COMMENTARY
Almeida et al. (1) have interrogated the genomes of two Pneumocystis species, Pneumocystis jirovecii and its sister Pneumocystis carinii, for genes known to be involved in sexual reproduction in the widely studied fission yeast Schizosaccharomyces pombe, with the hope that defining genetic pathways governing sexual reproduction in Pneumocystis will inform disease prevention strategies. Pneumocystis spp. cause host-specific lung infections in mammals, and sexual reproductive propagules appear to be the infectious stage of the life cycle (2). P. jirovecii, a genetically intractable obligate human pathogen, causes pneumonia in immunosuppressed individuals, with an estimated 400,000 life-threatening infections reported annually worldwide and a mortality rate of up to 80% (3). P. carinii inhabits the lungs of rats (4).
P. carinii and relatives were long thought to be protozoan parasites until molecular phylogenetic analysis (1988) clearly placed them within the ascomycetes (5, 6), together with baker’s yeast (Saccharomyces cerevisiae), the human pathogen Candida albicans (in the subphylum Saccharomycotina), the human pathogen Coccidioides immitis, pricey European truffles (Tuber spp.) and morels (Morchella spp.), and familiar contemporary genetic models, such as the saprobes Neurospora spp. and the destructive cereal pathogens Cochliobolus heterostrophus and Fusarium graminearum (all Pezizomycotina). Although in the same phylum, Pneumocystis is only distantly related to these other fungi. In fact, it is associated with a diverse group of ancient lineages at the base of the ascomycete phylogenetic tree collectively known as the Taphrinomycotina (7, 8). The Taphrinomycotina include, in addition to the Pneumocystis mammalian pathogens, Taphrina deformans, a dimorphic plant pathogen that causes leaf curl disease of peach, and S. pombe, used in the fermentation of millet beer and a genetic model second only to S. cerevisiae (9, 10). Molecular requirements for S. pombe sexual reproduction were elucidated more than 25 years ago (11).
Unlike S. pombe and T. deformans, Pneumocystis species are obligate pathogens and thus cannot be cultured. This element complicates the study of Pneumocystis biology, including its possible sexual cycle, and is challenging from a clinical perspective, because sex is thought to play a crucial role in the survival of Pneumocystis. Only the cysts, which are considered to be asci containing the sexual spores, are infectious and able to spread to new hosts (2). Despite the crucial potential importance of sex to the epidemiology of Pneumocystis pneumonia, little is known about molecular mechanisms associated with this developmental pathway in Pneumocystis. Earlier studies hinting at a sexual lifestyle include a report on the possible observation of synaptonemal complexes (12), a report identifying conserved mating and meiotic genes that are functional when heterologously expressed in S. pombe mutants (13), and evidence that the meiotic recombinase Dmc1 is expressed in cysts (14). The study by Almeida et al. (1) offers significant insight into the mechanism by which sexual reproduction might occur in Pneumocystis.
Almeida et al. (1) queried genome sequences of P. jirovecii, P. carinii, and their relative T. deformans with genes known to be involved in sexual reproduction in S. pombe and identified candidate homologs. Mating in S. pombe is controlled by the single mating type locus mat1 and is successful when strains of opposite mating type, designated P and M, pair. P and M cells differ in gene content at mat1 (15, 16). Furthermore, as with the budding yeast, S. cerevisiae (17, 18), S. pombe has, in addition to the active mat1 mating type locus, two linked but silent mating type loci, one containing the P and the other the M gene content. By programmed interconversion, one of the silent copies can change places with the active copy at the mat1 locus, leading to “switching” of cell type. Thus, homothallism in both yeasts refers to a change in mating type in some of the cells within a culture of a formerly uniform mating type, followed by mating of “switched” cells with “unswitched” cells within the culture, culminating in the production of sexual spores. This type of homothallism with mating type switching has not been described in Pezizomycotina to date.
Given that Pneumocystis is related to S. pombe, one might expect these fungi to have similar mating systems, but this is not what Almeida et al. (1) found. Instead, they detected a single mating type locus in the two Pneumocystis species, one or two loci in T. deformans (short contig sequences make linkage uncertain), and no silent loci (Fig. 1). This configuration indicates that these fungi are unable to switch mating type using an S. pombe-type mechanism. Also, the Pneumocystis and Taphrina mating type loci contain both P and M mating type genes, an arrangement, denoted as primary homothallism, known to enable selfing in Pezizomycotina. Where it has been examined carefully in Pezizomycotina, all instances of primary homothallism arose from a genetic recombination event (and loss in some cases) between heterothallic relatives.
FIG 1 .
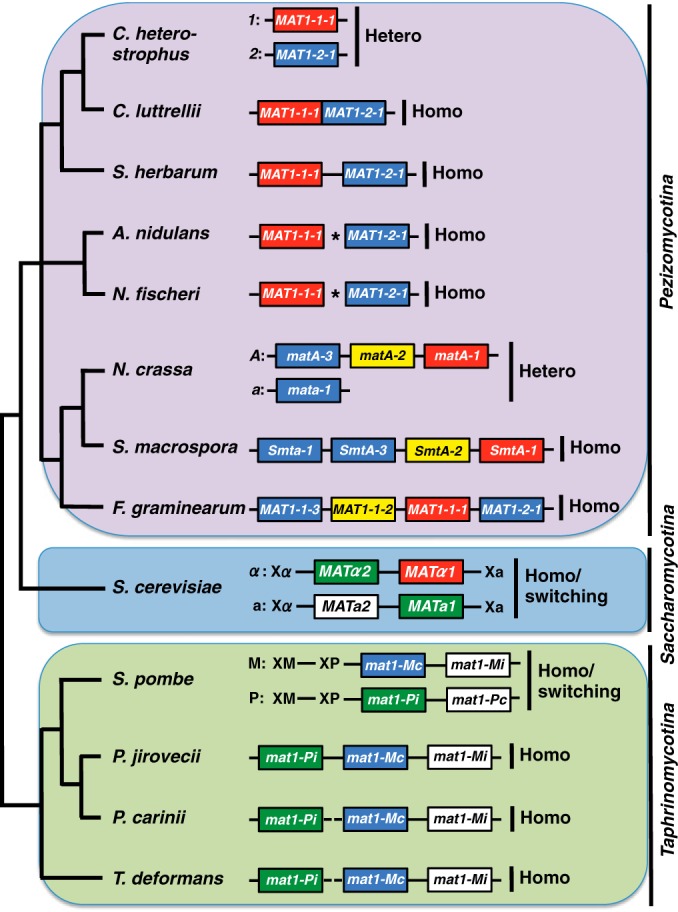
Mating type locus organization in select species mentioned in the text. The phylogenetic relationship of the species is given on the left, and mating type arrangements are on the right. Boxes correspond to mating type genes, and gene names are within the boxes. Red boxes encode alpha1 (α1) domain proteins, blue boxes high-mobility-group (HMG) box proteins, yellow boxes amphipathic alpha-helix proteins, green boxes homeobox proteins, and white boxes proteins with other or unknown domains. An “X” signifies a silent mating type locus. Horizontal lines between the boxes correspond to noncoding regions or non-mating type genes. Dashed lines indicate unknown DNA sequence, and an asterisk between boxes means that mating type genes are unlinked. For species with more than one mating type allele, both alleles, including allele designations, are provided. Gene diagrams are not to scale. “Hetero” stands for heterothallic, “Homo” for primary homothallic, and “Homo/switching” for homothallic by switching. Ascomycete subphyla are indicated by vertical lines on the right. For references, see the text. The phylogenetic topology is based on the work of Schoch et al. (42). S. herbarum, Stemphylium herbarum; S. macrospora, Sordaria macrospora; N. crassa, Neurospora crassa.
Evidence for primary homothallism is new to the Taphrinomycotina, but homothallism was inferred previously in population genetics studies which demonstrated widespread clonality in P. jirovecii (19, 20). As noted above, primary homothallism has been observed in the largest group of ascomycetes, the Pezizomycotina. Examples include the mostly saprobic, but sometimes opportunistic, human pathogens Aspergillus nidulans (21, 22) and Neosartorya fischeri (23, 24) and various plant pathogens and saprobes. One of the best-studied examples is Cochliobolus spp. All heterothallic Cochliobolus species have a single mating type locus (MAT1), with a single gene, either MAT1-1 or MAT1-2, and only isolates that differ at MAT1 are able to mate (Fig. 1). Like Pneumocystis, the primary homothallic Cochliobolus species have both mating type genes in their genomes, generally arranged side by side at a single locus (25). Functional analyses involving swapping of heterothallic for homothallic MAT genes and of homothallic for heterothallic MAT genes demonstrate that mating lifestyle can be altered by an exchange of MAT genes. Heterothallic C. heterostrophus can be rendered homothallic by introduction of the homothallic Cochliobolus luttrellii MAT genes, and homothallic C. luttrellii can be rendered heterothallic by introduction of the heterothallic C. heterostrophus MAT genes (25, 26). We note that homothallism without switching using silent mating type cassettes has also been described in the Saccharomycotina. Examples include strains of predominantly heterothallic C. albicans that become capable of self-mating through alterations in pheromone signaling (27) and the recently described novel switching mechanism in Hansenula polymorpha, in which only one of two linked MAT1 and MAT2 genes is expressed in a single nucleus (28, 29). Homothallism in H. polymorpha is achieved by a chromosomal inversion of the MAT region.
How do primary homothallic fungi evolve? The origin of the mating type gene arrangement in Pneumocystis and Taphrina is unknown, because these are the first and only MAT configurations described, but there is evidence in the Pezizomycotina that primary homothallic species originated from heterothallic ancestors by means of recombination between DNA motifs shared between opposite mating type alleles (25, 30–33). Opposite mating type alleles differ in DNA sequence, but when, for example, both MAT1-1 and MAT1-2 of heterothallic C. heterostrophus are aligned with the fused MAT1-1/MAT1-2 sequence of primary homothallic C. luttrellii, all sequences are identical across an 8-nucleotide stretch that, in C. luttrellii, is located at the fusion junctions between opposite mating type alleles. This suggests that recombination between MAT genes of a Cochliobolus heterothallic ancestor resulted in the fused mating type arrangement found in C. luttrellii today. For some primary homothallic representatives, no recombination sites have been identified, and in some, only one MAT gene is present (e.g., Neurospora africana has only matA [MAT1], while Huntiella moniliformis has only MAT2) (30, 34–41). In the genomes of other primary homothallic ascomycetes and possibly in T. deformans, the opposite mating type alleles are unlinked. This configuration can be explained by hypothesizing that heterothallic MAT genes are first linked by recombination and then rendered unlinked via a double-strand break between the linked MAT genes and a chromosomal translocation event. Examples include A. nidulans (22), N. fischeri (24), and possibly one species of Cochliobolus (25).
How the primary homothallic mating type arrangement in Pneumocystis evolved is unknown. The P and M mat1 genes are present on the same Pneumocystis chromosome; thus, recombination between the mat genes in an as-yet-undiscovered heterothallic ancestor is the most likely mechanistic scenario. It is curious, however, that the Pneumocystis mating type genes are more closely related to their homologs in T. deformans than to homologs in S. pombe (1) (Fig. 1), because Pneumocystis is more closely related to S. pombe than to T. deformans (7, 10). This suggests that the mating type arrangement of Pneumocystis and T. deformans may have evolved following the separation of these two lineages and then was transferred horizontally from one lineage to the other, as demonstrated for Stemphylium MAT genes (30). Alternatively, the mating type arrangement of Pneumocystis may have evolved before the separation of the Taphrinomycotina lineages. The S. pombe silent mating type cassettes used to effect switching and homothallism may have been acquired later (Fig. 1).
In conclusion, the evidence generated by Almeida et al. (1) and the body of genetic and phylogenetic evidence from the study of other ascomycete species, strongly suggest that the examined Pneumocystis species are primary homothallic species. Whether this is the case for all Pneumocystis species and how primary homothallism evolved in Pneumocystis and the Taphrinomycotina, in general, require additional studies. Given the clinical importance of Pneumocystis and the plant-pathogenic nature of Taphrina, molecular understanding of their reproductive strategies and evolutionary trajectory may have substantial practical implications.
The views expressed in this Commentary do not necessarily reflect the views of this journal or of ASM.
Footnotes
Citation Inderbitzin P, Turgeon BG. 2015. Pondering mating: Pneumocystis jirovecii, the human lung pathogen, selfs without mating type switching, in contrast to its close relative Schizosaccharomyces pombe. mBio 6(3):e00583-15. doi:10.1128/mBio.00583-15.
REFERENCES
- 1.Almeida JMGCF, Cissé OH, Fonseca Á, Pagni M, Hauser PM. 2015. Comparative genomics suggests primary homothallism of Pneumocystis species. mBio 6(1):e02250-14 doi: 10.1128/mBio.02250-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez A, Halliez MCM, Aliouat EM, Chabé M, Standaert-Vitse A, Fréalle E, Gantois N, Pottier M, Pinon A, Dei-Cas E, Aliouat-Denis C-M. 2013. Growth and airborne transmission of cell-sorted life cycle stages of Pneumocystis carinii. PLoS One 8:e79958. doi: 10.1371/journal.pone.0079958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Frenkel JK. 1976. Pneumocystis jiroveci n. sp. from man: morphology, physiology, and immunology in relation to pathology. Natl Cancer Inst Monogr 43:13–30. [PubMed] [Google Scholar]
- 5.Aliouat-Denis C-M, Martinez A, Aliouat EM, Pottier M, Gantois N, Dei-Cas E. 2009. The Pneumocystis life cycle. Mem Inst Oswaldo Cruz 104:419–426. doi: 10.1590/S0074-02762009000300004. [DOI] [PubMed] [Google Scholar]
- 6.Edman JC, Kovacs JA, Masur H, Santi DV, Elwood HJ, Sogin ML. 1988. Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature 334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Leigh JW, Brinkmann H, Cushion MT, Rodriguez-Ezpeleta N, Philippe H, Lang BF. 2009. Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Mol Biol Evol 26:27–34. doi: 10.1093/molbev/msn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berbee ML, Taylor JW. 2001. Fungal molecular evolution: gene trees and geologic time, p 229–245. In McLaughlin L (ed), The mycota VII, part B. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 9.Hartmeier W, Reiss M. 2002. Production of beer and wine, p 49–65. In Osiewacz HD (ed), The mycota X industrial applications. Springer Verlag, Berlin, Germany. [Google Scholar]
- 10.Sugiyama J, Hosaka K, Suh S-O. 2006. Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas. Mycologia 98:996–1005. doi: 10.3852/mycologia.98.6.996. [DOI] [PubMed] [Google Scholar]
- 11.Kelly M, Burke J, Smith M, Klar A, Beach D. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J 7:1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto Y, Yoshida Y. 1984. Sporogony in Pneumocystis carinii: synaptonemal complexes and meiotic nuclear divisions observed in precysts. J Protozool 31:420–428. doi: 10.1111/j.1550-7408.1984.tb02989.x. [DOI] [PubMed] [Google Scholar]
- 13.Burgess JW, Kottom TJ, Limper AH. 2008. Pneumocystis carinii exhibits a conserved meiotic control pathway. Infect Immun 76:417–425. doi: 10.1128/IAI.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kutty G, Achaz G, Maldarelli F, Varma A, Shroff R, Becker S, Fantoni G, Kovacs JA. 2010. Characterization of the meiosis-specific recombinase Dmc1 of Pneumocystis. J Infect Dis 202:1920–1929. doi: 10.1086/657414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen O, Egel R. 2007. The mat genes of Schizosaccharomyces pombe: expression, homothallic switch, and silencing, p 143–157. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in fungi. ASM Press, Washington, DC. [Google Scholar]
- 16.Klar AJ. 2007. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet 41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- 17.Herskowitz I. 1989. A regulatory hierarchy for cell specialization in yeast. Nature 342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- 18.Haber JE. 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteves F, Gaspar J, Tavares A, Moser I, Antunes F, Mansinho K, Matos O. 2010. Population structure of Pneumocystis jirovecii isolated from immunodeficiency virus-positive patients. Infect Genet Evol 10:192–199. doi: 10.1016/j.meegid.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Matos O, Esteves F. 2010. Pneumocystis jirovecii multilocus gene sequencing: findings and implications. Future Microbiol 5:1257–1267. doi: 10.2217/fmb.10.75. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell RG, Chaplin AJ, Mackenzie DWR. 1987. Emericella nidulans in a maxillary sinus fungal mass. Med Mycol 25:339–341. doi: 10.1080/02681218780000401. [DOI] [PubMed] [Google Scholar]
- 22.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S JE, Birren BW. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 23.Lonial S, Williams L, Carrum G, Ostrowski M Jr, McCarthy PPM. 1997. Case report Neosartorya fischeri: an invasive fungal pathogen in an allogeneic bone marrow transplant patient. Bone Marrow Transplant 19:753–755. doi: 10.1038/sj.bmt.1700715. [DOI] [PubMed] [Google Scholar]
- 24.Rydholm C, Dyer PS, Lutzoni F. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot Cell 6:868–874. doi: 10.1128/EC.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yun SH, Berbee ML, Yoder OC, Turgeon BG. 1999. Evolution of fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci U S A 96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S-W, Yun S-H, Lee T, Turgeon BG. 2011. Altering sexual reproductive mode by interspecific exchange of MAT loci. Fungal Genet Biol 48:714–724. doi: 10.1016/j.fgb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Alby K, Bennett RJ. 2011. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci U S A 108:2510–2515. doi: 10.1073/pnas.1017234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maekawa H, Kaneko Y. 2014. Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha. PLoS Genet 10:e1004796. doi: 10.1371/journal.pgen.1004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson SJ, Byrne KP, Wolfe KH. 2014. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc Natl Acad Sci U S A 111:E4851–E4858. doi: 10.1073/pnas.1416014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. 2005. Lateral transfer of mating system in Stemphylium. Proc Natl Acad Sci U S A 102:11390–11395. doi: 10.1073/pnas.0501918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inderbitzin P, Shoemaker RA, O’Neill NR, Turgeon BG, Berbee ML. 2006. Systematics and mating systems of two fungal pathogens of opium poppy: the heterothallic Crivellia papaveracea with a Brachycladium penicillatum asexual state and a homothallic species with a B. papaveris asexual state. Can J Bot 84:1304–1326. doi: 10.1139/b06-067. [DOI] [Google Scholar]
- 32.Chitrampalam P, Inderbitzin P, Maruthachalam K, Wu B-M, Subbarao KV. 2013. The Sclerotinia sclerotiorum mating type locus (MAT) contains a 3.6-kb region that is inverted in every meiotic generation. PLoS One 8:e56895. doi: 10.1371/journal.pone.0056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woudenberg JHC, De Gruyter J, Crous PW, Zwiers L-H. 2012. Analysis of the mating-type loci of co-occurring and phylogenetically related species of Ascochyta and Phoma. Mol Plant Pathol 13:350–362. doi: 10.1111/j.1364-3703.2011.00751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glass NL, Smith ML. 1994. Structure and function of a mating-type gene from the homothallic species Neurospora africana. Mol Gen Genet 244:401–409. doi: 10.1007/BF00286692. [DOI] [PubMed] [Google Scholar]
- 35.Glass NL, Metzenberg RL, Raju NB. 1990. Homothallic Sordariaceae from nature: the absence of strains containing only the a mating type sequence. Exp Mycol 14:274–289. doi: 10.1016/0147-5975(90)90025-O. [DOI] [Google Scholar]
- 36.Pöggeler S, Risch S, Kück U, Osiewacz HD. 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beatty NP, Smith ML, Glass NL. 1994. Molecular characterization of mating-type loci in selected homothallic species of Neurospora, Gelasinospora and Anixiella. Mycol Res 98:1309–1316. doi: 10.1016/S0953-7562(09)80304-3. [DOI] [Google Scholar]
- 38.Yun SH, Arie T, Kaneko I, Yoder OC, Turgeon BG. 2000. Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fungal Genet Biol 31:7–20. doi: 10.1006/fgbi.2000.1226. [DOI] [PubMed] [Google Scholar]
- 39.Lepoint P, Renard ME, Legrève A, Duveiller E, Maraite H. 2010. Genetic diversity of the mating type and toxin production genes in Pyrenophora tritici-repentis. Phytopathology 100:474–483. doi: 10.1094/PHYTO-100-5-0474. [DOI] [PubMed] [Google Scholar]
- 40.Yun S-H, Yoder OC, Turgeon BG. 2013. Structure and function of the mating-type locus in the homothallic ascomycete, Didymella zeae-maydis. J Microbiol 51:814–820. doi: 10.1007/s12275-013-3465-2. [DOI] [PubMed] [Google Scholar]
- 41.Wilson AM, Godlonton T, van der Nest MA, Wilken PM, Wingfield MJ, Wingfield BD. 2015. Unisexual reproduction in Huntiella moniliformis. Fungal Genet Biol, in press. [DOI] [PubMed] [Google Scholar]
- 42.Schoch CL, Sung G-H, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JZ, Arzanlou M, Sybren de Hoog G, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh S-O, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O’Donnell K, Sipman H, Rogers JD, Shoemaker RA. 2009. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol 58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]


