Abstract
HIV vaccine research increasingly uses polychromatic flow cytometry as a tool to monitor T cell responses. The use of this technology allows for the analysis of highly defined subsets of cells with unique phenotypes and functions. Ultimately, such studies may identify surrogate markers of protection from disease progression. However, this powerful technology comes with a number of technical hurdles, and there is a need to standardize the assays and protocols used in clinical trial monitoring. Here an optimized protocol, with variations for specific circumstances, is presented. This protocol covers the analysis of multiple cytokines, cell surface markers, and other functional markers such as perforin, CD107, and CD154. While the protocol can be adapted to various numbers of fluorescence parameters, optimized panels of 8–10 colors are presented.
Keywords: Antigen-specific, intracellular staining, multicolor, polychromatic, fixation, permeabilization, AIDS vaccine research
1. Introduction
HIV vaccine researchers have been among the first to adopt polychromatic flow cytometry (more than four colors) as a tool to dissect T cell responses to HIV infection and to HIV vaccines. There is little doubt that a successful HIV vaccine will need to induce a strong cellular immune response, as well as neutralizing antibodies (1). However, the nature of a protective cellular immune response to HIV is only beginning to be elucidated, largely by studies in the SIV model (2–8), and by examination of HIV+ long-term nonprogressors (9 –11). The ability of polychromatic flow cytometry to interrogate multiple subsets of immune cells for their functional capacities makes it a powerful tool for discovering potential surrogates of protection. In fact, it could be argued that this technology provides the most in-depth view currently possible into the workings of the human immune system at a cellular level.
What are the potential surrogates of T cell-based protection from HIV progression? Obviously, no definitive answers are yet available, as the only information comes from animal models or correlative studies in humans. However, current evidence suggests that HIV+ nonprogressors maintain proliferative capacity of their HIV-specific CD4+ and CD8+ T cells (9, 10), at least in part by maintenance of the ability to produce IL-2 (10–12). In fact, recent studies using polychromatic flow cytometry suggest that a greater proportion of HIV-specific CD8+ T cells in nonprogressors are “multifunctional” (11), being able to produce several cytokines (e.g., IFNγ, TNFα, IL-2, and MIP-1β) and to degranulate (as evidenced by cell-surface CD107 expression).
Phenotypic studies in SIV models suggest that protected animals maintain T cells of a “central memory” phenotype (CD28+, CD95 +) in contrast to nonprotected animals (7, 8). HIV+ progressors are known to show altered differentiation of HIV-specific T cells, as seen by staining for markers such as CCR7, CD62L, CD27, CD28, CD45RA, and CD127 (13–19). Very recent work indicates that upregulation of a death receptor, PD-1, on HIV-specific T cells may lead to dysfunction of those cells and consequent disease progression (20, 21). And extensive literature has correlated an elevated expression of activation markers such as CD38 and HLA-DR on CD8+ T cells with poor prognosis [reviewed in (22)].
Some researchers may believe that functions (e.g., cytokines and degranulation capacity) are more important than phenotypes (e.g., memory/effector markers) in categorizing antigen-specific T cells. But the emergence of markers such as PD-1 suggests that cell-surface proteins may hold important prognostic value as well. Thus, researchers find themselves in the position of wanting to monitor an ever-growing number of phenotypic and functional markers; and polychromatic flow cytometry is the best available tool to meet that goal.
1.1. Instrumentation Considerations
Polychromatic flow cytometry has been made practical only recently by the availability of commercial instruments with digital signal processing and detectors for up to 18 colors. Digital processing is important in that it allows for more precise calculation of optical spillover between detectors, and thus more precise compensation than was possible with analog systems (23,24). Equally important, software routines are now available that will automate the compensation process when presented with a set of single-color fluorescent controls. This makes it possible to collect data in eight or more colors with nearly the same ease as traditional two to four color experiments, despite an exponentially more complex spillover matrix.
Much of the instrumentation for polychromatic flow cytometry is customized, which means that each user’s system can have different lasers and optical filters. This in turn can alter the efficiency of detection of particular fluorochromes, such that the same antibody panel on one instrument will not yield identical results on another instrument. Some degree of standardization, or at least awareness of these variables, is beginning to occur, with the emergence of groups sharing their experiences (see for example http://maeckerlab.typepad.com). A minimum level of instrument standardization will need to be defined in order to achieve comparable results with polychromatic antibody panels on different instruments. Such standardization is a prerequisite to doing multicenter trials in which polychromatic flow cytometry will be performed at more than one site; and it is certainly a prerequisite to comparing results from different vaccine trials.
Even with two identical cytometers, setup of the instrument is still a variable that can vastly alter results. There has been recent progress in automated software routines using standardized particles to create optimal instrument setups (e.g., CST, BD Biosciences). For users not equipped with such automated setup paradigms, manual procedures need to be defined that optimize photomultiplier tube (PMT) voltage gains (25), as well as optimize instrument performance in general (26). Finally, monitoring of certain cytometer parameters over time is an important quality control tool to ensure consistent data and to anticipate and/or identify potential problems that might require service.
1.2. Reagent Considerations
The mere possession of an instrument capable of detecting, for example, 12 fluorescence parameters, does not guarantee success in 12-color flow cytometry. Choice of fluorochromes, antibody specificities, and the combination of these into an optimized reagent panel, are important considerations. In many cases, increasing a panel by one additional fluorochrome detracts so severely from the resolution sensitivity in other detectors, that it is not warranted. This occurs because of fluorescence spillover, the fact that each fluorochrome contributes some signal to neighboring detectors as well as to its primary detector. Because the optical spectrum is limited, the addition of new fluorescent reagents becomes more and more difficult without creating severe spillover problems, as the number of fluorochromes in an experiment increases. This is clearly a case where more is not always better.
Selection of fluorochromes and antibody conjugates based on brightness and minimal spillover has been recently reviewed (24). In general, one should start by selecting a set of fluorochromes that offer the greatest brightness, within the constraints of the user’s instrument, while minimizing spectral overlaps between detectors. Suggestions of fluorochrome sets to use for common instrument configurations are given in Table 25.1.
Table 25.1.
Suggested fluorochrome configurations
| Laser | 6-color | 8-color | 10-color | 12-color |
|---|---|---|---|---|
| 488 nm | FITC | FITC | FITC | FITC |
| PE | PE | PE | PE | |
| 488 nm or 532 nm | PE-Texas Red, PE-Alexa 594 or PE-Alexa 610 | PE-Texas Red, PE-Alexa 594 or PE-Alexa 610 | ||
| PerCP-Cy5.5 | PerCP-Cy5.5 | PerCP-Cy5.5 | PerCP-Cy5.5 | |
| PE-Cy7 | PE-Cy7 | PE-Cy7 | PE-Cy7 | |
| APC | APC | APC | APC | |
| 633 nm | Alexa 680 or Alexa 700 | Alexa 680 or Alexa 700 | ||
| APC-Cy7 | APC-Cy7 | APC-Cy7 | APC-Cy7 | |
| Pacific Blue | Pacific Blue | Pacific Blue | ||
| 405 nm | AmCyan | AmCyan | AmCyan | |
| Qdot 655 | ||||
| Qdot 705 |
Next, one should assign antibody specificities to particular fluorochromes by matching the dimmest specificities with the brightest fluorochromes. Further adjustments should then be made to minimize potential spillover issues. For example, two fluorochromes with significant spectral overlap might be used to identify non-overlapping cell populations, thereby negating their spillover. Conversely, one should avoid compromising a reagent for which high sensitivity is required by having a reagent in a neighboring detector that brightly stains the same cell population. For example, use of CD8 APC-Cy7 with anti-IL-2 APC is a potential problem, if IL-2 is to be detected on CD8+ cells. In this case, not only does APC-Cy7 cause spillover into the APC detector, but the tandem dye can also degrade, resulting in false positive signals in APC (24). Among tandem dyes, APC-Cy7, followed by PE-Cy7, are most sensitive to such degradation, which is catalyzed by light, increased temperature, and exposure to fixative (24). Finally, all of the above considerations need to be tempered by what antibody conjugates are commercially available or can be made by the investigator.
Once an antibody panel is selected, titration of certain reagents is often required to achieve an optimal signal:noise ratio. While many reagents are sold pre-titered, this does not always mean that the specified titer is optimal in a given application. Since polychromatic experiments already compromise sensitivity due to spillover between detectors, the need for optimal signal:noise is critical. It should also be noted that the optimal titer for cell-surface staining with an antibody is often higher than that for intracellular staining (after fixation and permeabilization). For many antibody specificities, resolution is compromised so drastically by fixation and permeabilization that these antibodies need to be used prior to application of a fixative.
Given the many considerations in optimizing a reagent panel for polychromatic flow cytometry, it makes sense to take advantage, wherever possible, of panels already validated by others. Table 25.2 shows some staining panels successfully used in the author’s laboratory. These can be taken as a starting point, given that small variations are still likely to be successful, as opposed to starting from “scratch” in the design of a new panel.
Table 25.2.
Some suggested multicolor antibody panels
| 8-color1 | 10-color2 | 10-color3 |
|---|---|---|
| Anti-IFNγ FITC | CD27 FITC | Anti-IFN FITC |
| Anti-IL-2 PE | CD154 PE | Anti-IL-2 PE |
| CD107 PE-Alexa 610 | Anti-TNFα PE-Alexa 610 |
|
| CD28 PerCP-Cy5.5 | CD4 PerCP-Cy5.5 | CD28 PerCP-Cy5.5 |
| CD45RA PE-Cy7 | Anti-IFNγ PE-Cy7 | CD45RA PE-Cy7 |
| CD27 APC | Anti-IL-2 APC | CD27 APC |
| Anti-TNFα Alexa 700 | CD3 Alexa 700 | |
| CD8 APC-Cy7 | CD8 APC-Cy7 | CD8 APC-Cy7 |
| CD3 Pacific Blue | CD3 Pacific Blue | CD4 Pacific Blue |
| CD4 AmCyan | CD14 AmCyan | CD14 AmCyan |
Used in ref. (19).
Used for International Flow Cytometry School (IFCS) 2006, Florence, Italy. Note that readout of IL-2 on CD8+ cells could be compromised by the use of CD8 APC-Cy7 with anti-IL-2 APC (see Section 1.2).
Used for Multicolor ICS Users Group standardization studies (see also http://maeckerlab.typepad.com for additional panel suggestions and details).
The basics of intracellular cytokine staining have been reviewed elsewhere (27, 28), and tips for optimizing protocols have also been recently published (29). The main focus of the protocol presented here is to show how intracellular cytokine staining can be adapted to a polychromatic format, which may include readout not only of phenotypic markers, but also of functional markers such as CD107 or CD154, in addition to cytokines. Variables in the stimulation and processing steps that apply to these markers and others are summarized in Table 25.3.
Table 25.3.
Procedural variables for different functional markers
| Variable | Covered by this protocol IL-2, IL-4, IL-5, IL-102, IL-13, IFNγ, MIP-1β, TNFα | CD107, CD154 | Not covered1 TGFβ |
|---|---|---|---|
| Stimulation conditions | 6–12 h | 5–6 h in the presence of staining antibodies for these markers | 16–24 h in serum-free medium3 |
| Secretion inhibitor | brefeldin A | monensin4 | monensin |
| Fixation/permeabilization system | FACS Lysing Solution, FACS Permeabilizing Solution 25 | FACS Lysing Solution, FACS Permeabilizing Solution 25 | Cytofix, Cytoperm6 |
This marker would be difficult to combine with those in the first two columns, mainly due to the longer optimal stimulation time.
We have performed IL-10 staining with some positive results under the listed conditions, but have not attempted to optimize conditions for IL-10 detection.
Serum-free medium (e.g., AIM V, Invitrogen, Grand Island, NY) produces much stronger TGFβ responses, presumably because serum contains free TGFβ that blocks staining for this marker.
CD107 and CD154 are taken up by endocytic vescicles and degraded. Monensin blocks this degradation by preventing acid-ification of these vescicles. When doing combined assays with cytokines, a monensin+brefeldin A combination is recommended (see Note 5).
BD Biosciences, San Jose, CA. Note that the Cytofix/Cytoperm system (BD Biosciences, San Diego, CA) is also used successfully for these markers by many investigators.
BD Biosciences, San Diego, CA.
2. Materials
2.1. Reagents
Freshly isolated or cryopreserved PBMC, isolated by Ficoll gradient centrifugation or via Cell Preparation Tubes (CPT; BD Vacutainer, Franklin Lakes, NJ) or equivalent.
RPMI-1640 medium with 20 mM HEPES, 10% fetal bovine serum, and antibiotic/antimycotic solution (cRPMI-10, components from Sigma Chemical Co., St. Louis, MO).
Stimulation antigens, e.g., peptide mixes [see ref. (30)], or SEB as a positive control. Optional: Preconfigured plates containing lyophilized stimulation reagents and secretion inhibitor(s) can be purchased [BD Lyoplate, BD Biosciences, San Jose, CA (31)].
Recommended: costimulatory antibodies to CD28 and CD49d, 0.1 mg/mL each in sterile PBS (FastImmune, BD Biosciences).
Brefeldin A, 5 mg/mL in DMSO (Fast Immune, BD Biosciences); or brefeldin A+monensin, 2.5 mg/mL each in 50% DMSO+50% methanol. For the latter stock, combine brefeldin A and monensin (Golgistop, BD Biosciences) 1:1. Aliquot and store both stocks at −20°C.
EDTA, 20 mM in PBS (pH 7.4) (FastImmune, BD Biosciences)
Cell fixation reagent, e.g., BD FACS Lysing Solution (BD Biosciences) or equivalent.
Cell permeabilizing reagent, e.g., BD FACS Permeabilizing Solution 2 (BD Biosciences) or equivalent.
Fluorescent-labeled antibodies (see for example Table 25.2). Optional: Preconfigured plates containing lyophilized antibody cocktails [BD Lyoplate, BD Biosciences (31)].
Wash buffer: 0.5% bovine serum albumin +0. 1% NaN3 in PBS.
Recommended: BD CompBeads [anti-mouse Ig κ, anti-rat Ig κ, or anti-rat/hamster Ig κ (BD Biosciences)], for creating single-color compensation controls.
Optional: To reduce biohazard potential, or if samples will be stored > 24 h prior to acquisition: 1% paraformaldehyde in PBS [dilute 10% paraformaldehyde (EM Science, Gibbstown, NJ) 1:10 in PBS]; or BD Stabilizing Fixative (BD Biosciences).
2.2. Equipment
96-well conical bottom polypropylene plates with lids [e.g., BD Falcon (Bedford, MA) or equivalent] (see Note 1).
12-channel aspiration manifold with 7-mm prongs (V&P Scientific, San Diego, CA).
Plate holders for table-top centrifuge [e.g., Sorvall Instruments (Newtown, CT)].
Polychromatic flow cytometer with digital signal processing, e.g., BD LSR II (BD Biosciences) or Dako Cyan ADP (Dako Corporation, Fort Collins, CO).
Optional: 96-well plate loader for flow cytometer.
3. Methods
3.1. Sample Collection
For fresh PBMC (see Note 2): Resuspend at 5 × 106 to 1 × 107 viable lymphocytes/mL in warm (37 °C) cRPMI-10 (see Note 3).
For cryopreserved PBMC (see Note 4): Thaw briefly in a 37 °C water bath, then slowly dilute up to 10 mL with warm (37 °C) cRPMI-10 and centrifuge for approximately 7 min at 250 × g. Resuspend in a small volume of warm cRPMI-10, perform a viable cell count, and dilute to a final concentration of 5 × 106 to 1 × 107 viable lymphocytes/mL (see Note 3).
Add 200 μL of cell suspension per well to a 96-well plate (see Section 2.2.1 for appropriate plates). For cryopreserved PBMC, incubate at 37 °C for 6–18 h prior to stimulation (see Note 4).
3.2. Cell Activation
For assays not involving CD107 or CD154: Thaw an aliquot of 5 mg/mL brefeldin A stock (see Note 5). Dilute 1:10 in sterile PBS to make a 50× working stock.
For assays measuring CD107 and/or CD154: Thaw an aliquot of 2.5 mg/mL brefeldin A+2.5 mg/mL monensin stock (see Note 5). Dilute 1:10 in sterile PBS to make a 50× working stock.
For assays using preconfigured lyophilized stimulation reagents in plates: Add 200 μL of cell suspension directly to the appropriate wells, let sit for a few minutes, then pipet up and down thoroughly to mix. Skip to step 3.2.7.
Resuspend peptides or peptide mixes in DMSO at a concentration of 500 μg/mL/peptide or greater (see Note 6). Store resuspended peptides in aliquots at −80 °C. Dilute peptide stocks in sterile PBS, if necessary, to achieve a 50× working stock that is between 50 and 100 μg/mL/peptide (when diluted 1:50, this will yield a final concentration of 1–2 μg/mL/peptide).
Prepare a 50× SEB stock of 50 μg/mL in sterile PBS. Store this stock at 4 °C (aliquoting is not necessary).
-
For each stimulation condition, prepare a “master mix” of the 50× working stocks and costimulatory antibodies as follows:
4μL/well peptides, SEB (positive control), or PBS (negative control).
4μL/well brefeldin A or brefeldin A+monensin.
4μL/well CD28 + CD49d Ab stock (see Note 7).
Pipet 12 μL of the appropriate master mix into each well containing cells. Mix by gently pipetting.
For assays involving CD107 and/or CD154, also add the recommended titer of the antibody conjugate(s) to each well. Minimize exposure to light, particularly for tandem dye conjugates (see Note 8).
Incubate covered plate for 6–12 h at 37 °C (see Notes 9 and 10).
3.3. Sample Processing
To halt activation and detach adherent cells, add 20 μL per well of 20 mM EDTA in PBS and mix by pipetting.
Incubate 15 min at room temperature, then mix again by vigorous pipetting to fully resuspend adhered cells.
Centrifuge plate at 250 × g for 5 min. Aspirate supernatant with 7 mm vacuum manifold (see Note 11).
-
For assays using amine-reactive dye for staining non-viable cells: Resuspend the amine dye at optimum concentration in PBS (usually around 2. 5 μg/mL, but this should be determined for individual lots of dye). Resuspend each well with 100 μL of this solution, incubate 20 min at room temperature, then add 100 μL wash buffer, and wash as in step 3.3.3 above.
For assays using liquid reagents and cell-surface markers other than CD3, CD4, and CD8: Resuspend each well in 100 μL wash buffer and add optimal titers of all Abs to cell-surface markers (see Note 12), incubate 30–60 min at room temperature, then add 100 μL wash buffer, and wash as in step 3.3.3 above.
For assays using preconfigured lyophilized staining reagents and cell-surface staining Abs: Resuspend the appropriate wells of the surface Ab plate with 50 μL of wash buffer. Let sit for a few minutes, then pipet up and down thoroughly to mix. Transfer the solution to appropriate wells of the cell plate, incubate 30–60 min at room temperature in the dark, then add 100 μL wash buffer, and wash as in step 3.3.3 above.
Resuspend cell pellets with 100 μL of 1× BD FACS Lysing Solution per well. Incubate at room temperature for 10 min (see Notes 13 and 14).
Add 100 μL wash buffer to each well, then centrifuge plate at 500 × g for 5 min (see Note 15). Aspirate supernatant with 7 mm vacuum manifold.
Resuspend cell pellets with 200 μL of 1× BD FACS Permeabilizing Solution 2 per well. Incubate at room temperature for 10 min (see Note 14).
Centrifuge plate at 500 × g for 5 min (see Note 15). Aspirate supernatant with 7 mm vacuum manifold.
Add 200 μL wash buffer to each well, and wash as in step 3.3.8 above.
Again add 200 μL wash buffer to each well, and wash as in step 3.3.8 above.
For assays using liquid reagents: Resuspend pellet in 100 μL wash buffer and add optimal titers of all Abs to intracellular markers. Incubate in the dark at room temperature for 60 min, mixing by pipetting or gentle agitation every 15–20 min.
For assays using preconfigured lyophilized intracellular staining reagents: Resuspend the appropriate wells of the intracellular Ab plate with 50 μL of wash buffer. Let sit for a few minutes, then pipet up and down thoroughly to mix. Transfer the solution to the appropriate wells of the cell plate, and incubate at room temperature in the dark for 60 min, mixing by pipetting or gentle agitation every 15–20 min.
Add 200 μL of wash buffer to each well, and wash as described in Sample Processing step 8 above.
Again add 200 μL wash buffer to each well, and wash as described in Sample Processing step 8 above.
Resuspend pellets with 150 μL wash buffer. Store at 4 °C in the dark until ready for data acquisition, which should be performed within 24 h. Optional: resuspend pellets with 150 μL of 1% paraformaldehyde in PBS or BD Stabilizing Fixative (see Note 16).
3.4. Data Acquisition and Analysis
-
First determine optimal PMT settings for the instrument and reagent panel in question. If automated software for this purpose is not available, follow guidelines as described in reference (24). In brief:
Establish minimum baseline PMT settings for the instrument by acquiring a set of dim particles at various voltages, and choosing the lowest voltage for each PMT for which the CV of these particles is minimized.
Run a sample stained with the full reagent cocktail in question, and adjust baseline PMT voltages as needed so that events are mostly above zero but do not register in the highest fluorescence channel.
Create a set of compensation controls consisting of single-stained cells or beads (see Note 17). Acquire these controls and use the software’s automated algorithm to calculate compensation (see Note 18).
Create a template for acquisition that displays the relevant parameters in the test samples in the form of dot plots. This template need not be the same as that used for analysis, i.e., it does not need to specify all gates or regions of interest. In fact, a simplified acquisition template will allow faster processing of data. However, the template should show any gates used to define the saved population of cells or the stopping criteria (e.g., CD3+ cells).
Set an appropriate threshold, usually on FSC, to eliminate small debris, and set the stopping and storage criteria. It is usually safest to store all events (rather than a gated subset) to allow exploration of all data on analysis. However, sometimes a threshold or gate on CD3+ cells may be employed in order to reduce file sizes (see Note 19). When using a plate loader, be sure to set a stopping criterion based on time, so that samples with insufficient cells will not run dry.
Acquire data from a fully stained sample to verify that the settings chosen are appropriate. If any changes to PMT voltages are made, be sure to re-run compensation samples and recalculate compensation based on the new voltages.
Record data from samples.
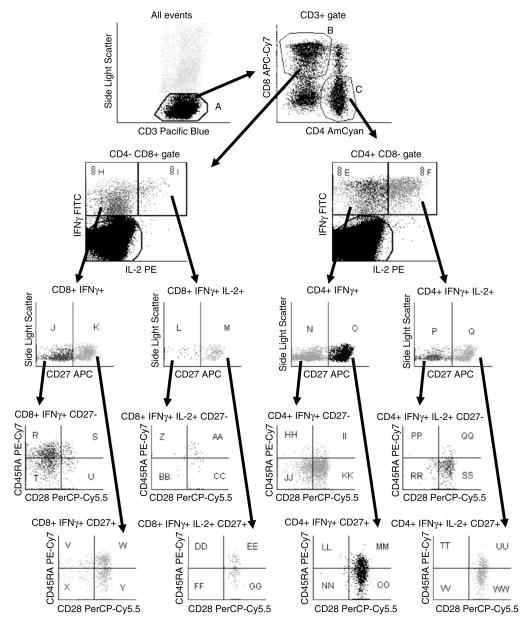
Analyze data using the acquisition software or compatible third-party software. Be sure to define all regions of interest and report the desired statistics on these (see Note 20 and Fig. 25.1). Where possible, use a batch analysis function to analyze all samples from a given experiment or study and export the statistical data to a spreadsheet (see Note 21).
For large studies, it is helpful to create a database to accept the statistical output files from batch analysis. This database can then be queried to create data tables from subsets of the data, allowing rapid graphing, statistical analysis, background subtraction, conversion to absolute counts, etc.
Fig. 25.1.
Gating scheme for eight-color ICS study [reproduced from reference (19)]. An acquisition threshold was used to exclude CD3-negative cells. Dynamic gates were used for defining CD3+, CD4+, and CD8+ lymphocytes, tethered regions were used to define the cytokine-positive cells, and conventional (static) regions or quadrants were used to define all other (rare) subsets. Note that all possible combinatorial subsets of CD27, CD28, and CD45RA were reported for each subset of cytokine-positive cells, resulting in a set of 32 populations whose frequencies describe a response “fingerprint” for that sample.
Acknowledgments
Details of this protocol were optimized by Laurel Nomura and Maria Suni (BD Biosciences).
Footnotes
Plates vs. tubes: Cells can also be stimulated in 15 mL conical polypropylene tubes, with staining in 12 × 75 mm polystyrene tubes (BD Falcon). However, plates are preferred for ease of handling multiple samples, and results for human PBMC are equivalent to those in tubes (32).
Fresh PBMC: If PBMC are not to be cryopreserved, they should ideally be prepared on the day of blood draw, then either stimulated the same day, or rested at 37 °C in cRPMI-10 overnight and stimulated the following day. Overnight resting at 37 °C increases the staining intensity of cytokines, but the effect is more pronounced with cryopreserved samples. Overnight shipping of whole blood or PBMC at ambient temperatures can cause a variable decrease in cell function and should be avoided if possible, though shipping PBMC is preferable to shipping whole blood.
Higher cell concentrations (1 × 107/mL, 2 × 106/well) should be used when possible, especially when response levels are low and/or there are many cell subsets to enumerate.
Cryopreserved PBMC: If cells cannot be stimulated within 24 h of blood draw, they should be cryopreserved by a validated protocol (33). Upon thawing, recoveries of > 60% and viabilities of > 80% should be obtained to minimize loss of functional responses. The method of thawing is equally as important as that of cryopreservation (33). Thawed cells should be rested in cRPMI-10 for 6–18 h at 37 °C to maximize cytokine staining intensity (32). Some cell loss may occur during this period, so plating a slight excess of cells is desirable. Alternately, the cells can be rested in bulk (e.g., in a slanted 15 mL conical polypropylene tube), then recounted and resuspended at the desired concentration after resting.
Brefeldin A vs. monensin: Secretion of most cytokines of interest (IFNγ, IL-2, etc.) is best inhibited by brefeldin A at 10 μg/mL cells. However, CD107 and CD154 are transiently expressed on the cell surface. Therefore, staining Abs to CD107 and/or CD154 are added to the stimulation culture to bind the antigen(s) as soon as they are expressed. Monensin increases the intensity of staining under these conditions by preventing the acidification and degradation of lysosomal vesicles that contain the recycled CD107 and CD154. Thus, for combined cytokine and CD107 or CD154 detection, 5μg/mL each of brefeldin A and monensin is recommended.
Peptide mixes: Peptide mixes can be prepared and lyophilized as premixed pools of up to several hundred peptides (30). These can then be resuspended in DMSO at high concentration per peptide, avoiding DMSO toxicity. The total concentration of DMSO in the assay should be kept at < 0. 5%.
Costimulatory antibodies: Antibodies to CD28 and CD49d can increase the cytokine response to protein antigens, peptides, and SEB by amplifying the signal for low-affinity T cells (34). In occasional donors, they increase cytokine production in the absence of antigen (TNFα is usually most affected).
Adding staining Abs during stimulation: As described in Note 5, staining Abs to CD107 and CD154 are best added during stimulation, to capture the transiently expressed antigen. Fluorochrome conjugated Abs are sensitive to light exposure, so they should be handled in low light and, once added, the samples should be incubated in the dark. Certain tandem dyes such as APC-Cy7 and PE-Cy7 are particularly sensitive to light and temperature (24) and are not optimal choices for use in stimulation cultures.
Stimulation time: A minimum of 5–6 h allows adequate detection of most proinflammatory cytokines like IFNγ, TNFα, and IL-2 (35). Increasing the time of incubation (in the presence of brefeldin A) increases cytokine staining intensity, but is not recommended for CD107 or CD154. For whole proteins requiring intracellular processing, a preincubation of 2 h prior to adding brefeldin A and/or monensin is recommended (35). CD8 responses to whole protein antigens can sometimes be detected, and are increased with longer incubation in antigen alone, but not in all donors (36).
Automating incubation times: A programmable heat block, incubator, or water bath can be used to time activation, cooling the samples to 4–18 °C at the end of a specified period at 37°C, and holding them for later processing.
A fixed-length vacuum manifold helps achieve consistent washing without undue cell loss in microtiter plates. Because of the small wash volume, a sufficient number of washes and efficient removal of supernatant are essential.
CD3, CD4, and CD8 can be stained either before or after fixation and permeabilization. Down-modulation of these antigens occurs to a variable degree depending upon the stimulus. Cells that have down-modulated these antigens can be better detected by intracellular staining (postfixation and permeabilization) (30), although the overall staining intensity is usually decreased. Most other cell-surface antigens are optimally stained before fixation.
Freezing of activated samples: Samples can be frozen at −80 °C directly in FACS Lysing Solution (35,37). This allows for samples to be sent to another laboratory for processing, or for longitudinal samples to be accumulated for batch processing. Lysed whole blood should be washed once prior to freezing.
Fixation and permeabilization steps: Solutions for these steps should be stored and used at 22–25 °C. FACS Lysing Solution simultaneously lyses erythrocytes and fixes leukocytes. While erythrocyte lysis is not required for PBMC samples, fixation is still helpful to prevent cell loss prior to permeabilization.
Centrifugation speed: All centrifugation post-fixation should be done at higher g force (500 × g) due to increased cell buoyancy.
Use of paraformaldehyde is only helpful when samples are stored for more than 24 h prior to acquisition, or to ensure neutralization of potentially biohazardous samples. In addition to subtle effects on cell scatter and fluorescence, storage in paraformaldehyde can cause degradation of tandem dyes such as APC-Cy7 and PE-Cy7. An alternative fixative is available that protects these tandems from degradation (BD Stabilizing Fixative, BD Biosciences), but it is not compatible with AmCyan staining.
Compensation controls: Where possible, anti-immunoglobulin coated capture beads (BD Biosciences) are preferred as compensation controls, because they provide a bright and homogeneous population of events stained with the antibody conjugate of interest. Ideally, the same lot of antibody should be used for compensation as is used in the experiment. In practice, however, this is only important for certain tandem conjugates, such as APC-Cy7 and PE-Cy7. The compensation controls should ideally be treated identically to the experimental samples in terms of fixation, etc., although this too is only important for the above tandem dyes.
When to apply compensation: While compensation can be calculated and changed at any time by software packages such as FloJo (TreeStar, Ashland, OR) or FACSDiva (BD Biosciences), it is helpful to perform compensation before sample acquisition, so that any setup problems can be more readily detected.
Number of events to collect: Because multiparameter ICS assays tend to divide responding populations of cells into ever-smaller subsets, it is important to process and collect enough cells per sample to allow statistically significant differences between samples to be detected. The number of events required will depend upon the anticipated levels of responses and background, as well as the number of subsets of responding cells being identified. Statistical tools for sample size calculation can be found at http://maeckerlab.typepad.com.
Gating of down-modulated cells: Be sure that gates set on CD3, CD4, and CD8 parameters include dim-positive cells, since down-modulation of these markers occurs with activation. When using dynamic gating (see Note 21), set the region size to the maximum value possible without causing inclusion of neighboring populations. Some donors have a significant population of CD4 + CD8dim T cells. This population contains a disproportionate number of cells specific for chronic antigens such as CMV and HIV, and should be included in the CD4+ T cell gate to avoid under-reporting of responses.
Batch analysis: Dynamic gating tools such “Snap-To” gates in FACS Diva (BD Biosciences) can be used to accommodate staining differences between samples for populations such as CD3+, CD4+, and CD8+ cells (see Fig. 25.1). This in turn allows use of a single analysis template and batch analysis across multiple samples in an experiment or study. However, dynamic gates are not always useful for rare populations, and their specifications (size and movement) need to be adjusted for the data set being analyzed. Batch analysis and dynamic gating thus do not replace the need for visual inspection of all data.
References
- 1.Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don’t know, what we should know. Nat Med. 2004;10:806. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- 2.Hel Z, Nacsa J, Tryniszewska E, et al. Containment of simian immunodefi-ciency virus infection in vaccinated macaques: correlation with the magnitude of virus-specific pre- and postchallenge CD4(+) and CD8(+) T cell responses. J Immunol. 2002;169:4778. doi: 10.4049/jimmunol.169.9.4778. [DOI] [PubMed] [Google Scholar]
- 3.Mooij P, Nieuwenhuis IG, Knoop CJ, et al. Qualitative T-helper responses to multiple viral antigens correlate with vaccine-induced immunity to simian/human immunodeficiency virus infection. J Virol. 2004;78:3333. doi: 10.1128/JVI.78.7.3333-3342.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer JD, Maciag PC, Parkinson R, et al. Rhesus macaques with high levels of vaccine induced IFN-gamma producing cells better control viral set-point following challenge with SIV239. Vaccine. 2005 doi: 10.1016/j.vaccine.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y, Schmitz JE, Acierno PM, et al. Dysfunction of simian immun-odeficiency virus/simian human immunod-eficiency virus-induced IL-2 expression by central memory CD4+ T lymphocytes. J Immunol. 2005;174:4753. doi: 10.4049/jimmunol.174.8.4753. [DOI] [PubMed] [Google Scholar]
- 6.Acierno PM, Schmitz JE, Gorgone DA, et al. Preservation of Functional Virus-Specific Memory CD8+ T Lymphocytes in Vaccinated, Simian Human Immunodeficiency Virus-Infected Rhesus Monkeys. J Immunol. 2006;176:5338. doi: 10.4049/jimmunol.176.9.5338. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Schmitz JE, Buzby AP, et al. Virus-specific cellular immune correlates of survival in vaccinated monkeys after simian immunodeficiency virus challenge. J Virol. 2006 doi: 10.1128/JVI.01458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letvin NL, Mascola JR, Sun Y, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migueles SA, Laborico AC, Shupert WL, et al. HIV-specific CD8(+) T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 10.Boaz MJ, Waters A, Murad S, et al. Presence of HIV-1 Gag-specific IFN-gamma + IL-2+ and CD28 + IL-2 + CD4 T cell responses is associated with nonprogression in HIV-1 infection. J Immunol. 2002;169:6376. doi: 10.4049/jimmunol.169.11.6376. [DOI] [PubMed] [Google Scholar]
- 11.Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T-cells. Blood. 2006;107:4781. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Younes SA, Yassine-Diab B, Dumont AR, et al. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198:1909. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G, Shankar P, Lange C, et al. CD8 T cells specific for human immunodeficiency virus, Epstein-Barr virus, and cytomegalovirus lack molecules for homing to lymphoid sites of infection. Blood. 2001;98:156. doi: 10.1182/blood.v98.1.156. [DOI] [PubMed] [Google Scholar]
- 14.Champagne P, Ogg GS, King AS, et al. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410:106. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 15.Harari A, Rizzardi GP, Ellefsen K, et al. Analysis of HIV-1- and CMV-specific memory CD4 T-cell responses during primary and chronic infection. Blood. 2002;100:1381. doi: 10.1182/blood-2001-11-0080. [DOI] [PubMed] [Google Scholar]
- 16.Ellefsen K, Harari A, Champagne P, et al. Distribution and functional analysis of memory antiviral CD8 T cell responses in HIV-1 and cytomegalovirus infections. Eur J Immunol. 2002;32:3756. doi: 10.1002/1521-4141(200212)32:12<3756::AID-IMMU3756>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Appay V, Dunbar PR, Callan M, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 18.Paiardini M, Cervasi B, Albrecht H, et al. Loss of CD127 expression defines an expansion of effector CD8+ T cells in HIV-infected individuals. J Immunol. 2005;174:2900. doi: 10.4049/jimmunol.174.5.2900. [DOI] [PubMed] [Google Scholar]
- 19.Nomura LE, Emu B, Hoh R, et al. IL-2 production correlates with effector cell differentiation in HIV-specific CD8+ T cells. AIDS Res Ther. 2006;3:18. doi: 10.1186/1742-6405-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day CL, Kaufmann DE, Kiepiela P, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 21.Trautmann L, Janbazian L, Chomont N, et al. Upregulation of PD-1 expression on HIV-specific CD8(+) T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 22.Savarino A, Bottarel F, Malavasi F, et al. Role of CD38 in HIV-1 infection: an epiphenomenon of T-cell activation or an active player in virus/host interactions? Aids. 2000;14:1079. doi: 10.1097/00002030-200006160-00004. [DOI] [PubMed] [Google Scholar]
- 23.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry. 2001;45:194. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 24.Maecker HT, Frey T, Nomura LE, et al. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62:169. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
- 25.Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A. 2006 doi: 10.1002/cyto.a.20333. [DOI] [PubMed] [Google Scholar]
- 26.Perfetto SP, Ambrozak D, Nguyen R, et al. Quality assurance for polychromatic flow cytometry. Nat Protocols. 2006;1:1522. doi: 10.1038/nprot.2006.250. [DOI] [PubMed] [Google Scholar]
- 27.Maecker HT. In: Flow Cytometry Protocols. Hawley TS, Hawley RG, editors. Humana Press; Totowa, NJ: 2004. p. 95. [Google Scholar]
- 28.Maecker HT. In: Cancer Vaccine Protocols. Kieber-Emmons T, editor. Humana Press; Totowa, NJ: 2007. [Google Scholar]
- 29.Lamoreaux LL, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protocols. 2006;1:1507. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- 30.Maecker HT, Dunn HS, Suni MA, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 31.Maecker HT, Rinfret A, D’Souza P, et al. Standardization of cytokine flow cytometry assays. BMC Immunol. 2005;6:13. doi: 10.1186/1471-2172-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suni MA, Dunn HS, Orr PL, et al. Performance of plate-based cytokine flow cytometry with automated data analysis. BMC Immunology. 2003;4:9. doi: 10.1186/1471-2172-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Disis ML, dela Rosa C, Goodell V, et al. Maximizing the retention of antigen specific lymphocyte function after cryopreservation. J Immunol Methods. 2006;308:13. doi: 10.1016/j.jim.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Waldrop SL, Davis KA, Maino VC, et al. Normal human CD4+ memory T cells display broad heterogeneity in their activation threshold for cytokine synthesis. J Immunol. 1998;161:5284. [PubMed] [Google Scholar]
- 35.Nomura LE, Walker JM, Maecker HT. Optimization of whole blood antigenspecific cytokine assays for CD4(+) T cells. Cytometry. 2000;40:60. doi: 10.1002/(sici)1097-0320(20000501)40:1<60::aid-cyto8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 36.Maecker HT, Ghanekar SA, Suni MA, et al. Factors affecting the efficiency of CD8+ T cell cross-priming with exogenous antigens. J Immunol. 2001;166:7268. doi: 10.4049/jimmunol.166.12.7268. [DOI] [PubMed] [Google Scholar]
- 37.Nomura LE, DeHaro ED, Martin LN, et al. Optimal preparation of rhesus macaque blood for cytokine flow cytometric analysis. Cytometry. 2003;53A:28. doi: 10.1002/cyto.a.10038. [DOI] [PubMed] [Google Scholar]