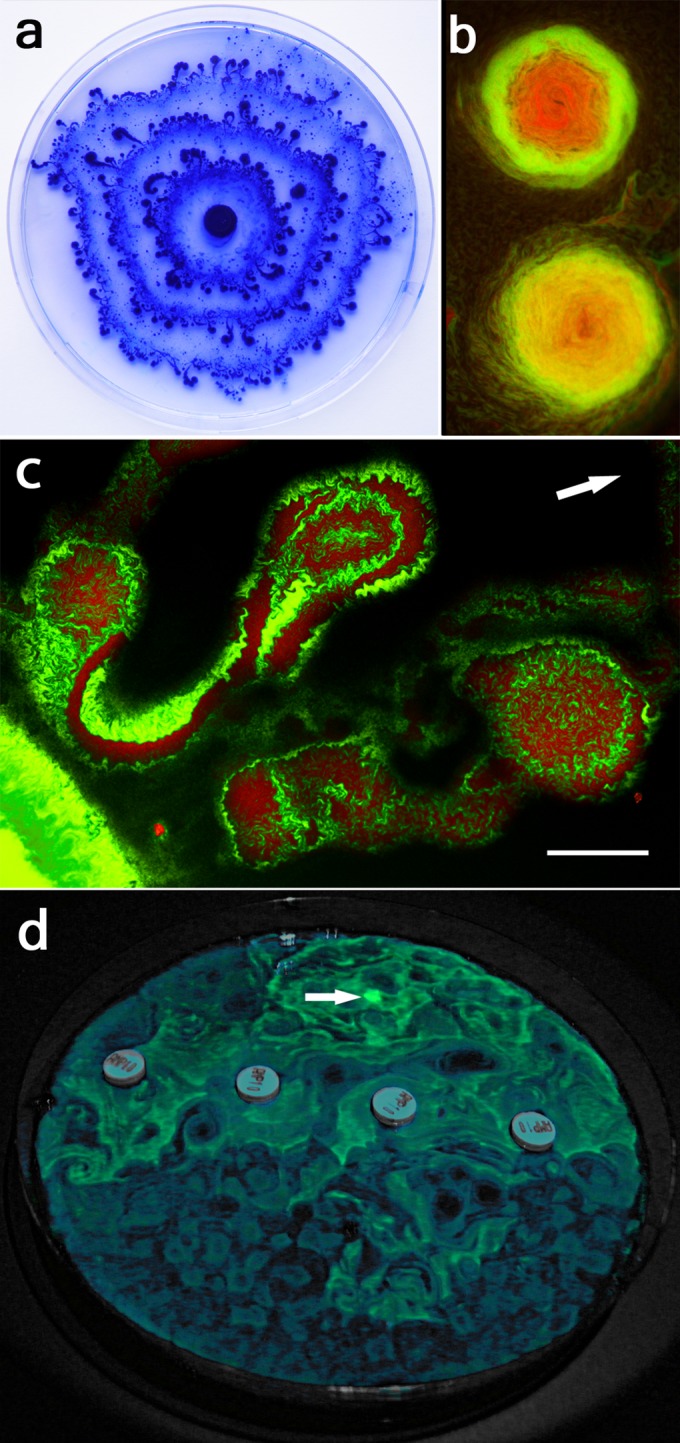
Colony and microcolony imaging of cargo transport. (a) A periodically expanding colony composed of swarming P. vortex and Ampr E. coli cargo imaged after incubation for 72 h at 37°C on a 14-cm-diameter MH agar plate with 200 µg ml−1 Amp. The plate was stained with Coomassie blue to enhance contrast. (b) Imaging by fluorescence microscopy of two moving peripheral colonies (rotating and moving progressively outwards from the inoculation point, in an experiment identical to that shown in panel a), using hexidium iodide to identify P. vortex (red) and GFP expression for E. coli (green). (c) Imaging by fluorescence microscopy of microcolony transport of a Ctxr strain of Enterobacter aerogenes GA2 (stained by Syto 9 [green]) by P. vortex (stained by hexidium iodide [red]) over agar containing 3 µg ml−1 Ctx. The arrow shows the overall direction of transport at a rate of ~3 mm h−1. (d) Transport of a consortium of P. vortex and Ampr E. coli over an MH agar plate with a barrier of Amp created by four Neosensitabs (see Fig. S2 in the supplemental material). Imaging is of E. coli via a blue light LED to visualize GFP expressed by the cargo strain. The scale bar in panel c (300 μm) corresponds to 3.8 cm (panel a), 200 µm (panel b images), and 1.9 cm for the cell in panel d.
The strain VL001 was able to swim but not swarm. To clarify the role of cargo motility, the experiments described above were repeated using a nonflagellated, Ampr E. coli cargo strain (VL003). On MH with Amp, the nonmotile Ampr strain was able to assist P. vortex in spreading toward the edges of the plates. This indicated that motility of the cargo was not required and P. vortex was providing the driving force for the dispersal process.
Fluorescence microscopy was used to observe the moving microcolonies, after allowing the bacteria to swarm across regions of agar impregnated with fluorogenic dyes to allow distinction of the two species by selective staining (hexidium iodide for P. vortex [red fluorescence] and Syto 9 for cargo [green]) or by using green fluorescent protein (GFP)-expressing cargo. Microcolonies (hundreds of microns across) at the periphery of an expanding colony contained both cargo and transporter strains (Fig. 1b). Cargo bacteria were patterned by the transporting P. vortex (Fig. 1b and c). On MH agar plates with 200 µg ml−1 Amp, the colony was composed of concentric rings of bacteria (Fig. 1a). Colony expansion was periodic (Fig. 2), with active swarming alternating with periods of little progress. The periods of reduced expansion apparently coincided with detoxification of the antibiotic by the BL-expressing cargo, followed by swarming when the BLA remaining at the colony periphery was no longer harmful to P. vortex. This conclusion was supported by the observation that after a pause of the peripheral swarming cells, adding additional Amp (to a local concentration of >200 µg ml−1) at the edge of the colony delayed the reinitiation of swarming by 1 to 2 h. Further, addition of the BL inhibitor clavulanic acid (>0.1 µg ml−1) also delayed the initiation of swarming for 3 to 4 h. In contrast to the Ampr (ampicillin-resistant) cargo strain VL001, an Amps (sensitive) strain of E. coli (VL002; see Table S1 in the supplemental material) was unable to facilitate colonization of MH agar plates containing 200 µg ml−1 Amp when coinoculated with P. vortex. Additionally, the periodicity of swarming decreased with higher concentrations of ampicillin in the plate (see Fig. S1 in the supplemental material). Taken together, these observations suggest that the time taken to detoxify the antibiotic was the limiting factor in colony expansion.
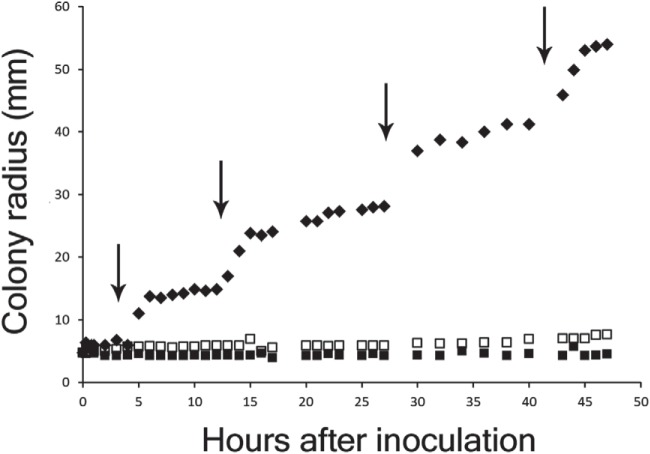
FIG 2 .
Periodicity of P. vortex swarming with Ampr E. coli cargo in the presence of Amp. One or both strains were inoculated in the center of MH agar plates containing 200 µg ml−1 Amp, followed by incubation at 37°C and tracking the expansion of the resulting colony by automated imaging. White squares, E. coli inoculated alone; black squares, P. vortex alone; black diamonds, both strains. Coinoculation of P. vortex and E. coli allowed a colony to form with a concentric ring architecture (Fig. 1a). At the expanding edge of such a colony, periods of rapid advancement (indicated by arrows) alternated with phases of less active progress (plateaus).
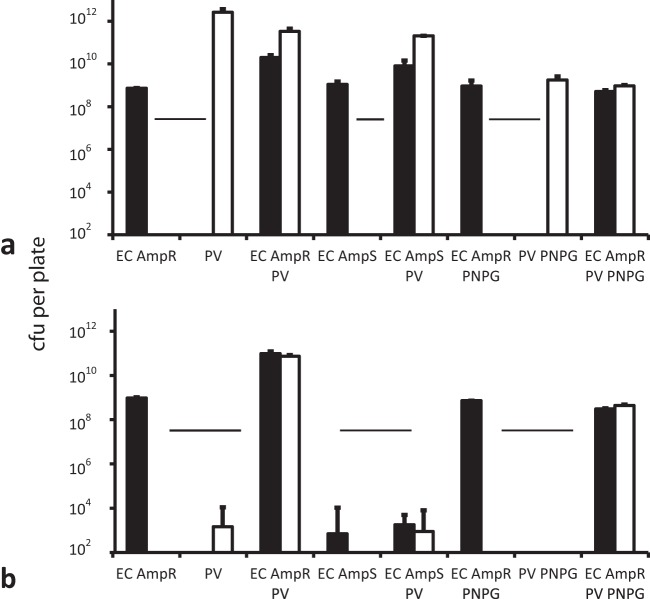
Changes in populations during the interaction of swarming P. vortex cells with Ampr or Amps cargo strains of E. coli were quantified by selective viable counts. When inoculated alone on MH agar without Amp, both E. coli strains expanded their populations by nearly 2 orders of magnitude by static growth (Fig. 3a). On the same medium, a monoculture of P. vortex achieved a major population expansion (over 4 orders of magnitude) by swarming and growth. When each of the three strains was inoculated as a monoculture on plates containing Amp, the Ampr E. coli strain expanded its population due to local growth, while the viability of P. vortex and Amps E. coli strains declined (Fig. 3b). When the Ampr cargo strain was coinoculated with P. vortex in the presence of the swarm inhibitor 0.5 mM p-nitrophenylglycerol (PNPG) (9, 21), both species gained moderately from localized growth. In this situation, P. vortex benefited from close proximity to the antibiotic-degrading E. coli. When E. coli Ampr was coinoculated with P. vortex under swarming conditions on medium containing Amp, both species expanded their population to a similar extent, around 3 orders of magnitude (Fig. 3b). The E. coli Amps strain could not support the same population expansion when coinoculated with P. vortex on medium containing Amp (Fig. 3b). Finally, when inoculated together without Amp (P. vortex and Amps or Ampr E. coli), the cargo still gained from the association: the colony expanded and E. coli was transported and grew in new regions of the agar. However, Ampr E. coli gained less from being a cargo, relative to the situation in the presence of Amp (population increase almost 2,000-fold with Amp and only 400-fold without), and P. vortex gained more from the situation (population increase 1,500-fold with Amp and 7,000-fold without).
FIG 3 .
Quantification of the effects of transport on population changes in both transport and cargo strains in the presence and absence of Amp. Selective viable counts (black bars, E. coli; white, P. vortex) were used to determine transporter and cargo CFU after inoculation on 14-cm-diameter MH agar plates with transporter (P. vortex [PV]) and/or cargo (E. coli [EC]) bacteria, with the change in viable count assessed by selective plating after 72 h at 37°C. PNPG, addition of the swarming inhibitor p-nitrophenylglycerol (0.5 mM). The broken horizontal line indicates the inoculum level (5 × 107 CFU) for each species. (a) Plates without antibiotic; (b) plates with 200 µg ml−1 Amp.
Local analysis of cargo distribution during transport with and without a BLA.
The distributions of the cargo and transporter strains were analyzed by isolating cells from different locations within the spreading colonies. Inoculated plates were sampled after 50 to 72 h, capturing >107 CFU, and the species composition was determined by selective viable counts. The expanding colony, typically 6 cm in radius, was divided into three sampling zones: the periphery of the colony (up to 1 cm from the edge), the immediate interior (1 to 2 cm from the edge), and the central region (>2 cm from the edge, including the inoculation site), as shown in Fig. S2 in the supplemental material. This analysis was particularly revealing for the situation in which the cargo and transporter strains were coinoculated on MH agar without Amp. Moving colonies at the periphery of the colony contained no detectable cargo bacteria. Both cargo and transporter bacteria were present within the colony interior (see Fig. S2). In contrast, analysis of the species distribution after coinoculation on MH agar with Amp revealed that the cargo strain was highly represented in all zone samples, including the edge of the colony. These data, combined with selective viable counts from entire plates (Fig. 3), indicated that the presence of Amp favored the transport of the E. coli Ampr strain and that this selection pressure had the greatest effect at the colony edge. The peripheral colonies lacking cargo in the absence of Amp were smaller, faster, and more highly dispersed than the interior cargo-carrying colonies (see Table S2 in the supplemental material). Microscopy showed that the peripheral colonies were derived from single cells and small groups of P. vortex cells that made an early escape from the central mass. Analysis of expanding colonies at earlier stages in growth indicated that the first wave of moving colonies could be detected after 20 min to 7 h, and these colonies (>90%) lacked cargo. A first scenario was created, where the transporter and cargo initiated swarming in a region of the plate and then the consortium was forced to encounter localized Amp after swarming 4 cm (Fig. 1d; see also Fig. S3 in the supplemental material). Tracking the moving colonies indicated that those lacking cargo (judged by microscopy imaging via GFP and confirmed by viable plating) reached the region of antibiotic first (by 1 to 2 h) and that these then failed to grow. After 3 to 6 h, the region was occupied by larger swarming colonies that contained cargo and which were able to grow.
In a second scenario, serial culture on agar plates was used to assess the robustness of the cargo-transporter system. Inoculations with P. vortex and a cargo strain were made on one edge of a 14-cm-diameter MH agar plate, and >108 CFU were isolated from the far edge of the plate after 36 to 80 h, to compare plates with and without Amp (see Fig. S3 in the supplemental material). When Amp was absent, it took 1 to 4 transfers (10 replicates; average, 1.8 transfers) to eliminate the cargo, as judged by selective plating. When Amp was present, the system was stable, as after >20 transfers the cargo continued to be highly represented (30 to 65% of the population by viable count; n = 6). A third scenario was created in which exposure to Amp was periodic: the consortium initiated transport in the absence of Amp, then passed through a region of agar containing Amp and was recovered from the far side of this chemical barrier (Fig. 1d; see also Fig. S3). As with the situation with Amp present throughout the agar plate, the cargo was stably maintained (>20 transfers; n = 6). This suggests that intermittent exposure to a BLA is sufficient to maintain the association of the Ampr cargo and P. vortex.
Comigration of P. vortex with E. coli Ampr on plates with Amp led to a stable mixed population, a system that corrected for initial imbalances in the inoculum. When the cargo was inoculated in as little as a 1:1,000 minority (5 × 107 CFU P. vortex, 5 × 104 CFU of Ampr E. coli) or in up to a 20-fold excess (109 CFU E. coli), the result after 72 h was a final population close to parity (with up to a 3-fold excess of cargo or transporter). This, along with the long-term robustness of the system, suggests that population levels move rapidly toward equilibrium as long as the selection pressures for both colony expansion and Amp resistance are maintained. The exception was at high cargo ratios (>20-fold cargo excess; >109 CFU), a situation in which swarming usually failed to initiate due to cargo overgrowth. However, in the cases (<20%) where swarming occurred when there was a 20-fold cargo excess, the consortium formed a stable and apparently balanced population after 72 h.
Transport is mediated by a phenotypic subpopulation of P. vortex.
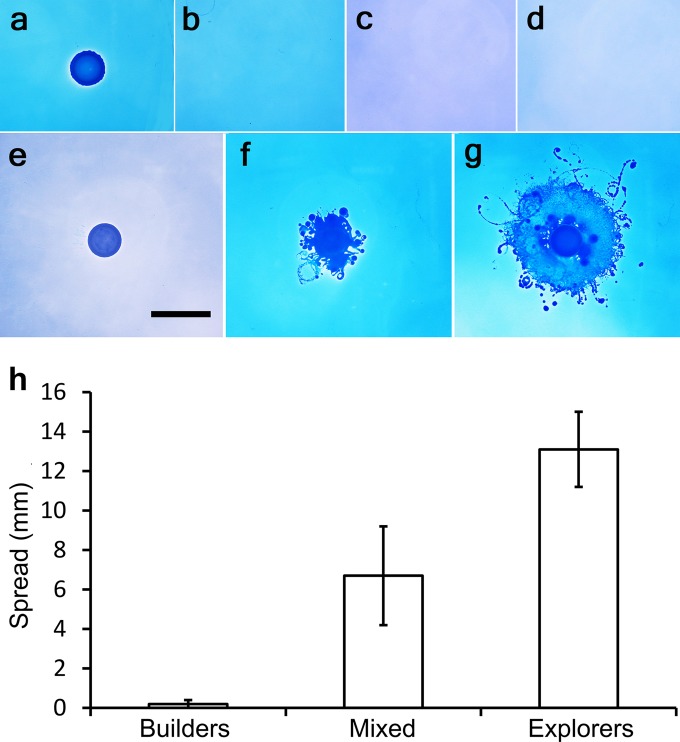
Previous studies have shown that an isogenic P. vortex colony contains two subpopulations: explorers with elevated antibiotic resistance and enhanced motility, and more rapidly dividing but less motile builders that form the majority of the colony interior (17). These two phenotypic variants of isogenic cells were enriched separately to >99% purity (see Fig. S4 in the supplemental material) and tested for their ability to transport an E. coli cargo (compared to the mixed population, which was ~30% explorers) (17). Transport assays were run for 12 h after inoculation with E. coli (Ampr) on LB agar plates containing 100 µg ml−1 Amp. This allowed the majority of the population to be maintained as the inoculated morphotype (17). Neither species could disperse or grow without the cargo—the limited refractory nature of explorers to antibiotics was not sufficient to support swarming on this medium (Fig. 4b to d). The explorer-enriched subpopulation mediated transport and the builders did not (Fig. 4). During transport, flagella create tangled networks, apparently wrapping around the cargo (10). One of the differences between the subpopulations is a higher expression of flagella in explorers versus builders (17). Thus, a possible explanation of the explorers’ ability to transport cargo could be more flagella enhancing one or both of motility and cargo capture.
FIG 4 .
Investigation of the P. vortex subpopulation responsible for cargo transport. Explorers and builders from a P. vortex mixed culture were enriched, and the subpopulations as well as the mixed culture were coinoculated with Ampr E. coli, plated on LB agar with Amp (100 µg ml−1), and incubated for 12 h. (a to d) Controls. Each culture was plated on LB with 100 µg ml−1 Amp, as follows: Ampr E. coli (a), mixed culture (b), builders (c), and explorers (d). P. vortex (mixed culture, builders, and explorers) plated alone on Amp plates showed no growth, while Ampr E. coli managed to grow but did not spread. (e to g) Coinoculation results for the Ampr E. coli with P. vortex builders (e), a mixed culture (f), and explorers (g), respectively. The scale bar in panel e (10.6 mm) also applies to panels a to g. (h) Spreading of E. coli combined with P. vortex (either purified builders or explorers or a mixed culture).
Cargo bacteria with higher BLA resistance support more effective dispersal on BLA-containing plates.
An isogenic collection of cargo strains, differing in their levels of BLA resistance, were tested for their ability to complement P. vortex when swarming in the presence of cefotaxime (Ctx). These strains expressed a TEM-1 type BL, with the encoding gene containing mutations leading to the production of enzymes with different levels of activity (22), and consequently had a wide range of resistance to Ctx, with MICs from 0.03 to 512 µg ml−1. P. vortex was highly sensitive to this BLA (MIC, 0.06 µg ml−1) and could not grow or swarm in the presence of Ctx. As in previous experiments, E. coli alone could grow on MH agar with Ctx if the MIC was sufficient, but could not swarm. Coinoculation of different cargo strains with P. vortex on Ctx plates and incubation for 72 h indicated that the higher the MIC of the cargo, the more effectively the consortium grew and dispersed (Fig. 5a). A higher resistance level was as beneficial to the transporting P. vortex as was the BL-producing strain of E. coli, as long as the two strains had the potential to form moving consortia (Fig. 5a to c). The benefits in terms of population expansion and dispersal were greater than for the E. coli in monoculture (Fig. 5b and c). The consortia appeared more responsive to an increase in MIC of the cargo than E. coli alone, when the MIC was higher than the concentration of Ctx in the agar (Fig. 5b and c). This may be because in the static situation, E. coli only detoxifies local antibiotic, and once that is achieved it gains little additional benefit from a more effective BL. In the moving consortia, the leading edges are expanding into new territory containing fresh antibiotic. This has the potential to happen rapidly—P. vortex can transport cargo at a rate of >1 cm h−1—so we suggest that there is a greater selection pressure for rapid degradation of the target antibiotic.
FIG 5 .
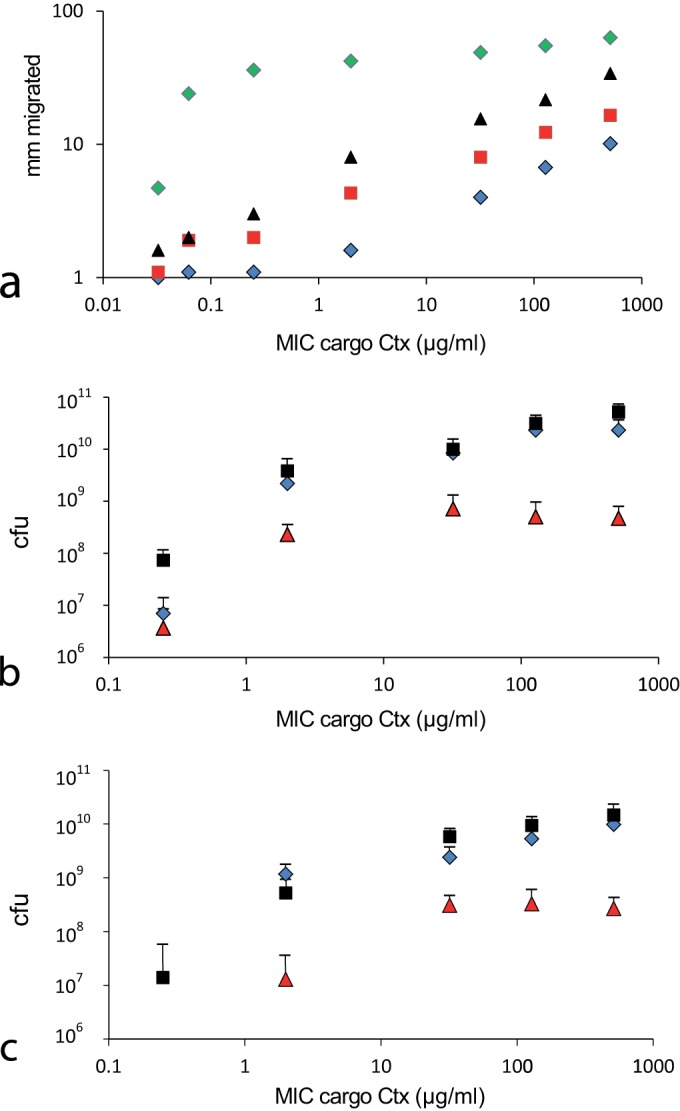
Relationship between the MIC of Ctx for the cargo bacteria and how this affects transport by P. vortex in the presence of this BLA. (a) Migration of a consortium of P. vortex coinoculated with E. coli strains with different levels of resistance to Ctx due to mutations within a TEM-1 type BL. Green diamonds, 0.3 µg ml−1 Ctx in MH agar; black triangles, 1 µg ml−1 Ctx; red squares, 10 µg ml−1 Ctx; blue diamonds, 100 µg ml−1 Ctx. The migration distance from the inoculation point on MH agar plates containing the antibiotic was measured after 72 h and plotted against the MIC of the cargo E. coli. (b) Quantification of cargo and transporter populations after coculture for 72 h on 14-cm-diameter MH agar plates containing 1 µg ml−1 Ctx, plotted against the MIC of Ctx for the cargo bacterium. Red triangles, E. coli strain plated alone; black squares, CFU for E. coli strain plated with P. vortex; blue diamonds, CFU for P. vortex plated with E. coli. (c) Quantification of cargo and transporter bacteria on MH agar containing 10 µg ml−1 Ctx plotted against the MIC of Ctx for the cargo bacterium. Red triangles, E. coli strain plated alone; black squares, CFU for E. coli strain plated with P. vortex; blue diamonds, CFU for P. vortex plated with E. coli. Due to widespread cell death, data points related to the viability of bacterial swarms with cargo bacteria on low-MIC Ctx agar were too small to be reliably quantified and were not plotted.
P. vortex has superior transport capacity compared to other swarming bacteria.
Other swarming bacteria were tested for their ability to transport Ampr cargo. Vegetative and swarmer cells of Amps strains of Proteus mirabilis failed to transport Ampr E. coli, as judged by swarm expansion and GFP visualization by fluorescence microscopy. These experiments were performed on MH Eiken agar (0.6% to 1.8%, wt/vol) plates containing 40 to 200 µg ml−1 Amp. Up to 72 h after inoculation, no expansion of the colony was observed beyond a few millimeters from the inoculation point. Ampr strains of P. mirabilis swarmed, but again, transport was not found. Similarly, a clinically derived Amps strain of Enterobacter aerogenes was able to swarm on MH agar (0.8%, wt/vol) but could not transport the same cargo. Other Paenibacillus spp. swarmed on MH agar. A collection of these were tested for their ability to employ Ampr cargo to disperse and grow on MH agar containing Amp (100 µg ml−1). Coinoculation with the cargo strain allowed localized growth of both Paenibacillus spp. and E. coli within the inoculation region, indicating that the former could benefit from a nearby BL-producing strain. Paenibacillus alvei, Paenibacillus polymyxa, and Paenibacillus glucanolyticus had minor transport capacities. Limited transport by Paenibacillus lautus was possible, as judged by the expansion of colonies on Amp-containing plates (Fig. 6) and by the distribution of the cargo strain, assessed by fluorescence microscopy. Paenibacillus dendritiformis has two phenotypic variants on agar. These morphotypes segregated stably, with rare interconversion, as the T (tip-splitting) and C (chiral) morphotypes, with different colony morphologies (23). The C morphotype transported most effectively. None of the Paenibacillus spp. tested could match the transport capacity of P. vortex. Further, the intricate patterning that P. vortex created with the cargo organism (Fig. 1b and 7) was absent with other Paenibacillus spp. The effectiveness of transport by P. vortex could be related to its swarming strategy, which involves multilayers of cells in which cargo microorganisms become stably entrapped by P. vortex, apparently within a network of flagella (10). Modeling of P. vortex “traffic systems” during cargo transport (24) supports physical connections between formed transporters in the context of a highly organized traffic network of cells (25) in which individuals maintain the same neighbors over a period of at least minutes.
FIG 6 .
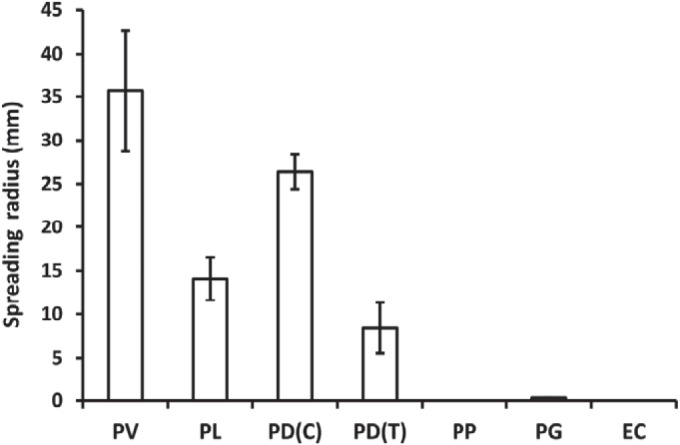
Transport by different Paenibacillus strains of Ampr E. coli in the presence of Amp. MH agar plates containing 200 µg ml−1 Amp were inoculated with a combination of Ampr E. coli and a strain of Paenibacillus, or in control experiments with individual strains. All of the Paenibacillus spp. were able to swarm, but none spread when inoculated alone, due to BLA sensitivity. PV, P. vortex plus E. coli V001; PL, P. lautus DSMZ3035 plus E. coli V001; PD(C), P. dendritiformis C454 (C morphotype) plus E. coli V001; PD(T), P. dendritiformis T454 (T morphotype) plus E. coli V001; PP, P. polymyxa DSMZ36 plus E. coli V001; PG, P. glucanolyticus 5162 plus E. coli V001; EC, E. coli V001 alone.
Clinical isolates expressing ESBLs gain from transport.
To investigate whether the P. vortex transport system can aid the spread of antibiotic-resistant pathogens, a collection of clinical isolates (see Table S3 in the supplemental material) of Enterobacteriaceae, both sensitive and resistant to cephalosporins, including Ctx (the resistant group was classified as ESBL producers), were tested as cargo organisms. This collection of resistant strains all complemented P. vortex in colonizing 14-cm MH agar plates containing 3 µg ml−1 Ctx (see Table S3). Ctxr clinical isolates of E. coli, nonmotile, capsulated Klebsiella pneumoniae (26), and Enterobacter aerogenes were all transported by P. vortex, and those that had appropriate BLs allowed transport and cocolonization of plates containing BLAs (Fig. 1c and 7; see also Table S3). Sensitive strains did not complement swarming of P. vortex on Ctx-containing agar. The motility of cargo bacteria was tested in the absence of P. vortex, and most were found to be nonswarming. One E. coli strain (see Table S3) was able to swarm under the test conditions. Even for this strain, the presence of P. vortex was still an advantage in the spreading of the consortium.
Environmental isolates expressing BLs gain from transport.
To test the ecological relevance of a transport strategy involving BLA-resistant cargo, bacteria isolated from arable, industrial, and woodland soils were screened for isolates capable of complementing P. vortex, i.e., allowing the latter to swarm in a sustained fashion on MH agar plates containing either Ctx (3 µg ml−1) or the carbapenem BLA meropenem (Mer; 2 µg ml−1). Candidate cargo strains recovered from the edge of MH agar plates containing Ctx or Mer were purified and retested by combining them with fresh P. vortex. This resulted in the isolation of Gram-negative bacterial strains which acted as useful cargo and so enhanced the spread of P. vortex on plates containing one of these BLAs. Identification by 16S sequencing revealed that most of the Ctxr isolates were pseudomonads with Ctx MICs of >8 µg ml−1 and Stenotrophomonas maltophilia (see Table S4 in the supplemental material). The S. maltophilia isolates were resistant to imipenem but not when this BLA was supplemented with EDTA. This suggested that the primary BL that was expressed was a metallo-BL (27). Therefore, non-TEM-1-type BLs can complement P. vortex swarming on BLA. The Mer screen resulted in a different range of isolated bacteria, predominantly Flavobacterium spp. All of these groups of bacteria have been encountered within hospitals and identified as causing opportunistic infections that are particularly hard to eradicate due to their broad spectrum of antibiotic resistance (28, 29). All isolates yielded satellite colonies when plated with a BL-sensitive strain of E. coli (VL002), indicating that they could degrade BLAs. The isolation of BLA-resistant cargo organisms directly from the soil in these experiments suggests that P. vortex can employ environmental bacteria as useful cargo.
DISCUSSION
When Paenibacillus vortex swarms, it can carry other microorganisms for many centimeters. The cargo microorganisms may be useful in solving problems that P. vortex cannot overcome alone. In this work, we have described a widely applicable selective system for looking at cargo-transporter interactions, one that requires dispersal across an agar plate in the presence of an antibiotic for the transporter and cargo to maximize their populations. We used BLAs to prevent P. vortex from growing or swarming alone and BL-expressing cargo unable to disperse. Together, a consortium of the two strains could colonize a BLA-containing agar plate to mutual benefit as long as the selective pressure was maintained. Colonies of BL-expressing bacteria can protect nearby sensitive strains, i.e., the phenomenon of satellite colonies. The situation we have described here is more dynamic and with reciprocal benefits during expansion of a mixed population. In other ways, P. vortex gained more from the relationship. The cargo bacterium benefited by dispersal, but alone it would have grown near the inoculation site, albeit to a lesser extent than if transported by P. vortex. The latter, when inoculated onto Amp alone, suffered a population decline due to the effects of the antibiotic. P. vortex was inoculated alone and with Ampr cargo on Amp-containing plates. When inoculated alone, the viability of P. vortex declined by over 4 orders of magnitude; with cargo assistance, P. vortex gained ~108-fold CFU/ml (Fig. 3). Ampr cargo gained 100-fold CFU/ml in the presence of the transporter.
A swarming colony of P. vortex contains a highly flagellated subpopulation (explorers) that localize toward the periphery and help to expand the colony, with builders (a less motile but rapidly dividing phenotype) dominating the interior (17). A P. vortex colony can be considered a temporarily multicellular organism with differentiation and task distributions (30–33). Only explorers mediate transport; therefore, we conclude that the explorers are specialized in movement, both movement of their own cells (motility) and of others (transport), the latter being a type of bacterial task distribution not previously reported. Further, of the Paenibacdillus spp. tested, only P. vortex created intricate patterns with the cargo. P. vortex operates in a self-organizing system, but we have shown here that when it is swarming it also can act as an “other-organizing” system (Fig. 1c and 7). Ecosystem patterning is of interest at a macroscopic level (34) and also within a developmental context, i.e., how different cell types organize to form tissues (35). P. vortex appears to provide an important model for studying how cells form more complex structures, as a system with two phenotypes with different spatial distributions (explorers and builders) and the explorer subpopulation organizing other bacteria.
In the absence of antibiotics, the Ampr cargo was still transported by P. vortex, but it was absent from the colony periphery. The faster, smaller lead colonies running ahead of the rest on MH agar did not contain cargo. In this situation, the cargo was transported by a second wave of larger, slower-moving colonies lagging behind the lead elements. This contrasted with the results of the experiment when Amp was present and where the cargo was invariably found at the edge. This has an interesting consequence: swarming colonies solely composed of P. vortex get a chance to explore and exploit new territory first in the absence of selection pressure for the presence of potentially competing cargo bacteria. However, if the lead elements cannot thrive, the second rank carrying the cargo may catch up and allow the swarm to do better. In this sense, P. vortex appears to exploit the cargo bacteria. The latter appear to be treated as an equal partner only when needed. This can be considered a bet-hedging strategy: hitchhikers are only given the best ride when useful to P. vortex.
An important question is how the degree of antibiotic resistance of the cargo affects the survival of both cargo and transporter in the presence of a BLA. A higher level of BL resistance not only promoted local cargo survival but also facilitated dispersal by P. vortex (Fig. 5). High levels of antibiotic resistance made little difference to the growth of the E. coli strain on Ctx alone when the MIC was higher than the concentration of the BLA. A static colony need only survive local BLA, and once this is done it only needs to degrade BLA on the edge of the growing colony. In contrast, a cargo strain expressing a BL can be moved rapidly by P. vortex, in excess of 1 cm h−1. In that situation, the demands for rapid degradation of the BLA are likely to be higher, as the moving consortium encounters fresh antibiotic continuously. Modeling studies support the role of antibiotic gradients in promoting the evolution of resistance in bacteria (36). Transport of cargo bacteria may be one situation where such a selection pressure is generated.
Because clinical BLA resistance is a major issue, we tested isolates obtained from human infections (expressing ESBLs) as cargo. The BL-expressing strains, even an independently swarming strain, showed enhanced dispersal when combined with the more vigorously swarming P. vortex on BLA-containing plates. Additionally, a screening of soil bacteria was undertaken to isolate cargo bacteria which complemented P. vortex when coinoculated on MH agar, also containing BLAs. This resulted in the isolation of a collection of Gram-negative, BLA-degrading bacteria, including ones with BLA resistance profiles commonly found in opportunistic hospital infections (29). Further, when presented with a mixed cargo population (sensitive and resistant E. coli) in the presence of a BLA, P. vortex retained the resistant strain more so than the sensitive strain. Therefore, from a mixed population, the most useful strain (to P. vortex) was retained. This suggests that P. vortex may play a role in the (re)distribution of clinically relevant BLA-resistant bacteria in the environment. BLs are the enzymes responsible for the most prominent and one of the most clinically relevant antibiotic-degrading activities. However, other antibiotic classes, both natural and semisynthetic, can be metabolized by bacteria from the environment, and other interspecies interactions can affect antibiotic survival (37, 38). Therefore, it is possible that other antibiotic-protecting bacteria may be productively transported by P. vortex. Recently, P. vortex was shown to disperse pathogenic species of Xanthomonas spp. on tomato leaves (39), increasing infection. In this case, the less motile “cargo” stimulated P. vortex motility with unknown volatiles, suggesting a role of interspecies communication in forming swarming consortia. Different cargo microorganisms may bring other “skills” which P. vortex lacks, and P. vortex may transport such microorganisms under other types of selection pressures, driving associations in which both partners gain an advantage. Range expansion is a powerful driver in evolution (40) and here may promote interactions between the sessile and motile microbiota. Further work could productively explore the genetic basis of this phenomenon, in both cargo and transporter, and the evolutionary and ecological importance of such relationships, i.e., the extent to which swarming bacteria act as moving ecosystems. Long-term coevolution experiments of transporter and cargo combined under different selection pressures may prove particularly useful in constructing stable or metastable synthetic ecologies (41). Such constructed or artificially evolved consortia are likely to be valuable in examining cooperation, competition, and cheating at a fundamental level (42).
MATERIALS AND METHODS
Strains and culture conditions.
The strains used in this study are summarized in Table S1 in the supplemental material. P. vortex and other Paenibacillus species (P. dendritiformis, P. polymyxa, P. glucanolyticus, P. alvei, and P. lautus) were grown, as previously described, on MH agar (1% [wt/vol] Eiken agar) (8), Luria broth agar, or peptone agar (17), aerobically at 20 to 37°C. Cargo strains were grown on MH agar at 20 to 37°C. Proteus mirabilis was grown on MH agar (0.8 to 1.5% [wt/vol] Eiken agar) at 37°C (43).
Cargo transport assays.
For cargo transport assays, 5 × 107 cells of each species (P. vortex or another candidate transporter and/or cargo organism) were inoculated into the center of an MH agar plate (1% [wt/vol] Eiken agar) supplemented with appropriate antibiotics and incubated under aerobic conditions at 20 to 37°C. Cargo transport assays with the Ctx series of E. coli strains (see Table S1 in the supplemental material) were performed in the same way with 50 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) to induce the TEM-1 BL (22). Selective plating of wild-type P. vortex was on MH agar containing 1 µg ml−1 kanamycin and 0.5 mM of the swarming inhibitor PNPG with incubation at 20°C for 25 h. Selective plating of P. vortex Rifr was on MH agar with 3 µg ml−1 rifampin (Rif) and 1 mM PNPG (8, 21), and plates were incubated at 20°C for 25 h. Selective plating to determine the viable count of E. coli was on MH agar supplemented with 1 µg ml−1 vancomycin with incubation at 37°C for 18 h.
Passage to extinction experiments.
To assess the robustness of long-term transporter-cargo interactions, coinoculations were made as described above, but at 1 cm from the edge of a 14-cm-diameter MH agar plate containing appropriate antibiotics (see Fig. S2 in the supplemental material). Recovery of swarming bacteria was achieved by harvesting >108 CFU from the opposite side of the plate. These samples were subjected to selective viable counts to assess the numbers of cargo/transporter organisms and reinoculation on a new plate to repeat the process. The number of passages required to eliminate the cargo was assessed. In a variation of this assay, four antibiotic-containing Neosensitabs (Roscoe Diagnostics, Taastrup, Denmark) were placed between inoculation and recovery points to create a continuous region of antibiotic-impregnated agar that had to be traversed to reach the recovery point.
Enrichment and assays with P. vortex subpopulations.
Both explorer and builder subpopulations were enriched as previously described (17). Briefly, enrichment for builders was by growth in 2 ml LB overnight at 37°C. The overnight culture was diluted 1:100 into fresh LB and grown for 4 h; serial cultivation was repeated for three more passages. For explorers, cultures of P. vortex were cultured in LB containing 60 µg ml−1 kanamycin for 18 h. Verification of the two enrichment procedures is given in Fig. S3 in the supplemental material.
MIC determinations.
MIC values for antibiotics were determined on MH agar (1% [wt/vol] Eiken agar) containing a 2-fold dilution series of the appropriate antibiotic, using EUCAST breakpoints and procedures to determine sensitivity and resistance (Clinical breakpoints for bacteria, version 3.1, 11 February 2013; http://www.Eucast.org/clinical_breakpoints/).
Staining and imaging of swarming cells.
Staining cells with the fluorogenic dyes hexidium iodide and Syto 9 was also used to distinguish P. vortex from Gram-negative cargo bacteria. To stain moving colonies, 4-mm-diameter areas of agar 1 cm ahead of the moving colony were impregnated with 30 µM Syto 9 and 15 µM hexidium iodide (Life Technologies, Bleiswijk, Netherlands) in a 10-µl aliquot of sterile water and allowed to dry into the agar. Bacteria were stained as they swarmed into this region. Imaging was done with a BX-41 fluorescence microscope equipped with a Kappa 8 b, black and white charge-coupled-device camera and 4× and 10× Fluorotar objective lenses (8). Swarming on MH agar plates was monitored with a Dino-Lite AM7031MT USB microscope with Dinocapture software (version 1.3.6K) and by imaging plates on a heated slide warmer at 37°C through the agar. Illumination of whole plates to detect GFP expression was performed with a 470-nm LED (Thorlabs, Dachau, Germany). Calculations of swarm rates and colony diameters were made using the ImageJ software package, version 1.45s (44).
Isolation of environmental microorganisms.
Arable and woodland soil samples from locations within 1 km of Utrecht (Netherlands) or within 1 km of Rotterdam harbor (Netherlands) were screened for cargo bacteria that could assist P. vortex in colonizing plates containing BLAs (3 µg ml−1 Ctx or 2 µg ml−1 Mer). Soil (5 g) was soaked in 50 ml MH broth for 16 h. After settling, samples of the supernatant were mixed with equal volumes of P. vortex (108 CFU in a 50-µl inoculum) in MH broth and incubated at room temperature for 3 days on 14-cm-diameter MH agar plates containing the appropriate BLA. Where there was migration toward the plate edge, the bacteria were isolated and streaked to single colonies on MH agar containing the same antibiotic and 1 mM PNPG to inhibit swarming. Isolates that were not P. vortex (on the basis of swarming behavior and colony morphology) were identified by 16S sequencing (45). Purified strains were tested for antibiotic resistance (MIC, ability to swarm alone, and ability to complement P. vortex on MH agar containing Ctx or Mer, as appropriate). BL activity was assessed by nitrocefin hydrolysis (Fluka, St. Louis, USA). A general bioassay for the hydrolysis of Ctx was performed, based on the growth of sensitive satellite colonies around the resistant strain. An indicator strain, a BL-sensitive strain of E. coli expressing GFP (VL001), was spread plated (ca. 106 CFU per plate) on MH agar containing 3 µg ml−1 Ctx. The candidate BL-producing strains were then streaked on the same plates. Those isolates that promoted the growth of the indicator strains within a few millimeters, after 2 days at 25°C, were scored as BLA degrading (see Table S4 in the supplemental material).
SUPPLEMENTAL MATERIAL
Periodicity of colony patterning with transport of Ampr E. coli (VL001) by P. vortex on MH agar plates containing different concentrations of ampicillin. Each data point is the average of three measurements ± the standard deviation. For ampicillin concentrations below 10 µg ml−1, swarming was continuous. Download
Sampling from dispersing consortia of P. vortex and E. coli on 14-cm-diameter MH agar plates in the absence (Amp −) or the presence of 200 µg/ml (Amp +). For each sampling region, the percentage of cargo bacteria, determined by selective viable count, is given as the average ± the standard deviation (5 plates, 10 samples per zone per plate). The sampling zones were as follows: (A) edge of 6-cm-radius colony, sampling within 1 cm of the lead colonies; (B) immediate interior (1 to 2 cm from the edge); (C) interior, including the inoculation point. Download
Experimental setup for passage to extinction experiments on 14-cm-diameter MH agar plates. (A) Inoculation point for P. vortex and/or cargo bacteria. (B) Agar which may contain BLA or other antibiotics (for example, specifically targeting transporter or cargo bacteria). (C) Optional barrier of antibiotic-containing tablets. (D) Recovery point where bacteria derived from those bacteria inoculated on the plates in panel A were isolated and then reinoculated on a new plate, allowing serial culture under continuous (antibiotic on plate) or intermittent (antibiotic-containing tablet barrier; see Fig. 1d in the main text) antibiotic-mediated stress. Download
Explorers and builders after enrichment. The two phenotypic subpopulations that make up a spreading colony of P. vortex were enriched, as described previously (D. Roth et al., 2013). The purified subpopulations were subsequently used in transport assays (see Fig. 4 in the main text). Builders are characterized by decreased spreading abilities and increased growth rates compared to the mixed culture. Explorers have a decreased growth rate but increased swarming motility. To verify subpopulation enrichment (>98% purity) from a mixed population (30% explorers), equal numbers of cells were inoculated on LB 1.5% (wt/vol) agar. (a) Demonstration of the explorer phenotype compared to that of the mixed population after 15 h at 37°C, following staining with Coomassie blue. (b) Builders compared with mixed culture (by viable count) after 15 h at 37°C. Scale bar, 6 mm applied to the two panels comprising part a, and 125 µm when applied to either panel in part b. Download
Laboratory strains used in this study
Characteristics of transporting and nontransporting P. vortex colonies
Clinical strains tested as cargo
Summary of soil isolates that complement P. vortex in crossing BLA-containing agar (isolates were obtained from soil and identified by 16S rRNA sequencing)
ACKNOWLEDGMENTS
This work was funded in part by EU FP7 program EvoTAR (C.J.I.), the Tauber Family Funds, the Maguy-Glass Chair in Physics of Complex Systems at Tel Aviv University, by the NSF Center for Theoretical Biological Physics (grant no. PHY-1308264), and the Cancer Prevention and Research Institute of Texas at Rice University (E.B.J.).
The funders played no direct role in this work and were not involved in the decision to publish.
Thanks to Arjan van de Visser for strains and Rob Willems and Willem van Shaik for the supply of antibiotics.
Footnotes
Citation Finkelshtein A, Roth D, Ben Jacob E, Ingham CJ. 2015. Bacterial swarms recruit cargo bacteria to pave the way in toxic environments. mBio 6(3):e00074-15. doi:10.1128/mBio.00074-15.
REFERENCES
- 1.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barak JD, Gorski L, Liang AS, Narm KE. 2009. Previously uncharacterised Salmonella enterica genes required for swarming play a role in seedling colonization. Microbiology 155:3701–3709. doi: 10.1099/mic.0.032029-0. [DOI] [PubMed] [Google Scholar]
- 3.Mobley HL, Belas R, Lockatell V, Chippendale G, Trifillis AL, Johnson DE, Warren JW. 1996. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect Immun 64:5332–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venturi V, Bertani I, Kerényi A, Netotea S, Pongor S. 2010. Coswarming and local collapse: quorum sensing conveys resilience to bacterial communities by localizing cheater mutants by Pseudomonas aeruginosa. PLoS One 5:e9998. doi: 10.1371/journal.pone.0009998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, Ma Q, Yi H, Wang L, Song H, Yuan Y-J. 2011. Metabolome profiling reveals metabolic cooperation between Bacillus megaterium and Ketogulonicigenium vulgare during induced swarm motility. Appl Environ Microbiol 77:7023–7030. doi: 10.1128/AEM.05123-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowbotham TJ. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoeba. J Clin Pathol 33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brock DA, Douglas TE, Queller DC, Strassmann JE. 2011. Primitive agriculture in a social amoeba. Nature 469:393–398. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Jacob E, Cohen I, Golding I, Gutnick DL, Tcherpakov M, Helbing D, Ron IG. 2000. Bacterial cooperative organization under antibiotic stress. Physica A 282:247–282. doi: 10.1016/S0378-4371(00)00093-5. [DOI] [Google Scholar]
- 9.Ingham CJ, Ben Jacob E. 2008. Swarming and complex pattern formation in Paenibacillus vortex studied by imaging and tracking cells. BMC Microbiol 8:36. doi: 10.1186/1471-2180-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingham CJ, Kalisman O, Finkelshtein A, Ben-Jacob E. 2011. Mutually facilitated dispersal between the nonmotile fungus Aspergillus fumigatus and the swarming bacterium P. vortex. Proc Natl Acad Sci U S A 108:19731–19736. doi: 10.1073/pnas.1102097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy GB. 1950. A unit of penicillinase. Nature 166:740–741. doi: 10.1038/166740a0. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Bae IK, Lee SH. 2012. New definitions of extended-spectrum β-lactamase conferring worldwide emerging antibiotic resistance. Med Res Rev 32:216–232. doi: 10.1002/med.20210. [DOI] [PubMed] [Google Scholar]
- 13.Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, Le Roux F, Mincer T, Polz MF. 2012. Ecological populations of bacteria act as socially cohesive units of antibiotic resistance production and resistance. Science 337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 14.Vetsigian K, Jajoo R, Kishony R. 2011. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol 9:e1001184. doi: 10.1371/journal.pbio.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler MT, Wang Q, Harshey RM. 2010. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A 107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth D, Finkelshtein A, Ingham C, Helman Y, Sirota-Madi A, Brodsky L, Ben-Jacob E. 2013. Identification and characterisation of a highly motile and antibiotic refractory subpopulation involved in the expansion of swarming colonies of Paenibacillus vortex. Environ Microbiol 15:2532–2544. doi: 10.1111/1462-2920.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benisty S, Ben-Jacob E, Ariel G, Be’er A. 2015. Antibiotic-induced anomalous statistics of collective bacterial swarming. Phys Rev Lett 114:018105. doi: 10.1103/PhysRevLett.114.018105. [DOI] [PubMed] [Google Scholar]
- 20.Kim W, Surette MG. 2003. Swarming populations of Salmonella represent a unique physiological state coupled to multiple mechanisms of antibiotic resistance. Biol Proced Online 5:189–196. doi: 10.1251/bpo61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams FD. 1973. Abolition of swarming of Proteus by p-nitrophenyl glycerin: general properties. Appl Microbiol 25:745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schenk MF, Szendro IG, Krug J, de Visser JA. 2012. Quantifying the adaptive potential of an antibiotic resistance enzyme. PLoS Genet 8:e1002783. doi: 10.1371/journal.pgen.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirota-Madi A, Olender T, Helman Y, Brainis I, Finkelshtein A, Roth D, Hagai E, Leshkowitz D, Brodsky L, Galatenko V, Nikolaev V, Gutnick DL, Lancet D, Ben-Jacob E. 2012. Genome sequence of the pattern-forming social bacterium Paenibacillus dendritiformis C454 chiral morphotype. J Bacteriol 194:2127–2128. doi: 10.1128/JB.00158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shklarsh A, Finkelshtein A, Ariel G, Kalisman O, Ingham C, Ben-Jacob E. 2012. Collective navigation of cargo-carrying swarms. Interface Focus 2:786–798. doi: 10.1098/rsfs.2012.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ariel G, Shklarsh A, Kalisman O, Ingham C, Ben-Jacob E. 2013. From organized internal traffic to collective navigation of bacterial swarms. New J Phys 15:125019. doi: 10.1088/1367-2630/15/12/125019. [DOI] [Google Scholar]
- 26.Favre-Bonte S, Joly B, Forestier C. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect Immun 67:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page MI, Badarau A. 2008. The mechanisms of catalysis by metallo β-lactamases. Bioinorg Chem Appl doi: 10.1155/2008/576297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naas T, Bellais S, Nordmann P. 2003. Molecular and biochemical characterisation of a carbapenem-hydrolysing β-lactamase from Flavobacterium johnsoniae. J Antimicrob Chemother 51:267–273. doi: 10.1093/jac/dkg069. [DOI] [PubMed] [Google Scholar]
- 29.Higgins CS, Murtough SM, Williamson E, Hiom SJ, Payne DJ, Russell AD, Walsh TR. 2001. Resistance to antibiotics and biocides among non-fermenting gram-negative bacteria. Clin Microbiol Infect 7:308–315. doi: 10.1046/j.1198-743x.2001.00253.x. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Jacob E, Shmueli H, Shochet O, Tenenbaum A. 1992. Adaptive self-organization during growth of bacterial colonies. Physica A 187:378–424. doi: 10.1016/0378-4371(92)90002-8. [DOI] [Google Scholar]
- 31.Ben-Jacob E, Schochet O, Tenenbaum A, Cohen I, Czirók A, Vicsek T. 1994. Generic modelling of cooperative growth patterns in bacterial colonies. Nature 368:46–49. doi: 10.1038/368046a0. [DOI] [PubMed] [Google Scholar]
- 32.Czirók A, Ben-Jacob E, Cohen I, Vicsek T. 1996. Formation of complex bacterial colonies via self-generated vortices. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 54:1791–1801. doi: 10.1103/PhysRevE.54.1791. [DOI] [PubMed] [Google Scholar]
- 33.Sirota-Madi A, Olender T, Helman Y, Ingham C, Brainis I, Roth D, Hagi E, Brodsky L, Leshkowitz D, Galatenko V, Nikolaev V, Mugasimangalam RC, Bransburg-Zabary S, Gutnick DL, Lancet D, Ben-Jacob E. 2010. Complete genome of the Paenibacillus vortex reveals grounds for thriving in complex environments and for advanced social behavior. BMC Genomics 11:710. doi: 10.1186/1471-2164-11-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandermeer J, Yitbarek S. 2012. Self-organized spatial pattern determines biodiversity in spatial competition. J Theor Biol 300:48–56. doi: 10.1016/j.jtbi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Wennekamp S, Mesecke S, Nédélec F, Hiiragi T. 2013. A self-organization framework for symmetry breaking in the mammalian embryo. Nat Rev Mol Cell Biol 14:452–459. doi: 10.1038/nrm3602. [DOI] [PubMed] [Google Scholar]
- 36.Hermsen R, Deris JB, Hwa T. 2012. On the rapidity of antibiotic resistance evolution facilitated by a concentration gradient. Proc Natl Acad Sci U S A 109:10775–10780. doi: 10.1073/pnas.1117716109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. 2013. Salmonella typhimurium intercepts Escherichia coli signalling to enhance antibiotic tolerance. Proc Natl Acad Sci U S A 110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 39.Hagai E, Dvora R, Havkin-Blank T, Zelinger E, Porat Z, Schulz S, Helman Y. 2014. Surface-motility induction, attraction and hitchhiking between bacterial species promote dispersal on solid surfaces. ISME J 8:1147–1151 doi: 10.1038/ismej.2013.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datta MS, Korolev KS, Cvijovic I, Dudley C, Gore J. 2013. Range expansion promotes cooperation in an experimental microbial metapopulation. Proc Natl Acad Sci U S A 110:7354–7359. doi: 10.1073/pnas.1217517110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brenner K, Arnold FH. 2011. Self-organization, layered structure, and aggregation enhance persistence of a synthetic biofilm consortium. PLoS One 6:e16791. doi: 10.1371/journal.pone.0016791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eldar A. 2011. Social conflict drives the evolutionary divergence of quorum sensing. Proc Natl Acad Sci U S A 108:13635–13640. doi: 10.1073/pnas.1102923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budding AE, Ingham CJ, Bitter W, Vandenbroucke-Grauls CM, Schneeberger PM. 2009. The Dienes phemonenon: competition and territoriality in swarming Proteus mirabilis. J Bacteriol 191:3892–3900. doi: 10.1128/JB.00975-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingham CJ, Sprenkels A, Bomer JG, Molenaar D, van den Berg A, van Hycklama Vlieg JET, de Vos WM. 2007. The million well petri dish: highly subdivided microbial growth chip constructed on a porous ceramic support. Proc Natl Acad Sci U S A 46:18217–18222. doi: 10.1073/pnas.0701693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Periodicity of colony patterning with transport of Ampr E. coli (VL001) by P. vortex on MH agar plates containing different concentrations of ampicillin. Each data point is the average of three measurements ± the standard deviation. For ampicillin concentrations below 10 µg ml−1, swarming was continuous. Download
Sampling from dispersing consortia of P. vortex and E. coli on 14-cm-diameter MH agar plates in the absence (Amp −) or the presence of 200 µg/ml (Amp +). For each sampling region, the percentage of cargo bacteria, determined by selective viable count, is given as the average ± the standard deviation (5 plates, 10 samples per zone per plate). The sampling zones were as follows: (A) edge of 6-cm-radius colony, sampling within 1 cm of the lead colonies; (B) immediate interior (1 to 2 cm from the edge); (C) interior, including the inoculation point. Download
Experimental setup for passage to extinction experiments on 14-cm-diameter MH agar plates. (A) Inoculation point for P. vortex and/or cargo bacteria. (B) Agar which may contain BLA or other antibiotics (for example, specifically targeting transporter or cargo bacteria). (C) Optional barrier of antibiotic-containing tablets. (D) Recovery point where bacteria derived from those bacteria inoculated on the plates in panel A were isolated and then reinoculated on a new plate, allowing serial culture under continuous (antibiotic on plate) or intermittent (antibiotic-containing tablet barrier; see Fig. 1d in the main text) antibiotic-mediated stress. Download
Explorers and builders after enrichment. The two phenotypic subpopulations that make up a spreading colony of P. vortex were enriched, as described previously (D. Roth et al., 2013). The purified subpopulations were subsequently used in transport assays (see Fig. 4 in the main text). Builders are characterized by decreased spreading abilities and increased growth rates compared to the mixed culture. Explorers have a decreased growth rate but increased swarming motility. To verify subpopulation enrichment (>98% purity) from a mixed population (30% explorers), equal numbers of cells were inoculated on LB 1.5% (wt/vol) agar. (a) Demonstration of the explorer phenotype compared to that of the mixed population after 15 h at 37°C, following staining with Coomassie blue. (b) Builders compared with mixed culture (by viable count) after 15 h at 37°C. Scale bar, 6 mm applied to the two panels comprising part a, and 125 µm when applied to either panel in part b. Download
Laboratory strains used in this study
Characteristics of transporting and nontransporting P. vortex colonies
Clinical strains tested as cargo
Summary of soil isolates that complement P. vortex in crossing BLA-containing agar (isolates were obtained from soil and identified by 16S rRNA sequencing)