ABSTRACT
Recent studies strongly suggest that the gene expression sustaining both normal and pathogenic bacterial growth is governed by the structural dynamics of the chromosome. However, the mechanistic device coordinating the chromosomal configuration with selective expression of the adaptive traits remains largely unknown. We used a holistic approach exploring the inherent relationships between the physicochemical properties of the DNA and the expression of adaptive traits, including virulence factors, in the pathogen Dickeya dadantii (formerly Erwinia chrysanthemi). In the transcriptomes obtained under adverse conditions encountered during bacterial infection, we explored the patterns of chromosomal DNA sequence organization, supercoil dynamics, and gene expression densities, together with the long-range regulatory impacts of the abundant DNA architectural proteins implicated in pathogenicity control. By integrating these data, we identified transient chromosomal domains of coherent gene expression featuring distinct couplings between DNA thermodynamic stability, supercoil dynamics, and virulence traits.
IMPORTANCE
We infer that the organization of transient chromosomal domains serving specific functions acts as a fundamental device for versatile adjustment of the pathogen to environmental stress. We believe that the identification of chromosomal “stress-response” domains harboring distinct virulence traits and mediating the cellular adaptive behavior provides a breakthrough in understanding the control mechanisms of bacterial pathogenicity.
INTRODUCTION
Organization of the bacterial nucleoid structure is a compromise between the requirements of about thousand-fold compaction of the DNA and unobstructed accessibility of the chromosomal loci to molecular machines governing the essential cellular functions of transcription, replication, and repair. This implies a high degree of structural organization of the chromosome. In this regard, instrumental clues were provided by recent studies demonstrating that distinct genetic loci occupy specific positions in the cell according to their physical arrangement in the chromosome (1–3). Furthermore, in Gammaproteobacteria, the order of genes was found highly conserved along the chromosomal replication origin-to-terminus (OriC-Ter) axis (4). In addition, a limited diffusion of the mRNAs from their chromosomal transcription sites was observed, suggesting a physical proximity of the sites of transcription and translation (5, 6). The corollary is that transcription factors, especially the highly abundant nucleoid-associated proteins (NAPs) diffusing from their production sites may exert graded long-range effects on gene expression both laterally and also along the OriC-Ter axis of the plectonemically interwound bacterial chromosome (4).
Bacterial NAPs both constrain the DNA supercoils facilitating the chromosome compaction and also act as global transcriptional regulators (7, 8). The composition of NAPs changes in a growth-phase-dependent manner and the dynamic alteration of chromosomal supercoil density during bacterial growth involve cross talk between the NAPs and the DNA topoisomerases (9–11), which is continuously adjusted to the metabolic state of the cell (10, 12, 13). Under the influence of environmental factors, instant changes of global supercoiling can mediate the genomic transcriptional response to a challenge (14–17). This adaptive response is optimized by cooperative binding effects of the NAPs, interacting in a quasi-continuous manner with hundreds of genomic sites of various affinities and stabilizing distinct supercoil structures in the chromosome (18–20).
In pathogenic bacteria, understanding of the concerted rearrangements of gene activities during transition from a saprophytic to pathogenic lifestyle is a fundamental problem. In principle, the various stages of the infection process can be conceived as an array of successive environmental challenges to which the bacteria need to adapt. Most of the hostile conditions encountered by bacteria within the host, including acidic and oxidative stresses, induce changes of DNA topology (21). Variation of the DNA supercoiling state in response to changing environmental conditions may serve as a signal triggering the virulence program and coordinating the global gene expression during the infection process (22, 23). Indeed, DNA topology has been shown to control expression of various virulence genes, including the fim operon in Escherichia coli and Klebsiella pneumoniae, the type III secretion system (TTSS) of Shigella flexneri (24), and the pathogenicity islands SPI-1 and SPI-2 of Salmonella enterica (25–27). In Salmonella, the DNA topology and factor for inversion stimulation (FIS)—an abundant NAP transiently expressed during early exponential growth (28)—were shown to cooperate in optimizing the virulence factor production at different infection stages. During the early stages, the TTSS that is encoded by the SPI-1 system is induced by increased negative DNA supercoiling under conditions encountered in the lumen of the mammalian gut. In contrast, both the SPI-2-encoded TTSS required for the survival of the bacterium in the hostile environment of the macrophage and also the SPI-2 gene promoters are activated under conditions of DNA relaxation found in the macrophage. Such distinct dependence on the state of DNA topology appears to be a key discriminator between these two sets of virulence factors, securing that each is produced in the correct environment and otherwise repressed.
Our recent study implicated cross talk between the DNA topology and two abundant NAPs, FIS and H-NS (i.e., heat-stable nucleoid structuring protein), in optimizing the virulence program of the plant pathogen Dickeya dadantii (21, 29–32). Expression of the pel genes essential for the virulence was found strongly decreased when the chromosomal DNA was relaxed by treatment with the DNA gyrase inhibitor novobiocin or application of environmental (oxidative and acidic) stresses, whereas FIS and H-NS distinctly modulated the response of the pel promoters to supercoiling both in vivo and in vitro. Accordingly, the virulence of both the hns and fis mutants was strongly attenuated (30, 31, 33). However, pathogenicity of D. dadantii is a multifactorial process involving numerous virulence genes scattered around the chromosome, and how their expression is coordinated during pathogenic growth remains an open question. In this study, we adopt a holistic approach integrating the data on the DNA sequence organization, supercoil dynamics, and functional expression of the D. dadantii chromosome. For this purpose, we on the one hand experimentally modulated the superhelical density of the DNA in exponentially growing D. dadantii wild-type (WT) and mutant cells lacking FIS and H-NS—the two “domainin” NAPs implicated in forming topological domains in the bacterial chromosome (34). On the other hand, we imitated the hazardous impacts of the environmental stress in batch cultures. Using transcriptomics analyses, we integrated the data on the physicochemical, topological, functional, and regulational properties of the expressed DNA sequences (35), enabling us to distinguish chromosomal domains coherently responding to stress and harboring distinct virulence determinants. We infer that transient chromosomal domains organized by distinct couplings of the physicochemical and functional properties of the DNA sequence mediate the versatile and selective expression of virulence traits facilitating the adaptation of the pathogen.
RESULTS
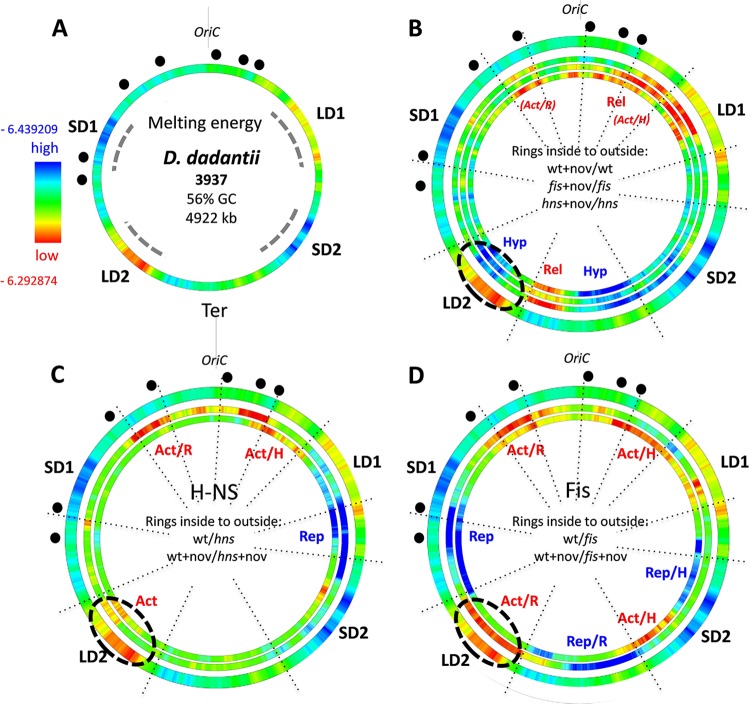
In all of the gammaproteobacterial genomes, the gene order relative to OriC is highly conserved (4) and this organization is paralleled by the average underlying physical properties of the circular DNA genome. In E. coli, it was observed that gene expression correlates with gradients of DNA supercoiling and average negative melting energy (−ΔG) extending from OriC to Ter along both replichores (4, 35). This OriC-Ter gradient of the DNA melting energy distribution is characteristic of Gammaproteobacteria in general (35). However, in the D. dadantii genome, four extended regions of high and low DNA thermodynamic stability (approximate high and low average GC content) are apparent, being organized almost symmetrically around the Ori and Ter ends of the chromosome on the left and right replichores (SD1, LD1, SD2, and LD2 in Fig. 1A). These distinct regions thus separate, as it were, the Ori and Ter ends from the rest of the chromosome (or vice versa), whereby each of the two regions of high and low thermodynamic stability face each other diagonally across the OriC-Ter axis. Similar extended regions of high and low DNA thermodynamic stability were observed in other Dickeya species, although in different spatial arrangements (see Fig. S1 in the supplemental material). This common organizational feature prompted us to ask whether the distinct genomic pattern of DNA thermodynamic stability in D. dadantii is pertinent to the organization of the gene expression patterns, as recently observed in E. coli (35).
FIG 1 .
Spatial wheels of the genomic supercoiling response of transcription. (A) Averaged distributions of negative melting energy in the genome of D. dadantii. Red and blue indicate low and high average negative melting energy, respectively. The average negative melting energy range for D. dadantii (high, −6.439209; low, −6.292874) was calculated for 500-kb windows; a much wider variation range is observed for individual genes. SD1/SD2 and LD1/LD2 represent, respectively, the two high and two low negative melting energy chromosomal regions, with extensions indicated by dashed lines. The origin of replication (OriC) is indicated. The locations of rrn genes are marked by black dots. (B) Genomic wheels showing the effect of novobiocin addition (+nov) in wild type (wt + nov/wt; 1,401 genes), fis (fis + nov/fis; 1,113 genes) and hns (hns + nov/hns; 1,105 genes) mutants arranged from inside to outside. Only the significant differentially expressed genes (P < 0.05; FC, >1.2) were considered. The genes activated and inactivated on DNA relaxation are denoted as the rel and hyp genes, respectively. The outer ring is the same as in panel A. The three inner rings show gene expression density distributions (red, high; blue, low). High gene density (red) indicates a higher proportion of genes upregulated in the first versus the second sample (wt + nov versus wt), whereas low gene density (blue) indicates a higher proportion of genes downregulated in the first versus the second sample. Blue and red in the rings indicate the significance of change (Z scores of >3). (C) Comparison of gene densities in the wild type and hns mutant under conditions of physiological levels of superhelicity (innermost ring) and DNA relaxation by novobiocin (middle ring). The outer ring is the same as in panel A. (D) Comparison of gene densities in the wild type and fis mutant under conditions of physiological levels of superhelicity (innermost ring) and DNA relaxation (middle ring). The outer ring is the same as in panel A. “Act” and “Rep” indicate regions that are, respectively, activated or repressed under the given conditions (Act/H, activated at physiological supercoiling levels; Act/R, activated on DNA relaxation; Rep/H, repressed at physiological supercoiling levels; Rep/R, repressed on DNA relaxation). For all analyzed parameters, the scanning window size was 500 kb, shifting by 4 kb and resulting in 1,230 overlapping windows.
Supercoiling response.
To assess the impact of the distinct genomic sequence organization of D. dadantii on the supercoiling response of the bacterial transcriptome, we induced chromosomal DNA relaxation by adding the DNA gyrase inhibitor novobiocin to the wild-type and fis and hns mutant strains grown in the exponential phase. From these experiments, we derived a large pool of genes (n = 2,169) that were significantly activated or repressed (either directly or indirectly) in response to DNA relaxation. These “supercoiling response” gene sets were analyzed in terms of their density distributions in the genome (see Materials and Methods), conceivably reflecting the supercoil dynamics of the D. dadantii chromosome. Comparisons of the supercoiling responses within each genetic background (intrastrain comparisons) revealed consistent spatial genomic patterns (Fig. 1B), although among these three expression profiles, only about 50% of the genes were similar (see Fig. S2 in the supplemental material). Overall we observed that the chromosomal Ori end was more activated and the Ter end more repressed in all three genetic backgrounds. More specifically, the DNA relaxation by novobiocin treatment repressed transcription (blue color) in two adjacent regions around the terminus (indicated as Hyp in Fig. 1B), one of which partially overlapped with LD2 (indicated by a dashed ellipse), whereas the region in between (indicated as Rel) was activated (shown in red). Furthermore, in wild-type cells, an extended region around OriC showed patches of activation by DNA relaxation as well, whereas in fis and hns mutants this Ori proximal activation appeared delimited to the region of the right replichore overlapping with LD1. While the Hyp and Rel clusters of supercoiling sensitivity could not be readily assigned to the genomic regions of high and low DNA thermodynamic stability, they were largely delimited to the chromosomal sectors between SD1 and LD1 in the Ori end and between LD2 and SD2 in the Ter end, whereas the chromosomal sectors between SD1 and LD2 on the left and LD1 and SD2 on the right chromosomal arm were devoid of any extended supercoiling-sensitive regions.
We next investigated the impacts of FIS and H-NS on the transcript profiles comparing the wild-type and mutant cells grown both under the normal supercoiling regimen and under conditions of DNA relaxation by novobiocin (interstrain comparisons). Again, regional differences of the expressed gene density were observed, consistent with the proposed long-range effects of the abundant NAPs on transcription (8). Overall both of the NAPs activated (red) the Ori end of the chromosome, but the effect varied, depending on the supercoiling regimen. The region on the left side of OriC was more strongly activated by both FIS and H-NS on DNA relaxation (Act/R in Fig. 1C and D). Accordingly, in the intrastrain comparisons, the relaxation of the DNA strongly activated this region in the wild type but not in mutant cells lacking the NAPs (red color of the OriC-proximal region labeled “Act/R” in Fig. 1B), suggesting that both NAPs preferentially activate this region on DNA relaxation. Conversely, a confined region in the vicinity of LD1 was preferentially activated by both NAPs at the normal physiological level of negative superhelicity (Act/H in Fig. 1C and D), and accordingly, in the intrastrain comparisons, activation of this region by DNA relaxation was least pronounced in the wild type (compare the density of red color at Act/H between the three rings in Fig. 1B), suggesting that the NAPs prevent the activation of this region on DNA relaxation. Thus, long-range effects of the NAPs distinctly modulate the supercoiling response on both sides of OriC. Furthermore, H-NS and FIS, respectively, repressed, albeit to different extents depending on the supercoiling regimen, the chromosomal regions between LD1 and SD2 on the right and between SD1 and LD2 on the left replichore (indicated as Rep in Fig. 1C and D). Notably, while this repression by H-NS and FIS was supercoiling dependent, showing, respectively, extension and shrinkage on DNA relaxation, these repressed regions showed the least conspicuous differences in the intrastrain comparisons (see the same regions in Fig. 1B). In addition, FIS repressed the chromosomal Ter end between LD2 and SD2 on DNA relaxation (Rep/R in Fig. 1D) and activated the flanking region at normal physiological negative superhelicity (Act/H to the right of Rep/R in Fig. 1D). Notably, in wild-type cells, this region also showed a stronger response to superhelicity than the fis mutant in the intrastrain comparisons, indicating that FIS is modulating the supercoiling response intensity of the Ter end (Fig. 1B). Finally, under conditions of DNA relaxation, FIS activated transcription of an extended region overlapping the thermodynamically labile region LD2, whereas H-NS activated transcription at the left edge of LD2 independent of the supercoiling regimen (Act and Act/R in Fig. 1C and D, respectively, indicated by the dashed ellipse).
Thus, while the regions showing explicit supercoiling sensitivity are roughly delimited by SD1 and LD1 at the Ori end and SD2 and LD2 at the Ter end, the regions in between (that is the regions between SD1 and LD2 and LD1 and SD2 on the left and right replichores, respectively) are repressed by FIS and H-NS, respectively, in a supercoiling-dependent manner. Taken together, these data strongly suggest that the chromosomal supercoiling response is largely coordinated by the primary sequence organization of the genomic DNA and the long-range binding effects of the NAPs in the genome.
Stress response.
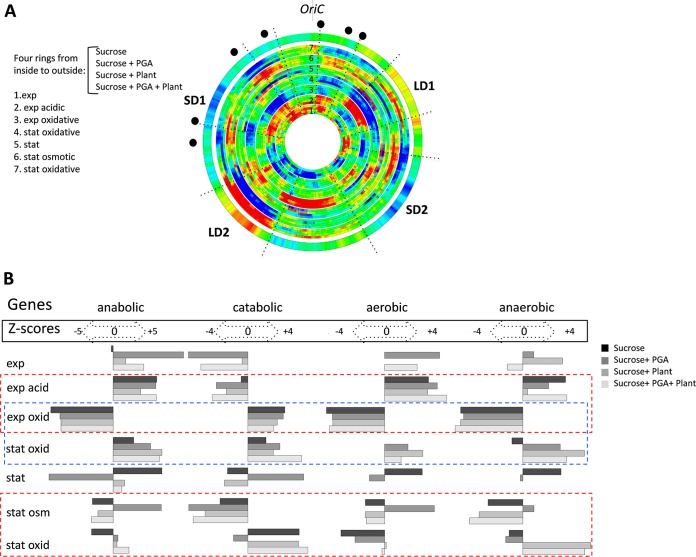
What is the relevance of these organizational features to the genomic stress response during the pathogenicity process? To answer this question, we derived transcript profiles from D. dadantii cells grown under adverse conditions of environmental stress normally encountered in the course of the infection process (see Fig. S3 in the supplemental material). We applied three distinct kinds of environmental stress (acidic, oxidative, and osmotic) during two distinct stages of growth (exponential and early stationary phases) to cells grown in four different growth media, such that for each type of stress, we ultimately obtained four biological replicates. On the chromosomal wheel shown in Fig. 2A, the expression patterns are arranged according to the applied stress in all four media (bundled in four rings corresponding to the four media used) in a sequential order (from inside to outside the wheel), corresponding to the assumed temporal order of the growth environments encountered during the infection process (indicated by the numbers 1 to 7). Thus, each pattern is compared to the preceding one to reveal the changes of stress response expected during the process of pathogenesis, as illustrated in Fig. S3. Among these, conditions 4 and 7 are identical as both correspond to oxidative stress applied in the stationary phase. However, in one case we compare the effects of the oxidative stress in stationary and exponential phases (comparison 4), whereas in another we compare the effect of oxidative stress in the stationary phase to that of the osmotic shock in the stationary phase (comparison 7). The latter comparison was carried out since the phenolic compounds released on degradation of plant tissues at the advanced stages of infection can induce oxidative stress, although D. dadantii is subject to oxidative stress primarily during the early stages of infection. Overall, we observed a remarkable variation of the gene expression densities in response to stress demonstrating chromosomal segments encompassing extended clusters of coherently activated (red) or repressed (blue) genes (Fig. 2A). Interestingly, the densities varied mostly coherently irrespective of the medium composition (see, e.g., the ring bundles 6 and 7 encompassing the LD2 region), but we observed also medium-dependent variation (see, e.g., the ring bundles 5, 6, and 7 encompassing the LD1 region). A similar pattern of gene expression density variation was observed when the chromosomal wheel was assembled by comparing each kind of stress applied either in the exponential or stationary phase to corresponding untreated samples (see Fig. S4 in the supplemental material).
FIG 2 .
Spatial genomic wheels of the D. dadantii transcriptional response to stress. (A) Gene density distributions. The wheels corresponding to each single condition are arranged in bundles of four rings (due to the four different media used: sucrose, sucrose-polygalacturonate [i.e., PGA, a pectin derivative], sucrose-plant, and sucrose-plant-PGA). The conditions (listed on the left side of the panel) are ordered from inside to outside and numbered from 1 to 7. Each of the consecutive four-ring bundles in the wheel (from inside to outside) shows the change in the density of differentially expressed genes (red, high; blue, low) compared to the preceding condition. The different stress conditions are arranged according to their assumed occurrence during the infection. The first four-ring bundle (exp, exponential phase) is compared to the stationary-phase (stat) expression. Only the significant differentially expressed genes (P < 0.05; FC, >1.2) are compiled in this representation. The outer ring shows the averaged distributions of negative melting energy in the genome. Locations of rrn genes are marked by black dots. The colors in the rings (blue and red) indicate the significance of the change (Z scores of >3). (B) Distribution of the different functional groups of genes in the stress-response patterns. The conditions are indicated in the leftward column and arranged according to the wheels in panel A. The four remaining columns represent the significant changes in the anabolic, catabolic, aerobic, and anaerobic genes for each condition. The corresponding Z scores (both positive and negative) are indicated for each functional group. Positive and negative Z scores indicate the increase and decrease of each particular functional group in the corresponding pattern. Functional gene groups were taken from the respective GO branches provided by the KEGG database. The dashed rectangles indicate the comparisons between different stresses (red) and between different growth phases with oxidative stress (blue).
When we inspected the functional content of the expressed sequences, we again observed a variable pattern (Fig. 2B). Coordinated switches of large functional classes of the anabolic, catabolic, aerobic, and anaerobic genes occurred both with a single stress during growth phase transitions (blue dashed rectangle) and also with different kinds of stress applied to cells in the same growth phase (red dashed rectangles). We infer that during the stress response, the changing dynamics of chromosomal gene expression densities are associated with coordinated switches of the metabolic profile.
Determination of the chromosomal “stress-response” domains.
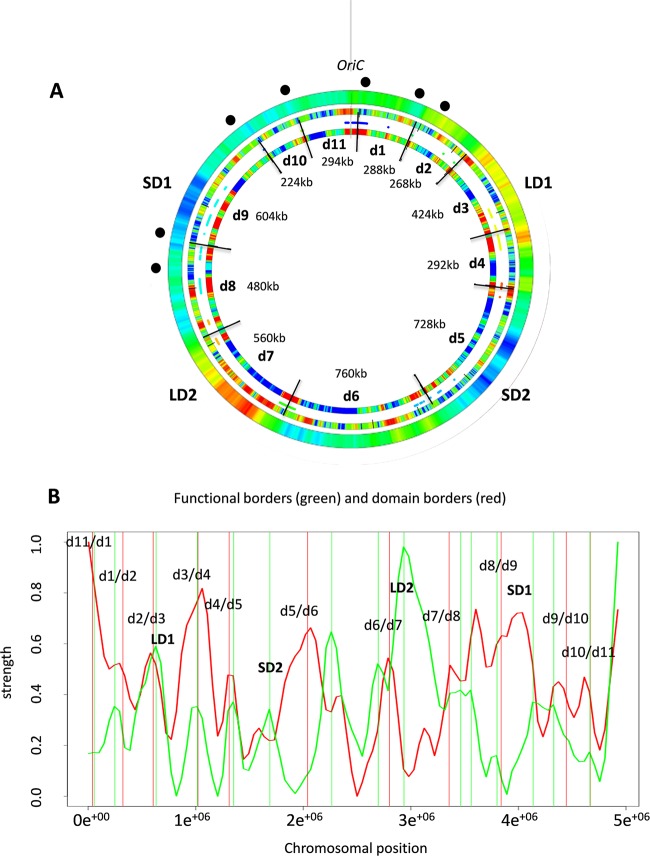
To gain insight into the device coordinating the patterns of the chromosomal gene expression density, we adopted a recently established holistic approach (35). In the entire stress-response transcriptome, we integrated different kinds of parameters determined for the expressed gene sequences, including (i) expression density, (ii) average negative melting energy, (iii) preferred superhelicity regimen of transcription, and (iv) response to the NAPs. All of these features collectively we denote here as the DNA sequence parameters. Each of these parameters formed distinct patterns in the global expression profiles analyzed under the different growth conditions. We correlated pairwise the spatial distributions of all these parameters in the genome to detect their statistically significant boundaries. For this analysis, we used the parameter distributions observed both in the wheels assembled according to the sequence of stress conditions encountered during the infection process (Fig. 2A), as well as the wheels assembled by comparisons of the stress conditions with untreated samples (see Fig. S4 in the supplemental material). The result of this analysis is shown in Fig. 3. (For full analysis of these parameters and their correlations in expression patterns see Materials and Methods and Fig. S5 to S7 in the supplemental material). We thus approximated 11 statistically significant boundaries delimiting the chromosomal “stress-response” domains numbered d1 to d11 (innermost ring in Fig. 3A). Four of these transient domains encompass the four regions of distinct thermodynamic stability observed in the D. dadantii genome. For example, d3 largely contains LD1, d5 contains SD2, d7 contains LD2, and d9 contains SD1 with the adjacent OriC proximal region, whereas d6 encompasses the Ter end of the chromosome. In the chromosomal half encompassing OriC, the “stress-response” domains are both smaller and more frequent, whereas many of the boundaries are located in close proximity to the rRNA operons (e.g., d8/d9, d9/d10, d10/d11, d11/d1, and d1/d2).
FIG 3 .
Stress-response domains. (A) Compilation of the spatial boundaries based on pairwise correlations of different DNA sequence parameters in D. dadantii genome. The outermost ring shows the average negative melting energy distributions in the genome. The locations of rrn genes are marked by black dots. The middle ring represents the static GO tree functional boundaries. Red indicates a substantial change in the functional composition of genes in the genome (35). The innermost ring shows the detected 11 stress-response boundaries delimiting 11 domains (d1 to d11). Red indicates the statistically significant change in the expressed DNA sequence parameter values. (B) Comparison of the functional borders (green vertical lines) and domain borders (red vertical lines). The change of function (green curve) and the correlation of domain borders in the expression profiles (red curve) are depicted in arbitrary units. The coincidence of both borders was tested by a random model. Only 2.4% of randomly chosen border positions show a smaller median nearest-neighbor distance between both sets of borders.
To relate these stress-response domains to genetic function, we next determined the distribution of the functionally distinct classes of metabolic genes compiled in the Gene Ontology (GO) data set (see Materials and Methods) and mapped them on the D. dadantii genome as described previously (35). The result showed that functionally related genes are separated by boundaries on the chromosome (indicated by red in the middle ring in Fig. 3A), some of which either closely match (e.g., d2/d3, d3/d4, and d4/d5), or are in the vicinity of (e.g., d6/d7, d7/d8, d9/d10, d10/d11, and d11/d1) the statistically determined borders of structural domains (Fig. 3A and B). The domain d7 encompassing LD2 is particularly enriched in functionally distinct gene classes. The observed modular organization of the functionally related genes and the transient structural domains emerging in response to stress in the chromosome suggested that the structural boundaries could delimit biologically relevant functions. However, can these transient structural-functional domains appearing in response to stress be related to the virulence program?
Coordination of the virulence program.
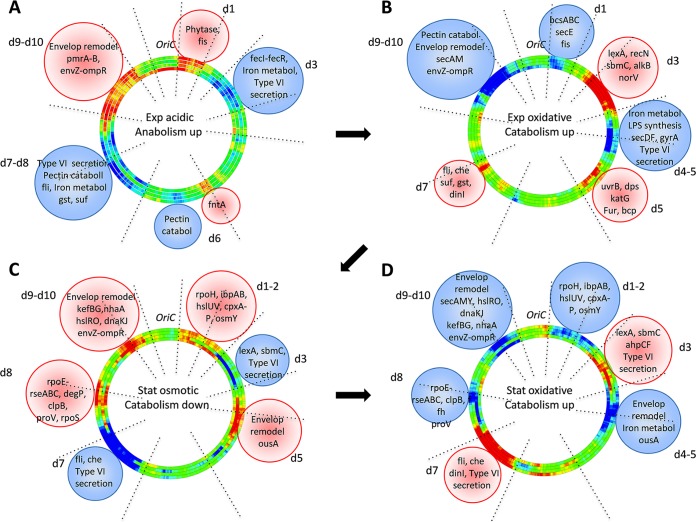
To answer this question, we analyzed the expression of different adaptation and virulence traits in the identified stress-response domains under various stress conditions. (For the list of the relevant genes in chromosomal domains, see Table S1 in the supplemental material.) The results of this analysis are shown in Fig. 4. Activation and repression of the stress-response domains are indicated on the chromosomal wheels by red and blue, respectively (as in Fig. 2A). The relevant genes in the domains are written beneath the domains harboring them (encircled within spheres of corresponding color). During acidic stress, the activated domain d1, delimited by boundaries coinciding with strong ribosomal operons (Fig. 3A), expresses phytase, which degrades phytic acid, providing access to plant phosphorus, and also expresses fis, which boosts ribosome production and growth (Fig. 4A), consistent with activation of anabolism at this stage (Fig. 2B; see Fig. S3 in the supplemental material). The activated domains d9 and d10 express genes involved in modification of lipopolysaccharides (LPS) and outer membrane permeability, leading to increased resistance to antimicrobial peptides produced by plants (36). At the same time, the type VI system secreting the Rhs proteins coordinating the multicellular behavior (37), the genes involved in pectin catabolism (38), the flagellar operon required for motility (39), and the genes involved in iron metabolism (40), are all repressed (blue color of the domains d3, d6, and d7-d8). Repression of iron metabolism under conditions of acidic stress could be explained by the increased solubility of this oligonucleotide element, which is toxic for the cells at high concentration. This pattern is fully consistent with the notion that at this stage, the cells actively multiply, presumably preparing to the stage of colonization during the late asymptomatic stage. Colonization occurs under conditions of oxidative stress (see Fig. S3) and is associated with downregulation of anabolism (see Fig. 2B). Under oxidative stress applied to exponentially growing cells, the domains (d1 and d9-d10) active under acidic stress conditions, are repressed, whereas d5, as well as d3 and d7, which are repressed during acidic stress, are activated (Fig. 4B). In these activated domains, the expression of the genes coding for protective and SOS response proteins (norV, recN, alkB, lexA, and sbmC in d3; katG, dps, bcp, and uvrB in d5; and gst and dinI in d7) is induced, whereas in the repressed domain d1, the expressions of both the growth stimulator gene fis and the cellulose biosynthesis operon bcsABC involved in bacterial aggregation and biofilm formation (not required during the stage of colonization) are inhibited. Also the iron metabolism in d4 is repressed, as iron can facilitate the generation of reactive oxygen species (ROS) via the Fenton reaction (41). Accordingly, in d5 a repressor of iron assimilation fur is activated (see Table S1), and also the suf genes involved in iron sulfur cluster assembly and repair, as well as the flagellar and chemotaxis operons required for plant invasion are induced in d7. Importantly, while the flagellum expression facilitates apoplast invasion during the colonization stage, the cell-wall-degrading enzymes are not produced (42). Indeed, the sec genes (secE in d1, secDF in d4, and secAM in d9) required for translocation of proteins across the inner membrane are repressed, and, therefore, the pectinases and cellulases cannot be secreted. Accordingly, in d11 the quorum sensing genes vfmNOP, controlling the production of plant cell-wall-degrading enzymes, are also repressed (43). Furthermore, the activation of sbmC expression (gyrase inhibitor) in d3 and the repression of gyrA (gyrase subunit A) in d4 at this stage are fully consistent with DNA relaxation observed under oxidative stress (21). Importantly, at this stage, bacteria either survive and the infection process continues, or they die, leading to abortive infection (42). If the infection progresses, bacteria enter the stationary phase and the cell-wall-degrading enzymes (pectinases PelA, -E, and -D in d8 and PelB and -C in d9, cellulase CelZ in d7, and proteases PrtA, -B, -C, and -G in d6) are produced (see Table S1). Maceration of plant tissues and cell lysis occur, and consequently, the release of vacuolar contents in apoplast creates osmotic shock conditions for the bacteria.
FIG 4 .
Stress-response domains coordinate the virulence program. The genomic wheels (represented as four-ring-bundles as in Fig. 2A) in panels A, B, C, and D show the expression densities (red, high; blue, low) of the genes in the stress-response domains. The relevant genes in the domains are written beneath the domains harboring them (encircled within spheres of the corresponding color). (A) Acidic stress applied during the exponential phase. (B) Oxidative shock applied during the exponential phase. (C) Osmotic shock applied during the stationary phase. (D) Oxidative shock applied during the stationary phase.
After colonization and during the osmotic shock in the early symptomatic phase (see Fig. S3 in the supplemental material), the cells are not expected to be motile, and indeed, we observed a repression of the flagellar and chemotaxis operons in d7 (Fig. 4C). The type VI secretion in d7 and d3 is also repressed, whereas in the activated domains d1 to 2, d5, d8, and d9-d10, the genes involved in the cell envelope stress (rpoE, rseABC, degP, and cpxP-cpxRA), heat shock response (rpoH, ibpAB, dnaKJ, clpB, hslUV, and hslRO), general stress response regulation (rpoS), and osmoprotection (osmY, ousA, proVWX, kefBG, and nhaA) are induced. Catabolism is downregulated (Fig. 2B), and at the same time, the gyrase inhibitor gene sbmC is inhibited, consistent with the high global negative superhelicity observed under conditions of osmotic shock (44).
Finally, during oxidative stress in the stationary phase (Fig. 4D) corresponding to the late symptomatic stage of infection (see Fig. S3 in the supplemental material), catabolism is upregulated (Fig. 2B), and the responding chromosomal domains are identical to those of the osmotic shock response, but the pattern of activation and repression is inverted. Indeed, at this stage, the cells are expected to be motile, as is the case during oxidative stress in the exponential phase (compare to Fig. 4B), and again, activation of the gyrase inhibitor sbmC (in d3) at this stage could facilitate global DNA relaxation.
From these analyses, we infer that the virulence program is coordinated by specific constellations of the stress-response domains that are temporarily activated or repressed during the adaptation of D. dadantii cells to changing environmental conditions.
Various combinations of DNA sequence parameters and impacts of the NAPs coordinate the gene expression in the domains.
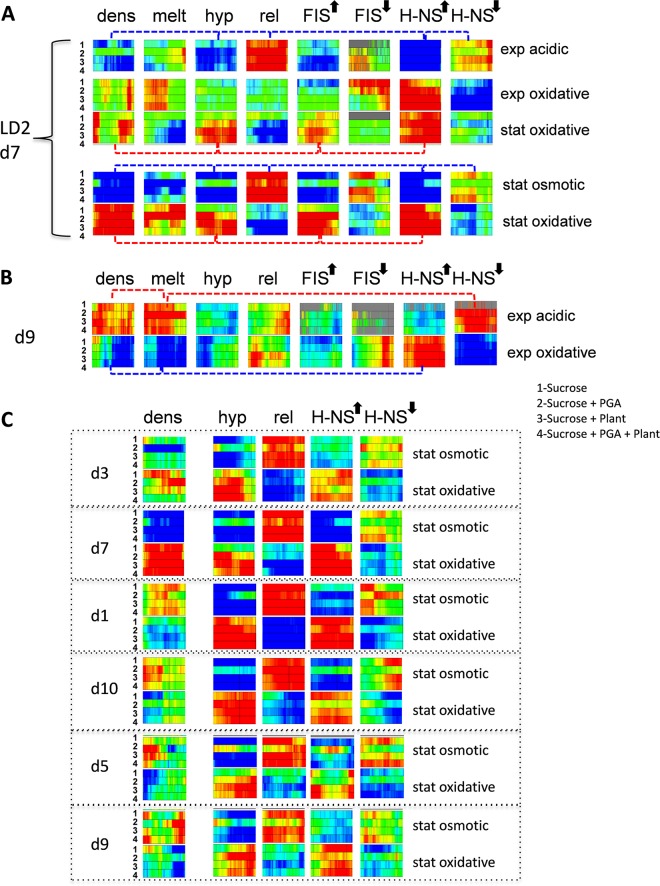
Whereas the coordinated behavior of the stress-response domains under changing environmental conditions is manifest in changing gene expression densities, our results strongly suggest that in turn, the changing gene densities in the domains are correlated with distinct constellations of the DNA sequence parameters. We illustrate this fundamental principle with the example of the GC-poor chromosomal region LD2 (largely coinciding with d7 [Fig. 3]). In Fig. 5A, the domain LD2/d7 is represented in form of squares composed of four rectangles corresponding to four different growth media numbered 1 to 4. The squares are arranged in five rows and eight columns, corresponding to the kind of applied stress and the analyzed parameter, respectively. The five stress conditions are arranged according to their assumed occurrence during the infection. The changes of color in each row of the squares indicate the difference (where red represents increase, blue decrease, and green no change) from the previous row (except for the acidic and osmotic stresses, which are compared to those with untreated cells harvested during exponential and stationary growth, respectively).
FIG 5 .
Combinations of DNA sequence parameters coordinate the gene expression densities. (A) Stress response of the domain LD2/d7. The domain is represented in the form of squares composed of four rectangles corresponding to four different growth media numbered 1 to 4 (listed on the right side of the panel). The squares are arranged in five rows corresponding to different stress conditions (arranged according to their assumed occurrence during the infection) and eight columns corresponding to the analyzed parameters. The upward and downward arrows indicate the genes, respectively, activated or repressed by the NAPs. The changes of color in each row of the squares indicate the difference (red, increase; blue, decrease; green, no change) from the previous row (except for the acidic and osmotic stresses, which are compared to those of untreated cells). The dashed connector lines indicate the combinations of parameters leading to an increased expressed gene density (red) or decreased gene density (blue). (B) Response of the chromosomal domain d9 to the acidic and oxidative stresses applied in the exponential phase. The representation is as in panel A. Couplings of the distinct DNA sequence parameters are indicated by the dashed red (for acidic stress) and blue (for oxidative stress) connectors. (C) Response of the chromosomal domains to the osmotic and oxidative stresses during the stationary phase. The representation is as in panel A. Note that the changes of gene expression density in response to oxidative stress in the two domains associated with DNA of low thermodynamic stability LD2/d7 and LD1/d3 are opposite to those observed in the OriC proximal domains d1 and d10 or the domains associated with high thermodynamic stability, SD1/d5 and SD2/d9. Note also that the response of supercoiling-sensitive genes (hyp and rel), as well as H-NS-activated (upward arrow) and -repressed (downward arrow) genes, is uniform throughout all domains. For details see the text.
We observed that the gene expression density in LD2/d7 was low with both the acidic and osmotic shocks and increased in response to oxidative stress applied in the stationary phase (compare the color code in the “dens” column). Moreover, there was an inherent coupling between the changes in gene expression density in LD2/d7 and the other measured parameters. For example, low expression density was coupled, albeit to different degrees, to certain parameters of the expressed sequences: abundance of genes activated by DNA relaxation (red in the “rel” column), scarcity of genes activated by high negative superhelicity (blue in the “hyp” column), and by H-NS (blue in the entry for “H-NS” with an upward arrow), as well as relative abundance of genes (predominance of red) that are normally repressed by H-NS (H-NS with a downward arrow). Thus, although there is a variation in the penetrance of each parameter in the particular pattern as judged by the variation in the color intensity (full red and blue indicate Z scores of >3 and >−3, respectively [see Materials and Methods]), the patterns of variation are very similar (coupled parameters are connected by blue dashed lines; see also Fig. S5 in the supplemental material). This similarity of patterns suggests that on average under both the acidic stress in exponential phase and osmotic stress in stationary phase, the low expression density in LD2/d7 is associated with derepression by H-NS (albeit to different extents) of the DNA relaxation-dependent genes and concomitant repression of the genes activated by DNA supercoiling and H-NS. Conversely, increased expression density in LD2/d7 under oxidative stress in the stationary phase was coupled to high abundance of the genes transcribed at normal superhelical density (“hyp” [red]) and albeit to different extent, an abundance (red) of the H-NS- and FIS-activated genes (coupling indicated by red dashed connectors). This suggests that under oxidative stress in the stationary phase, the increased expression density in LD2/d7 results from increased transcription of genes activated by both H-NS and FIS at physiological levels of superhelical density. Notably, we observed that LD2 is largely repressed by DNA relaxation (Fig. 1B), and indeed under oxidative stress, the rel genes (i.e., genes activated by DNA relaxation) are underrepresented (Fig. 5A, the predominantly blue color of the “rel” column in the “stat. oxidative” rows). However, we also observed that FIS counteracts the inactivation of LD2 by DNA relaxation (Act/R at LD2 in Fig. 1D). This suggests that the genes activated by FIS in LD2/d7 are essentially the genes rescued by FIS (either directly or indirectly, as the FIS concentration is assumed to be very low in stationary phase) from inactivation on global DNA relaxation (45, 46) induced by oxidative stress. In addition, the transcriptional response to oxidative stress in the stationary phase shows a preference for sequences with high average negative melting energy (predominance of blue in the “melt” column) compared to those in the exponential phase, which are nevertheless of lower average negative melting energy (red in the “melt” column) compared to sequences activated by the osmotic shock. We infer that the transcriptional response of LD2/d7 to environmental stress is both variable and selective, being mediated by distinct couplings of the DNA sequence parameters in response to different kinds of stress. Figure 5B illustrates the same principle in the example of domain d9 associated with the region of high thermodynamic stability SD1 (Fig. 3).
Pearson’s correlation analyses support this notion (see Fig. S5 in the supplemental material). Overall we find high positive correlation coefficients for the hyp- and FIS-activated genes (0.9) and reasonable correlations for FIS-repressed and H-NS-activated genes (0.7), FIS-activated and H-NS-repressed genes (0.5), as well as the FIS- and H-NS activated genes and high negative melting energy of sequences (0.6 for both). Furthermore, in the domains d3 and d7 associated with DNA of low thermodynamic stability (respectively, LD1 and LD2 [Fig. 3]), we observed that application of oxidative stress in the stationary phase increased, albeit to different degrees, the gene expression density compared to that under osmotic stress (“dens” column in Fig. 5C). In contrast, under the same conditions in the OriC-proximal domains d1 and d10, as well as in d5 and d9, associated, respectively, with thermodynamically stable DNA regions SD1 and SD2, the gene expression density rather tends to decrease (see the “dens” column for corresponding domains). Despite the apparently opposite changes of expression density on application of oxidative stress (compared to osmotic shock) in the stationary phase, in all analyzed domains, the activities of the genes transcribed at high superhelical density and DNA relaxation increased and decreased, respectively (“hyp” and “rel” columns in Fig. 5C). In addition, all of these distinct domains demonstrated increased levels of H-NS-activated genes (predominance of red in the “H-NS” column with upward arrow) and low levels of genes repressed by H-NS (predominance of the blue color in the “H-NS” column with downward arrow). We thus infer that in different domains, similar kinds of stress can induce similar couplings of the expressed DNA sequence parameters leading to opposite changes of gene expression density.
Taken together, our results suggest that in D. dadantii, the peculiar DNA sequence parameters in conjunction with regulatory effects of abundant NAPs coordinate the changes of gene expression density, forming transient structural-functional domains in response to environmental stress.
DISCUSSION
In this study, we explored the determinants of the genomic transcriptional response in adaptation of the pathogenic bacterium D. dadantii to adverse conditions normally encountered during plant infection. Environmental stress is known to elicit strong supercoiling responses (47–49), and therefore, to facilitate our investigations, we initially identified the supercoiling-sensitive pools of genes under conditions of DNA relaxation. By mapping the supercoiling-sensitive patterns in the D. dadantii genome, we detected extended chromosomal regions selectively responding to alterations of supercoiling in wild-type, fis, and hns mutant cells. The boundaries of the detected supercoiling domains are not precise, but their organization is fairly uniform in all three backgrounds. These supercoiling-sensitive gene clusters are enriched in the chromosomal Ori and Ter ends and are delimited by regions of distinct thermodynamic stability SD1/LD1 and SD2/LD2, respectively (Fig. 1B). We also detected distinct regulatory effects of FIS and H-NS in the Ori and Ter ends of the chromosome under different supercoiling regimens (Fig. 1C and D), whereby most of the activated genes were organized between SD1 and LD1 on both sides of the origin. Furthermore, the extended regions between SD1 and LD2 on the left and SD2 and LD1 on the right arm of the chromosome were devoid of extended supercoiling-sensitive clusters and repressed by FIS and H-NS, respectively. From our data, we infer that the supercoiling response of the chromosomal transcription is largely coordinated by the primary sequence organization of the DNA and the long-range NAP binding effects.
Detection of the “stress-response” domains in the D. dadantii chromosome.
To test the biological relevance of the observed supercoiling-dependent patterns, we next determined the chromosomal gene expression densities in the transcript profiles of the D. dadantii cells grown under various stress conditions. We found that different types of stress distinctly affected both the gene expression densities and the metabolic profile (Fig. 2). Notably, the composition of the growth medium did not substantially affect the expression patterns, except for the minimal medium with polygalactonurate (PGA) in the absence of the plant extract (see Fig. S5 in the supplemental material), where the highest expression of pel genes, the major virulence factors of D. dadantii, was observed (50). One possible explanation is that since transcription, translation, and insertion into the membrane are concurrent (51), the coupled massive expression and secretion of pectinases in this medium could impose physical constraints on the chromosomal dynamics due, for example, to locus repositioning toward the membrane.
To understand the organization of the stress-induced gene expression patterns, we correlated the spatial distributions of DNA sequence parameters in the genome using an established holistic approach (35). These statistical analyses revealed 11 chromosomal boundaries delimiting 11 domains. Importantly, many of these dynamic boundaries closely coincided with the genomic location of strong rRNA operons, and many were found in the vicinity of the boundaries separating functionally distinct genes (Fig. 3). The identified domains highlight distinct characteristic combinations of DNA sequence parameters and harbor different virulence determinants. We found that on the one hand, particular domains respond to different kinds of stress by utilizing distinct couplings of DNA sequence parameters leading to opposite changes in the gene expression densities, and on the other hand, we observed that different domains highlighting similar couplings of DNA sequence parameters can respond to the same kind of applied stress by an increased or decreased gene expression density. This versatility of response likely depends on the thermodynamic properties of the DNA in the domains (Fig. 5). We thus infer that the chromosome of D. dadantii is organized in distinct stress-response domains with characteristic supercoil dynamics expressing virulence determinants that are used selectively and transiently during the adaptation of cells to environmental challenge (Fig. 4). Overall, this organization is consistent with previously observed macrodomains (52), extended spatial gene clusters (53, 54), and especially, the transient structural-functional domains detected in the E. coli chromosome (35, 55). Given that the detection of the gene expression density domains involved normalization by coherence of transcription (i.e., uniformity in the direction of the transcriptional response) rather than assessments of transcriptional strength, our finding that many domain boundaries are coinciding with strongly transcribed loci (such as the ribosomal operons [Fig. 3 and 4B]) is in keeping with previous observations of highly transcribed loci separating long regions of low transcription in E. coli and Salmonella enterica serovar Typhimurium (56). We propose that the domain boundaries may represent topologically sensitive DNA regions presumably associated with strongly transcribed units. In E. coli, such boundaries are assumed to act as supercoil traps driving, according to the physiological state, the morphological alterations of the bacterial chromosome (57, 58). We infer that D. dadantii takes advantage of DNA topology fluctuations during infection to orchestrate, with the help of the NAPs, the sequential establishment of the virulence and survival factors.
Properties of the stress-response domains.
Importantly, both in E. coli and D. dadantii, the transient organization of chromosomal domains is paralleled by the average underlying physical properties of the circular genomic DNA. In E. coli, the functional significance of the gradients of gene expression and of DNA thermodynamic stability have been related to the Ori-Ter gradient of average DNA negative superhelicity from high to low in both replichores (4, 35, 58). In contrast, in D. dadantii the genomic distribution of DNA melting energy shows four regions of distinct sequence composition organized almost symmetrically around the OriC and Ter ends of the chromosome (Fig. 1A). We propose that, whereas in E. coli laboratory strains the sequence organization is largely streamlined by the requirements of the fast replication process, the more complex pattern observed in D. dadantii reflects specialization selected for by a constitutive requirement to cope with a range of variable stresses and host defense mechanisms. The corollary to this argument is that the genomes of the pathogens with a versatile lifestyle might require a higher structural variability, allowing flexible regulation of spatially separated domains in the chromosome. This requirement is fully consistent with the distinct sequence organization of the bacterial pathogenicity islands assumed to be subject to horizontal gene transfer during microbial evolution (59). Notably, the low average melting energy region LD2/d7 implicated in D. dadantii pathogenicity is shown to have acquired AT-rich DNA sequences by horizontal gene transfer (60). Furthermore, our analyses show that the thermodynamically distinct structural regions identified in D. dadantii are conserved, albeit in a distinct spatial arrangement, in other Dickeya species (see Fig. S1 in the supplemental material), corroborating their functional importance. These signatures went unnoticed until this study, thus underscoring the predictive value of our approach.
In conclusion, we present the first evidence for the determinative role of the physicochemical properties of the primary DNA sequence in organizing chromosomal stress-response domains, which harbor specific virulence/adaptation determinants and react distinctly to adverse conditions encountered by the infecting bacteria and ultimately determine their phenotypic behavior (Fig. 4; see Fig. S3 in the supplemental material). We found that selective induction of these domains involves a cross talk between DNA supercoiling and the abundant NAPs, FIS and H-NS (and most likely, also the other abundant NAPs not studied here). The stress-response domains thus appear as modular structural-functional entities coordinating the genetic expression of the chromosome and sustaining the bacterial pathogenic growth. Overall, the organizational properties of at least some of them closely resemble those of the horizontally acquired genomic pathogenicity islands, suggesting variations on a common theme. It is obvious that the role of the discovered transient structural-functional domains in the bacterial pathogenic behavior needs to be explored further, including, for example, deliberate changes of their boundaries and chromosomal coordinates, as well as exchanges of the virulence and adaptation determinants between the individual domains.
MATERIALS AND METHODS
Bacterial strains, cell growth, and stress conditions.
D. dadantii strain 3937, isolated from Saintpaulia, and its hns (31) and fis (30) derivatives were used to analyze the global gene expression of cells grown in vitro under different stress conditions that mimic the hostile environment encountered by the bacteria in the plant host. A multifactorial design covering 32 different experimental conditions with two biological replicates each was used to study the transcriptome by pangenomic microarrays. Wild-type cells were cultivated in four different growth media: M63 supplemented with 0.2% (wt/vol) sucrose as the carbon source, with or without 0.2% (wt/vol) polygalacturonate (PGA, a pectin derivative) and with or without 1% (vol/vol) Saintpaulia leaf extracts. (The leaf extract was 100 g leaves in 1 liter M63 boiled for 20 min and sterilized by filtration through a 0.45-µm-pore filter.) PGA induces the synthesis of pectinases, which are the essential virulence factors of D. dadantii, whereas supplementation with leaf extracts mimics the nutrient conditions encountered by bacteria in the plant. Cells were grown in these four media until the exponential or early stationary phases. The aliquots were then transferred to four flasks: one of them was kept as a control, and the three others were subjected to the different stresses as follows. (i) Acidic stress was induced by 15 min of incubation in the presence of 30 mM malic acid, resulting in a final pH of 5.0. (ii) Oxidative stress was induced by 15 min of incubation in the presence of 100 µM H2O2. (iii) Osmotic stress was induced by 15 min of incubation in the presence of 300 mM NaCl. The cells were then harvested for RNA extraction. For DNA relaxation, wild-type, hns, and fis strains were grown in M63 medium supplemented with 0.2% (wt/vol) sucrose until the early exponential phase (optical density at 600 nm [OD600] of 0.2), and then aliquots were transferred to two flasks: one of them was kept as a control, and the second was treated for 15 min with novobiocin to a final concentration of 100 µg/ml. At this concentration, novobiocin has no impact on D. dadantii growth (21).
RNA extraction.
RNAs from D. dadantii were extracted as previously described (61) from cultures grown to the early exponential phase (OD600 of 0.2) and to the early stationary phase (OD600 of 1.2 for cells grown in M63-sucrose, OD600 of 1.5 in M63-sucrose-plant extract medium, and OD600 of 1.9 for cells grown in M63-sucrose-PGA medium or M63-sucrose-PGA-plant extract medium). The different OD600s retained for the various culture media correspond to a similar growth stage (i.e., the transition from the late exponential phase to the early stationary phase). Complementation with PGA, plant extract, or both PGA and plant extract does not modify the bacterial growth rate, but it does increase the final biomass. Isolated RNA was quantified spectrophotometrically using an ND 2000 Nanodrop spectrophotometer, visualized on an agarose gel for quality, and stored at −80°C until further use. The absence of genomic DNA contamination was checked by PCR. Four target genes were selected to assess the impact of novobiocin and the stresses in quantitative reverse transcription (RT-qPCR) experiments as previously described in reference 61 (see Fig. S8 in the supplemental material). The primers used for these RT-PCR experiments are as follows: 16S RNA, forward, GATCATGGCTCAGATTGAACG, and reverse, AGTTATCCCCCTCCATCAGG; gyrB, forward, AGTATTAAAAGGGCTGGATGC, and reverse, ACCGACACCGAGTTATCAGC; pelE, forward, AGCGAATTCAAAGCAGCACT, and reverse GGCGTTTCGATGTACAGGTT; and asr, forward, GCTCTGGGTCTGTCCTCTGT, and reverse, CTGAGCTTTCTGCGTTGC.
DNA microarray experiment and data analysis.
The microarrays used in this study were custom designed and produced by Roche NimbleGen, Inc. (Madison, WI) based on the annotated sequence of D. dadantii, available at GenBank accession no. CP002038, which comprises 4,597 coding sequences (CDSs) (62). The 4-plex expression microarrays consist of 60-mer oligonucleotides, triplicated in three blocks on the array (5 oligonucleotides per CDS). For microarray analyses, cDNA was synthesized, labeled, and hybridized by Roche NimbleGen, Inc. Microarrays comprising 32 conditions with 2 replicates each as well as the 12 novobiocin arrays were normalized using quantile normalization and further analyzed using the R limma package. Significantly differentially expressed genes were identified using a P value of <0.05 and fold change (FC) of >1.2.
For all parameters analyzed in this study, the scanning window size was 500 kb, shifting by 4 kb and resulting in 1,230 overlapping windows. In order to evaluate the spatial distributions of DNA sequence parameters (gene expression density, average negative melting energy, preferred superhelicity regimen [hyp or rel], spatial orientation in the genome, distance to origin, response to FIS, and response to H-NS), we determined Z scores. For gene density, the ratio of up- and downregulated genes was used to generate Z scores, again taking randomly chosen up- and downregulated gene sets as reference. The average melting energy was calculated using the parameters of SantaLucia (63). The difference of the average melting energy within the window in up- and downregulated genes was compared to the difference of melting energy of randomly chosen up- and downregulated genes. For other parameters, the frequency ratio of the feature in the up- and downregulated genes was compared to the frequency ratios of randomly chosen up- and downregulated gene sets of the respective size.
Variation of expression patterns with changing conditions was determined by first normalizing the gene expression pattern of each gene in the range from 0 to 1. For each condition-dependent variation pattern of gene groups (e.g., hyp or rel), we applied the average normalized expression of all genes in this group. In the case of melting energy and distance to origin, we applied the average melting energy/distance to origin of all genes, weighted by their normalized expression. Functional gene groups were taken from the respective GO branches provided by the KEGG database.
Determination of structural-functional domains.
In order to obtain structural domains, we investigated the correlation of the feature dynamics (gene expression density, hyp genes, rel genes, leading-strand preference, and average negative melting energy) under the growth conditions analyzed. For a pair of features, we first computed for each window a Pearson correlation of the window feature Z scores. Taking all windows together, we obtained a feature correlation ring for each pair of features. The term “ring” is used here to describe the set of all windows (500-kb size, 4-kb shift) with the contained values. In the next step, we determined common changes of correlations in all correlation rings (comprising all combinations of features). To determine the change of correlation, i.e., the spatial borders of a certain correlation, we compare the correlation value of two windows that have a distance of 20 windows (20*4 kb = 80 kb). The more the values differ, the stronger is the border character of this region (a fast change of correlation within 80 kb). Hence we use the difference to detect the presence of a border. For each ring, we computed the absolute difference of feature correlations within a 20-window span (|correlation [−10] − correlation [+10]|) to get rings of correlation change. These represent the spatial change of correlation of a pair of features and therefore the change of realized combination of the two features. Then, we summed up the correlation change rings window-wise for all feature correlation rings and obtained a ring summing up the changes. Hence, proximal correlation changes of different feature combinations lead to a large sum. Finally we computed correlation change Z scores by applying the same procedure to randomly rotated correlation change rings. Windows with Z scores of ≥3, indicating a change in feature usage, with less than a 30-window distance (120 kb) were clustered, and the center of the cluster was considered a domain border.
For the determination of functional domains, we investigated the number of genes assigned to GO groups at level 4 in the metabolic function branch (4). We compared the distribution of genes among GO groups in two windows at a center distance of 250 kb using the root mean square of the group size difference. Prior to the comparison, the number of genes in each group was normalized by the total number of genes assigned to all groups in a window to avoid biases introduced by regions enriched in genes with unknown function. High values can be interpreted as a change of the functional composition of genes in the two windows. These peaks were compared to the positions of the structural domains. For each position in one set of borders (e.g., functional borders), the nearest neighbor to a border of the other set (e.g., domain borders) was determined. For a robust result, we took the median of the derived distances to the nearest neighbors of all borders. In 10,000 repetitions of this procedure, 98.0% of the random borders showed a larger median nearest neighbor than the actual border positions, indicating a tight connection between the functional and structural domain borders.
Microarray data accession numbers.
The microarray data from this study are compiled in the EBI Gene Expression (ArrayExpress) database (http://www.ebi.ac.uk/arrayexpress/) and are accessible through accession no. E-MTAB-541 and E-MTAB-2597.
SUPPLEMENTAL MATERIAL
(A) Synteny of the Dickeya dadantii genome and the genomes of other Dickeya species. Pairwise alignments of genomes were generated using Mauve. Colored blocks outline the conserved regions of the genomic sequence that align. The colored bars inside the blocks are related to the level of sequence similarities. The thermodynamically distinct structural regions identified in D. dadantii (LD1, LD2, SD1, and SD2) are conserved, albeit in a distinct spatial arrangement, in other Dickeya species. (B) Averaged distributions of negative melting energy in the genomes of various Dickeya species. Red and blue indicate low and high average negative melting energy, respectively. Calculations were done as in the legend to Fig. 1. Download
Venn diagrams of the intrastrain comparisons of supercoiling response genes. The genes active at high physiological supercoiling levels are indicated as “up” and the genes activated on DNA relaxation by novobiocin as “down.” (A) Comparison of the wild-type cells with the fis mutant. (B) Comparison of the wild-type cells with the hns mutant. (C) Comparison of the fis and hns mutants. Download
Sketch of the infection process by D. dadantii. The Dickeya pathogenicity program comprises asymptomatic (A) and symptomatic (B) phases. In the asymptomatic phase during plant penetration and colonization, the bacteria experience an acidic stress and an oxidative stress, respectively. During the symptomatic phase, bacteria experience osmotic and oxidative stresses. During these stresses, DNA topology is changed as indicated in panel C. For acidic and oxidative stresses, DNA was isolated from the exponential phase. For NaCl stress, DNA was isolated from the stationary phase. The lists of the up- and downregulated genes at each stage are provided in Table S1 in the supplemental material and discussed in reference 22. Download
Spatial genomic wheels of the D. dadantii transcriptional response to stress compared to that of untreated cells. The wheels corresponding to each single condition are arranged in bundles of four rings (due to the four different media used: sucrose, sucrose-polygalacturonate [i.e., PGA, a pectin derivative], sucrose-plant, and sucrose-plant-PGA). The conditions (listed on the left side of the panel) are ordered from inside to outside and numbered from 1 to 8. Each of the consecutive four-ring-bundles in the wheel (from inside to outside) shows the change in the density of differentially expressed genes (red, high; blue, low) compared to that in the untreated cells. The first four-ring bundle (exp, exponential phase) is compared to the stationary-phase (stat) expression. The four-ring bundle no. 5 corresponds to the stationary phase compared to the exponential phase. Only the significant differentially expressed genes (P < 0.05; FC, >1.2) are compiled in this representation. The outer ring shows the averaged distributions of negative melting energy in the genome. The locations of rrn genes are marked by black dots. Download
Compilation of the variations of expressed gene parameters with growth condition. Graphical representation of the variation of analog DNA parameters with changing growth conditions assembled along the x axis: 1, stationary phase; 2, exponential phase; 3, exponential-phase cells under acidic stress; 4, exponential-phase cells under oxidative stress; 5, stationary-phase cells under oxidative stress; 6, stationary phase (same as 1); 7, stationary-phase cells under osmotic stress; 8, stationary-phase cells under oxidative stress (same as 5). The analyzed parameter is indicated above each graph, together with the range of variation indicated in brackets. The y axis in panels A, B, C, D, E, and F shows the relative expression within the given variation range normalized to 0 to 1 for each separate group. (A) Relative expression of genes upregulated by FIS. (B) Relative expression of genes downregulated by FIS. (C) Relative expression of genes upregulated by H-NS. (D) Relative expression of genes downregulated by H-NS. (E) Relative expression of genes active at physiological supercoiling levels. (F) Relative expression of genes activated on DNA relaxation. (G) Expression weighted average negative melting energy of all genes. The variation range is indicated in brackets above the graph. (H) Expression weighted leading strand utilization calculated for all genes. (I) Expression weighted relative distance from the origin of replication calculated for all genes. The order of the growth conditions was retained in accordance with the fluctuation of the conditions encountered by D. dadantii during the infection process (see Fig. S3 in the supplemental material). Download
Comparison of the parameter curve shapes using Pearson’s correlation coefficient. Perfect positive and negative correlation in the matrix is color coded (red, 1; blue, −1). As expected, we observed that the hyp and rel genes were anticorrelated, as indicated by the negative value (−0.7) in the intersection square. We also found a strong anticorrelation for H-NS-activated and -repressed genes (−0.9) and a less strong one for FIS-activated and -repressed genes (−0.6). Furthermore, positive correlation was observed for the FIS-activated genes and hyp genes (0.9), whereas both of these anticorrelated with leading-strand bias (−0.7). Note that correlations (both positive and negative) are observed between particular gene groups and the distance to the origin, suggesting a spatial organization. Overall, the genes repressed by H-NS were closer to the origin (−0.6), as opposed to the genes activated by H-NS and those repressed by FIS (0.9 and 0.7, respectively). The proximity to the origin correlated also with increasing average negative melting energy (−0.6), consistent with observations in E. coli (39). Download
Spatial genomic wheels of the D. dadantii transcriptional response to stress. The wheels corresponding to each single condition are arranged in bundles of four rings (due to the four different media used: sucrose, sucrose-PGA, sucrose-plant, and sucrose-plant-PGA). The conditions (listed on the right side of the figure) are ordered from inside to outside and numbered from 1 to 7. Each of the consecutive four-ring bundles in the wheel (from inside to outside) shows the change in the parameter value (red, increase; blue, decrease) compared to the preceding condition. The different stress conditions are arranged according to their assumed occurrence during the infection. The first four-ring bundle (exp, exponential phase) is compared to the stationary-phase (stat) expression. Only the significant differentially expressed genes (P < 0.05; FC, >1.2) are compiled in this representation. (A) Gene density distributions (red, high; blue, low). (B) Melting energy distributions (color inverted: red, low; blue, high). (C) Leading-strand bias (red, high; blue, low). (D) Distribution of hyp genes (red, high; blue, low). (E) Distribution of the FIS-activated genes (red, high; blue, low). (F) Distribution of the H-NS-activated genes (red, high; blue, low). (G) Distribution of rel genes (red, high; blue, low). (H) Distribution of the FIS-repressed genes (red, high; blue, low). (I) Distribution of the H-NS-repressed genes (red, high; blue, low). The three outer rings are the same as in Fig. 3. The 11 significant boundaries delimiting the stress-response domains are indicated by black lines. Download
Expression profiles of four genes selected for their differential responses to stresses. RT-qPCR assays were performed on gyrB, pelE, asr, and 16S RNA genes under the experimental conditions previously tested in microarrays (with two biological replicates). One microgram of total RNA was used in each reaction. The transcript levels were normalized to those of the WT strain grown exponentially in sucrose medium without stress. The expression profiles of other genes (ahpC induced under oxidative stress and proV induced under osmotic stress) were already reported (61). Download
Features and expression of selected adaptation/virulence genes involved in various stages of infection.
ACKNOWLEDGMENTS
This work was supported by grants from the French Agence National de la Recherche (ANR), ANR Blanc Régupath 2007, and Bonus Qualité Recherche (BQR) INSA de Lyon 2012, and a research grant from the Deutsche Forschungsgemeinschaft to G.M. Exchanges between the French and German teams were supported by CNRS Programme International de Cooperation Scientifique (PICS 2009).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Servane Le Guillouzer for help with RNA extraction for novobiocin experiments.
Footnotes
Citation Jiang X, Sobetzko P, Nasser W, Reverchon S, Muskhelishvili G. 2015. Chromosomal “stress-response” domains govern the spatiotemporal expression of the bacterial virulence program. mBio 6(3):e00353-15. doi:10.1128/mBio.00353-15.
Contributor Information
Ivan Junier, Centre for Genomic Regulation (CRG), Barcelona, Spain.
Ronald K. Taylor, Dartmouth Medical School.
REFERENCES
- 1.Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. 2010. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci U S A 107:4991–4995. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. 2006. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol 62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- 3.Umbarger MA, Toro E, Wright MA, Porreca GJ, Baù D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, Shapiro L, Dekker J, Church GM. 2011. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell 44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobetzko P, Travers A, Muskhelishvili G. 2012. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc Natl Acad Sci U S A 109:E42–E50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhlman TE, Cox EC. 2012. Gene location and DNA density determine transcription factor distributions in Escherichia coli. Mol Syst Biol 8:610. doi: 10.1038/msb.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. 2006. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep 7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimsky S, Travers A. 2011. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr Opin Microbiol 14:136–141. doi: 10.1016/j.mib.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Balke VL, Gralla JD. 1987. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol 169:4499–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider R, Travers A, Muskhelishvili G. 1997. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol 26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 11.Weinstein-Fischer D, Altuvia S. 2007. Differential regulation of Escherichia coli topoisomerase I by Fis. Mol Microbiol 63:1131–1144. doi: 10.1111/j.1365-2958.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 12.van Workum M, van Dooren SJ, Oldenburg N, Molenaar D, Jensen PR, Snoep JL, Westerhoff HV. 1996. DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol Microbiol 20:351–360. doi: 10.1111/j.1365-2958.1996.tb02622.x. [DOI] [PubMed] [Google Scholar]
- 13.Travers A, Muskhelishvili G. 2005. DNA supercoiling—a global transcriptional regulator for enterobacterial growth? Nat Rev Microbiol 3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh LS, Burger RM, Drlica K. 1991. Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J Mol Biol 219:443–450. doi: 10.1016/0022-2836(91)90185-9. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh LS, Rouviere-Yaniv J, Drlica K. 1991. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. J Bacteriol 173:3914–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenschein N, Geertz M, Muskhelishvili G, Hütt MT. 2011. Analog regulation of metabolic demand. BMC Syst Biol 5:40. doi: 10.1186/1752-0509-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. 2004. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol 5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dame RT. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol 56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 19.Maurer S, Fritz J, Muskhelishvili G. 2009. A systematic in vitro study of nucleoprotein complexes formed by bacterial nucleoid-associated proteins revealing novel types of DNA organization. J Mol Biol 387:1261–1276. doi: 10.1016/j.jmb.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 20.Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee AS, Luscombe NM. 2012. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res 40:3524–3537. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouafa ZA, Reverchon S, Lautier T, Muskhelishvili G, Nasser W. 2012. The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res 40:4306–4319. doi: 10.1093/nar/gks014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reverchon S, Nasser W. 2013. Dickeya ecology, environment sensing and regulation of virulence programme. Environ Microbiol Rep 5:622–636. doi: 10.1111/1758-2229.12073. [DOI] [PubMed] [Google Scholar]
- 23.Hérault E, Reverchon S, Nasser W. 2014. Role of the LysR-type transcriptional regulator PecT and DNA supercoiling in the thermoregulation of pel genes, the major virulence factors in Dickeya dadantii. Environ Microbiol 16:734–745. doi: 10.1111/1462-2920.12198. [DOI] [PubMed] [Google Scholar]
- 24.Dorman CJ, Porter ME. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol 29:677–684. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 25.Dorman CJ, Corcoran CP. 2009. Bacterial DNA topology and infectious disease. Nucleic Acids Res 37:672–678. doi: 10.1093/nar/gkn996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron AD, Dorman CJ. 2012. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet 8:e1002615. doi: 10.1371/journal.pgen.1002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron AD, Stoebel DM, Dorman CJ. 2011. DNA supercoiling is differentially regulated by environmental factors and FIS in Escherichia coli and Salmonella enterica. Mol Microbiol 80:85–101. doi: 10.1111/j.1365-2958.2011.07560.x. [DOI] [PubMed] [Google Scholar]
- 28.Ball CA, Osuna R, Ferguson KC, Johnson RC. 1992. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol 174:8043–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lautier T, Blot N, Muskhelishvili G, Nasser W. 2007. Integration of two essential virulence modulating signals at the Erwinia chrysanthemi pel gene promoters: a role for Fis in the growth-phase regulation. Mol Microbiol 66:1491–1505. doi: 10.1111/j.1365-2958.2007.06010.x. [DOI] [PubMed] [Google Scholar]
- 30.Lautier T, Nasser W. 2007. The DNA nucleoid-associated protein Fis co-ordinates the expression of the main virulence genes in the phytopathogenic bacterium Erwinia chrysanthemi. Mol Microbiol 66:1474–1490. doi: 10.1111/j.1365-2958.2007.06012.x. [DOI] [PubMed] [Google Scholar]
- 31.Nasser W, Faelen M, Hugouvieux-Cotte-Pattat N, Reverchon S. 2001. Role of the nucleoid-associated protein H-NS in the synthesis of virulence factors in the phytopathogenic bacterium Erwinia chrysanthemi. Mol Plant Microbe Interact 14:10–20. doi: 10.1094/MPMI.2001.14.1.10. [DOI] [PubMed] [Google Scholar]
- 32.Nasser W, Reverchon S. 2002. H-NS-dependent activation of pectate lyases synthesis in the phytopathogenic bacterium Erwinia chrysanthemi is mediated by the PecT repressor. Mol Microbiol 43:733–748. doi: 10.1046/j.1365-2958.2002.02782.x. [DOI] [PubMed] [Google Scholar]
- 33.Duprey A, Reverchon S, Nasser W. 2014. Bacterial virulence and Fis: adapting regulatory networks to the host environment. Trends Microbiol 22:92–99. doi: 10.1016/j.tim.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Hardy CD, Cozzarelli NR. 2005. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol 57:1636–1652. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- 35.Sobetzko P, Glinkowska M, Travers A, Muskhelishvili G. 2013. DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle. Mol Biosyst 9:1643–1651. doi: 10.1039/c3mb25515h. [DOI] [PubMed] [Google Scholar]
- 36.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koskiniemi S, Lamoureux JG, Nikolakakis KC, T’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol 50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 39.Antúnez-Lamas M, Cabrera-Ordóñez E, López-Solanilla E, Raposo R, Trelles-Salazar O, Rodríguez-Moreno A, Rodríguez-Palenzuela P. 2009. Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937). Microbiology 155:434–442. doi: 10.1099/mic.0.022244-0. [DOI] [PubMed] [Google Scholar]
- 40.Franza T, Expert D. 2013. Role of iron homeostasis in the virulence of phytopathogenic bacteria: an “a la carte” menu. Mol Plant Pathol 14:429–438. doi: 10.1111/mpp.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 42.Lebeau A, Reverchon S, Gaubert S, Kraepiel Y, Simond-Côte E, Nasser W, Van Gijsegem F. 2008. The GacA global regulator is required for the appropriate expression of Erwinia chrysanthemi 3937 pathogenicity genes during plant infection. Environ Microbiol 10:545–559. doi: 10.1111/j.1462-2920.2007.01473.x. [DOI] [PubMed] [Google Scholar]
- 43.Nasser W, Dorel C, Wawrzyniak J, Van Gijsegem F, Groleau MC, Déziel E, Reverchon S. 2013. Vfm a new quorum sensing system controls the virulence of Dickeya dadantii. Environ Microbiol 15:865–880. doi: 10.1111/1462-2920.12049. [DOI] [PubMed] [Google Scholar]
- 44.Meury J, Kohiyama M. 1992. Potassium ions and changes in bacterial DNA supercoiling under osmotic stress. FEMS Microbiol Lett 78:159–164. [DOI] [PubMed] [Google Scholar]
- 45.Rochman M, Aviv M, Glaser G, Muskhelishvili G. 2002. Promoter protection by a transcription factor acting as a local topological homeostat. EMBO Rep 3:355–360. doi: 10.1093/embo-reports/kvf067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auner H, Buckle M, Deufel A, Kutateladze T, Lazarus L, Mavathur R, Muskhelishvili G, Pemberton I, Schneider R, Travers A. 2003. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J Mol Biol 331:331–344. doi: 10.1016/S0022-2836(03)00727-7. [DOI] [PubMed] [Google Scholar]
- 47.Weinstein-Fischer D, Elgrably-Weiss M, Altuvia S. 2000. Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol Microbiol 35:1413–1420. doi: 10.1046/j.1365-2958.2000.01805.x. [DOI] [PubMed] [Google Scholar]
- 48.Karem K, Foster JW. 1993. The influence of DNA topology on the environmental regulation of a pH-regulated locus in Salmonella typhimurium. Mol Microbiol 10:75–86. doi: 10.1111/j.1365-2958.1993.tb00905.x. [DOI] [PubMed] [Google Scholar]
- 49.Ni Bhriain N, Dorman CJ, Higgins CF. 1989. An overlap between osmotic and anaerobic stress responses: a potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol Microbiol 3:933–942. doi: 10.1111/j.1365-2958.1989.tb00243.x. [DOI] [PubMed] [Google Scholar]
- 50.Reverchon S, Expert D, Robert-Baudouy J, Nasser W. 1997. The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi. J Bacteriol 179:3500–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Libby EA, Roggiani M, Goulian M. 2012. Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc Natl Acad Sci U S A 109:7445–7450. doi: 10.1073/pnas.1109479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. 2004. Macrodomain organization of the Escherichia coli chromosome. EMBO J 23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marr C, Geertz M, Hütt MT, Muskhelishvili G. 2008. Dissecting the logical types of network control in gene expression profiles. BMC Syst Biol 2:18. doi: 10.1186/1752-0509-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scolari VF, Bassetti B, Sclavi B, Lagomarsino MC. 2011. Gene clusters reflecting macrodomain structure respond to nucleoid perturbations. Mol Biosyst 7:878–888. doi: 10.1039/c0mb00213e. [DOI] [PubMed] [Google Scholar]
- 55.Ma Q, Yin Y, Schell MA, Zhang H, Li G, Xu Y. 2013. Computational analyses of transcriptomic data reveal the dynamic organization of the Escherichia coli chromosome under different conditions. Nucleic Acids Res 41:5594–5603. doi: 10.1093/nar/gkt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Booker BM, Deng S, Higgins NP. 2010. DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol Microbiol 78:1348–1364. doi: 10.1111/j.1365-2958.2010.07394.x. [DOI] [PubMed] [Google Scholar]
- 57.Berger M, Farcas A, Geertz M, Zhelyazkova P, Brix K, Travers A, Muskhelishvili G. 2010. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep 11:59–64. doi: 10.1038/embor.2009.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Travers AA, Muskhelishvili G. 2013. DNA thermodynamics shape chromosome organization and topology. Biochem Soc Trans 41:548–553. doi: 10.1042/BST20120334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 60.Ferrandez Y, Condemine G. 2008. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol Microbiol 69:1349–1357. doi: 10.1111/j.1365-2958.2008.06366.x. [DOI] [PubMed] [Google Scholar]
- 61.Hommais F, Zghidi-Abouzid O, Oger-Desfeux C, Pineau-Chapelle E, Van Gijsegem F, Nasser W, Reverchon S. 2011. lpxC and yafS are the most suitable internal controls to normalize real time RT-qPCR expression in the phytopathogenic bacteria Dickeya dadantii. PLoS One 6:e20269. doi: 10.1371/journal.pone.0020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glasner JD, Yang CH, Reverchon S, Hugouvieux-Cotte-Pattat N, Condemine G, Bohin JP, Van Gijsegem F, Yang S, Franza T, Expert D, Plunkett G III, San Francisco MJ, Charkowski AO, Py B, Bell K, Rauscher L, Rodriguez-Palenzuela P, Toussaint A, Holeva MC, He SY, Douet V, Boccara M, Blanco C, Toth I, Anderson BD, Biehl BS, Mau B, Flynn SM, Barras F, Lindeberg M, Birch PR, Tsuyumu S, Shi X, Hibbing M, Yap MN, Carpentier M, Dassa E, Umehara M, Kim JF, Rusch M, Soni P, Mayhew GF, Fouts DE, Gill SR, Blattner FR, Keen NT, Perna NT. 2011. Genome sequence of the plant-pathogenic bacterium Dickeya dadantii 3937. J Bacteriol 193:2076–2077. doi: 10.1128/JB.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.SantaLucia J. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A 95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Synteny of the Dickeya dadantii genome and the genomes of other Dickeya species. Pairwise alignments of genomes were generated using Mauve. Colored blocks outline the conserved regions of the genomic sequence that align. The colored bars inside the blocks are related to the level of sequence similarities. The thermodynamically distinct structural regions identified in D. dadantii (LD1, LD2, SD1, and SD2) are conserved, albeit in a distinct spatial arrangement, in other Dickeya species. (B) Averaged distributions of negative melting energy in the genomes of various Dickeya species. Red and blue indicate low and high average negative melting energy, respectively. Calculations were done as in the legend to Fig. 1. Download
Venn diagrams of the intrastrain comparisons of supercoiling response genes. The genes active at high physiological supercoiling levels are indicated as “up” and the genes activated on DNA relaxation by novobiocin as “down.” (A) Comparison of the wild-type cells with the fis mutant. (B) Comparison of the wild-type cells with the hns mutant. (C) Comparison of the fis and hns mutants. Download
Sketch of the infection process by D. dadantii. The Dickeya pathogenicity program comprises asymptomatic (A) and symptomatic (B) phases. In the asymptomatic phase during plant penetration and colonization, the bacteria experience an acidic stress and an oxidative stress, respectively. During the symptomatic phase, bacteria experience osmotic and oxidative stresses. During these stresses, DNA topology is changed as indicated in panel C. For acidic and oxidative stresses, DNA was isolated from the exponential phase. For NaCl stress, DNA was isolated from the stationary phase. The lists of the up- and downregulated genes at each stage are provided in Table S1 in the supplemental material and discussed in reference 22. Download
Spatial genomic wheels of the D. dadantii transcriptional response to stress compared to that of untreated cells. The wheels corresponding to each single condition are arranged in bundles of four rings (due to the four different media used: sucrose, sucrose-polygalacturonate [i.e., PGA, a pectin derivative], sucrose-plant, and sucrose-plant-PGA). The conditions (listed on the left side of the panel) are ordered from inside to outside and numbered from 1 to 8. Each of the consecutive four-ring-bundles in the wheel (from inside to outside) shows the change in the density of differentially expressed genes (red, high; blue, low) compared to that in the untreated cells. The first four-ring bundle (exp, exponential phase) is compared to the stationary-phase (stat) expression. The four-ring bundle no. 5 corresponds to the stationary phase compared to the exponential phase. Only the significant differentially expressed genes (P < 0.05; FC, >1.2) are compiled in this representation. The outer ring shows the averaged distributions of negative melting energy in the genome. The locations of rrn genes are marked by black dots. Download
Compilation of the variations of expressed gene parameters with growth condition. Graphical representation of the variation of analog DNA parameters with changing growth conditions assembled along the x axis: 1, stationary phase; 2, exponential phase; 3, exponential-phase cells under acidic stress; 4, exponential-phase cells under oxidative stress; 5, stationary-phase cells under oxidative stress; 6, stationary phase (same as 1); 7, stationary-phase cells under osmotic stress; 8, stationary-phase cells under oxidative stress (same as 5). The analyzed parameter is indicated above each graph, together with the range of variation indicated in brackets. The y axis in panels A, B, C, D, E, and F shows the relative expression within the given variation range normalized to 0 to 1 for each separate group. (A) Relative expression of genes upregulated by FIS. (B) Relative expression of genes downregulated by FIS. (C) Relative expression of genes upregulated by H-NS. (D) Relative expression of genes downregulated by H-NS. (E) Relative expression of genes active at physiological supercoiling levels. (F) Relative expression of genes activated on DNA relaxation. (G) Expression weighted average negative melting energy of all genes. The variation range is indicated in brackets above the graph. (H) Expression weighted leading strand utilization calculated for all genes. (I) Expression weighted relative distance from the origin of replication calculated for all genes. The order of the growth conditions was retained in accordance with the fluctuation of the conditions encountered by D. dadantii during the infection process (see Fig. S3 in the supplemental material). Download
Comparison of the parameter curve shapes using Pearson’s correlation coefficient. Perfect positive and negative correlation in the matrix is color coded (red, 1; blue, −1). As expected, we observed that the hyp and rel genes were anticorrelated, as indicated by the negative value (−0.7) in the intersection square. We also found a strong anticorrelation for H-NS-activated and -repressed genes (−0.9) and a less strong one for FIS-activated and -repressed genes (−0.6). Furthermore, positive correlation was observed for the FIS-activated genes and hyp genes (0.9), whereas both of these anticorrelated with leading-strand bias (−0.7). Note that correlations (both positive and negative) are observed between particular gene groups and the distance to the origin, suggesting a spatial organization. Overall, the genes repressed by H-NS were closer to the origin (−0.6), as opposed to the genes activated by H-NS and those repressed by FIS (0.9 and 0.7, respectively). The proximity to the origin correlated also with increasing average negative melting energy (−0.6), consistent with observations in E. coli (39). Download
Spatial genomic wheels of the D. dadantii transcriptional response to stress. The wheels corresponding to each single condition are arranged in bundles of four rings (due to the four different media used: sucrose, sucrose-PGA, sucrose-plant, and sucrose-plant-PGA). The conditions (listed on the right side of the figure) are ordered from inside to outside and numbered from 1 to 7. Each of the consecutive four-ring bundles in the wheel (from inside to outside) shows the change in the parameter value (red, increase; blue, decrease) compared to the preceding condition. The different stress conditions are arranged according to their assumed occurrence during the infection. The first four-ring bundle (exp, exponential phase) is compared to the stationary-phase (stat) expression. Only the significant differentially expressed genes (P < 0.05; FC, >1.2) are compiled in this representation. (A) Gene density distributions (red, high; blue, low). (B) Melting energy distributions (color inverted: red, low; blue, high). (C) Leading-strand bias (red, high; blue, low). (D) Distribution of hyp genes (red, high; blue, low). (E) Distribution of the FIS-activated genes (red, high; blue, low). (F) Distribution of the H-NS-activated genes (red, high; blue, low). (G) Distribution of rel genes (red, high; blue, low). (H) Distribution of the FIS-repressed genes (red, high; blue, low). (I) Distribution of the H-NS-repressed genes (red, high; blue, low). The three outer rings are the same as in Fig. 3. The 11 significant boundaries delimiting the stress-response domains are indicated by black lines. Download
Expression profiles of four genes selected for their differential responses to stresses. RT-qPCR assays were performed on gyrB, pelE, asr, and 16S RNA genes under the experimental conditions previously tested in microarrays (with two biological replicates). One microgram of total RNA was used in each reaction. The transcript levels were normalized to those of the WT strain grown exponentially in sucrose medium without stress. The expression profiles of other genes (ahpC induced under oxidative stress and proV induced under osmotic stress) were already reported (61). Download
Features and expression of selected adaptation/virulence genes involved in various stages of infection.