Abstract
Islet transplantation is considered to be a curative treatment for type 1 diabetes mellitus. However, disruption of the extracellular matrix (ECM) leads to β-cell destruction and graft dysfunction. In this study, we developed a functionalized self-assembling peptide, KLD-F, with ECM mimic motifs derived from fibronectin and collagen IV, and evaluated its effect on β-cell function and proliferation. Atomic force microscopy and rheological results showed that KLD-F could self-assemble into a nanofibrous scaffold and change into a hydrogel in physiological saline condition. In a three-dimensional cell culture model, KLD-F improved ECM remodeling and cell-cell adhesion of INS-1 β-cells by upregulation of E-cadherin, fibronectin, and collagen IV. KLD-F also enhanced glucose-stimulated insulin secretion and expression of β-cell function genes, including Glut2, Ins1, MafA, and Pdx-1 in INS-1 cells. Moreover, KLD-F promoted proliferation of INS-1 β-cells and upregulated Ki67 expression by mediating cell cycle progression. In addition, KLD-F improved β-cell function and proliferation via an integrin/focal adhesion kinase/extracellular signal-regulated kinase/cyclin D pathway. This study highlights the fact that the β-cell-ECM interaction reestablished with this functionalized self-assembling peptide is a promising method to improve the therapeutic efficacy of islet transplantation.
Keywords: extracellular matrix, self-assembling peptide, islet transplantation, β-cell proliferation, insulin secretion
Introduction
Diabetes mellitus is a chronic metabolic disease caused by insufficient β-cell mass, which leads to hyperglycemia and serious complications, such as diabetic nephropathy, neuropathy, and retinopathy.1–3 There is increasing evidence that islet transplantation is an effective treatment for type 1 diabetes.4 Currently, a major problem in islet transplantation is the high rate of primary dysfunction and early islet destruction.5 Therefore, research efforts have focused on developing more efficient strategies to improve β-cell function and achieve a higher survival rate after transplantation.
It has been reported that the extracellular matrix (ECM) plays a critical role in maintaining β-cell function and viability.6,7 In native pancreatic islets, β-cells are surrounded by various ECM substrates, including collagen I/IV, laminin, and fibronectin, which support cell structure, mediate cell adhesion, and stimulate cell surface receptors to influence biological processes.6 During islet isolation and transplantation, enzymatic digestion and mechanical agitation disrupt cell-ECM and cell-cell contacts, causing loss of islet integrity and viability, resulting in matrix signaling-related cell apoptosis (anoikis) and graft failure.8 In contrast, coculture of islets with ECM components such as collagen IV and fibronectin can maintain β-cell function and viability in vitro,9,10 and improve islet graft survival and long-term blood glucose control in vivo.11 Therefore, regulation of the cellular microenvironment using ECM materials may be a useful strategy to improve the success of islet transplantation.
The self-assembling peptide (SAP) is a designed bio-material that typically consists of alternating hydrophilic and hydrophobic amino acids.12,13 SAP can self-assemble into cross-linked nanofibers and form a hydrogel rapidly in response to stimulation with physiological saline. SAP has been widely used in many biomedical applications, including three-dimensional (3D) cell culture, tissue engineering, and drug delivery.12,14,15 SAP can be designed with functional motifs, and therefore provides a superior microenvironment for specific applications. Recent studies have reported that functionalized SAP enhances angiogenesis, liver regeneration, skin cell proliferation, and the neural stem cell phenotype.16–19 In a previous study, we found that SAP enhanced islet viability and function in vitro,20 and identified functionalized SAP as a potential scaffold for islet transplantation.
To improve the ECM-cell adhesion and performance of β-cells, we specifically designed a functionalized KLD12 (KLD-F) with ECM mimic motifs driven by collagen IV and fibronectin (Figure 1A), which are abundant in natural islets. Fibronectin mimic motif (GGRGDSP) contains a well-defined cell-adhesion motif of RGD, which is located at the tenth type III repeating domain of fibronectin.21 The collagen IV mimic motif (GEFYFDLRLKGDK) contains a triple helical sequence present in the α1 chain of collagen IV.22 These motifs have been widely used in tissue engineering, and can promote various biological activities, including cell adhesion, migration, and ECM production via binding to cell surface receptors.22
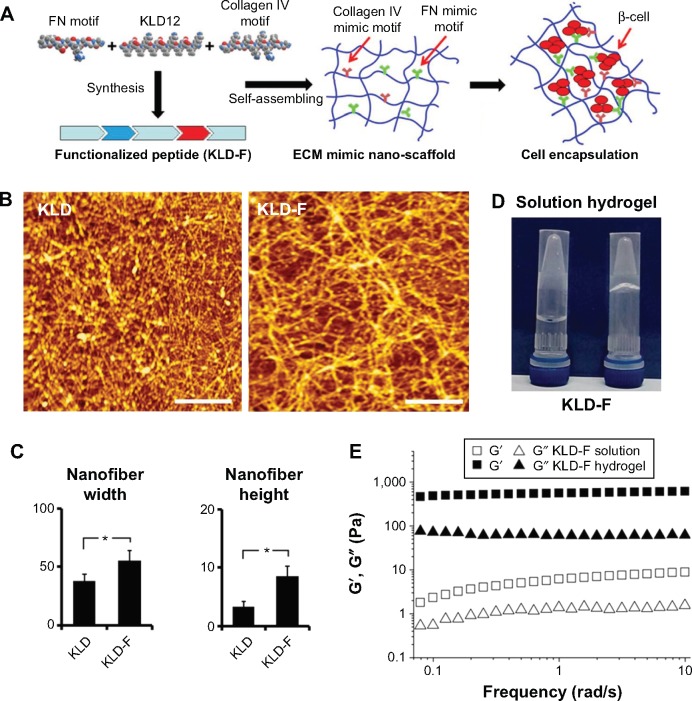
Figure 1.
Molecular design and characteristics of functionalized self-assembling peptide.
Notes: (A) Design and synthesis of KLD-F with FN and collagen IV mimic motifs, with KLD-F self-assembling into a nanoscale scaffold for encapsulating INS-1 β-cell. (B) Atomic force micrographs of KLD12 and KLD-F (0.1 mg/mL, bar 500 nm). (C) Width and height of KLD and KLD-F nanofibers (*P<0.05). (D) Photograph of KLD-F solution and hydrogel (10 mg/mL). (E) Rheological properties of KLD-F solution and hydrogel (Pa).
Abbreviations: ECM, extracellular matrix; FN, fibronectin.
In this study, we conjugated KLD12 with collagen IV and fibronectin mimic motifs, and investigated its morphological and rheological properties. We also evaluated the effect of functionalized SAP on insulin secretion and cell proliferation in a 3D culture of INS-1 β-cells, and explored the molecular mechanism of ECM-cell cross-talk mediated by functionalized SAP.
Materials and methods
SAP design and synthesis
SAPs, including KLD12 (Ac-KLDLKLDLKLDL-CONH2), KLD12-fn (Ac-KLDLKLDLKLDLGGRGDSP-CONH2), and KLD12-collagen IV (Ac-KLDLKLDLKLDLGGGE-FYFDLRLKGDK-CONH2), were commercially synthesized by Shanghai Biotech Bioscience and Technology Co Ltd (Shanghai, People’s Republic of China). Each peptide was purified to higher than 95% by high-performance liquid chromatography (Shimadzu, Kyoto, Japan). The lyophilized peptide powder was dissolved at 10 mg/mL in sterile water and used as a stock solution. KLD-F was prepared by mixing a stock solution of KLD12, KLD-fn, and KLD12-collagen IV at a volume ratio of 6:2:2, followed by sonication for 20 minutes and storage at 4°C for further use.
Atomic force microscopy
The morphology of SAP was observed by atomic force microscopy (SPI4000 Probe Station, Seiko Instruments Inc, Chiba, Japan) with a tapping mode. For preparation of the sample, 10 μL of diluted peptide solution was evenly placed on a freshly cleaved mica substrate. The mica surface was then rinsed with Milli-Q water to remove unattached peptide and air-dried. The atomic force microscopy test was performed at room temperature and images were acquired with a resolution of 512×512 pixels. The typical scan parameters were set as follows: scan speed 1.00 Hz, amplitude 1–1.2 V, and integral and proportional gains of 0.2–0.3 and 0.03–0.04, respectively.
Rheological analysis
Rheological analysis of the peptide was performed using a rheometer (AR2000, TA Instruments, New Castle, DE, USA) with a 20 mm diameter and a 1° steel cone with a 25 μm truncation gap. In brief, 70 μL of the sample solution was placed on the plate of the rheometer, and gelation of the peptide was induced by addition of phosphate-buffered saline (PBS). After equilibrating for 1 minute, the excess PBS solution was removed. The frequency sweep tests of storage (G′) and loss (G″) modulus were measured at 37°C with parameters of strain 0.5% and a frequency range of 0.01–100 rad/sec.
INS-1 cell culture
A rat insulinoma (INS-1) β-cell line was cultured in Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 U/mL streptomycin, 1 mM sodium pyruvate (Sigma-Aldrich, St Louis, MO, USA), and 50 μm β-mercaptoethanol (Sigma-Aldrich) in an atmosphere of 5% CO2 at 37°C. For 3D culture of SAP hydrogel, cells at 80% confluence were harvested from a cell culture flask by trypsinization, washed in PBS, and resuspended in 20% sucrose (w/v, 584.2 mM, dissolved in sterile water). The cell suspension was gently mixed with an equal volume of peptide solution (10 mg/mL), after which the mixture was pipetted into six-well plates and immediately equilibrated with culture medium to induce gel formation. The culture medium in the peptide group was changed after 30 minutes, and every 3 days thereafter.
Glucose-stimulated insulin secretion test
The cells were collected and washed with Krebs-Ringer bicarbonate (KRB, pH 7.2) buffer containing 1% bovine serum albumin. Cells were first incubated in KRB with low glucose (2 mM) at 37°C for 1 hour. The buffer was then removed and replaced with fresh KRB containing high glucose (20 mM) for 1 hour. The insulin concentration in KRB was measured using a commercial enzyme-linked immunosorbent assay kit for rat insulin (R&D Systems, Minneapolis, MN, USA). The glucose stimulation index (SI) was calculated as the insulin level in high glucose condition/insulin level in low glucose condition. After treatment with 20 mM glucose, the cells were collected and disrupted by sonication. The intracellular insulin level was measured and normalized to the DNA content of each sample to eliminate the influence of cell numbers.
Cell proliferation assay
Cell proliferation was evaluated using Cell Counting Kit-8 (CCK-8) and 5-bromo-2-deoxyuridine (BrdU) assays. For the CCK-8 assay, cells were seeded in a 96-well plate and treated with different materials. Next, 10 μL of CCK-8 solution (Dojindo, Kumamoto, Japan) were added to each well. After incubation at 37°C for 1.5 hours, the medium was removed and its absorbance at 450 nm was measured using a microplate reader (BioTek Instruments Inc, Winooksi, VT, USA). For the BrdU enzyme-linked immunosorbent assay (Abcam, Cambridge, MA, USA), cells were replated in a 96-well plate after treatment. BrdU solution was then added into each well, with incubation at 37°C for 6 hours. After fixation and denaturation, the cells were incubated with primary anti-BrdU antibody, followed by secondary horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G antibody. TMB peroxidase substrate was added to the plate for reaction with peroxidase, and its absorbance at 450 nm was recorded. The glucose concentration of the culture medium at day 0 (Glud0) and day 3 (Glud3) was measured in each group, and glucose consumption (%) was calculated as (Glud0 – Glud3)/Glud0*100%.
Cell cycle analysis
After the experimental treatment, the cells were harvested, washed with cold PBS, and fixed in 70% ethanol. The cell samples were washed, resuspended in PBS, and then incubated with propidium iodide (100 μg/mL, Sigma-Aldrich) and RNase (10 μg/mL, Sigma-Aldrich) solution at room temperature for 30 minutes in the dark. The stained cell samples were analyzed using a FC500 flow cytometer (Beckman Coulter, Brea, CA, USA), and the percentage of cells in each cell cycle phase was calculated. All cell samples were measured in triplicate. The proliferation index was calculated as (S + G2/M)/(G1 + S + G2/M) and the S-phase cell fraction as S/(G1 + S + G2/M).
Real-time polymerase chain reaction analysis
Total RNA was extracted from the cells by Trizol Reagent (Gibco, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA was quantified using a microspectrophotometer (NanoDrop 2000, Thermo Fisher Scientific Inc, Waltham, CA, USA). Complementary DNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). The primers were synthesized by Invitrogen Inc (Waltham, MA, USA), and their sequences are listed in Table S1. Polymerase chain reaction (PCR) reactions were carried out on a CFX96 real-time PCR detection system (Bio-Rad) with SYBR Green Supermix (SsoFast EvaGreen, Bio-Rad). Real-time PCR data were analyzed using Bio-Rad CFX Manager software, and relative changes in the mRNA level were calculated by the delta-delta Ct method with β-actin as the internal reference gene.
Immunocytochemistry
The cells were fixed with 4% paraformaldehyde in PBS for 15 minutes at room temperature, and permeabilized with 0.3% Triton X-100 in PBS for 10 minutes. After blocking with 1% bovine serum albumin and 0.3% Triton X-100 in PBS for 30 minutes, the cells were incubated with diluted primary antibodies to E-cadherin (1:100, BD Biosciences, Franklin Lakes, NJ, USA), fibronectin (1:100, BioWorld, Dublin, OH, USA), collagen IV (1:100, Abcam), insulin (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, USA), Pdx-1 (1:100, Abcam), and Ki67 (1:100, Santa Cruz, USA) overnight at 4°C, followed by incubation with fluorescein isothiocyanate or tetramethylrhodamine-conjugated secondary antibody (1:200, Millipore, Billerica, MA, USA) at 37°C for 1 hour. After staining with 4,6-diamidino-2-phenyllindile (Sigma-Aldrich) and washing with PBS, images of the stained cells were acquired using a fluorescent microscope (IX71, Olympus, Tokyo, Japan). The percentage of Ki67-positive cells in total cells present in the micrographs was analyzed using ImageJ software.
Western blot analysis
Cells were collected and lysed in cold RIPA buffer, and the protein concentration was determined using a bicinchoninic acid assay kit (Pierce Biotechnology, Waltham, MA, USA). Protein samples were electrophoresed on 10% sodium dodecyl sulfate polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (PVDF, Merck Millipore). The PVDF membrane was blocked in 5% nonfat milk buffer and then incubated with primary antibodies against E-cadherin (1:500, BD Biosciences), fibronectin (1:500, BioWorld), Pdx-1 (1:500, Abcam), Ki67 (1:500, Santa Cruz), extracellular signal-regulated kinase (ERK) 1/2 (1:500, Abcam), p-ERK1/2 (1:500, Cell Signaling Technology, Danvers, MA, USA), focal adhesion kinase (FAK, 1:500, Cell Signaling Technology), p-FAK (1:500, Cell Signaling Technology), Akt (1:500, Cell Signaling Technology), p-Akt (1:500, Cell Signaling Technology), integrin α5 (1:500, Abcam), integrin β1 (1:500, Abcam), cyclin D1 (1:500, Santa Cruz), p27 (1:500, Cell Signaling Technology), and β-actin (1:1,000, Santa Cruz) overnight at 4°C. The PVDF membrane was washed in PBST and incubated with horseradish peroxidase-conjugated secondary antibodies (1:2,000, Santa Cruz) at 37°C for 1 hour. The target protein band on the PVDF membrane was observed using an electrochemiluminescence kit (Pierce).
Statistical analysis
Quantitative data are presented as the mean ± standard deviation, and analyzed using Statistical Package for the Social Sciences version 11.5 software (SPSS Inc, Chicago IL, USA) with one-way analysis of variance using the Bonferroni post hoc test. P<0.05 was considered to be statistically significant.
Results
Morphologic and rheological properties of functionalized SAP
As shown in Figure 1B, KLD-F could self-assemble via cross-linked nanofibers, with some bulges and aggregates, and the width and height of the KLD-F nanofibers were higher than those of the KLD nanofibers (Figure 1C). After addition of PBS, the KLD-F solution rapidly changed to an obvious hydrogel (Figure 1D). In rheological analysis (Figure 1E), the value of the storage modulus (G′, response to elasticity) and loss modulus (G″, response to viscosity) in KLD-F solution (5 mg/mL) was lower than 10 Pa, while the storage modulus of KLD-F increased significantly (500–600 Pa) after addition of PBS, suggesting formation of a strong hydrogel. These results indicate that KLD-F can form an elastic hydrogel with a 3D nanofiber structure.
Effect of KLD-F on ECM remodeling and cell-cell adhesion
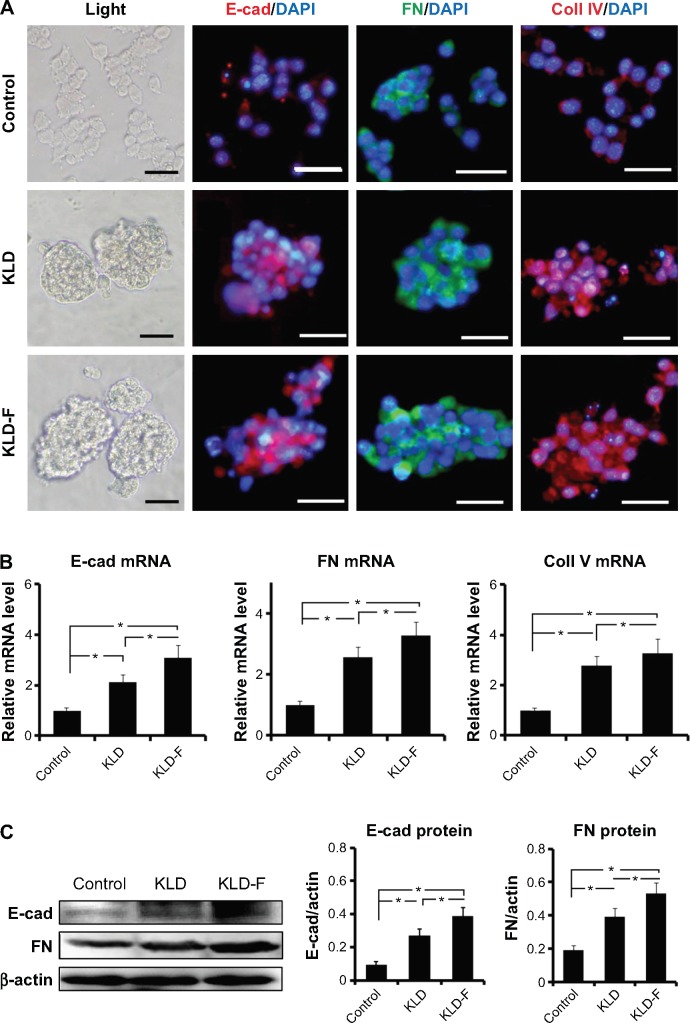
To investigate the effect of KLD-F on ECM remodeling, INS-1 β-cells were cultured and tested. In the control group (Figure 2A), two-dimensional culture of INS-1 β-cells as a monolayer on a TCP dish showed loose clusters with an irregular shape, while clear islet-like cell aggregates formed in 3D culture of the KLD and KLD-F hydrogel. In addition, immunofluorescence staining (Figure 2A), real-time PCR (Figure 2B), and Western blot (Figure 2C) results showed that the INS-1 cells in the KLD-F group expressed higher levels of E-cadherin, fibronectin, and collagen IV than the other groups. These results indicate that KLD-F was able to improve ECM remodeling of β-cells and cell-cell interaction.
Figure 2.
Functionalized self-assembling peptide enhanced ECM remodeling and cell-cell adhesion in INS-1 β-cells.
Notes: (A) Light and immunofluorescent micrographs of E-cad, FN, and Coll IV (bar 50 μm) after 3 days of culture. (B) Real-time polymerase chain reaction analysis of E-cad, FN, and Coll IV mRNA expression after 3 days of culture. (C) Western blot and quantitative analysis of E-cad and FN protein expression after 3 days of culture (*P<0.05).
Abbreviations: FN, fibronectin; E-cad, E-cadherin; Coll IV, collagen IV.
Effect of KLD-F on insulin secretion
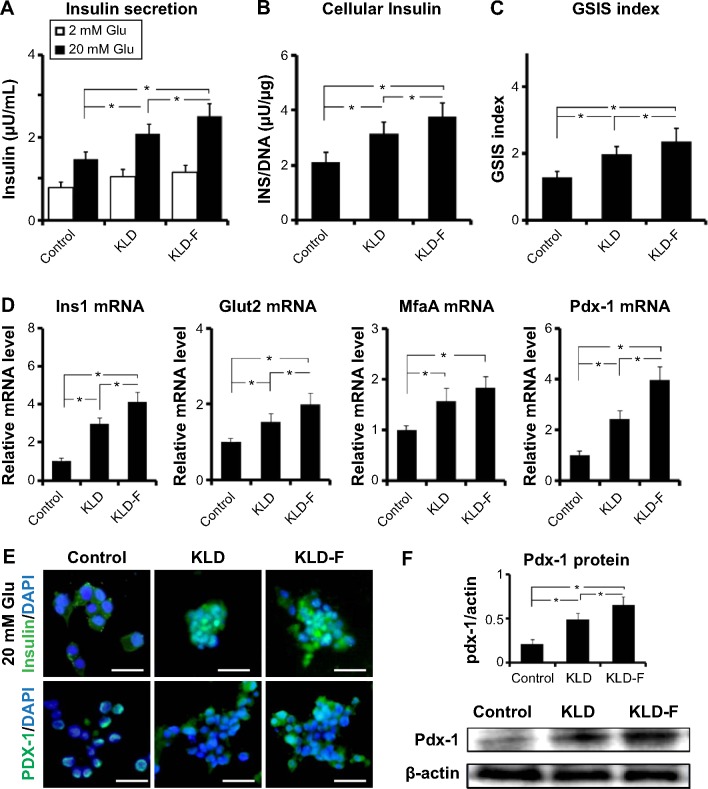
The insulin secretion of INS-1 cells was analyzed. Compared with the control and KLD groups, the KLD-F group showed increased insulin secretion (Figure 3A) and higher intracellular insulin content (Figure 3B) in response to 20 mM glucose, as well as an elevated SI (Figure 3C). Expression of β-cell function genes, including glucose transporter 2 (Glut2), insulin I (Ins1), v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A (MafA), pancreatic and duodenal homeobox 1 (Pdx-1, see Figure 3D), and Pdx-1 protein (Figure 3E), were also significantly upregulated in the KLD-F group. These results demonstrate that KLD-F enhanced β-cell function, and this effect may due to enhanced cell-ECM and cell-cell adhesion.
Figure 3.
Functionalized self-assembling peptide improved insulin secretion function in INS-1 β-cells.
Notes: (A) Insulin secretion, (B) intracellular insulin (under 20 mM glucose), and (C) GSIS index in the different groups after 3 days of culture. (D) Real-time polymerase chain reaction analysis of Glut2, Ins1, MfaA, and Pdx-1 mRNA expression after 3 days of culture. (E) Immunofluorescent staining for insulin (under 20 mM glucose condition) and Pdx-1 (bar 50 μm). (F) Western blot and quantitative analysis of PDX-1 protein level after 3 days of culture (*P<0.05).
Abbreviations: DAPI, 4,6-diamidino-2-phenyllindile; GSIS, glucose-stimulated insulin secretion; Glu, glucose.
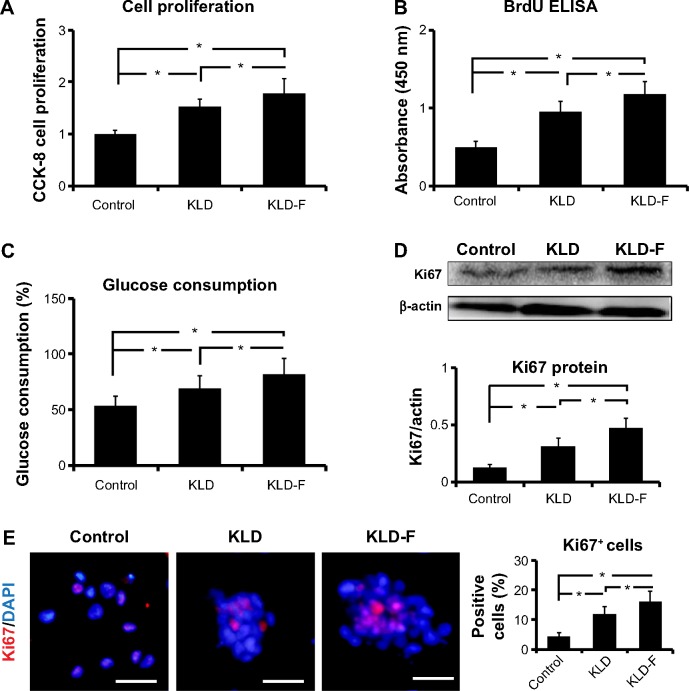
Effect of KLD-F on cell proliferation
Compared with the control and KLD groups, INS-1 cells in the KLD-F group showed significantly increased cell proliferation rates, which was determined by a CCK-8 assay (Figure 4A), BrdU enzyme-linked immunosorbent assay (Figure 4B), and glucose consumption assay (Figure 4C). In addition, the protein level of Ki67, a cellular marker for proliferation, was increased significantly in KLD-F group (Figure 4D). Immunofluorescence staining also showed that the KLD-F group had more Ki67-positive cells than the other groups (Figure 4E). These results indicate that the KLD-F hydrogel promoted cell proliferation as well as insulin secretion in INS-1 β-cells.
Figure 4.
Functionalized self-assembling peptide promoted cell proliferation in INS-1 β-cells.
Notes: (A) Cell proliferation rates detected by CCK-8 assay after 3 days of culture. (B) BrdU enzyme-linked immunosorbent assay of INS-1 cell proliferation after 3 days of culture. (C) Glucose consumption rates (%) in different groups after 3 days of culture. (D) Western blot and quantitative analysis of Ki67 protein expression after 3 days of culture. (E) Immunofluorescent staining of Ki67 (bar 50 μm) and quantitative analysis of Ki67-positive cells (*P<0.05).
Abbreviations: CCK-8, Cell Counting Kit-8; DAPI, 4,6-diamidino-2-phenyllindile; ELISA, enzyme-linked immunosorbent assay.
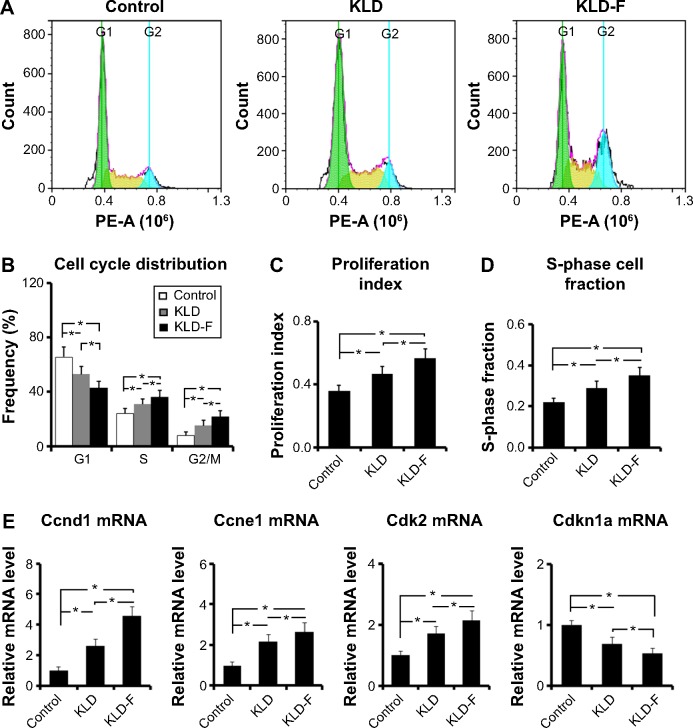
Effect of KLD-F on cell cycle distribution
To investigate the potential effect of KLD-F on the cell cycle, INS-1 cells in each group were analyzed by flow cytometry (Figure 5A). Compared with the other groups, the KLD-F group showed a reduced G1 phase population and an elevated S and G2/M phase population (Figure 5B), as well as an increased proliferation index and S-phase cell fraction (Figure 5C and D), suggesting enhanced cell division and DNA duplication on 3D culture in KLD-F hydrogel. Real-time PCR data showed that the KLD-F group had higher levels of Ccnd1, Ccne1, and Cdk2 (induction of cell cycle progression), and a lower level of Cdkn1a (inhibition of cell cycle progression) than the other groups (Figure 5E). These results suggest that the effect of KLD-F on promotion of β-cell proliferation may be via modulation of cell cycle progression.
Figure 5.
Functionalized self-assembling peptide induced cell cycle progression in INS-1 β-cells.
Notes: (A) Flow cytometry analysis of cell cycle. (B) G1, S, and G2/M phase distribution after 3 days of culture. (C) Proliferation index and (D) S-phase cell fraction detected by flow cytometry (*P<0.05). (E) Real-time polymerase chain reaction analysis of Ccnd1, Ccne1, Cdk2, and Cdkn1a mRNA levels after 3 days of culture (*P<0.05).
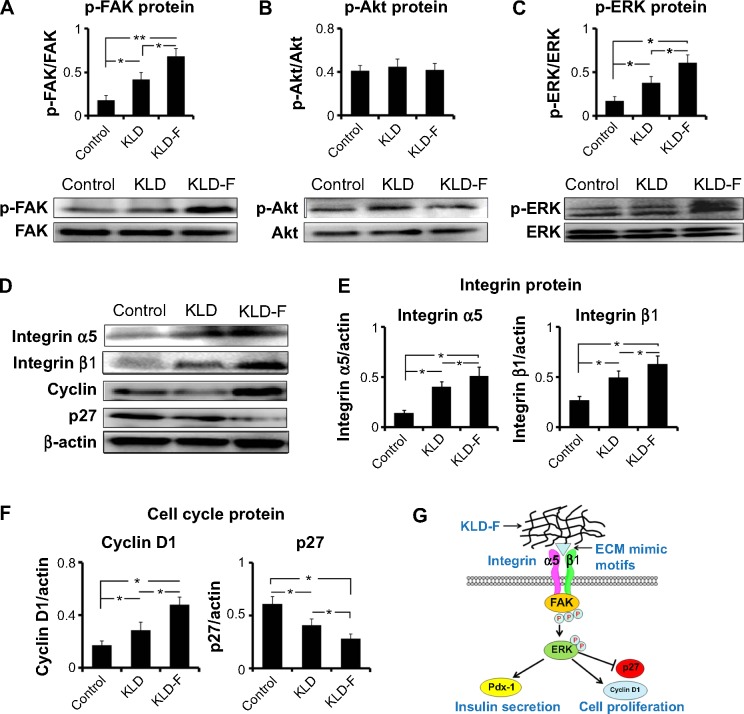
Effect of KLD-F on integrin/FAK/ERK/cyclin signaling
To explore the underlying mechanism of KLD-F on cell proliferation, expression of signaling molecules related to ECM-integrin and the cell cycle was analyzed. Although the total levels of FAK and ERK did not change, the level of p-FAK and p-ERK in the KLD-F group increased significantly (Figure 6A and C). There was no significant difference in total Akt or p-Akt between the groups (Figure 6B). INS-1 cells in the KLD-F group had a higher level of integrin α5 and β1 when compared to the control and KLD groups (Figure 6D and E). The level of key mediators in G1/S phase, including cyclin D1 and p27, was also analyzed. The data show that KLD-F group had elevated cyclin D1 and reduced p27 compared with the other groups (Figure 6D and F). These results suggest that KLD-F induces β-cell proliferation via an integrin α5/β1-FAK-ERK-cyclin D1/p27 pathway (Figure 6G).
Figure 6.
Mechanism of functionalized self-assembling peptide inducing β-cell proliferation.
Notes: (A–C) Western blot for p-FAK/FAK, p-Akt/Akt, and p-ERK/ERK after 1 day of culture, and quantitative analysis of protein level. (D–F) Western blot for integrin α5 and β1, cyclin D1, and p21 after 1 day of culture, and quantitative analysis of protein level (*P<0.05, **P<0.01). (G) Schematic map of signaling pathways. KLD-F binds to integrin α5 and β1, subsequently activating FAK/ERK and downstream cyclin D1 as well as inhibiting p27 signaling, finally promoting β-cell proliferation.
Abbreviations: ECM, extracellular matrix; FAK, focal adhesion kinase; ERK, extracellular signal-regulated kinase.
Discussion
Beta-cell death and dysfunction caused by disruption of ECM is a major problem in islet transplantation. Increasing evidence suggests that encapsulation of islets within an ECM scaffold is a promising strategy to improve β-cell survival and function.7 It is well documented that pancreatic cell lines have physiological properties similar to those of primary islets, and serve as a useful tool for studies of the function and molecular events in β-cells, thereby overcoming the limited availability of primary islets.23 In this study, we developed a functionalized SAP with mimic motifs driven by collagen IV and fibronectin, and evaluated its effect on function and proliferation in a 3D culture model of a rat INS-1 β-cell line.
To assess whether the linking of functional motifs to KLD12 affected formation of the nanostructure, atomic force microscopy and rheological analysis was performed. Our results showed that, under physiological saline conditions, KLD-F could self-assemble to form an elastic hydrogel consisting of cross-linked nanofibers. We next investigated the effect of KLD-F on cell morphology and ECM in a 3D cell culture model. Unlike the cell monolayer in two-dimensional culture, INS-1 cells formed islet-like aggregates in SAP hydrogel, suggesting a possible difference in ECM between two-dimensional and 3D culture. It has been reported that natural or artificial materials induced ECM secretion of islets.6 Consistent with these reports, we observed increased ECM protein, including collagen IV and fibronectin, as well as elevated E-cadherin in INS-1 cells cultured in the KLD-F hydrogel. E-cadherin is a Ca2+-dependent adhesion molecule, and plays an important role in cell growth and development.24 Cell adhesion via E-cadherin had been shown to have a distinct role in the regulation of cell-cell communication between β-cell within islets, with potential repercussions for insulin secretion.25 Overall, our results indicate that ECM remodeling and cell-cell adhesion of β-cells could be enhanced by KLD-F.
The ability of designed ECM materials to improve β-cell survival and function has been reported,7 and MIN6 β-cells cultured in gels containing collagen IV or laminin enhanced insulin release.10 Both islet structure and cell-cell interaction within islets are essential for the normal function of β-cells, and the dispersed islet is associated with a dramatic decline of glucose-induced insulin release.26 In this study, we observed an increased SI, increased insulin release, and increased intracellular insulin under high-glucose stimulation in INS-1 clusters in the KLD-F group, suggesting improvement of β-cell function by KLD-F. Expression of key genes controlling insulin biosynthesis and secretion, including Glut2, Ins1, MafA, and Pdx-1, was upregulated in the KLD-F group. Pdx1 is a key transcription factor mediating the development and function of β-cells, and affects insulin secretion via generation of nicotinamide adenine dinucleotide phosphate, mobilization of intracellular Ca2+, and mitochondrial biosynthesis.27 These results suggest that KLD-F can improve β-cell function, and that this may due to the enhanced cell-cell and ECM-cell interaction.
The effect of KLD-F on β-cell proliferation was also analyzed. BrdU is a uridine derivative that can be incorporated into DNA during the S-phase of cell cycle as a substitute for thymidine, and serves as a common marker for cell proliferation.28 In this study, BrdU and CCK-8 results demonstrated that INS-1 β-cells in the KLD-F group had higher proliferation rates than the other groups. The glucose consumption rate of INS-1 cells in the KLD-F group was also increased, corresponding to an increased number of living cells. Ki67 is a general marker for cell proliferation, and is highly expressed during the active phases of the cell cycle.28 We observed increased numbers of Ki67+ cells and elevated Ki67 protein in INS-1 clusters in the KLD-F group, suggesting a potential relationship between cell-cell interaction and proliferation. A previous study had reported that cell-cell contact positively regulated proliferation in endothelial cells.29 Although the exact mechanism is not clear, our results indicate that KLD-F promoted β-cell proliferation via enhancing cell-cell and ECM-cell interaction, which may have an influence on the cell cycle.
The cell cycle is a series of events leading to replication and division of the cell into two equal copies (proliferation). In eukaryotes, the cell cycle is divided into G0 (gap 0), G1 (gap 1), S (synthesis), G2 (gap 2), and M (mitosis) phase. DNA synthesis and cell division occur during S, G2, and M phase.30 INS-1 cells in the KLD-F group had a reduced G1 phase population and elevated S and G2/M phase populations, as well as an increased proliferation index and S-phase cell fraction, indicating that KLD-F induced cell cycle progression in β-cells. In addition, the mRNA level of signaling molecules that induce cell cycle progression, including Ccnd1 (cyclin D1), Ccne1 (cyclin E1), and Cdk2 (CDK2), were upregulated, while the mRNA level of the cell cycle inhibitor Cdkn1a (p21) was reduced in the KLD-F group. These results indicate that KLD-F induced β-cell proliferation by regulating cell cycle and its related pathways.
ECM-cell interaction is essential for cell proliferation, and the response of cells to ECM is mainly mediated by cell surface adhesion receptors such as integrins.31 The composition of integrins on islets is complex, including α3, α5, αv, β1, β3, and β5.6 Integrin β1 is the major receptor of type I and IV collagen, and has been shown to regulate development and function of the pancreas.32 Integrin α5β1 is the primary receptor for fibronectin and is involved in cell adhesion, migration, and ECM formation.33 We observed increased integrin α5 and β1 in the KLD-F group, suggesting that KLD-F can interact with multiple integrins and thereby trigger downstream pathways in the β-cell. FAK is a tyrosine kinase that plays a key role in regulating intracellular signals in response to ECM stimuli.34 ECM-integrin interaction leads to phosphorylation of FAK, and further activates the mitogen-activated protein kinase pathways. Mitogen-activated protein kinases, including ERK, play an important role in transition of extracellular signals to the nucleus.35 Upon activation, ERK translocates from the cytoplasm to the nucleus, where it phosphorylates various transcription factors that regulate cell cycle progression.36 We observed increased p-FAK and p-ERK in INS-1 cells in the KLD-F group, providing evidence for the synergistic activation of FAK/ERK signals by KLD-F. Cyclin D is a key mediator of G1/S phase transition, which can be activated by ERK to form an active cyclin D/CDK4/6 complex, thereby driving entry of the cell into the next phase of the cell cycle.37 In contrast, p27 is an important inhibitor of the cell cycle, which restricted G1/S phase transition by inactivation of cyclin E/CDK2 and the cyclin A/CDK2 complex.34 INS-1 cells in the KLD-F group had increased cyclin D1 as well as reduced p27, which may be caused by activation of the FAK/ERK pathways by KLD-F. Therefore, our results indicate a possible mechanism whereby KLD-F interacted with integrin α5/β1 and led to activation of the FAK-ERK pathways, which in turn induced cell cycle progression by upregulation of cyclin D1 and inhibition of p27 signaling.
Naturally derived polymers often contain undefined residual factors and substances, which may induce an immune response or side effects in clinical therapies. SAP is made of natural amino acids and can be synthesized commercially with high purity to solve these problems. SAP had been widely used in various cell culture and animal experiments, and rarely showed any toxic or immune response.12,13 Our results suggest that functionalized SAP is a promising scaffold for clinical islet transplantation. In our next experiment, we are planning to encapsulate primary islets in SAP hydrogel and transplant them into diabetic monkeys.
Conclusion
In conclusion, we designed a functionalized SAP with ECM mimic motifs derived from collagen IV and fibronectin. KLD-F can self-assemble into an elastic hydrogel consisting of cross-linked nanofibers. Islet-like cell aggregates formed in 3D culture of INS-1 β-cells in KLD-F hydrogel. INS-1 cells in the KLD-F group had higher levels of E-cadherin, fibronectin, and collagen IV, suggesting enhanced β-cell remodeling and cell-cell adhesion. INS-1 cells in the KLD-F group showed enhanced insulin secretion, and increased expression of Glut2, Ins1, MafA, and Pdx-1. In addition, KLD-F promoted β-cell proliferation by mediating cell cycle progression. Further, we found that KLD-F improved β-cell function and proliferation via an integrin α5/β1-FAK/ERK-cyclin D1/p27 pathway. Our study indicates that ECM-cell interaction is critical for β-cells, and functionalized SAP with ECM mimics is a promising method to improve β-cell function and proliferation for islet transplantation.
Supplementary material
Table S1.
Primer sequence for real-time polymerase chain reaction
| Gene | Primer sequence |
|---|---|
| E-cad | Forward: 5′-AAGACAGAGCGGAACTAT-3′ |
| Reverse: 5′-ATCATCCTCAGCATCAGT-3′ | |
| Fibronectin | Forward: 5′-GTCACCTACAACATCATA-3′ |
| Reverse: 5′-TACAGTAACAACCTCTTC-3′ | |
| Collagen IV (Col4a1) | Forward: 5′-TGAGAAGAACATAGTGAT-3′ |
| Reverse: 5′-TTAACAATACAACAGGAG-3′ | |
| Ins1 | Forward: 5′-CAATCATAGACCATCAGCAAGC-3′ |
| Reverse: 5′-AGAAACCACGTTCCCCAC-3′ | |
| Glut2 | Forward: 5′-CACATCCTACTTGGCCTATCTG-3′ |
| Reverse: 5′-TCAGTGCCCCTTAGTCTTTTC-3′ | |
| MafA | Forward: 5′-GTCTTCAGGGTCGCCGTCTAG-3′ |
| Reverse: 5′-GAGGTTGGGACGCAGAACTG-3′ | |
| Pdx-1 | Forward: 5′-CCCGAGCTTCTGAAAACTTTG-3′ |
| Reverse: 5′-CTTTTCATTGTCCTCAGTTGGG-3′ | |
| Ccnd1 | Forward: 5′-CTTCAGCAAGGAGGAGGTCATC-3′ |
| Reverse: 5′-GCGTAGCCGCGGTTCTT-3′ | |
| Ccne1 | Forward: 5′-GTCTTCAGGGTCGCCGTCTAG-3′ |
| Reverse: 5′-GAGGTTGGGACGCAGAACTG-3′ | |
| Cdk2 | Forward: 5′-GACTGATGTTGTTGACAGCCA-3′ |
| Reverse: 5′-ATGCTTAGGCATAACGCACTAGGTT-3′ | |
| Cdkn1a | Forward: 5′-GACTGATGTTGTTGACAGCCA-3′ |
| Reverse: 5′-ATGCTTAGGCATAACGCACTAGGTT-3′ | |
| β-actin | Forward: 5′-GGAGATTACTGCCCTGGCTCCTA-3′ |
| Reverse: 5′-GACTCATCGTACTCCTGCTTGCTG-3′ |
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31200754) and the China Postdoctoral Science Foundation (2012M511931).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2008;S1:S55–S60. doi: 10.2337/dc08-S055. [DOI] [PubMed] [Google Scholar]
- 2.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Wang D, Chen Y, et al. 1H NMR-based metabonomic analysis of serum and urine in a nonhuman primate model of diabetic nephropathy. Mol Biosyst. 2013;9:2645–2652. doi: 10.1039/c3mb70212j. [DOI] [PubMed] [Google Scholar]
- 4.Bruni A, Gala-Lopez B, Pepper AR, Abualhassan NS, Shapiro AJ. Islet cell transplantation for the treatment of type 1 diabetes: recent advances and future challenges. Diabetes Metab Syndr Obes. 2014;7:211–223. doi: 10.2147/DMSO.S50789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korsgren O, Lundgren T, Felldin M, et al. Optimising islet engraftment is critical for successful clinical islet transplantation. Diabetologia. 2008;51:227–232. doi: 10.1007/s00125-007-0868-9. [DOI] [PubMed] [Google Scholar]
- 6.Stendahl JC, Kaufman DB, Stupp SI. Extracellular matrix in pancreatic islets: relevance to scaffold design and transplantation. Cell Transplant. 2009;18:1–12. doi: 10.3727/096368909788237195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng JY, Raghunath M, Whitelock J, Poole-Warren L. Matrix components and scaffolds for sustained islet function. Tissue Eng Part B Rev. 2011;17:235–247. doi: 10.1089/ten.TEB.2011.0004. [DOI] [PubMed] [Google Scholar]
- 8.Thomas FT, Contreras JL, Bilbao G, Ricordi C, Curiel D, Thomas JM. Anoikis, extracellular matrix, and apoptosis factors in isolated cell transplantation. Surgery. 1999;126:299–304. [PubMed] [Google Scholar]
- 9.Davis NE, Beenken-Rothkopf LN, Mirsoian A, et al. Enhanced function of pancreatic islets co-encapsulated with ECM proteins and mesenchymal stromal cells in a silk hydrogel. Biomaterials. 2012;33:6691–6697. doi: 10.1016/j.biomaterials.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber LM, Hayda KN, Anseth KS. Cell-matrix interactions improve beta-cell survival and insulin secretion in three-dimensional culture. Tissue Eng Part A. 2008;14:1959–1968. doi: 10.1089/ten.tea.2007.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yap WT, Salvay DM, Silliman MA, et al. Collagen IV-modified scaffolds improve islet survival and function and reduce time to euglycemia. Tissue Eng Part A. 2013;19:2361–2372. doi: 10.1089/ten.tea.2013.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Zhao X. Design of self-assembling peptides and their biomedical applications. Nanomedicine (Lond) 2011;6:1621–1643. doi: 10.2217/nnm.11.142. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, Zhang S. Molecular designer self-assembling peptides. Chem Soc Rev. 2006;35:1105–1110. doi: 10.1039/b511336a. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Zhang L, Yang Z, Zhao X. Controlled release of paclitaxel from a self-assembling peptide hydrogel formed in situ and antitumor study in vitro. Int J Nanomedicine. 2011;6:2143–2153. doi: 10.2147/IJN.S24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Song H, Zhang L, Xu H, Zhao X. Self-assembly-peptide hydro-gels as tissue-engineering scaffolds for three-dimensional culture of chondrocytes in vitro. Macromol Biosci. 2010;10:1164–1170. doi: 10.1002/mabi.200900450. [DOI] [PubMed] [Google Scholar]
- 16.Bradshaw M, Ho D, Fear MW, Gelain F, Wood FM, Iyer KS. Designer self-assembling hydrogel scaffolds can impact skin cell proliferation and migration. Sci Rep. 2014;4:6903. doi: 10.1038/srep06903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Wang X, Horii A, et al. In vivo studies on angiogenic activity of two designer self-assembling peptide scaffold hydrogels in the chicken embryo chorioallantoic membrane. Nanoscale. 2012;4:2720–2727. doi: 10.1039/c2nr00001f. [DOI] [PubMed] [Google Scholar]
- 18.Cheng TY, Wu HC, Huang MY, Chang WH, Lee CH, Wang TW. Self-assembling functionalized nanopeptides for immediate hemostasis and accelerative liver tissue regeneration. Nanoscale. 2013;5:2734–2744. doi: 10.1039/c3nr33710c. [DOI] [PubMed] [Google Scholar]
- 19.Koutsopoulos S, Zhang S. Long-term three-dimensional neural tissue cultures in functionalized self-assembling peptide hydrogels, matrigel and collagen I. Acta Biomater. 2013;9:5162–5169. doi: 10.1016/j.actbio.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Y, Cong C, Zhang J, et al. Self-assembling peptide nanofiber as potential substrates in islet transplantation. Transplant Proc. 2008;40:2571–2574. doi: 10.1016/j.transproceed.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Mardon HJ, Grant KE. The role of the ninth and tenth type III domains of human fibronectin in cell adhesion. FEBS Lett. 1994;340:197–201. doi: 10.1016/0014-5793(94)80137-1. [DOI] [PubMed] [Google Scholar]
- 22.Yamada Y, Kleinman HK. Functional domains of cell adhesion molecules. Curr Opin Cell Biol. 1992;4:819–823. doi: 10.1016/0955-0674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 23.Skelin M, Rupnik M, Cencic A. Pancreatic beta cell lines and their applications in diabetes mellitus research. ALTEX. 2010;27:105–113. doi: 10.14573/altex.2010.2.105. [DOI] [PubMed] [Google Scholar]
- 24.Bhatt T, Rizvi A, Batta SP, Kataria S, Jamora C. Signaling and mechanical roles of E-cadherin. Cell Commun Adhes. 2013;20:189–199. doi: 10.3109/15419061.2013.854778. [DOI] [PubMed] [Google Scholar]
- 25.Rogers GJ, Hodgkin MN, Squires PE. E-cadherin and cell adhesion: a role in architecture and function in the pancreatic islet. Cell Physiol Biochem. 2007;20:987–994. doi: 10.1159/000110459. [DOI] [PubMed] [Google Scholar]
- 26.Kelly C, McClenaghan NH, Flatt PR. Role of islet structure and cellular interactions in the control of insulin secretion. Islets. 2011;3:41–47. doi: 10.4161/isl.3.2.14805. [DOI] [PubMed] [Google Scholar]
- 27.Brissova M, Blaha M, Spear C, et al. Reduced PDX-1 expression impairs islet response to insulin resistance and worsens glucose homeostasis. Am J Physiol Endocrinol Metab. 2005;288:E707–E714. doi: 10.1152/ajpendo.00252.2004. [DOI] [PubMed] [Google Scholar]
- 28.Muskhelishvili L, Latendresse JR, Kodell RL, Henderson EB. Evaluation of cell proliferation in rat tissues with BrdU, PCNA, Ki-67(MIB-5) immunohistochemistry and in situ hybridization for histone mRNA. J Histochem Cytochem. 2003;51:1681–1688. doi: 10.1177/002215540305101212. [DOI] [PubMed] [Google Scholar]
- 29.Nelson CM, Chen CS. Cell-cell signaling by direct contact increases cell proliferation via a PI3K-dependent signal. FEBS Lett. 2002;514:238–242. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- 30.Pu QQ, Streuli CH. Integrin control of cell cycle: a new role for ubiquitin ligase. Bioessays. 2002;24:17–21. doi: 10.1002/bies.10049. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz MA, Assoian RK. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J Cell Sci. 2001;114:2553–2560. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- 32.Diaferia GR, J Jimenez-Caliani AJ, Ranjitkar P, et al. Beta1 integrin is a crucial regulator of pancreatic beta-cell expansion. Development. 2013;140:3360–3372. doi: 10.1242/dev.098533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson EE, Foty RA, Corbett SA. Fibronectin matrix assembly regulates alpha5beta1-mediated cell cohesion. Mol Biol Cell. 2004;15:973–981. doi: 10.1091/mbc.E03-07-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Boudreau NJ, Jones PL. Extracellular matrix and integrin signalling: the shape of things to come. Biochem J. 1999;339(Pt 3):481–488. [PMC free article] [PubMed] [Google Scholar]
- 36.Chambard JC, Lefloch R, Pouyssegur J, Lenormand P. ERK implication in cell cycle regulation. Biochim Biophys Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primer sequence for real-time polymerase chain reaction
| Gene | Primer sequence |
|---|---|
| E-cad | Forward: 5′-AAGACAGAGCGGAACTAT-3′ |
| Reverse: 5′-ATCATCCTCAGCATCAGT-3′ | |
| Fibronectin | Forward: 5′-GTCACCTACAACATCATA-3′ |
| Reverse: 5′-TACAGTAACAACCTCTTC-3′ | |
| Collagen IV (Col4a1) | Forward: 5′-TGAGAAGAACATAGTGAT-3′ |
| Reverse: 5′-TTAACAATACAACAGGAG-3′ | |
| Ins1 | Forward: 5′-CAATCATAGACCATCAGCAAGC-3′ |
| Reverse: 5′-AGAAACCACGTTCCCCAC-3′ | |
| Glut2 | Forward: 5′-CACATCCTACTTGGCCTATCTG-3′ |
| Reverse: 5′-TCAGTGCCCCTTAGTCTTTTC-3′ | |
| MafA | Forward: 5′-GTCTTCAGGGTCGCCGTCTAG-3′ |
| Reverse: 5′-GAGGTTGGGACGCAGAACTG-3′ | |
| Pdx-1 | Forward: 5′-CCCGAGCTTCTGAAAACTTTG-3′ |
| Reverse: 5′-CTTTTCATTGTCCTCAGTTGGG-3′ | |
| Ccnd1 | Forward: 5′-CTTCAGCAAGGAGGAGGTCATC-3′ |
| Reverse: 5′-GCGTAGCCGCGGTTCTT-3′ | |
| Ccne1 | Forward: 5′-GTCTTCAGGGTCGCCGTCTAG-3′ |
| Reverse: 5′-GAGGTTGGGACGCAGAACTG-3′ | |
| Cdk2 | Forward: 5′-GACTGATGTTGTTGACAGCCA-3′ |
| Reverse: 5′-ATGCTTAGGCATAACGCACTAGGTT-3′ | |
| Cdkn1a | Forward: 5′-GACTGATGTTGTTGACAGCCA-3′ |
| Reverse: 5′-ATGCTTAGGCATAACGCACTAGGTT-3′ | |
| β-actin | Forward: 5′-GGAGATTACTGCCCTGGCTCCTA-3′ |
| Reverse: 5′-GACTCATCGTACTCCTGCTTGCTG-3′ |