Abstract
Purpose
We sought to determine whether dysregulation of the RB tumor suppressor pathway was associated with improved response to neoadjuvant chemotherapy in breast cancer.
Experimental Design
An RB-loss signature was used to analyze the association between pathway status and pathological complete response in gene expression datasets encompassing three different neoadjuvant regimens. Parallel immunohistochemical analysis of the RB-pathway was performed on pretreatment biopsies to determine the association with pathological response to neoadjuvant chemotherapy.
Results
An RB loss gene expression signature was asssociated with increased pathological complete response in datasets from breast cancer patients treated with FAC (p<0.001), T/FAC (p<0.001) and TA (p<0.001) neoadjuvant therapy encompasssing approximately 1,000 patients. The association with improved response to neoadjuvant chemotherapy was true in both ER-positive and ER-negative breast cancer. Elevated expression of p16ink4a is associated with the RB-loss of signature (R=0.493–0.5982), and correspondingly p16ink4a mRNA levels were strongly associated with pathological complete response in the same data sets analyzed. In an independent cohort, immunohistochemical analyses of RB and p16ink4a revealed an association of RB loss (p=0.0018) or elevated p16ink4a (p=0.0253) with pathological complete response. Additionally, by Miller-Payne and Clinical-Pathologic Scoring (CPS) analyses, RB-deficient tumors experienced an overall improved response to neoadjuvant chemotherapy.
Conclusion
Disruption of the RB-pathway as measured by several independent methods was associated with improved response to neoadjuvant chemotherapy. The RB-pathway status was relevant for pathological response in both ER-positive and ER-negative breast cancer with similar results observed with multiple chemotherapy regimens. Combined, these data indicate that RB-status is associated with the response to neoadjuvant chemotherapy in breast cancer and could be employed to inform treatment.
INTRODUCTION
In recent years, there has been an emphasis on breast cancer treatment that is directed at altered regulatory pathways in the tumor. For example, the presence of Her2 overexpression/amplification is associated with sensitivity to trastuzumab and other agents that target the receptor, and is therefore used clinically to direct therapy (1). While such markers are useful, additional undefined pathways contribute to the response to commonly used therapies. For example, in ER-positive breast cancer, while a substantial fraction of patients respond to endocrine therapy, there are clearly determinants beyond ER and Her2 that contribute to therapeutic sensitivity (2). In triple negative breast cancer, there are no established markers predicting chemotherapy response, though clearly there is heterogeneity in the response (3–5). One pathway that is emerging as a potential determinant of therapeutic response is the retinoblastoma (RB) tumor suppressor pathway (6–8).
The RB-pathway is disrupted in a large fraction of human cancers via distinct mechanisms (9). There is evidence for RB genetic inactivation and histological loss in some breast cancers (10–14). Additionally, the RB protein can be inactivated by post-translational modifications that are the subject of oncogenic (e.g. cyclin D1 overexpression/amplification) or tumor suppressive (e.g. p16ink4a silencing) alterations. These events compromise the activity of RB as a transcriptional repressor, and dysregulate cell cycle progression (10, 15). Historically, loss of RB function has been associated with aberrant proliferation contributing to cancer etiology and disease progression. However, RB also plays a critical role in the response to many anti-proliferative stresses that are engaged by therapeutic agents (6, 7). For example, preclinical studies have shown that RB loss compromises DNA damage checkpoints, leading to enhanced cell death in simple isogenic models treated with chemotherapy (16, 17). These findings suggest that distinct alterations in the RB-pathway could modulate the response to commonly used therapeutic regimens.
Breast cancer treatment typically involves a multidisciplinary approach combining surgery with systemic therapies. While historically surgery has preceded treatment with adjuvant therapy, there has been a significant increase in neoadjuvant therapy (18, 19). Multiple studies have shown that the response to neoadjuvant therapy, is effective at predicting the ultimate course of tumor behavior and specific determinants of that response are being sought. Importantly, pathological response in neoadjuvant studies reveals direct tumor response to a given therapy, independent of overall prognostic variables that can confound analyses of survival. In this study, we retrospectively analyzed the association of RB function with response to neoadjuvant chemotherapy in multiple distinct cohorts. We demonstrate that RB deficiency is associated with improved pathological response and could thus be used to identify those patients who will most benefit from neoadjuvant chemotherapy treatment.
MATERIALS AND METHODS
Neoadjuvant Breast Cancer Datasets
Raw microarray data files and sample annotation for datasets GSE20194, GSE20271, GSE22093, GSE23988, and GSE25066 were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). Information regarding the characteristic of the patients involved in these datasets is publicly available through GEO website. These datasets include gene expression data on pre-treatment tumor specimens, and the corresponding pathological response to three chemotherapy regimens: paclitaxel, 5-fluorouracil, cyclophosphamide and doxorubicin (TFAC), 5-fluorouracil, cyclophosphamide and doxorubicin (FAC) and taxane-anthracyline (TA). A summary of the patient cohorts and simple demographics, based on the information that is available on those datasets, are provided in supplemental Figure 1.
Dataset Normalization, RB-loss signature, and Cutpoints
Microarray sample CEL files were normalized against a reference dataset in Matlab version 2011b (The MathWorks, Natick, MA) using a modified version of the Robust Multichip Average (RMA) procedure previously described (15, 20). This procedure retains only those features included on the HGU133A microarray platform, for a total of 22277 probesets. Gene annotation for the 22277 probesets was obtained from the HGU133A annotations file version na32, dated June 9th, 2011, downloaded from the Affymetrix website (http://www.affymetrix.com/). Genes with multiple probesets were handled by averaging their rows together and scaling by the probeset with the largest standard deviation. For consistency with previous work on the RB-loss signature, transcript profiles were analyzed relative to values in the dataset analyzed in Ertel et al., 2010 (15). The genes making up the RB-loss signature are provided in supplemental Figure 2. The profiles of 137 RB-loss signature genes represented in the neoadjuvant datasets were centered around the median of the breast cancer dataset from Ertel et al. 2010, and averaged to obtain the RB-loss signature magnitude (15). In our previous study (15), X-Tile software was used to find thresholds for the RB-loss signature corresponding with optimal partitions in the breast cancer patient population, to define groups of low, intermediate, and high risk for 10-year recurrence. Those partitions were found at the 25th and 48th percentile in all patients, at the 29th and 60th percentile in ER-positive patients, and the 24th and 72nd percentile for ER-negative patients. While histological ER-status was available for all neoadjuvant treatment breast cancer microarray samples, a transcript-level-based ER status prediction was also computed from the ESR1 205225_at probeset, using a normalized RMA expression cutoff of 7.5, as described in previous work (15).
RB-loss Signature as a Predictor of Neoadjuvant Response
The RB-loss signature was evaluated as a predictor of neoadjuvant pathological response using several statistical tests for each neoadjuvant treatment. Initially, the RB-loss signature was used to rank the breast cancer samples from low to high signature expression. The Kolmogorov-Smirnov (KS) test was used to evaluate the null hypothesis that responders and non-responders would be evenly distributed across the continuous spectrum of RB-loss signature expression. In a similar manner, continuous values of the RB-loss signature were evaluated using Receiver Operating Characteristic (ROC) curves and area-under-the curve (AUC) values. Discrete analyses were also performed, using boxplots and two-sided, two-sample t-tests to evaluate differences between expression magnitudes in responders and non-responders. Additionally, the discrete RB-loss signature cutoffs were used to predict pathological response to neoadjuvant therapies. Breast Cancer samples were divided into high- and low- RB-loss signature expression, first based on a mean RB-loss signature magnitude cutoff, and then on high- and low- cutoffs that were previously established for optimal separation of short-term and long-term relapse-free-survival in an independent dataset (15). Significant differences in the distribution of responders and non-responders in the high- and low- RB-loss signature expressing groups were evaluated using the chi-squared test. Additionally, measures of test performances, including sensitivity, specificity and predictors values were calculated and are reported separately in supplemental Figure 3.
Neoadjuvant cohort and tissue staining
The TJU neoadjuvant cohort consists of 98 patients treated at the Thomas Jefferson University Hospital with neoadjuvant chemotherapy. The ER, PR, and Her2 status were determined in a clinical laboratory. The immunohistochemical (IHC) staining for p16ink4a and RB was performed and scored as previously described (21). The Miller-Payne and Clinical-Pathologic Scoring (CPS) analyses were performed by assessment of the pre and post-treatment specimens (22, 23). The Miller-Payne criteria takes into account the overall cellularity of the tumor, wherein grade 1 represent no reduction in cellularity, grade two represent a minor loss of tumor cells (up to 30%), grade 3 represents a 30–90% reduction in cellularity, grade 4 represents few distributed viable cells, and grade 5 represents no viable malignant cells. The CPS compares the pretreatment clinical tumor stage with the post treatment pathological evaluation. A low score is indicative of pathological response. The association between p16ink4a and Rb, measured by IHC, and pathological complete response was determined using an unpaired t-test. The association between RB-status with ER and Her2 status was also explored. Data analyses were performed in GraphPad Prism.
RESULTS
Gene Expression Signature of RB loss is associated with complete pathological response to FAC therapy
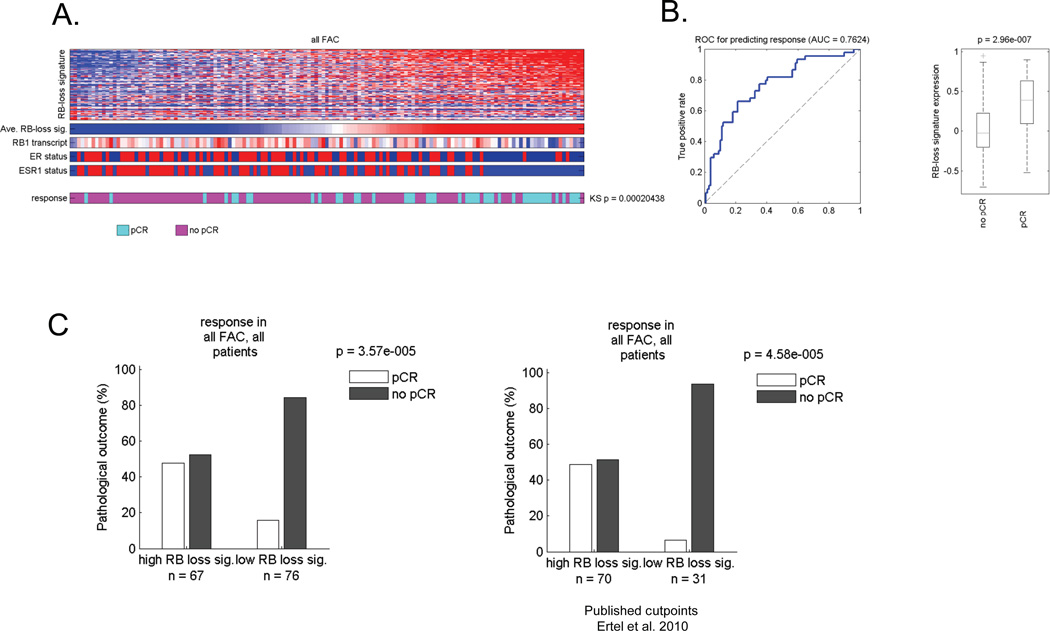
Preclinical studies have demonstrated that RB loss is associated with improved response to chemotherapeutic regimens (6, 7). Because the clinical response (e.g. overall survival or recurrence free survival) is modified by multiple prognostic variables, we focused on determining the predictive value of RB function on the pathological response to neoadjuvant chemotherapy. We employed a well-characterized gene expression signature of RB-loss, defined in preclinical models (15, 24, 25), to allow quantitative assessment of RB function across tumor specimens (15, 24, 25). This signature was employed to stratify of a cohort of 143 patients who were treated with neoadjuvant 5-fluorouracil/adriamycin/cytoxan (FAC) therapy at the MD Anderson Cancer Center (26–28). The signature demonstrated significant heterogeneity across tumors. In general, tumors with the highest level of the RB loss signature were ER-negative (Figure 1A). Using the non-parametric Kolmogorov-Smirnov (KS) statistic to test for an association across all tumors, the RB-loss signature associated with pathological complete response (pCR) and was highly significant (p=0.0002). By receiver operating curve (ROC) analyses, the area under the curve (AUC) for the RB-loss signature was 0.76, indicating relatively high sensitivity and specificity for predicting response (Figure 1B, left graph). Unbiased assessment of the RB-loss signature expression difference between cases with a pCR and other cases was similarly significant (Figure 1B, right graph). In this data set using either median signature value (Figure 1C, left panel) or previously determined high/low cutpoints (Figure 1C, right panel) the RB-loss signature was highly effective at determining tumors that experienced a pCR.
Figure 1. RB loss signature is associated with response to FAC neoadjuvant therapy.
(A) Tumors were clustered based on the relative level of the RB loss signature. The heat map depicts all of the genes within the signature. Below the bars denote the average RB loss signature value, the RB1 transcript, clinically defined ER status, and the expression of the ESR1 transcript. The bottom color bar provides the relationship to pathological complete response (pCR) vs residual or progressed disease (no pCR). The relationship of chemotherapy response to RB loss signature was determined using KS statistical modeling. (B) ROC analyses of the RB loss signature in this cohort was determined for predicting response (left panel) and the RB loss signature expression value as a function of no pCR vs. pCR was determined (right panel). (C) Bar graphs demonstrating the frequency of response based on median signature value (left panel) or a previously determined cutpoint (right panel).
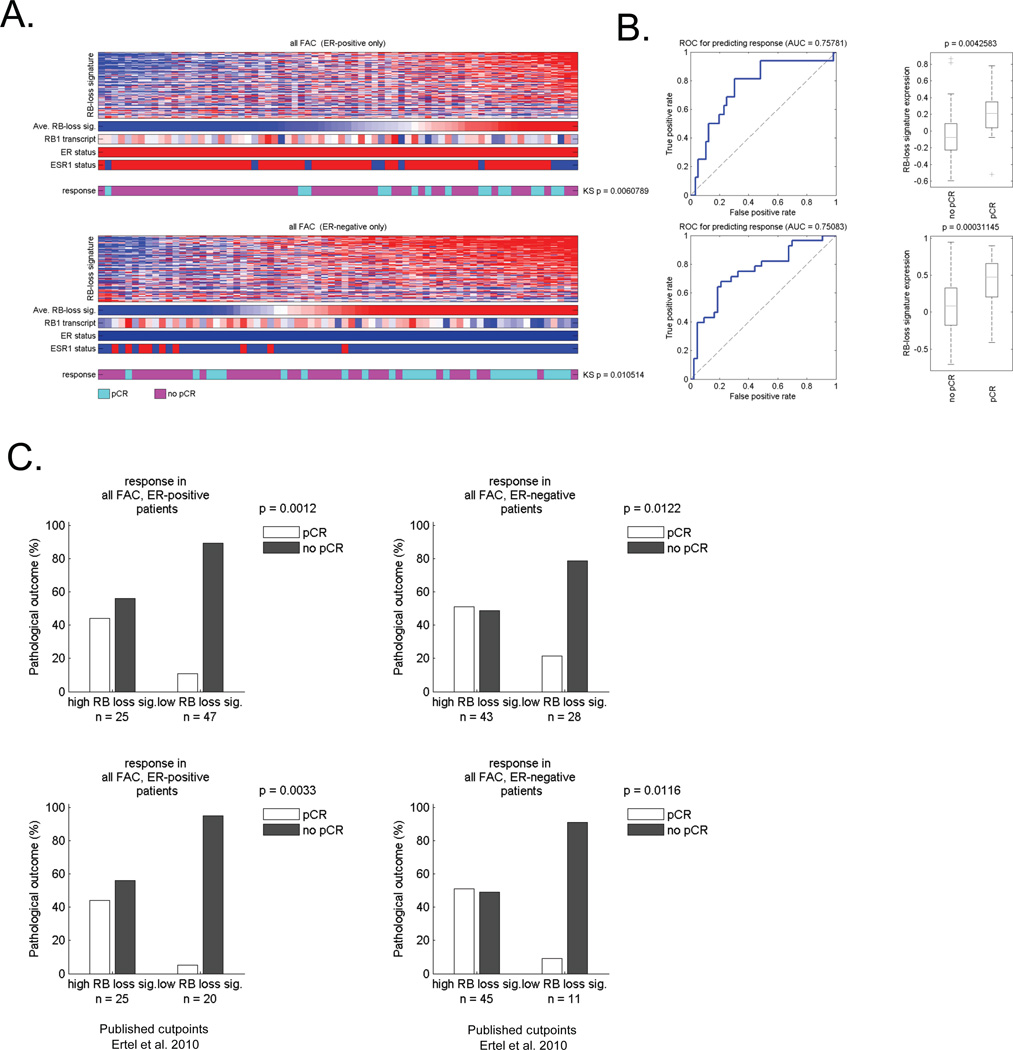
It is well known that ER-positive and ER-negative cancers exhibit a differential response rate to neoadjuvant chemotherapy that can be modified by Her2 status. In this cohort there were no Her2-positive tumors (supplemental Figure 1). However, the percent of pCR in ER-positive patients was ~15%, while in ER-negative patients was ~35%. To determine if the RB-loss signature had value in either tumor type, the ER-positive and negative cases were analyzed separately (Figure 2). These analyses showed that RB loss signature value was associated with pathological complete response in both ER-positive and ER-negative cases (Figure 2A). Interestingly the AUC was virtually identical irrespective of hormone receptor status (Figure 2B). Correspondingly, in ER-positive tumors exhibiting a high RB-loss signature had ~40% pCR, while those with low RB-loss signature had ~5% pCR. In ER-negative breast cancer, high RB-loss signature pCR frequency was ~50%, while low RB loss signature ~13%. These data were highly significant irrespective of cutpoint employed. These findings indicate that analyses of RB-status could be particularly useful in defining tumors with a particularly poor response to FAC neoadjuvant chemotherapy, and thereby enrich for responsive cases.
Figure 2. Signature of RB loss is associated with response to FAC neoadjuvant therapy in both ER-positive and ER-negative breast cancer.
(A) Tumors that were either clinically defined as ER-positive or ER-negative were clustered based on the relative level of the RB loss signature. The heat maps depicts all of the genes within the signature. The bars denote the average RB loss signature value, the RB1 transcript, clinically defined ER status, and the expression of the ESR1 transcript. The color bar provides the relationship to pathological complete response (pCR) vs residual or proressed disease (no pCR). The relationship of chemotherapy response to RB loss signature was determined using KS statistical modeling. (B) ROC analyses of the RB loss signature in either ER-positive or ER-negative cases was determined for predicting response (left panel) and the RB loss signature expression value as a function of no pCR vs. pCR was determined (right panel). (C) Bar graphs demonstrating the frequency of response of ER-positive and ER-negative cases based on median signature value (top panels) or a previously determined cutpoint (lower panels).
RB loss signature is associated with improved pathological response to other chemotherapy regimens
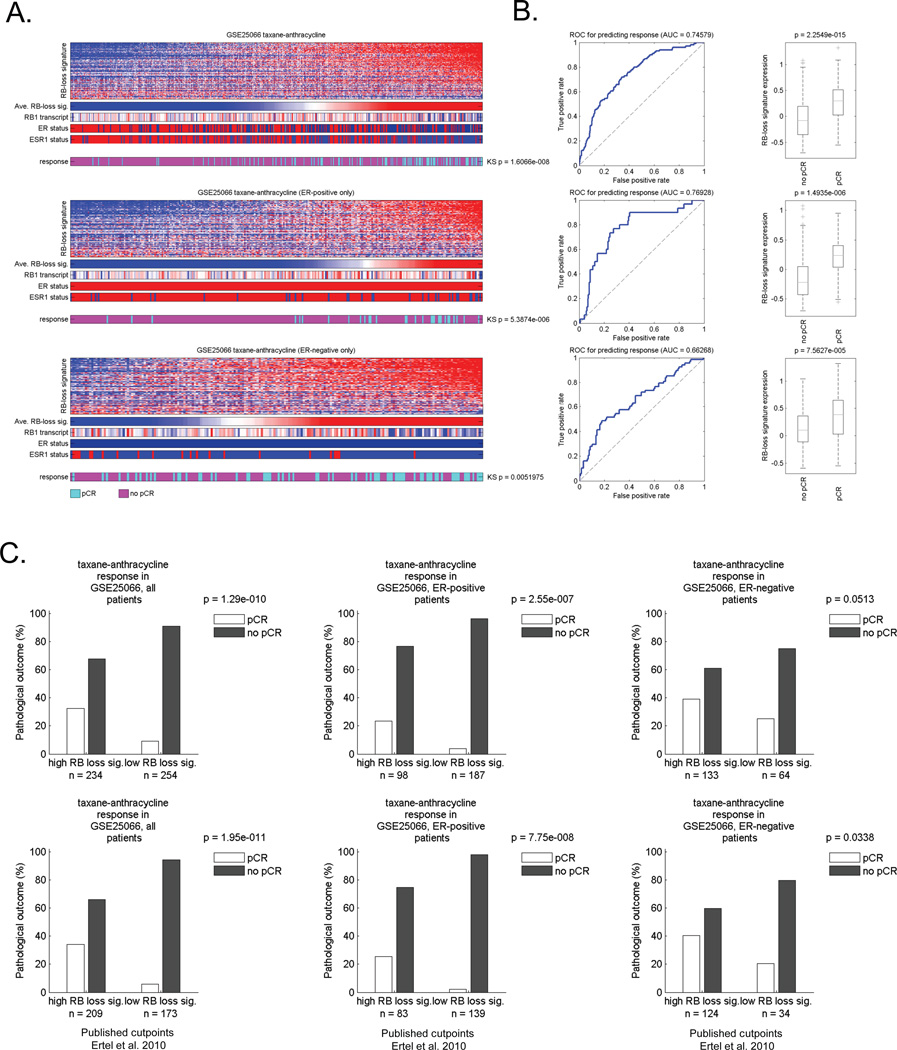
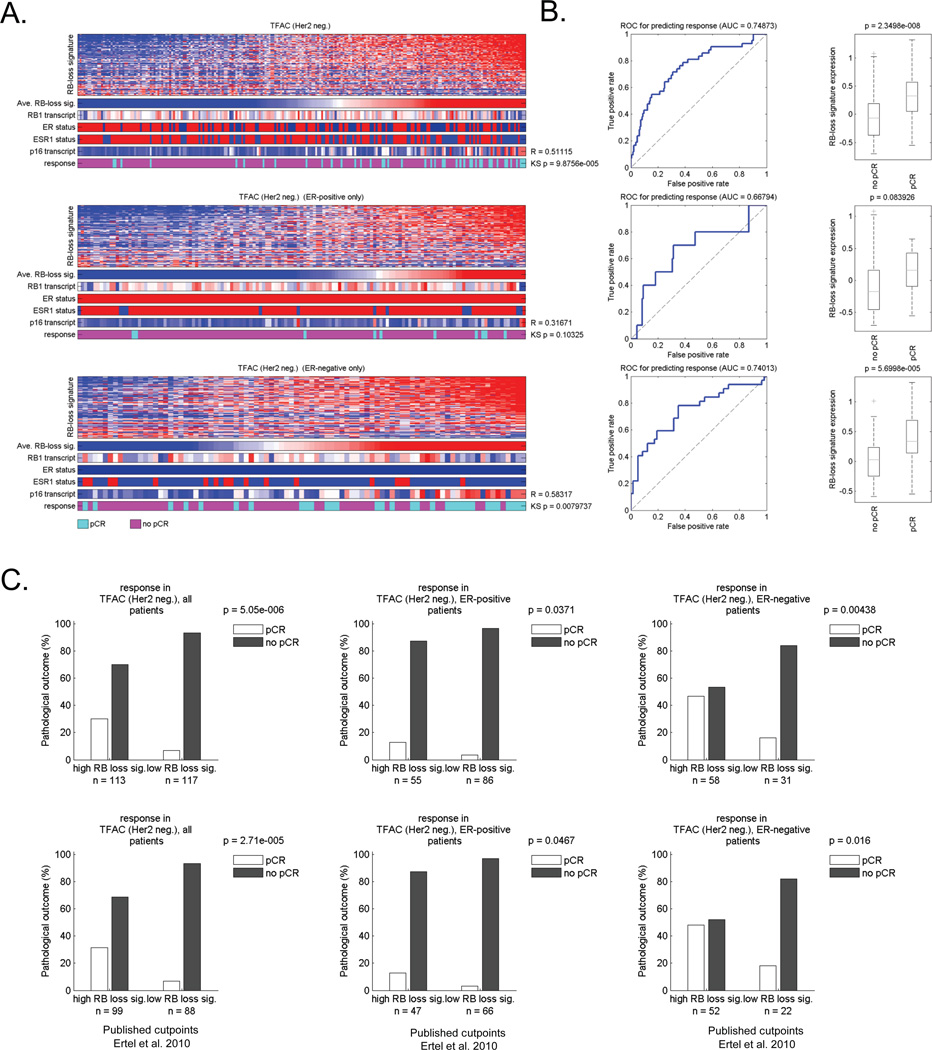
Since there are multiple forms of neoadjuvant chemotherapy used in the clinic, we also investigated cases that were treated with Taxane/Adriaymcin (TA) and Taxane/5-Fluorouracil/Adriamycin/Cytoxan (T/FAC) regimens (Figures 3 and 4, respectively) (26–28). In each cohort, the RB-loss signature was generally associated with pathological complete response (Figure 3A and 4A). The AUC was 0.75 for both TA and TFAC treatment (Figure 3B and 4B). These values were in very close agreement with that observed with FAC therapy, suggesting a wide-ranging impact of RB pathway dysfunction on the response to cytotoxic neoadjuvant chemotherapy regimens. Similarly, irrespective of ER-status, the RB loss signature was predictive for improved response to therapy (Figure 3B/C and 4B/C). Interestingly in the T/FAC cohort, there was a small subset of tumors which were Her2-positive (Supplemental Figures 1 and 5). Although the overall size of this subset was small, the RB loss signature had no predictive significance in these tumors (Supplemental Figure 5), suggesting that the predictive utility of RB-deficiency is most pronounced in luminal ER-positive and triple negative breast cancer.
Figure 3. The RB loss signature is associated with response to TA neoadjuvant therapy.
(A) Total tumors and ER-positive/ER-negative groupings were clustered based on the relative level of the RB loss signature. The heat map depicts all of the genes within the signature. Below the bars denote the average RB loss signature value, the RB1 transcript, clinically defined ER status, and the expression of the ESR1 transcript. Color bar provides the relationship to pathological complete response (pCR) vs residual or progressed disease (no pCR). The relationship of chemotherapy response to RB loss signature was determined using KS statistical modeling. (B) ROC analyses of the RB loss signature in this cohort was determined for all cases as well as by ER-status (left panel). The differentiation RB loss signature expression value was a function of no pCR vs. pCR was determined in total and ER-specific groups (right panel). (C) Bar graphs demonstrating the frequency of response based on median signature value (left panel) or a previously determined cutpoint (right panel).
Figure 4. The RB loss signature is associated with response to TFAC neoadjuvant therapy.
(A) Total tumors and ER-positive/ER-negative groupings were clustered based on the relative level of the RB loss signature. The heat map depicts all of the genes within the signature. Below the bars denote the average RB loss signature value, the RB1 transcript, clinically defined ER status, and the expression of the ESR1 transcript. Color bar provides the relationship to pathological complete response (pCR) vs residual or progressed disease (no pCR). The relationship of chemotherapy response to RB loss signature was determined using KS statistical modeling. (B) ROC analyses of the RB loss signature in this cohort was determined for all cases as well as by ER-stautus (left panel). The differentiation RB loss signature expression value was a function of no pCR vs. pCR was determined in total and ER-specific groups (right panel). (C) Bar graphs demonstrating the frequency of response based on median signature value (left panel) or a previously determined cutpoint (right panel).
Elevated p16ink4a expression correlates with the RB loss signature and is associated with improved pathological response
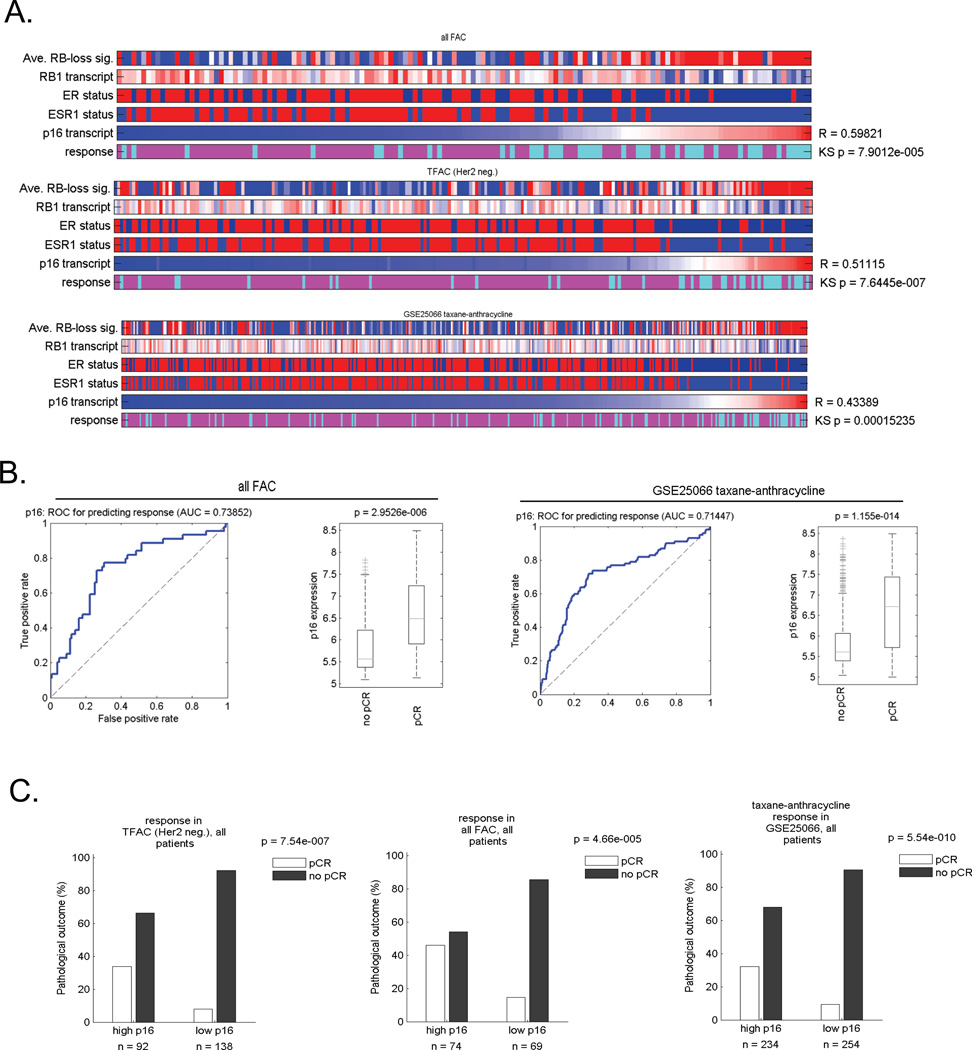
A key marker for RB pathway status in the clinic is the p16ink4a tumor suppressor (8, 24). In tumors that have lost RB function by deletion or viral oncoproteins, the expression of p16ink4a is highly elevated (8). Thus, p16ink4a staining is routinely used in the diagnoses of cervical and head and neck cancers that are human papilloma virus positive (8). Here we investigated the utility of p16ink4a as a single marker for neoadjuvant response. There was a significant correlation between RNA levels of the p16ink4a gene (CDKN2A) and the RB-loss signature (Figure 5A). Correspondingly, association between p16ink4a transcript level and pCR was significant based on testing with the KS statistic (Figure 5A). In ROC analyses, the AUC on the data sets was similar to that observed with the RB-loss signature (0.654 TFAC, 0.738 FAC, 0.7144 TA) and exhibited a similar enhanced expression in tumors that experienced a pCR (Figure 5B). The response rate of tumors with elevated expression approached that of RB deficient tumors (Figure 5C), and was relevant to both ER-positive and ER-negative cases (not shown). These combined studies suggest that p16ink4a levels and RB-status could be particularly relevant to the response of breast cancers to neoadjuvant chemotherapy.
Figure 5. The expression of CDKN2A is associated with RB deficiency and response to neoadjuvant therapy.
Cases treated with FAC, TA, and TFAC were clustered based on the relative expression of CDKN2A. The correlation between CDKN2A expression and the RB loss signature was determined (R-value shown). The RB1 expression, ER-status, ESR1 expression, and pathological response were shown. The statistical relationship with response was defined using the KS statistic. (B) ROC analyses of CDKN2A expression in these cohorts and the relationship relative expression levels in pCR vs. non pCR cases was determined. Data shown are for FAC and TA cases. (C) Bargraphs demonstrate the frequency of response based on the median CDKN2A expression value.
Immunohistochemical analyses of RB and p16ink4a demonstrate association with response to neoadjuvant chemotherapy
Gene expression profiling is a powerful approach for defining potential pathways involved in therapeutic response, but can be difficult to bring to the clinic (29). Additionally, it does not necessarily define the underlying lesion driving a particular gene expression signature, although the data with high levels of p16ink4a transcript support a direct role for the RB-pathway in response to neodjuvant chemotherapy. Therefore, we expanded our analysis by directly assessing the histological status of p16ink4a and RB protein in pretreatment biopsies from a cohort of patients treated with neoadjuvant chemotherapy (TJU cohort). Overall demographic and clinico-pathological features of this cohort are summarized in Table 1. RB negative tumors were statistically associated with ER negative status (P < 0.05) and higher nuclear grade (P < 0.05); however no association was found with Her2 status (Supplement Figure 4).
Table 1.
Clinical and pathological features of neoadjuvant cohort
| Factor | No. | % |
|---|---|---|
| Age, years | ||
| Median | 55 | |
| Range | 17–90 | |
| Clinical tumor size, cm | ||
| Median | 4.0 | |
| Mean | 4.3 | |
| Clinical stage at presentation | ||
| I | ||
| IIA | 1 | 1 |
| IIB | 25 | 26 |
| IIIA | 22 | 22 |
| IIIB | 20 | 20 |
| IIIC | 29 | 30 |
| 1 | 1 | |
| Pathologic tumor size, cm | ||
| Median | 0.9 | |
| Mean | 1.2 | |
| Range | 0–6 | |
| Post treatment stage | ||
| 0 | 20 | 20 |
| I | 31 | 31 |
| IIA | 19 | 19 |
| IIB | 9 | 9 |
| IIIA | 12 | 12 |
| IIIB | 0 | 0 |
| IIIC | 7 | 7 |
| ER status | ||
| Positive | 30 | 30 |
| Negative | 68 | 69 |
| PR status | ||
| Positive | 48 | 49 |
| Negative | 50 | 51 |
| HER2 status | ||
| Positive | 21 | 21 |
| Negative | 50 | 51 |
| Indeterminate | 10 | 10 |
| Unknown | 17 | 17 |
| Nuclear grade | ||
| 1 | 11 | 11 |
| 2 | 44 | 45 |
| 3 | 40 | 41 |
| Unknown | 3 | 3 |
| Neoadjuvant therapy | ||
| AC | 14 | 14 |
| ACT | 14 | 14 |
| AT | 8 | 8 |
| CMF | 34 | 35 |
| FAC | 7 | 7 |
| CT | 5 | 5 |
| Other | 9 | 9 |
| Unknown | 5 | 5 |
We found a clear reciprocal relationship between RB and p16ink4a tumors that exhibited low/absent p16ink4a staining were clearly positive for RB staining (Figure 6A). In contrast, tumors that exhibited high levels of p16ink4a were generally devoid of RB staining in the tumor compartment, although positive staining was apparent in stroma and lymphocytes (Figure 6B). Of the 97 cases evaluated, 26 were characterized by RB protein loss. The rate of pCR amongst these cases was 40.7% (Figure 6C, left panel). In contrast, in the remaining 71 cases that were clearly RB positive the pathological response rate was 12.8% (Figure 6C, left panel). One case exhibited mixed RB-positive and RB-negative portions of the tumor and was excluded from the analyses. In the case of p16ink4a there were 20 cases that exhibited robust staining (score 3), and the majority of these cases were RB deficient (not shown). The frequency of pathological complete response in this group was 38.1%, while in cases with intermediate and low p16ink4a staining (score 0–2) only 15.8% of tumors experienced a pathological complete response (Figure 6C, right panel). These data were significant in univariate analyses: p=0.0018 for RB loss and p=0.0253 for p16ink4a high.
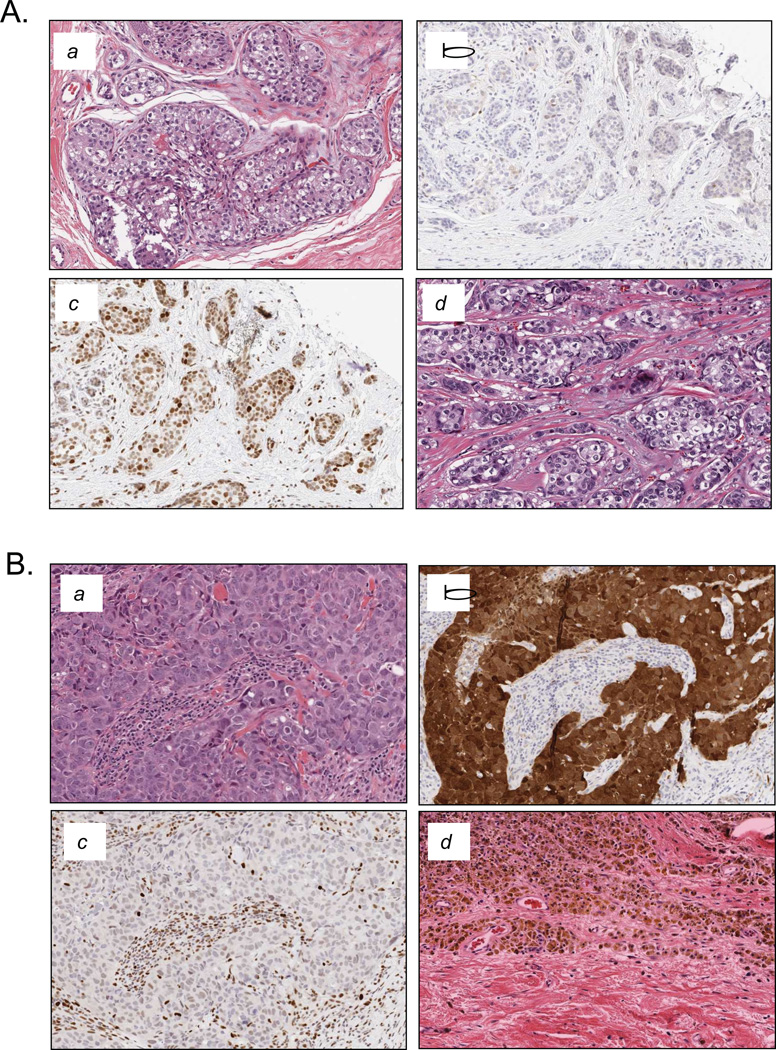
Figure 6. Histological analyses of RB and p16ink4a in neoadjuvant treated cases.
(A) Representative case that failed to respond to neoadjuvant chemotherapy. a. Pretreatment specimen stained with Hematoxylin/Eosin. b. Pretreatment specimen stained for p16ink4a expression with low staining in tumor. c. Pretreatment specimen stained for RB showing robust nuclear staining in tumor. d. Post treatment specimen showing residual disease. (B) Representative case that responded to neoadjuvant chemotherapy. a. Pretreatment specimen stained with Hematoxylin/Eosin. b. Pretreatment specimen stained for p16ink4a expression showing robust tumor specific staining. c. Pretreatment specimen stained for RB showing lack of staining in the tumor tissue (stroma/leukocytes stain positive for RB). d. Post treatment specimen showing complete pathological response. (C) Bar graphs depicting the response frequency stratified by RB status (left panel) or p16ink4a levels (right panel). (D) Scatter plots showing the Miller-Payne Score of individual cases in the cohort. Cases were stratified by RB status (left panel) or p16ink4a levels (right panel). (E) Scatter plots showing the CPS of individual cases in the cohort. Cases were stratified by RB status (left panel) or p16ink4a levels (right panel).
To determine if the sensitivity of tumors to chemotherapy was only reflective of complete response or if there was also a relationship with higher degrees of pathological response, the Miller-Payne and the Clinical-Pathologic Scoring (CPS) criteria were applied (22, 23). The Miller-Payne criteria takes into account the overall cellularity of the tumor, wherein grade 1 represent no reduction in cellularity, grade two represent a minor loss of tumor cells (up to 30%), grade 3 represents a 30–90% reduction in cellularity, grade 4 represents few distributed viable cells, and grade 5 represents no viable malignant cells. Amongst RB-negative cases, 13 of 25 (52%) experienced a grade 5 response, and the median response across all cases was 4 (average 4.2) (Figure 6D, left panel). In contrast, only 13 of 71 (18.3%) of the RB-positive cases experienced a grade 5 response, with a median response of 3 (average 3.2) (Figure 6D). Thus, the overall response to neoadjuvant chemotherapy, measured by Miller Payne criteria, was significantly higher in RB-negative tumors (p=0.0004). A similar improved response was observed with high p16ink4a (p=0.0063) cells. The CPS compares the pretreatment clinical tumor stage with the post treatment pathological evaluation. A low score is indicative of pathological response. For both RB-loss and p16ink4a high cases the median CPS was 1, while for RB-positive or p16ink4a intermediate/low tumors the median CPS was 2 (Figure 6E). Thus, by multiple different criteria dysregulation of the RB-pathway predicts improved response to neoadjuvant chemotherapy.
DISCUSSION
While molecular targeted therapies are generally considered the future of cancer treatment, current conventional cytotoxic chemotherapies can be quite effective in the treatment of breast cancer (5, 26). However, there are few accepted markers to define patients that will benefit from such therapies. Thus, there is significant concern that many patients are treated with therapies that have little benefit, with potential serious side effects. In the neoadjuvant setting, a pathological complete response is associated with long term durable response.; the CPS score can be employed to approximate the likelihood of 5 year disease free survival (23). Our data suggests that disruption of the retinoblastoma tumor suppressor pathway is a useful predictive marker of response to neoadjuvant chemotherapy. This was observed using three different approaches (RB-signature, p16ink4a levels, and RB histological levels), multiple chemotherapy regimens, different scoring criteria, and greater than 600 cases.
Since the RB-pathway can be disrupted by multiple mechanisms, a molecular profiling approach provides a means to quantitatively assess multiple modes of RB dysregulation (15). The RB-loss signature was specifically defined in preclinical models of RB manipulation, and represents a gene expression profile that reflects the functional inactivation of the protein as can occur through multiple mechanisms in cancer (15). The signature was associated with ER-negative disease, and pathological complete response. Because there is a general improved response to chemotherapy in ER-negative cases, it was important to independently evaluate RB-status with respect to ER-status. These data showed that in both ER-positive and ER-negative tumors RB loss was associated improved response to neoadjuvant therapy. In these analyses, we largely excluded Her2-positive cases due to the differential treatment for such tumors (i.e., incorporation of Her2-antagonists). Thus, our studies were generally applicable to luminal and triple negative breast cancer. Since there are many different neoaduvant chemotherapy regimens, used largely at physician discretion, any classifier of chemotherapeutic response must be applicable to a range of regimens. The RB-loss signature was relevant with multiple chemotherapy regimens, suggesting a potential general utility. However, it is possible that for certain regimens the RB-loss signature would not have utility. This was apparently the case in Her2-positive cancers, wherein RB-loss signature did not associate with response in a limited sub-type analyses. Furthermore, the association of the RB-loss signature with improved response was not applicable to all neoadjuvant regimens, since using the same methodology it was associated with poor response to neoadjuvant letrozole treatment (not shown). These data suggest that RB dysfunction could be a specific determinant of response to chemotherapy, and define ER-positive tumors that would experience little benefit from endocrine therapy and a substantial benefit from neoadjuvant chemotherapy.
Because diagnostic testing using a molecular signature in the clinical environment can be unwieldy (29), we evaluated whether surrogates of the RB-loss signature could be useful for predicting pathological response. Interestingly, Rb1 transcript levels are not a particularly useful determinant of RB status in tumors (15, 30). However, elevated p16ink4a transcript and protein levels are known to occur in multiple settings of RB loss (8). As shown here, p16ink4a mRNA levels were highly correlated with RB loss signature, and as a single marker was differentially associated with tumors experiencing a pathological response. The involvement of p16ink4a protein levels as a marker of response was also interrogated in a retrospective analyses of neoadjuvant cases. A high level of p16ink4a was associated with improved pathological response, whereas p16ink4a loss/low was associated with a worse response to neoadjuvant therapy. These studies are concordant with the observation in head and neck cancer that p16ink4a high tumors (those that have RB-pathway inactivated by HPV or other mechanisms) harbor an improved therapeutic response (31). Interestingly, the relatively few cases that had an intermediate level of p16ink4a staining experienced an intermediate pathological response rate between high and low or absent staining. As expected there was a significant inverse correlation between p16ink4a and RB status. This correlation was not absolute and several RB deficient tumors did not exhibit high levels of p16ink4a. The findings with elevated p16ink4a strongly supported that disruption of RB protein function is a key determinant of tumors that experience a pathological response to neoadjuvant therapy.
The preceding findings supported directly evaluating RB protein levels in clinical specimens. The staining for RB was extensively optimized and incorporated the use of positive/negative controls. In the cohort analyzed, RB loss was more prevalent in ER-negative cases (~50%), but also occurred in ER-positive cases (~18%). Histological RB loss was strongly associated with improved response to neoadjuvant chemotherapy as determined by frequency of pathological complete response. While the data were significant there is room to enhance the predictive value of RB loss, and presumably combining with other tumor suppressors (e.g. p53 and PTEN) or oncogenes (e.g. PIK3CA) the sensitivity and specificity could be enhanced. Importantly there was a striking agreement between the in silico profiling data and the direct analyses of RB by immunostaining. Interestingly, RB loss was a slightly better marker of response than p16ink4a, and defined cases with moderate or low staining for p16ink4a that exhibited a pathological complete response. Using CPS and Miller-Payne scoring systems, it was apparent that there was also general improvement in response to neoadjuvant therapy not solely at the level of complete response which again modestly outperformed p16ink4a staining. Together, these data indicate that the loss of RB, which occurs relatively frequently in locally advanced disease, could be a useful tool for defining patients that experience an improved response to neoadjuvant chemotherapy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Critical pathways involved in the response to chemotherapy in breast cancer have remained elusive. Hence, there are no commonly employed markers to define patients that will benefit from such treatment. In this study, a combination of gene expression profiling and direct histological analyses were performed to determine the relevance of the retinoblastoma tumor suppressor (RB) pathway to the pathological response to neo-adjuvant chemotherapy. Using multiple approaches and independent cohorts these analyses revealed that disruption of RB function is associated with improved response to multiple therapeutic regimens in both ER-positive and ER-negative breast cancer. Together, these data indicate that the loss of RB, which occurs relatively frequently in locally advanced disease, could be a useful tool for identifying patients with improved response to neoadjuvant chemotherapy.
REFERENCES
- 1.Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity. Nat Rev Clin Oncol. 2010;7:139–147. doi: 10.1038/nrclinonc.2009.234. [DOI] [PubMed] [Google Scholar]
- 2.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 3.Dawson SJ, Provenzano E, Caldas C. Triple negative breast cancers: clinical and prognostic implications. Eur J Cancer. 2009;45(Suppl 1):27–40. doi: 10.1016/S0959-8049(09)70013-9. [DOI] [PubMed] [Google Scholar]
- 4.Perez EA, Moreno-Aspitia A, Aubrey Thompson E, Andorfer CA. Adjuvant therapy of triple negative breast cancer. Breast Cancer Res Treat. 2010;120:285–291. doi: 10.1007/s10549-010-0736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 6.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008 doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knudsen ES, Wang JY. Targeting the RB-pathway in cancer therapy. Clin Cancer Res. 2010;16:1094–1099. doi: 10.1158/1078-0432.CCR-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witkiewicz AK, Knudsen KE, Dicker AP, Knudsen ES. The meaning of p16(ink4a) expression in tumors: functional significance, clinical associations and future developments. Cell Cycle. 2011;10:2497–2503. doi: 10.4161/cc.10.15.16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subhawong AP, Subhawong T, Nassar H, Kouprina N, Begum S, Vang R, et al. Most basal-like breast carcinomas demonstrate the same Rb-/p16+ immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol. 2009;33:163–175. doi: 10.1097/PAS.0b013e31817f9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosco EE, Knudsen ES. RB in breast cancer: at the crossroads of tumorigenesis and treatment. Cell Cycle. 2007;6:667–671. doi: 10.4161/cc.6.6.3988. [DOI] [PubMed] [Google Scholar]
- 13.Derenzini M, Donati G, Mazzini G, Montanaro L, Vici M, Ceccarelli C, et al. Loss of retinoblastoma tumor suppressor protein makes human breast cancer cells more sensitive to antimetabolite exposure. Clin Cancer Res. 2008;14:2199–2209. doi: 10.1158/1078-0432.CCR-07-2065. [DOI] [PubMed] [Google Scholar]
- 14.Trere D, Brighenti E, Donati G, Ceccarelli C, Santini D, Taffurelli M, et al. High prevalence of retinoblastoma protein loss in triple-negative breast cancers and its association with a good prognosis in patients treated with adjuvant chemotherapy. Ann Oncol. 2009;20:1818–1823. doi: 10.1093/annonc/mdp209. [DOI] [PubMed] [Google Scholar]
- 15.Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, et al. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–4163. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zagorski WA, Knudsen ES, Reed MF. Retinoblastoma deficiency increases chemosensitivity in lung cancer. Cancer Res. 2007;67:8264–8273. doi: 10.1158/0008-5472.CAN-06-4753. [DOI] [PubMed] [Google Scholar]
- 17.Bosco EE, Wang Y, Xu H, Zilfou JT, Knudsen KE, Aronow BJ, et al. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117:218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Generali D, Berruti A, Foroni C, Bazzola L, Andreis D, Allevi G, et al. Molecular oncology and the neoadjuvant setting: the perfect blend for treatment personalization and clinical trial design. J Natl Cancer Inst Monogr. 2011:67–70. doi: 10.1093/jncimonographs/lgr029. [DOI] [PubMed] [Google Scholar]
- 19.Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. 2010;17:R245–R262. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- 20.Ertel A, Tozeren A. Switch-like genes populate cell communication pathways and are enriched for extracellular proteins. BMC Genomics. 2008;9:3. doi: 10.1186/1471-2164-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witkiewicz AK, Rivadeneira DB, Ertel A, Kline J, Hyslop T, Schwartz GF, et al. Association of RB/p16-pathway perturbations with DCIS recurrence: dependence on tumor versus tissue microenvironment. Am J Pathol. 2011;179:1171–1178. doi: 10.1016/j.ajpath.2011.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogston KN, Miller ID, Payne S, Hutcheon AW, Sarkar TK, Smith I, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast. 2003;12:320–327. doi: 10.1016/s0960-9776(03)00106-1. [DOI] [PubMed] [Google Scholar]
- 23.Mittendorf EA, Jeruss JS, Tucker SL, Kolli A, Newman LA, Gonzalez-Angulo AM, et al. Validation of a novel staging system for disease-specific survival in patients with breast cancer treated with neoadjuvant chemotherapy. J Clin Oncol. 2011;29:1956–1962. doi: 10.1200/JCO.2010.31.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witkiewicz AK, Rivadeneira DB, Ertel A, Kline J, Hyslop T, Schwartz GF, et al. Association of RB/p16-Pathway Perturbations with DCIS Recurrence Dependence on Tumor versus Tissue Microenvironment. Am J Pathol. 2011;179:1171–1178. doi: 10.1016/j.ajpath.2011.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thangavel C, Dean JL, Ertel A, Knudsen KE, Aldaz CM, Witkiewicz AK, et al. Therapeutically activating RB: reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr Relat Cancer. 2011;18:333–345. doi: 10.1530/ERC-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 28.Tabchy A, Valero V, Vidaurre T, Lluch A, Gomez H, Martin M, et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin Cancer Res. 2010;16:5351–5361. doi: 10.1158/1078-0432.CCR-10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol. 2010;9:58–64. doi: 10.1038/nrclinonc.2011.125. [DOI] [PubMed] [Google Scholar]
- 30.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rischin D, Young RJ, Fisher R, Fox SB, Le QT, Peters LJ, et al. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol. 2010;28:4142–4148. doi: 10.1200/JCO.2010.29.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.