Abstract
Extracellular vesicles (EVs) are cell-secreted vesicles that range from 30–2000 nm in size. These vesicles are secreted by both normal and neoplastic cells. Physiologically, EVs serve multiple critical biologic functions, including cellular remodeling, intracellular communication, modulation of the tumor microenvironment and regulation of immune function. Because EVs contain genetic and proteomic contents that reflect the cell of origin, it is possible to detect tumor-specific material in EVs secreted by cancer cells. Importantly, EVs secreted by cancer cells transgress anatomic compartments and can be detected in the blood, cerebrospinal fluid, and other biofluids of cancer patients. In this context, there is a growing interest in analyzing EVs from the biofluid of cancer patients as a means of disease diagnosis and therapeutic monitoring. In this article, we review the development of EVs as a diagnostic platform for the most common form of brain cancer, glioblastoma, discuss potential clinical translational opportunities and identify the central challenges associated with future clinical applications.
Keywords: biomarker, exosomes, extracellular vesicle, glioma, microvesicle
Glioblastoma is the most common form of primary brain neoplasm. Despite aggressive surgical resection, chemotherapy and radiation therapy, the median survival for glioblastoma patients is approximately 14 months [1,2]. Optimal management of glioblastoma patients is limited by a lack of effective strategies for monitoring response to therapy [3]. In the current clinical practice, therapeutic responses are monitored through serial neurologic examinations and MRI studies. However, both forms of assessment are crude measures of the underlying disease status.
Monitoring the clinical status of the patient will detect disease progression only after sufficient tumor burden has accumulated to alter the neurologic examination. This level of disease burden occurs long after the tumor cells have acquired resistance to therapy. While MRI potentially shortens this delay, the resolution limit of MRI renders this modality insensitive. Current MRI has a resolution limit of approximately 2–3 mm [4]. Considering that the size of a tumor cell is on the order of 10 µm, tumor cells will undergo more than 20 rounds of exponential growth before detection is feasible by MRI. Moreover, modalities used to treat glioblastoma (radiation and chemotherapy) often induce changes in the brain that mimic those of tumor progression on MRI [5]. While advanced MR modalities have been developed to better detect tumor progression (reviewed in [6,7]), the time required for the scheduling, acquisition and interpretation of these studies further delay timely intervention. Though brain biopsy and histologic analysis can definitively evaluate disease progression, serial brain biopsy is both invasive and impractical given cumulative surgical risk [8].
The delay in the detection of therapeutic failure is particularly problematic since the standard chemotherapy used to treat glioblastoma, temozolomide, is a mutagenic agent [9]. Temozolomide induces tumor kill by alkylating the O6 position of the guanine nucleoside, resulting in stalled replication fork that ultimately triggers cell death [10,11]. However, in cells that have acquired resistance to temozolomide, the O6 methylated guanine can mispair with the thymine nucleoside, resulting in G:C → A:T transitions [12]. This mutagenesis is expected to promote the emergence of more aggressive clones [13], thereby limiting the efficacy of subsequent therapeutic regimens [14]. Thus, temozolomide treatment should be terminated as soon as resistance against this agent can be detected.
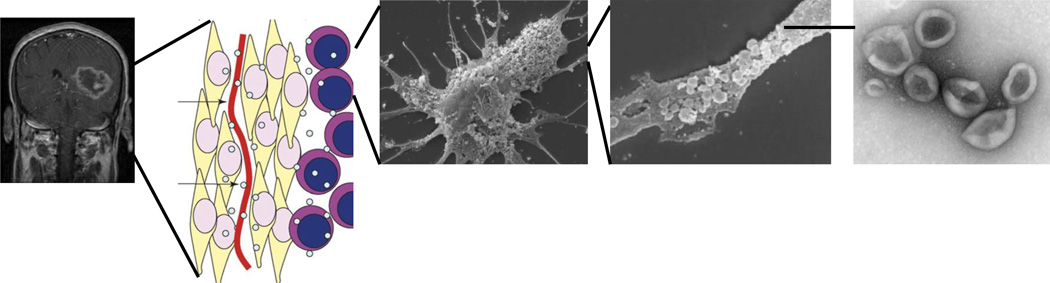
In these contexts, there is a critical need in the development of diagnostic tools that would afford timely assessment of disease burden and therapeutic response in glioblastoma patients. There is a growing interest in extracellular vesicles (EVs) as a biomarker platform in this development. Recent studies have demonstrated that glioblastoma cells secrete EVs that harbor tumor-specific mRNAs, miRNAs, and proteins (Figure 1) [15–18]. These EVs transgress multiple anatomic compartments and can be detected in the blood [18] and CSF [15] of glioblastoma patients. Pilot studies suggest that the analysis of EVs derived from the biofluid of glioblastoma patients may serve as a ‘liquid biopsy’ platform [19].
Figure 1. Extracellular vesicles (EV) as a diagnostic biomarker platform.
EVs are secreted by glioblastomas and carry tumor-specific genetic materials. (A) MRI demonstrating a glioblastoma in the left temporal region. (B) The secreted EVs transgress histo-anatomic boundaries of tumor cell (yellow), blood vessel (red) and stroma cell (purple). (C) Scanning electron microscopy image of a primary glioblastoma cell. Note the EVs on the cell surface. (D) Higher magnification showing the vesicles on the cell surface. (E) Electron micrograph of the EVs isolated from the biofluid of glioblastoma patients.
Adapted with permission from [18] © Macmillan Publishers Ltd. (2008).
Extracellular vesicles: overview of their biology, nomenclature & isolation
Biology
EVs play a key role in several biological processes. For the secreting cells, EVs facilitate cellular membrane remodeling, recycling and the removal of cellular components [20–24]. For instance, during reticulocyte maturation, the cell transforms from a spherical volume into a biconcave structure, a process that requires the removal of large amount of membrane and proteins from the cell surface. This removal is mediated through shedding of cellular membranes and components through EV secretion [23]. Similarly, EV secretion is the major mechanism by which eggs avoid polyspermy during fertilization. Mammalian fertilization is mediated through interaction between Izumo1, a sperm protein, and Juno, an egg surface receptor. Rapid shedding of Juno through EV secretion occurs rapidly after fertilization as to ensure that each egg fuses with a single sperm [21].
For the cells in the immediate vicinity of the EV-secreting cells, EVs mediate intercellular communication and transport membrane bound receptors, nucleic acids and other proteins to target cells [25–27]. For instance, oligodendrocytes and astrocytes secrete EVs that deliver specific proteins, mRNA and miRNA to surrounding neurons to provide trophic support. Similarly, neurons secrete EVs that are taken up by astrocytes and oligodendrocytes [25], thereby reciprocally influencing the functions of these cells.
In terms of microenvironment modulation, secreted EVs facilitate remodeling of the extracellular matrix [22,28–31]. Tumor microvesicles showed an enrichment of matrix degrading metalloproteases (MMP2, MMP9, MTI-MMP and urokinase-type plasminogen activator) [28–31]. These secreted EVs play critical roles in modulating the migration and invasion of cancer cells [32,33]. Moreover, EVs released by metastatic cells interact with bone marrow-derived cells to promote a pro-vasculogenic and pro-metastatic phenotype and to facilitate the establishment of the pre-metastatic niche [34].
Finally, EVs can play an important role in immunomodulation [22]. Depending on the cellular context, EVs can induce both immunostimulatory and immuno-suppressive effects [35–37]. Dendritic cells release EVs that augment immunologic responses by activating T cells [35] or transferring MHC molecules to other dendritic cells [36]. On the other hand, EVs isolated from tumor cells have been shown to induce the expansion of human T-regulatory cells and suppress the immune responses [37].
Nomenclature
EVs have been categorized into exosomes, microvesicles, apoptotic bodies, retroviral particles and other entities based on the mechanism of biogenesis, and an in-depth review of this topic can be found elsewhere [20]. In brief, exosomes refer to vesicles of 30–100 nm in size and arise from the endosomal network, a membranous compartment responsible for sorting various forms of intraluminal vesicles. Microvesicles refer to larger sized vesicles (50–2000 nm) that arise as a result of direct budding from the plasma membrane. Retrovirus-like particles are 90–100 nm vesicles that also arise from direct budding of the plasma membrane. In contrast to microvesicles, however, the mechanism of formation is driven by retroviral proteins, such as Gag [38]. Finally, apoptotic bodies refer to membrane-enclosed vesicles that are formed during apoptosis. These vesicles range between 50 and 4000 nm in size.
As above discussed, the terms ‘exosomes’, ‘microvesicles’, ‘apoptotic bodies’ and ‘retroviral particles’ each hold a unique significance based on their respective mechanisms of biogenesis. Unfortunately, surface markers that uniquely distinguish one EV sub-population from another have not yet been established. As such, it is the currently not possible to discriminate the mechanism of biogenesis when analyzing EVs derived from clinical specimens.
In this context, alternate nomenclatures have been loosely applied when discussing EVs isolated from clinical specimens. One nomenclature convention adopted was to name the vesicles based on the source of isolation. Terms such as oncosomes, prostatsomes and epididimosomes were coined [20]. EVs have also been classified based on size. In general, the term exosome refers to EVs <100 nm in size, whereas the term microvesicles refers to exosomes 100 nm–2 µm in size [20]. It is important to note that though there is some consensus in the literature regarding the size-based definitions, precise cut-offs are arbitrary and vary depending on the author. For the purposes of this review, we will refer to vesicles isolated from clinical specimens as EVs, while reserving the terms exosome and microvesicle for vesicles derived from model systems where biogenic mechanisms can be clearly defined.
Isolation & characterization
EVs have been isolated from blood, urine, CSF, lymphatics, tears, saliva and nasal secretions, ascites and semen. As indicated above, multiple methods have been used to isolate EVs. Some groups use differential centrifugation or density gradients to remove larger cellular debris and isolate EVs [15,39,40]. Other reported methods include chemical precipitation, the use of serial filters [41], EV-binding resins, immuno-isolation with magnetic beads [42,43], and microfluidic separations [17]. It is important to note that the biologic properties and contents of the EVs are greatly influenced by the method of isolation [44,45]. In this context, it is important to specify the method by which EVs are isolated in scientific communications.
Due to their small size, the analysis of EVs presents significant technical challenges. Large EVs (>300 nm) can be analyzed by flow cytometry [46]. Newer generation of cytometry instruments, including the Apogee (Apogee Flow Systems), Gallios (Beckman Coulter) and BD-Influx (Becton Dickinson) may be used to sort EVs down to 200 nm in size [47]. Current efforts are directed towards the development of cytometric instruments that can afford sorting of smaller particles.
Currently, EVs smaller than 200 nm are not amenable to flow cytometric analysis. The most frequently used descriptive analytic for these particles involve direct visualization using electron microscopy [15,48]. Nanoparticle tracking analysis is another method for quantifying the distribution of EVs and calculating particle size [49,50]. Nanoparticle tracking analysis uses a digital camera to capture the movement of EVs over a series of frames. The rate of the particle movement is then used to calculate particle size. Resistive pulse sensing is another method for quantifying EV size. Two cells are separated by a membrane containing a single pore. As EVs pass through the pore, a transient change in ionic current flow is detected and used to calculate the volume of the EV [51]. Other analytic techniques based on surface markers such as diagnostic MR, ELISA and western blotting have also been reported [20].
EV as a biomarker platform for glioblastoma
High-throughput genomic technologies have revolutionized our understanding of glioblastoma and identified novel biomarkers, with potential clinical application. In pre-clinical models, EVs isolated from glioblastoma cell lines contain tumor-specific mRNA and miRNAs [15,16,18]. Importantly, these markers can also be detected in EVs derived from the biofluid of glioblastoma patients. As such, the disappearance of these biomarkers from the patient’s biofluid may serve as proxy for therapeutic response. By the same rationale, re-emergence of these biomarkers may indicate disease recurrence. In the ensuing section, four studies describing the applications of this strategy will be reviewed.
EGFRvIII
The EGFRvIII mutation is found in 20–30% of all glioblastomas [52,53]. The EGFRvIII truncation mutant arises from the deletion of wild-type EGFR exons 2 through 7 [53]. EGFRvIII mutants express a constitutively active receptor that plays a significant role in glioblastoma pathogenesis and development [54]. Importantly, the EGFRvIII mutation is not found in normal tissue, thereby rendering EGFRvIII ideal as a tumor biomarker.
In a study by Skog et al., the EGFRvIII status of EVs derived from the sera of 30 glioblastoma patients were examined by qRT-PCR. EGFRvIII was found in the EV derived from five patients harboring EGFRvIII-expressing glioblastoma. Interestingly, two patients with biopsy specimen scoring negative for EGFRvIII harbored EGFRvIII in their sera EVs, suggesting the initial biopsy may have targeted a non-representative area in a histologically heterogeneous tumor. However, PCR contamination leading to a false-positive signal cannot be excluded given the study design. Of the patients who scored positive for EGFRvIII in sera EVs, longitudinal sera collection was performed in five of these patients. In all cases, EGFRvIII was undetectable after resection [18].
IDH1R132H
Mutations in isocitrate dehydrogenase 1(IDH1), a citric acid cycle enzyme, are found in approximately 10% of all glioblastomas and 80% of secondary glioblastomas [55]. The native function of the protein catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate [56]. The majority of IDH1 mutated tumors exhibit an arginine to histidine mutation (R132H) [55]. The resultant enzyme is deficient in its native function but harbor a new catalytic activity – the production of 2-hydroxyl-glutarate. The production of 2-hydroxylglutarate is associated with an altered chromatin state [57], which is thought to contribute to glioblastoma pathogenesis. However, the molecular mechanisms underlying this pathogenesis remain poorly understood. Despite a lack of clarity in these mechanisms, IDH1 mutation status has come to serve as a means of differentiating secondary glioblastomas (i.e., tumors that arise from lower grade gliomas) from primary glioblastomas that arise de novo.
A study by Chen et al. explored the hypothesis that mutant transcripts of IDH1 may be detected in EVs isolated from patient CSF [16]. The authors utilized highly sensitive digital PCR platform, with primers designed to specifically detect the mutated IDH1 sequence. Parallel experiments were performed to assess the IDH1 status of glioblastoma specimens with matched CSF collected from the same patient. While the authors were not able to detect mutant IDH1 transcript in the sera of patients afflicted with IDH1 mutated tumors, the detection of mutant IDH1 RNA transcript was achieved in five of eight CSFs derived from patients with IDH1 mutated tumors. Importantly, the absolute quantity of mutant IDH1 transcripts directly correlated with the tumor volume, suggesting the feasibility of using EV-based platform to assess glioblastoma disease burden.
miR-21
miRNAs are small RNA molecules that help regulate gene expression by silencing mRNA transcripts with partially complementary sequences. Given the inherent stability of miRNAs relative to mRNAs and their presence in EVs, EV miRNAs represent particularly attractive biomarker platform for ‘liquid biopsies’. miR-21 is a miRNA that is highly over-expressed in glioblastoma cells [58]. The expression of miR-21 in glioblastoma mediates several essential oncogenic functions, including suppression of apoptosis, growth proliferation and tolerance of DNA damages [59]. Akers et al. showed that miR-21 levels in EVs isolated from the CSF of glioblastoma patients were 10-fold higher than those isolated from controls. Importantly, using CSF-derived EV miR-21 levels, the authors were able to prospectively distinguish CSF derived from glioblastoma and non-oncologic patients. Furthermore, miR-21 levels in CSF EVs decreased by an order of magnitude after surgical resection of the glioblastoma in patients with available longitudinal follow-up [15].
Combined proteomic platform
Shao et al. developed a micro-fluidic chip technology that used magnetic nanoparticles to label EVs followed by analysis with a micro-nuclear MR system [17]. This technology allowed the detection of EV proteins with a sensitivity exceeding other protein detection methods (including western blotting and ELISA) by several orders of magnitude. The authors used this µNMR technology to measure EGFR, EGFRvIII, podoplanin and IDH1 R132H molecules in EV isolates from the sera of 24 glioblastoma patients and eight healthy volunteers. In this study, patients with higher levels of tumor-related molecules (such as EGFR, EGFRvIII and podoplanin) in their EVs were more likely to fail standard temozolomide/radiation treatment (p < 0.005).
Expert commentary
Significant strides have been made in the understanding of EVs and the role they play in cellular physiology and intercellular communications. The critical observation that glioblastoma cells secrete EV-containing genetic materials that mirror the intracellular tumor milieu has laid the foundation for EVs as a platform for ‘liquid biopsy’. With the uncovering of the genomic landscape of glioblastomas and the development of highly sensitive detection technologies, such as digital PCR, maturation in the clinical application of this platform is expected. Early reports suggest the EV platform for ‘liquid biopsy’ is promising. However, there are major challenges that require creative and thoughtful solutions before clinical translation. These issues are outlined below.
Since non-neoplastic cells secrete EVs and these cells outnumber neoplastic cells by several orders of magnitude in a patient, tumor-specific EVs remain a rarity in clinical samples. This rarity limits the sensitivity of EVs as a biomarker platform. While one method for bypassing this limitation is to analyze large volumes of the biofluid, such an approach may not be practical. As such, there is a critical need to identify reagents that enrich the yield of glioblastoma-specific EVs from smaller samples. To this end, two major advances are needed. First, surface markers that enhance the isolation in glioblastoma-specific EVs will need to be identified. Second, affordable technologies for the reliable and efficient sorting of EVs of all size ranges will need to be developed. If such technologies become widespread, they may also enhance communication and cooperation within the EV field by facilitating the development of standardized inter-institutional protocols.
It is also conceivable that certain genetic materials, such as miRNA, may be preferentially concentrated in glioblastoma EVs. If so, the analysis of these enriched genetic materials may further enhance the inherent sensitivity of the assay. Since the mechanisms responsible for transporting genetic materials from the cellular context into the EVs remain poorly understood, selection of the optimal assay for EV analysis will likely require empiric determination.
Quantitative analysis of the genetic contents of EVs is another issue that requires thoughtful consideration. Quantitative assessment of mRNA or miRNA expression in the cellular context typically involves normalization of the query gene to housekeeping genes, such as glyceraldehyde-3-phosphate dehydrogenase or 18S. These housekeeping genes are expressed at high levels, exhibit little cell-to-cell variation and serve as a proxy for the total number of cells analyzed. Unfortunately, the quantities of various housekeeping genes are rare and highly variable among EVs [15]. As such, random biases would be introduced if the query transcript level is normalized to an arbitrarily selected gene, such as glyceraldehyde-3-phosphate dehydrogenase. In this context, we favor PCR quantitation of the genetic material in absolute terms [15]. However, the question remains: should this absolute quantity be normalized to the total volume of the input biofluid, to the total protein content of the EVs, to the total number of EVs or to highly abundant transcripts in the EVs. Much work remains to be done to address this important question.
Another important issue that is at the core of any biofluid-based ‘liquid biopsy’ involves the dynamic nature of the biofluid. Like one’s blood pressure, samples of biofluid taken at any one moment may not be identical to those taken at another moment. The contents of the biofluid (CSF, blood or otherwise) are subject to changes related to natural circadian rhythm, food intake, medications, exercise and a plethora of other physiologic and iatrogenic factors. To further confound the matter, the method by which the biofluid is collected significantly impact the quality and quantity of EVs collected. Agitation of the collected fluid and the time interval between collection and processing of the samples are also equally important factors [47]. Given the inherent biologic complexity of the various biofluids, careful consideration should be given to the optimal method of collection – which will likely differ depending on the intended assay and biomarker.
While the initial studies of EV as a biomarker platform for glioblastomas show promise, validation through larger sample sets and prospective clinical trials are the necessary next steps. Given the rarity of glioblastomas, such efforts require multi-institutional collaborations and infrastructures. A pilot effort toward this end has been initiated by the Accelerated Brain Cancer Cure Foundation, an effort that involved 18 academic neurosurgical centers across the U.S. Undoubtedly, the optimal technical platform for EV analysis will continue to evolve during the course of this prospective study. As such, future clinical trials should be designed with an adaptive intent, with the possibility of comparing evolving methods of EV isolation and analysis. The trial design should also afford opportunities for comparing EV-based platforms to other established and emerging biomarker platforms such as circulating tumor cells [60] or cell-free DNA [61].
In conclusion, the concept of EV-based ‘liquid biopsy’ for monitoring disease status is promising. The preliminary reports exploring this concept with application to glioblastoma patients are compelling. Successful implementation of this platform can fundamentally alter the paradigm of care for glioblastoma patients and make meaningful gains in terms of improvement in clinical outcomes. Ultimately, thoughtful clinical trial design and technology advancements will be needed to harness the potential of EVs for this clinical translation.
Five-year view
Over the course of the next 5 years, priorities in EV research, as it pertains to biomarker development, should be placed on optimizing methods of isolation with the recognition that chemical and biologic content of clinical biofluids necessarily differ from those of cell culture supernatants. Optimal methods of isolation likely will require glioblastoma-specific EV markers as a function of the source of the biofluid. Given the spectrum of genetic materials present in biofluid EVs, the analytic of maximal utility remains a central question. Moreover, normalization and quantitation of genetic materials isolated from biofluid EV is an unresolved question. The solution to these challenging questions will likely involve empiric determination rather than theoretical considerations. Spontaneous fluctuation in the contents of patient biofluid as well as perturbation imposed by iatrogenic or iatrogenic poses another major challenge as they inevitably introduce ‘noise’ to the biomarker analytic. As such, efforts to mitigate these dynamic ‘noises’ will be needed, including defining strict collection protocol. With maturation of EV biomarker analytics in the upcoming years, there will be increasing need for clinical validation and comparison to other biomarker platforms, including circulating tumor cells. Thoughtful consideration in clinical trial design and multi-institutional collaboration are warranted in this regard. The challenges facing EV biomarker development are daunting but not insurmountable. Collegial and deliberate collaborative endeavors are undoubtedly required to address these central challenges.
Key issues.
Diagnosis and therapeutic monitoring remains a major challenge for neuro-oncologic diseases, such as glioblastomas.
Extracellular vesicles (EVs) are cell-secreted vesicles that are 30–2000 nm in size. EVs contain genetic and proteomic contents that reflect the cell of origin.
Glioblastoma cells secrete EVs that harbor tumor-specific mRNAs, miRNAs and proteins. These EVs transgress multiple anatomic compartments and can be detected in the blood and cerebrospinal fluid of glioblastoma patients.
Pilot studies have demonstrated that glioblastoma disease burden and therapeutic responses associate with protein, miRNA and mRNA profiles of EVs isolated from the biofluids of glioblastoma patients.
Optimization and validation of results provided by the pilot studies are needed to advance EV as a platform for glioblastoma biomarker development.
Acknowledgments
This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. see comment Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Preusser M, de Ribaupierre S, Wohrer A, et al. Current concepts and management of glioblastoma. Ann Neurol. 2011;70(1):9–21. doi: 10.1002/ana.22425. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen AG, Batchelor TT, Wen PY, et al. Response criteria for glioma. Nat Clin Pract Oncol. 2008;5(11):634–644. doi: 10.1038/ncponc1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahmathulla G, Marko NF, Weil RJ. Cerebral radiation necrosis: a review of the pathobiology, diagnosis and management considerations. J Clin Neurosci. 2013;20(4):485–502. doi: 10.1016/j.jocn.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Law M. Advanced imaging techniques in brain tumors. Cancer Imaging. 2009;9(Spec No A):S4–S9. doi: 10.1102/1470-7330.2009.9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemort M, Canizares-Perez AC, Van der Stappen A, Kampouridis S. Progress in magnetic resonance imaging of brain tumours. Curr Opin Oncol. 2007;19(6):616–622. doi: 10.1097/CCO.0b013e3282f076b2. [DOI] [PubMed] [Google Scholar]
- 8. Waters JD, Gonda DD, Reddy H, et al. Diagnostic yield of stereotactic needle-biopsies of sub-cubic centimeter intracranial lesions. Surg Neurol Int. 2013;4(Suppl 3):S176–S181. doi: 10.4103/2152-7806.110677. • The study described challenges associated with histologic diagnosis through brain biopsies.
- 9.Geiger H, Schleimer D, Nattamai KJ, et al. Mutagenic potential of temozolomide in bone marrow cells in vivo. Blood. 2006;107(7):3010–3011. doi: 10.1182/blood-2005-09-3649. [DOI] [PubMed] [Google Scholar]
- 10.Chen CC, Taniguchi T, D’Andrea A. The Fanconi anemia (FA) pathway confers glioma resistance to DNA alkylating agents. J Mol Med (Berl) 2007;85(5):497–509. doi: 10.1007/s00109-006-0153-2. [DOI] [PubMed] [Google Scholar]
- 11.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21(24):3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodell WJ, Gaikwad NW, Miller D, Berger MS. Formation of DNA adducts and induction of lacI mutations in Big Blue Rat-2 cells treated with temozolomide: implications for the treatment of low-grade adult and pediatric brain tumors. Cancer Epidemiol Biomarkers Prev. 2003;12(6):545–551. [PubMed] [Google Scholar]
- 13.Tian T, Olson S, Whitacre JM, Harding A. The origins of cancer robustness and evolvability. Integr Biol (Camb) 2011;3(1):17–30. doi: 10.1039/c0ib00046a. [DOI] [PubMed] [Google Scholar]
- 14.Silva AS, Gatenby RA, Gillies RJ, Yunes JA. A quantitative theoretical model for the development of malignancy in ductal carcinoma in situ. J Theor Biol. 2010;262(4):601–613. doi: 10.1016/j.jtbi.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 15. Akers JC, Ramakrishnan V, Kim R, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. doi: 10.1371/journal.pone.0078115. • The study demonstrated that miR-21 could be detected in extracellular vesicles (EVs) derived from the CSF of glioblastoma patients. Moreover, the level of miR-21 in CSF EV mirriored tumor burden.
- 16.Chen WW, Balaj L, Liau LM, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18(12):1835–1840. doi: 10.1038/nm.2994. • The study reported the development of a micro-fluidic chip technology for analyzing EVs in clinical patient glioblastoma specimens.
- 18. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. • Ground-breaking translational study that demonstrated that EVs isolated from the sera of glioblastoma patients could serve a liquid biopsy platform for detecting the EGFRvIII mutation.
- 19.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 20. Akers J, Gonda D, Kim R, et al. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113(1):1–11. doi: 10.1007/s11060-013-1084-8. • Important review that discussed the biogenesis of various forms of EVs and the issues related to EV nomenclature.
- 21.Bianchi E, Doe B, Goulding D, Wright GJ. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508(7497):483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonda DD, Akers JC, Kim R, et al. Neuro-oncologic applications of exosomes, microvesicles, and other nano-sized extracellular particles. Neurosurgery. 2013;72(4):501–510. doi: 10.1227/NEU.0b013e3182846e63. [DOI] [PubMed] [Google Scholar]
- 23. Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262(19):9412–9420. • Described the initial discovery that lead to the term ‘exosome’.
- 24.Ohshima K, Inoue K, Fujiwara A, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE. 2010;5(10):e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frohlich D, Fruhbeis C, Amphornrat J, et al. 2012 International Society for Extracellular Vesicles meeting. Gothenburg, Sweden: 2012. Crosstalk between neurons and glia involving exosomes as vesicular carriers of RNA and proteins [abstract. [Google Scholar]
- 26.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Zomer A, Vendrig T, Hopmans ES, et al. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010;3(5):447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angelucci A, D’Ascenzo S, Festuccia C, et al. Vesicle-associated urokinase plasminogen activator promotes invasion in prostate cancer cell lines. Clin Exp Metastasis. 2000;18(2):163–170. doi: 10.1023/a:1006778000173. [DOI] [PubMed] [Google Scholar]
- 29.Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55(7):808–818. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ginestra A, La Placa MD, Saladino F, et al. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res. 1998;18(5A):3433–3437. [PubMed] [Google Scholar]
- 31.Hakulinen J, Sankkila L, Sugiyama N, et al. Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. J Cell Biochem. 2008;105(5):1211–1218. doi: 10.1002/jcb.21923. [DOI] [PubMed] [Google Scholar]
- 32.Castellana D, Zobairi F, Martinez MC, et al. Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1–CX3CR1 axis. Cancer Res. 2009;69(3):785–793. doi: 10.1158/0008-5472.CAN-08-1946. [DOI] [PubMed] [Google Scholar]
- 33.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71(11):3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 34.Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Admyre C, Johansson SM, Paulie S, Gabrielsson S. Direct exosome stimulation of peripheral human T cells detected by ELISPOT. Eur J Immunol. 2006;36(7):1772–1781. doi: 10.1002/eji.200535615. [DOI] [PubMed] [Google Scholar]
- 36.Obregon C, Rothen-Rutishauser B, Gitahi SK, et al. Exovesicles from human activated dendritic cells fuse with resting dendritic cells, allowing them to present alloantigens. Am J Pathol. 2006;169(6):2127–2136. doi: 10.2353/ajpath.2006.060453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szajnik M, Czystowska M, Szczepanski MJ, et al. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PLoS One. 2010;5(7):e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pincetic A, Leis J. The mechanism of budding of retroviruses from cell membranes. Adv Virol. 2009;2009:6239691–6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raposo GA, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamparski HG, Metha-Damani A, Yao JY, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 42.Clayton A, Court J, Navabi H, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247(1–2):163–174. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 43.Koga K, Matsumoto K, Akiyoshi T, et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25(6A):3703–3707. [PubMed] [Google Scholar]
- 44.Rekker K, Saare M, Roost AM, et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47(1–2):135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 45.Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Rieseberg M, Kasper C, Reardon KF, Scheper T. Flow cytometry in biotechnology. Appl Microbiol Biotechnol. 2001;56(3–4):350–360. doi: 10.1007/s002530100673. [DOI] [PubMed] [Google Scholar]
- 47.Witwer KW, Buzas EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013:2. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 49.Soo CY, Song Y, Zheng Y, et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136(2):192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Pol E, Coumans F, Varga Z, et al. Innovation in detection of microparticles and exosomes. J Thromb Haemost. 2013;11(Suppl 1):36–45. doi: 10.1111/jth.12254. [DOI] [PubMed] [Google Scholar]
- 51.Roberts GS, Kozak D, Anderson W, et al. Tunable nano/micropores for particle detection and discrimination: scanning ion occlusion spectroscopy. Small. 2010;6(23):2653–2658. doi: 10.1002/smll.201001129. [DOI] [PubMed] [Google Scholar]
- 52.Aldape KD, Ballman K, Furth A, et al. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J Neuropathol Exp Neurol. 2004;63(7):700–707. doi: 10.1093/jnen/63.7.700. [DOI] [PubMed] [Google Scholar]
- 53.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci USA. 1990;87(21):8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16(6):748–754. doi: 10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 55.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 57.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 59.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balic M, Williams A, Lin H, et al. Circulating tumor cells: from bench to bedside. Annu Rev Med. 2013;64:31–44. doi: 10.1146/annurev-med-050311-163404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Mattos-Arruda L, Cortes J, Santarpia L, et al. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol. 2013;10(7):377–389. doi: 10.1038/nrclinonc.2013.80. [DOI] [PubMed] [Google Scholar]