Abstract
Interactions mediated by short linear motifs in proteins play major roles in regulation of cellular homeostasis since their transient nature allows for easy modulation. We are still far from a full understanding and appreciation of the complex regulation patterns that can be, and are, achieved by this type of interaction. The fact that many linear-motif-binding domains occur in tandem repeats in proteins indicates that their mutual communication is used extensively to obtain complex integration of information toward regulatory decisions. This review is an attempt to overview, and classify, different ways by which two and more tandem repeats cooperate in binding to their targets, in the well-characterized family of WW domains and their corresponding polyproline ligands.
Keywords: WW domain, protein domain repeats, peptide-mediated interactions, WW domain containing oxidoreductase, Yes-associated protein, cooperative binding, regulation of signaling
Introduction
The modular nature of protein interactions
Communication between proteins plays a major role in cellular homeostasis, as protein–protein interactions can create regulation of considerable complexity.1 To understand how these interactions are achieved, it is often helpful to look at protein partners not as single entities, each acting as an integrated whole, but rather as collections of smaller units, each with its specific role in interaction. These smaller units can be independently folding domains, short linear binding motifs,2 as well as the connecting regions that despite their lack of defined tertiary structure are increasingly recognized as important components of the protein and its function.3,4 Indeed, proteins have been shown to evolve by creating new combinations of existing self-autonomous domains—nature’s lego-building blocks5—rather than by the invention of novel domains6.
Linear motifs and their binding domains play a major role in cellular regulation—fine tuning is achieved by multiple interactions
Modular interactions between these building blocks are the driving force in the evolution and wiring of cell signaling circuits.7 Context-dependent, tunable interactions and switching behavior usually involve short linear motifs, post-translational modifications,8 and flexible linkers.9 A substantial fraction of protein–protein interactions is mediated by linear motifs or peptides (estimated around 40%10 or even higher11). It is therefore not surprising that interactions mediated by linear motifs and their corresponding binding domains, such as SH2, SH3, PDZ, and WW, are prominent in the regulation of pathways linked to serious diseases including several forms of cancer.
Cellular regulation and response to internal and external stimuli is directed by the combination, frequency, and intensity of linear-motif-mediated interactions between protein partners, which are easily modulated by post-translational modifications. Consequently, regulatory proteins, by means of the domains they contain, their local concentration, and post-translational modifications, compete with one another to determine the fates of their targets—to activate or to inhibit; to degrade or to sequester (see Figure 1). Concentration-dependent competition of targets for the regulatory protein can also help to determine which target will bind, and consequently which pathway will be activated (e.g. Kiel et al.12).
Figure 1.
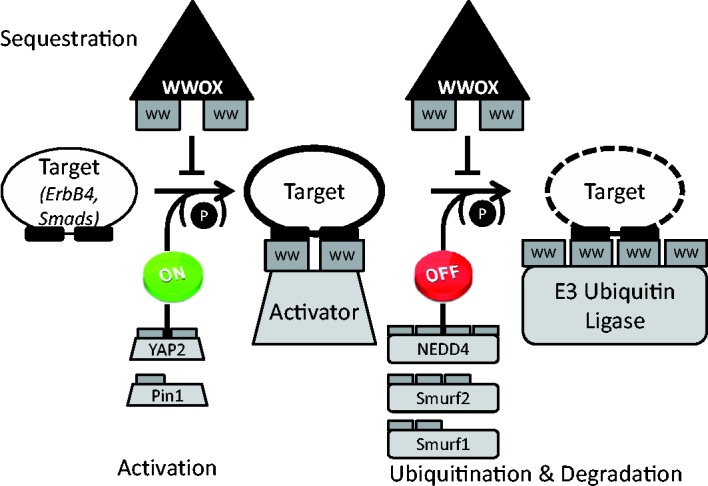
The life cycle of a regulatory protein is determined by WW domains. Proteins that contain (multiple) WW domains are involved in the tight regulation of both activation and clearance of major players in critical regulatory pathways. A simplified scheme of this process consists of the following steps: The target protein (white ellipse) is activated (bold outline) by binding to an activator protein (gray trapezoid), and subsequently targeted for degradation (dotted outline) by binding to an E3 ubiquitin ligase (gray rectangle).31,32,43 Both steps are regulated by competition with a sequestering protein (black triangle),20 as well as often by phosphorylation.32 All these interactions are mediated by interactions between WW domains (labeled dark gray rectangles) and the polyproline ligands on the target (black rectangles). The different targets, activators, and E3 ubiquitin ligases discussed in more detail in the text are shown, together with their number of WW domains. We note that interactions with different partners may involve different (numbers of) WW domains. See text for more details. (A color version of this figure is available in the online journal.)
Tandem repeated modules allow for fine-tuned regulation
Proteins involved in cellular regulation are often composed of multiple peptide binding domains of varying type and number. This multiplicity is usually matched by the presence of several binding motifs on corresponding target proteins. One particularly interesting case is the occurrence in a protein of tandem repeats of the same domain or the same linear motif. This review will discuss possible ways by which such domain and motif repeats cooperate to achieve regulations of increasing complexity. Our focus will be on the well-known prototype example: interaction and regulation mediated by the abundant WW domain and its polyproline peptide ligands.
WW domains: Short, versatile domains involved in a variety of interactions
With 35–40 residues, WW domains are among the smallest independently folding domains.13–15 As such, the WW domain has been the focus of many studies on protein structure stability and design.16–18 Nature also utilizes these domains extensively: WW domains have been found in many human proteins and play integral roles in systems connected to the appearance of Alzheimer’s, Huntington’s, muscular dystrophy, and cancer.19,20 In particular, loss-of-function mutations that disrupt WW domain–ligand interactions can lead to major complications, such as mental retardation that results from a point mutation in the WW domain of PQBP1 in Golabi–Ito–Hall syndrome21 and hypertension that results from a point mutation in the PPXY motif in the epithelial Na+ channel, ENaC, in Liddle syndrome.22 It is therefore not only interesting, but also very applicative to enhance our understanding of these domains and how they contribute to function within the full protein context.
WW domains fold into a three-stranded beta sheet that binds polyproline motifs. They have been divided into four main classes, corresponding to the polyproline motif with which they interact23: class 1 with PPxY motifs, class 2 with PPPL/R motifs, class 3 with (PxxGMxPP)*2 motifs, and class 4 with (pS/pT)P motifs (p = phosphorylation). Additional phosphorylation events regulate these interactions, in the WW domain (e.g. pY33 residue in the first WW domain of (WWOX) activates binding to p7324) as well as in binding motifs (PPxpY in WBP1 abolishes binding of YAP25). Here we will focus on the means by which WW domains are involved in manipulating cellular behavior. We first review studies of WW domains as autonomous units and then include the context imposed by neighboring WW domains occurring in tandem.
Isolated WW domain–peptide interactions are rather promiscuous
The most basic approach would assume an autonomous nature of domain-ligand binding even when removed from the context of their respective proteins. Several studies have attempted to evaluate the peptide-binding specificities of individual WW domains in humans and other organisms (using a wide range of assays including ELISA-like binding, mass spectrometry and pull-down, phage display, and peptide arrays).26–29 The most exhaustive study among these involved detailed mapping of interactions between all possible WW domains and corresponding peptide ligands in the human proteome.26 All predicted WW domains were cloned (a total of 65 clones) and every known consensus of peptide ligands (including flanking regions) was synthesized (around 2000 peptides), and each combination was matched in a quantitative ELISA-like binding assay, to be scored according to their determined binding affinity. Analysis of this experiment offers a glimpse into the indiscriminate nature of autonomous WW domain protein–peptide interactions. Among the roughly 1000 ligands that bound selectively to WW domains, only about one-third were qualified as “very selective” (i.e. binding to three or less of the WW domains), while two-thirds were ranked “moderately selective” (binding between four and 30 domains), and a revealing 77 peptide ligands displayed the ability to bind at least half of the identified WW domains. Conversely, among the 65 cloned WW domains, almost half bound up to 5% of the ligands, only a few bound to more than 25%, and none to more than 45% of the ligands.
As with any experiment that is done on a large scale and outside of its natural setting, results may not always parallel reality. For example, none of the 16 ligands identified in this study as binding the second WW domain in the WWOX protein could be recapitulated in physiological concentrations in a later study using mass spectrometry and phage display to identify binding partners to the first and second WW domains in WWOX.30
Putting domains back into context: How competition between WW domain containing proteins regulates cell proliferation
This considerable promiscuity among autonomous WW domain–peptide interactions indicates a rather non-specific way by which individual WW domains can bind to many different polyproline peptide partners, and vice versa, target proteins can bind to a variety of WW domains in different regulatory proteins—all depending on the specific temporal and spatial context. Unsurprisingly, WW proteins are important regulators in the activation and clearance of targets that play critical roles in a number of signaling pathways. This, however, also raises the question as to whether and how greater specificity and binding affinity are acquired in WW domain interactions. For this purpose, it is helpful to examine WW domains not only as autonomous units but also within their larger biological context (Figure 1).
Regulation of activation by WW domains
Take, for example, the well-studied dynamics between YAP and WWOX, two WW domain-containing proteins known to compete for interaction with several shared partners active in nuclear transcription control (Figure 1, left side).31 Most notable of these, ErbB4 undergoes cleavage in the cytoplasm and translocates to the nucleus where it acts as a transcription factor for the acceleration of cell proliferation. Binding of YAP with ErbB4 induces ErbB4 activity, in turn increasing cell proliferation. Conversely, binding of WWOX with ErbB4 inhibits this YAP–ErbB4 interaction, thus decreasing cell proliferation by sequestration of ErbB4 in the cytoplasm. A proper balance between YAP and WWOX binding with ErbB4 is therefore essential in maintaining a healthy rate of cell growth, as was established by the dose-dependent inhibition of YAP–ErbB4 transactivation upon introduction of increasing amounts of WWOX into the cell.31 This is a balance so important to cell stability that its disruption has been implicated in the appearance of tumor growth and cancer, in particular since the wwox gene is located in a fragile region.19,20
Regulation of clearance (degradation) by WW domains
The final stages of a target protein’s life cycle are also regulated by WW domains, here located in E3 ubiquitin ligases, the factors that lead to its degradation. Take, for example, the regulator proteins of the TGF-β and BMP signaling pathways.20,32 Pathway signaling is directly activated by R-Smads 1–3 and 5 and Co-Smad 4, and inhibited by I-Smads 6–7, all of which contain PPxY motifs. The opposing effects of the two Smad sets are coordinated by a common group of WW domains: in the sequester WWOX, the activators YAP and Peptidyl-prolyl cis-trans isomerase NIMA-interacting (Pin1), and finally in the E3 ubiquitin ligases SMAD ubiquitination regulatory factor (Smurf1/2) and E3 ubiquitin-protein ligase Nedd4-like (Nedd4L). Despite the antagonistic nature of regulatory and inhibitory Smads, their shared regulatory WW proteins act similarly toward both. That is to say, YAP exists simultaneously as an ON switch of both R- and I-Smads (Figure 1, center). Likewise, ubiquitin ligases Smurf1/2 and Nedd4L serve as OFF switches for several activator R-Smads, as well as inhibitor I-Smad7 (Figure 1, right side). WWOX on the other hand acts as mediator of the onset of the ON and OFF switches (Figure 1, left and right sides), since sequestration may prevent entry of its target proteins into the nucleus and thereby transcription factor activation, but also prevents degradation of the same target protein by competing with ubiquitin ligases such as Nedd4L. The differential regulation of R- and I-Smads is based on the details that distinguish their binding patterns: While the constitutive interactions with Inhibitory Smads are phosphorylation independent and involve only (or mainly) one WW domain (in YAP, Pin1, Nedd4L, and Smurf1/2), interactions with the Regulatory Smads are tightly regulated and predominantly involve the binding of tandem WW domain pairs to a more complex binding pattern of composite, phosphorylation-dependent (pS/pT)P-polyproline motifs.32
Strategies for fine-tuned regulation of WW domain–polyproline ligand interactions using tandem domains
It therefore appears that while in principle the competitions between different WW domain containing proteins for a common target described earlier (Figure 1) could be achieved by a single WW domain in each, it turns out that in fact many involve more than one domain (and consequently, often a more complex, composite binding motif on the partner32,33). Why? In this review we sketch out different possible ways by which WW tandem repeats have been used by nature to fine-tune regulation (Figure 2). Some have already been described in a previous review by Sudol in 2005.34 We start with strategies observed in the systems detailed earlier: First we describe the arguable simplest scenario of enhanced binding that is obtained by multiple, additive binding by tandem domains to polyproline motifs on a target, as originally suggested for YAP (Figure 2(a)). We then describe a scenario at the other extreme end, where the role of one WW domain in the tandem repeat has been reduced to stabilize—chaperone—the second domain, giving up its own binding ability to the target, as seen in WWOX (Figure 2(b)). Within the frame of these two extreme scenarios, we sketch out a broad range of possible ways of mutual influence of tandem domains toward overall binding affinity and specificity, and of different strategies for communication between WW domain repeats in Nedd4L and other ubiquitin ligases Su(dx), Smurf1, and Smurf2 (Figure 2(c) to (e)). Finally, we discuss what we can learn from solved structures of tandem WW domains, which provide a particularly useful source to demonstrate the rich variety of mutual influence of tandem WW domains (Figure 3).
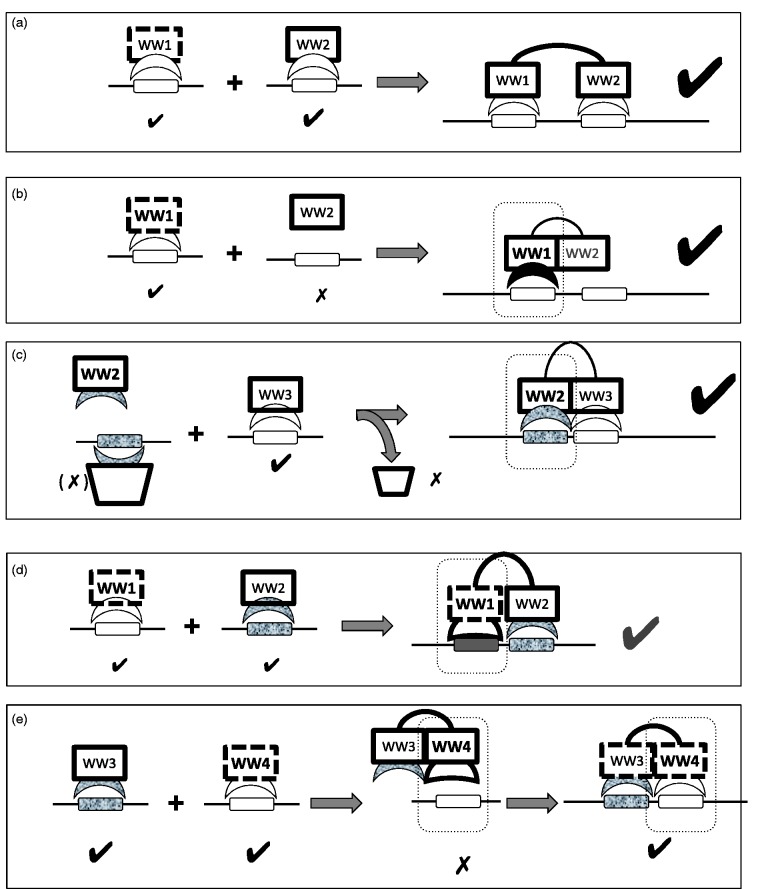
Figure 2.
Strategies for integration of input from different WW domains. Tandem WW domains in proteins can communicate in different ways during binding to their polyproline (and/or (pS/pT)P) ligand containing targets. This figure depicts the main strategies reported in literature and discussed in this review. In each example (except for (a)), the discussed WW domain is highlighted by a dotted rectangle and labeled in bold. For the WW domains, unstable domains are shown in dotted outline and the peptide binding site is indicated by a sickle. Different binding specificities are indicated by different fills (e.g. granite texture for Pin1-ligand binding to a (pS/pT)P ligand). Binding/nonbinding is shown by “v” and “x” signs, respectively. See text for more details. (a) Additive effect (as originally suggested for YAP2):39 Both tandem domains (e.g. WW1 and WW2 of YAP2) are independently capable of interaction with motifs on the target protein (e.g. ErbB4), but two such interactions significantly increase effective binding affinity. (b) Chaperone effect (e.g. WWOX):31,41 While only the first domain (e.g. WW1 of WWOX) is able to bind to the target (e.g. ErbB4), it is the second domain (WW2 of WWOX) that stabilizes this interaction to obtain the necessary binding affinity. The second domain has lost its ability to bind due to mutations at critical binding positions. (c) Binding-induced-binding (e.g. Nedd4L that outcompetes Pin1):32 In this case, the binding affinity of a weakly binding domain (e.g. WW2 domain of Nedd4L) is increased by positioning it next to its target peptide (e.g. on Smad2) after the first domain (e.g. WW3 domain of Nedd4L) has bound to an adjacent peptide motif. In this case, this binding event might outcompete an original, stronger interaction (trapezoid; e.g. with Pin1). (d) Modulation of binding conformation by adjacent WW domain (e.g. FBP11): An adjacent WW tandem domain (e.g. WW2 domain of FBP11) can change the stability as well as the dynamics of a WW domain (e.g. WW1 domain of FBP11),44 leading to changes in the target peptide bound conformation, and possibly in the binding specificity of the WW domain. (e) Integration of two binding events by competition between stabilization and binding (e.g. Su(dx)):46 A WW domain that can bind its polyproline target in isolation (e.g. WW4 domain of Su(dx)), loses this ability when connected to its tandem WW domain (e.g. WW3 domain of Su(dx)), due to stabilization of a binding-incompatible conformation. Upon binding of the tandem WW3 domain to its own polyproline target, the inhibition of the first is released. This results in an AND switch where full binding is only achieved when both WW domains are bound
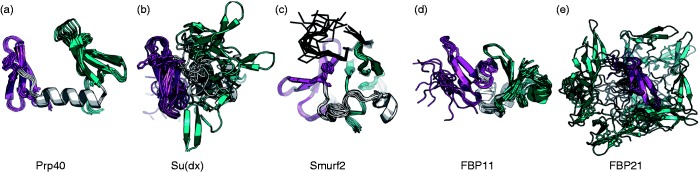
Figure 3.
Structures of tandem WW domains reveal a wide range of interdomain flexibility. A range of mobility between WW domains is observed in different solved tandem domain structures. (a) Yeast splicing factor Prp40 WW1–WW2 [Protein Data Bank (PDB54) id 1o6w45]. (b) E3 Ubiquitin ligase Su(dx) WW3–WW4 (PDB id 1tk746). (c) E3 Ubiquitin ligase Smurf2 WW3–WW4 bound to its peptide ligand (PDB id 2kxq33). (d) Human Prp40 homologs FBP11 (PDB id 2l5f) and (e) FBP21 (PDB id 2jxw40). The first WW domain, linker, and second WW domains are shown on the left in pink, in light gray, and to the right in cyan, respectively. The peptide is displayed as black trace (in (c)). Slight reorientations were performed to optimally display flexibility (in FBP21, the first domain is located in the center, in front of the rest). See text for more details. (A color version of this figure is available in the online journal.)
Increased binding affinity through joint binding of tandem WW domains (Figure 2(a))
At one end of the spectrum of tandem domain communication, we describe a scenario in which both tandem domains are independently capable of interacting with the target protein, and when both interactions occur simultaneously, the cooperative effect results in a significant jump in total affinity between the WW protein and its target. Confirmation of the occurrence in nature of this intuitive scenario has remained surprisingly elusive. Nevertheless, the mechanism has been recently observed and documented in a slightly modulated form in the case of the YAP2 (YAP1.2) isoform.35
NMR studies and molecular dynamics simulations have demonstrated that while the thermodynamic details differ between the two WW domains of YAP2—WW2 is structurally folded whereas WW1 undergoes substantially larger conformational changes upon binding,36,37 both tandem domains are capable of binding the target protein. Moreover, ITC measurements reveal that both YAP WW domains are able to interact with ErbB4 PPxY motifs in a physiologically relevant manner (most strongly with the third, PY3).37 Surprisingly, however, when the affinity of joint tandem domain binding is measured, a simple positive cooperative effect is not observed. Rather, a recent study by Schuchardt et al. reports that the presence of an additional WW domain alongside a functional, independently binding WW domain in YAP2 in fact has a negative effect on that domain’s binding with the ErbB4 PY3 target, along with other PPXY peptides.35 That is to say, independently binding WW1 and WW2 domains of the YAP2 isoform each bind the PY3 peptide of ErbB4 with reduced affinity when connected to their tandem domain (for this experiment, peptide binding ability of the added domain was abolished by targeted mutagenesis). Interestingly, a modulated form of positive cooperativity was instead observed within the context of this initial negative cooperativity, that is after an initial reduction of binding affinity within the tandem arrangement. In other words, after the presence of an additional WW domain causes an initial drop in its neighbor domain’s affinity for the target protein, simultaneous binding of the additional domain to an additional peptide motif restores binding affinity of the entire tandem domain (almost) to that of the isolated WW domain. This is, in short, a positive cooperative effect, albeit somewhat modulated for the purposes of controlling threshold levels in cell signaling. As such, it is similar to the positive cooperative effect observed for the WW3–WW4 tandem repeat of Su(dx), discussed below in the text and in Figure 2(e).38
While a straightforward positive cooperative effect between WW tandem domains remains yet to be found in nature, the inverse situation, that of two polyproline motifs in the target protein interacting with a WW domain for overall increased affinity, has indeed been observed. From the side of the peptide, we see a cooperative effect resulting in a sixfold increase in affinity when the YAP WW tandem domains interact with a dual PPxY peptide as compared to single PPxY peptides.36,39 Cooperativity has also been reported between tandem WW domains for the binding of FBP21 (Formin binding protein 21; WBP4) to splicing factor that interacts with polyglutamine binding protein 1 and PP1 (SIPP1) (WBP11) in the sub-spliceosomal complex. The two tandem group III WW domains in FBP21 bind to the group II and III adjacent polyproline motifs in SIPP1 with 60-fold enhancement compared to the individual domain–peptide interactions.40 This cooperativity is of functional importance, as removal of only one of the two WW domain–peptide interactions abolishes splicing.
Increased binding affinity by chaperoning activity of one WW domain on the tandem WW domain (Figure 2(b))
At the other end of the tandem domain communication spectrum, we again observe two domains that collaborate to modulate overall interaction affinity. This time, however, only one of the domains is capable of independently binding the target protein. Due to its structural instability, this first domain only weakly binds the target in isolation. The second domain does not interact with the target protein, but serves as a chaperone by binding the first domain, thus stabilizing the first domain’s structure and improving its interaction affinity with the target.
In the example of the two WW domains in WWOX, seen in both activation and clearance pathways earlier, only WW1 binds with any of the potential PPxY motifs on ErbB4 (again, most strongly to the isolated PY3 motif).31,41 Binding is—as if on purpose—abolished for WW2 by change of its sequence at two strategic positions: Y85 and E66 replace the generally conserved residues of WW1 at these positions (W44 and R25, respectively).41,42 Nevertheless, WW2 maintains a vital role in WWOX-ErbB4 binding. According to a recent study, WW2, the more thermally stable of the two WW domains of WWOX, remains structurally folded in isolation, whereas WW1 does not.41 Moreover, WW1 only adopts the standard WW-domain triple-stranded beta-fold when complexed with its chaperone WW2. And while WW1 can autonomously bind ErbB4 without the chaperone effect of WW2, WW1 interaction affinity is increased 2–3 fold when in the tandem WW1–WW2 module. Far from an isolated event, this pattern is repeated in WWOX binding with many of its partner PPxY ligands, among them p73,24 WBP1, and WBP2.42
Increased binding affinity through joint binding of tandem WW domains, in which binding of second domain depends on binding of first domain (Figure 2(c) to (e))
Above we presented two extremes on the spectrum of tandem domain communication. At one end, both domains are capable of rather independently binding the target protein. On the other end we again have two domains, the first of which independently binds the target with weak affinity, the second of which apparently never binds the target, but serves to stabilize and improve the binding affinity of the first. The following cases fall somewhere between these two extremes: while none of the WW domains have completely lost their binding ability, the mode of action of one WW domain (or both) is not restricted to its binding, but rather involves also regulation of binding of the tandem WW domain repeat.
WW domain with weak affinity can bind to its target once a neighboring WW domain–polyproline interaction has been established (Figure 2(c))
Here, it is the actual binding event of one WW domain that triggers conformational and dynamic changes in the neighboring WW domain, leading to changes in structure and affinity of its own interaction. This scenario is observed in the example of Nedd4L binding to Smad2/3 described in the clearance pathway earlier (Figure 1, right side) that leads to removal of the Pin1 activator.32 Phosphorylation plays a decisive role in this sequence of events: single phosphorylation (by Cdk8/9 in this case) creates a new binding site for the class 4 WW domain of the activator Pin1, turning Smad2/3 activity ON. The OFF switch is however much more intricate: The phosphorylation above serves also as a recognition signal for another kinase, GSK3, which subsequently phosphorylates a nearby PPXY motif, thereby creating a nearby target site recognized by the WW3 domain of Nedd4L. Importantly, binding of this WW3 domain juxtaposes the preceding Nedd4L WW2 domain to the location of the first motif, thereby increasing significantly its effective binding affinity. Consequently, binding of the Nedd4L WW2 domain releases Pin1 to turn OFF activation. Thus, nearby linear motif ligands on the target might interfere with each other to modulate nearby binding and activity. A similar sequential mechanism has been suggested for the binding of Smurf1/2 to a composite (pS/pT)P-polyproline motif in Regulatory Smads 1 and 2 (as well as to Inhibitory Smad7, in which case a slightly different motif containing negative charges removes the dependency on phosphorylation, see above).33 Here again, a WW domain is capable of binding Smad1/2/7 in isolation, but collaboration between tandem domains promotes joint, switchable binding in the case of regulatory Smads 1 and 2. This assertion has been contested by a different study of Smurf that suggests an alternative mechanism, in which the sole role of the first WW domain lies in its ability to multimerize,43 a strategy discussed in more detail below. Whatever the mechanism of cooperativity, the different studies agree that the tandem domains of Smurf1/2 bind better to the Smad7 PY peptide than the isolated domains.
Modulation of binding properties of WW domain by its tandem WW domain repeat (Figure 2(d) and (e))
Results from NMR experiments on the FBP11 protein suggest that while the isolated WW1 domain of FBP11 can bind polyprolines in two distinct conformations, where each binds the leucine residue in a different pocket (e.g. XP or XP2), the relative preference for these binding conformations is dramatically changed in the tandem WW1–WW2 construct due to addition of the WW2 domain that influences the flexibility of its binding groove44 (Figure 2(d)). Cooperative effects between ligand binding of neighboring domains WW3 and WW4 have been demonstrated also in the suppressor of deltex Su(dx) (homolog of human Nedd4 ubiquitin ligase) using a combination of ITC and NMR experiments.38 In this protein, domain–domain association competes with proper folding and ligand binding (Figure 2(e)). While the isolated WW4 domain can bind its ligand Notch, in the tandem construct WW3 domain stabilizes WW4 and by this prevents binding to its ligand. It is only upon binding of WW3 with its own ligand, e.g. a WBP1 peptide, that WW4 is released from this interdomain association to adapt a ligand binding conformation and to bind to the Notch polyproline motif. Thus, binding of WW4 to Notch is regulated by binding of WW3 to its own, distinct ligand. As described earlier, a similar scenario has been suggested recently for YAP2, in which binding of the tandem WW domain to a ligand recovers binding ability of its neighbor WW domain (with the difference that both bind to the same PY3 peptide ligand).35
Solved structures of WW domain tandem repeats highlight the variability of interaction patterns between the tandem domains and the crucial role played by the connecting linker (Figure 3)
As we have seen earlier, a wide spectrum of tandem domain communication strategies has been interpreted from experimental data regarding several high profile regulatory proteins containing multiple WW domains. However, little is known of the structural dynamics that determine exactly how two domains in a protein will interact to result in the regulatory scenarios described earlier.
Structural work on different tandem WW domain repeats has revealed early on that in addition to the individual WW domains, linker length as well as flexibility can significantly influence binding and activity. As the review by Sudol on tandem WW domains34 pointed out, comparison of solved structures of WW tandem domains, WW1–WW2 in the yeast splicing factor Prp4045 and WW3–WW4 in Su(dx),46 revealed two rather different mechanisms of cooperation between the domains, aimed to serve a rather different function: The helical, rigid linker (12 residues long) between WW1 and WW2 domains in Prp40 is responsible for a fixed, distant orientation of these domains, allowing (in principle completely independent) binding of different targets in a precise spatial orientation for the precise orchestration of the splicing machinery45 (Figure 3(a)). In contrast, the helical, but more flexible and longer linker (20 residues long) between WW3 and WW4 domains of Su(dx) allows for a wider, albeit still restricted, range of relative orientations between the two domains—helpful for the interaction of E3 ubiquitin ligases with a wide array of diverse proteins46 (Figure 3(b)). A still more rigid mode of association of WW domain tandem repeats has recently been observed in Smurf2, where the two associate to create one stable domain upon ligand binding33 (Figure 3(c); an alternative mechanism of action that involves homomultimerization has also been proposed for Smurf,43 see “Dimerization via WW—WW domain interactions—Another strategy to increase local concentration of polyproline binding domains” section). We note that linker flexibility per se is not necessarily conserved: Comparison of the structure of the tandem WW domain region of Prp40 above with two additional solved structures of human Prp40 homologs shows three distinct patterns of flexibility and WW domain interaction: while Prp40 includes a rigid helical linker of 12 residues that rigidly positions the two WW domains far apart from each other (Figure 3(a)), a slightly longer, still helical linker in FBP11 results in a relatively rigid, but interacting conformation (Figure 3(d)), enabling the mutual influence on the dynamics of neighboring WW tandem domains described earlier. In contrast, the linker between the tandem WW domain repeats in FBP21 is also 12 residues long, but does not form a helix, resulting in a high degree of flexibility and a large variation of WW domain orientations40 (Figure 3(e)). On the contrary, its high degree of flexibility has indeed been shown to be crucial for forming a tight interaction with SIPP1 and consequently for proper splicing, as described earlier.40 In YAP2 and medaka TAZ2 isoforms, even higher degree of flexibility between the WW1 and WW2 tandem domains is obtained by yet longer and less structured linkers (35 and 45 residues long, respectively).36 If, however, direct interaction between the domains is needed for the cooperative activity, a longer linker might actually decrease overall affinity, as exemplified in the case of the two Smurf1 isoforms that differ by linker length: It is only the short isoform that shows increased overall binding affinity to Smad7.33
Beyond tandem pairs
Increased robustness thanks to multiplicity of domains
Until now, we have described how tandem WW domain pairs communicate for the purpose of raising binding affinity and regulating interactions with a target protein. Frequently, these collaborative efforts are necessary to surpass a given threshold for functional binding, which allows to effectively integrate information from various sources to produce a desired result. This is because individual domains often bind independently with only marginal, nonfunctional affinity. Multiplicity of domains, however, may also provide redundancy, and thus increase robustness. Whereas generally the same two domains are coordinated to interact with a target protein, if the affinity threshold for effective functional binding is low enough, alternative matchups of domains may be sufficient to maintain functionality.
Nedd4L, the ubiquitin ligase observed in the Smad regulatory pathway discussed earlier, displays a high degree of flexibility and resourcefulness when interacting with an alternate target protein, Arrestin-Related Domain-Containing Protein-3 (ARRDC3).47 Affinity measurements of all combinations between the four WW domains of Nedd4L and the two corresponding PPxY motifs in ARRDC3 determined that tandem WW domains show higher affinity for the C-terminal domain of ARRDC3. Individually, WW3 binds with the highest affinity to both PPxY motifs, most probably due to its induced fit upon binding. WW2 and WW4 display lower levels of affinity to either motif, and WW1 interacts very weakly with both. Two pairings of WW domains—WW3–WW4 and WW3–WW2—were shown to interact strongly with ARRDC3. However, interaction between regulator and target proteins was not completely nullified until three of the four WW domains were abolished. That is to say, while only two WW domains bind with the target protein at any given time, interaction continued until WW2, WW3, and WW4 were all mutated. Thus, it is not only the multiplicity in WW domains utilized during interaction, but also the number of WW domains available for interaction, that can add to the strength of a regulator protein.
A similar study has tested the ability of different ligands to bind to each of the four WW domain repeats in the protein Itch (named AIP4 in that study).48 Ligands such as p68 were found to bind each of the WW domains with similar affinity, indicating synergistic binding as described earlier. However, other ligands such as EWS did show specificity for only one specific domain, indicating that Itch might serve as a scaffold to recruit specific ligands to specific domains. Therefore, the very same WW domain repeats in a protein can integrate information in different ways, depending on the ligands involved.
Multivalency and avidity as major players in the game of binding affinity and cellular regulation
We have considered the multiplicity of tandem WW domains in boosting interaction affinity with target proteins, but we have yet to discuss the reverse, that is the multiplicity of polyproline motifs for the same purpose.
The examples provided previously relate mainly to the mutual effect of tandem WW domains on binding to a site with one or two corresponding linear motifs. As described earlier, the strong binding of FBP21 to SIPP1 is achieved by a cooperative effect of two low-affinity WW domain–polyproline ligand interactions.40 Cooperativity effects seem however not to be the whole story—more and more evidence is accumulating that it is rather avidity that plays an important role in many multivalent interactions. In the case of FBP21, binding affinity indeed increases upon providing a second binding site on the target peptide, but importantly, further addition of up to four binding sites on the ligand increases effective binding affinity even more.49 Paramagnetic relaxation enhancement experiments in that study also suggest a dynamic equilibrium between at least two inverse binding orientations. It is therefore not the number of bound, but the number of locally available sites, i.e. multivalency, that determine strong effective binding affinity. These additional sites, and possible orientations, are thought to overall increase the local concentration of the partner, and by this to both enhance the effective on-rate and reduce the off-rate. This highlights the importance of local effective concentration of a partner for strong and biologically relevant binding.
Dimerization via WW—WW domain interactions—Another strategy to increase local concentration of polyproline binding domains
The local concentration of peptide binding sites might as well be modulated by WW domain-mediated homo-multimerization. Three separate studies have reported ability of WW domains to homo-multimerize.43,50,51 Specific to WW tandem pairs, the WW2 domain of Sav1,50 as well as WW1 of Smurf1 and its homolog WW2 in Smurf2,43 have been shown to undergo functional homo-multimerization. In conflict with Chong et al.’s earlier assertion that these Smurf WW domains participate in Smad7 interface binding,33 Aragon et al.43 claim they are in fact non-binding members of their respective tandem pair/triplet, but rather undergo homo-multimerization to increase the effective binding affinity of Smurf1/2 for the Smad7 peptide motif (via the neighboring Smurf1 WW2 and Smurf2 WW3 domains, respectively).
Intentional impairment of stability or binding ability of WW domains might allow for refined regulation of interactions
The same WW domains and WW domain proteins are present in many pathways of varying objectives within the cell, and it appears that full-strength interactions between proteins and their targets are not always ideal. We observe that several of the discussed WW domains in this review include sequence variation at critical sites for both folding stability and binding. As an example, WW2 domain in WWOX contains two such deviations from the general consensus (Y85 and E66) that abolish binding. Indeed, when these residues are reverted to the consensus (E66R/Y85W), this domain can again bind to its ligand peptides in ErbB4, WBP1, and WBP2.37,41,42 Moreover, in Smurf1 WW1 and Smurf2 WW2 domains replacement of a conserved tryptophan with tyrosine reduces stability (e.g. W257Y in the second WW repeat of Smurf2) as well as binding.33 Consequently, both are necessary for functional binding of the target Smad protein, leaving the interaction subject to possible control by contextual effects such as specific phosphorylation at either the peptide-binding or WW domain–domain interfaces.33 The WW domain is among the smallest folding domains and therefore often only marginally stable. As such, this domain is particularly amenable to regulation by structural destabilization and directed re-stabilization by contextual effects, e.g. neighboring tandem WW domain repeats. This sort of self-deregulation is another means of improving the protein’s versatility and suitability for regulation by external factors depending upon changing conditions. This is also reflected by the fact that positive cooperativity in peptide binding has so far only been reported in a background in which the ability of a WW domain to bind its peptide target within the context of the tandem domain is reduced compared to its binding as a free domain (see above).35,38 This could very well be necessary for maintaining a similar threshold of binding affinity that will elicit the functional outcome for both the case of single WW as well as tandem WW domains that distinguish, e.g. different isoforms.
Differential expression of isoforms with different numbers of WW domain repeats and linker lengths
Indeed, differential expression of isoforms is another way to modulate interaction of a tandem WW-domain protein with its respective targets. For example, YAP occurs in several isoforms that are expressed under different conditions: the YAP1 isoform contains only one WW domain, and previous findings have shown that YAP1 is a weaker binder, and therefore weaker coactivator, of ErbB4 than YAP2,52 and that it can bind only to I-Smad7 but not to R-Smads.43 Isoforms may also differ in binding affinity due to difference in linker length, as described earlier for Smurf1 binding to Smad7.33
Before we summarize our review, we would like to emphasize that the transient, often very weak nature of peptide-mediated interactions such as those discussed here between WW domains and their polyproline peptide partners makes accurate experimental characterization particularly challenging, and experimental noise might obscure the details, or even miss important interactions. Consequently, part of the proposed scenarios might undergo reinterpretation in the future. One illustration of this possibility is in the varying interpretations of data regarding Smurf1/2 binding to Smad7. While one group interpreted their experimental NMR and fluorescence binding results to conclude that for Smurf2, a tandem WW domain pair cooperatively binds Smad7,33 another group followed up with their own NMR and ITC experiments, only to propose that one WW domain in both Smurf1/2 tandem pairs does not actually bind the target protein at all, but rather serves to facilitate protein homodimerization.43 This dispute not only demonstrates the potential for conflicting determinations based on experimental data, but also opens the door for identifying further domain-communication strategies, in this case dimerization of domains. This leaves us with the need to constantly improve experimental approaches of improved accuracy and scope, but also to develop accurate simulation techniques that will provide structural models of tandem domain interactions to verify the uncertainty of experiment-based interpretation. In addition, it is important to remember that most of the experiments are performed in vitro, and it is not always clear whether the same experiments in a corresponding in vivo setting would provide equivalent results, and more so, to what extent the reported differences in binding affinities indeed translate to different functional outcomes in the cell.
Future outlook
We have presented a variety of well-studied systems in which WW domains in tandem were shown to play a range of different roles. How general then are these? Can we find additional examples of regulation using tandem domains? A simple inspection of WW domain-containing proteins in the human proteome (considering 55 proteins with bona fide WW domains reported previously53) reveals indeed a wide variation both in the number of WW domain repeats (around half contain more than one WW domain), as well as in the connecting linker lengths. Notably, such variation is also observed among different isoforms of a same protein (15 proteins are expressed as isoforms that differ in the number of WW domain repeats). The discovery of yet novel ways of regulating WW domain-mediated interactions is thus just around the corner. Further considerable accumulation of more experimental data on contextual effects on specific WW domain–polyproline mediated interactions and their classification will open up the horizon toward a golden era of significant advance in our understanding of integration strategies achieved by tandem repeats of WW domains as well as their ligands in particular, as well as peptide-binding domains in general. It is indeed a WoW34 era of tandem research!
Acknowledgements
This work was funded by the European Research Council under the ERC Grant Agreement No. 310873 to OSF.
Authors’ Contributions
EJD and OSF devised and wrote the manuscript. VFY contributed bioinformatics support. SRB collected relevant experimental data from the literature.
References
- 1.Lim WA, Lee CM, Tang C. Design principles of regulatory networks: searching for the molecular algorithms of the cell. Mol Cell 2013; 49: 202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Kruger D, Grebnev G, Kuban M, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res 2014; 42: D259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. Classification of intrinsically disordered regions and proteins. Chem Rev 2014; 114: 6589–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trudeau T, Nassar R, Cumberworth A, Wong ET, Woollard G, Gsponer J. Structure and intrinsic disorder in protein autoinhibition. Structure 2013; 21: 332–41. [DOI] [PubMed] [Google Scholar]
- 5.Das S, Smith TF. Identifying nature’s protein Lego set. Adv Protein Chem 2000; 54: 159–83. [DOI] [PubMed] [Google Scholar]
- 6.Vogel C, Morea V. Duplication, divergence and formation of novel protein topologies. Bioessays 2006; 28: 973–8. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem 2006; 75: 655–80. [DOI] [PubMed] [Google Scholar]
- 8.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science 2003; 300: 445–52. [DOI] [PubMed] [Google Scholar]
- 9.Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr Opin Struct Biol 2012; 22: 378–85. [DOI] [PubMed] [Google Scholar]
- 10.Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr Opin Biotechnol 2008; 19: 344–50. [DOI] [PubMed] [Google Scholar]
- 11.London N, Raveh B, Schueler-Furman O. Druggable protein-protein interactions—from hot spots to hot segments. Curr Opin Chem Biol 2013; 17: 952–9. [DOI] [PubMed] [Google Scholar]
- 12.Kiel C, Verschueren E, Yang JS, Serrano L. Integration of protein abundance and structure data reveals competition in the ErbB signaling network. Sci Signal 2013; 6: ra109–ra109. [DOI] [PubMed] [Google Scholar]
- 13.Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett 1995; 369: 67–71. [DOI] [PubMed] [Google Scholar]
- 14.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci 1994; 19: 531–3. [DOI] [PubMed] [Google Scholar]
- 15.Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol 1996; 65: 113–32. [DOI] [PubMed] [Google Scholar]
- 16.Piana S, Sarkar K, Lindorff-Larsen K, Guo M, Gruebele M, Shaw DE. Computational design and experimental testing of the fastest-folding beta-sheet protein. J Mol Biol 2011; 405: 43–8. [DOI] [PubMed] [Google Scholar]
- 17.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature 2005; 437: 579–83. [DOI] [PubMed] [Google Scholar]
- 18.Araya CL, Fowler DM. Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol 2011; 29: 435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardenswartz A, Aqeilan RI. WW domain-containing oxidoreductase’s role in myriad cancers: clinical significance and future implications. Exp Biol Med (Maywood) 2014; 239: 253–63. [DOI] [PubMed] [Google Scholar]
- 20.Aldaz CM, Ferguson BW, Abba MC. WWOX at the crossroads of cancer, metabolic syndrome related traits and CNS pathologies. Biochim Biophys Acta 2014; 1846: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ, et al. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem 2010; 285: 19391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu C, Pribanic S, Debonneville A, Jiang C, Rotin D. The PY motif of ENaC, mutated in Liddle syndrome, regulates channel internalization, sorting and mobilization from subapical pool. Traffic 2007; 8: 1246–64. [DOI] [PubMed] [Google Scholar]
- 23.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 2000; 14: 231–41. [PubMed] [Google Scholar]
- 24.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han SY, Melino G, Huebner K, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci USA 2004; 101: 4401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, Kelly JW, Sudol M. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem 1997; 272: 17070–7. [DOI] [PubMed] [Google Scholar]
- 26.Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L, et al. A map of WW domain family interactions. Proteomics 2004; 4: 643–55. [DOI] [PubMed] [Google Scholar]
- 27.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol 2005; 25: 7092–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otte L, Wiedemann U, Schlegel B, Pires JR, Beyermann M, Schmieder P, Krause G, Volkmer-Engert R, Schneider-Mergener J, Oschkinat H. WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci 2003; 12: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S. Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol 2006; 7: R30–R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abu-Odeh M, Bar-Mag T, Huang H, Kim T, Salah Z, Abdeen SK, Sudol M, Reichmann D, Sidhu S, Kim PM, et al. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J Biol Chem 2014; 289: 8865–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res 2005; 65: 6764–72. [DOI] [PubMed] [Google Scholar]
- 32.Aragon E, Goerner N, Zaromytidou AI, Xi Q, Escobedo A, Massague J, Macias MJ. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev 2011; 25: 1275–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong PA, Lin H, Wrana JL, Forman-Kay JD. Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity. Proc Natl Acad Sci USA 2010; 107: 18404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudol M, Recinos CC, Abraczinskas J, Humbert J, Farooq A. WW or WoW: the WW domains in a union of bliss. IUBMB Life 2005; 57: 773–8. [DOI] [PubMed] [Google Scholar]
- 35.Schuchardt BJ, Mikles DC, Hoang LM, Bhat V, McDonald CB, Sudol M, Farooq A. Ligand binding to WW tandem domains of YAP2 transcriptional regulator is under negative cooperativity. FEBS J. 2014; 281: 5532–51. [DOI] [PMC free article] [PubMed]
- 36.Webb C, Upadhyay A, Giuntini F, Eggleston I, Furutani-Seiki M, Ishima R, Bagby S. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway. Biochemistry 2011; 50: 3300–9. [DOI] [PubMed] [Google Scholar]
- 37.Schuchardt BJ, Bhat V, Mikles DC, McDonald CB, Sudol M, Farooq A. Molecular basis of the binding of YAP transcriptional regulator to the ErbB4 receptor tyrosine kinase. Biochimie 2014; 101: 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jennings MD, Blankley RT, Baron M, Golovanov AP, Avis JM. Specificity and autoregulation of Notch binding by tandem WW domains in suppressor of Deltex. J Biol Chem 2007; 282: 29032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald CB, McIntosh SK, Mikles DC, Bhat V, Deegan BJ, Seldeen KL, Saeed AM, Buffa L, Sudol M, Nawaz Z, et al. Biophysical analysis of binding of WW domains of the YAP2 transcriptional regulator to PPXY motifs within WBP1 and WBP2 adaptors. Biochemistry 2011; 50: 9616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Beullens M, Zhang J, Zhou Y, Nicolaescu E, Lesage B, Hu Q, Wu J, Bollen M, Shi Y. Structure and function of the two tandem WW domains of the pre-mRNA splicing factor FBP21 (formin-binding protein 21). J Biol Chem 2009; 284: 25375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuchardt BJ, Bhat V, Mikles DC, McDonald CB, Sudol M, Farooq A. Molecular origin of the binding of WWOX tumor suppressor to ErbB4 receptor tyrosine kinase. Biochemistry 2013; 52: 9223–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald CB, Buffa L, Bar-Mag T, Salah Z, Bhat V, Mikles DC, Deegan BJ, Seldeen KL, Malhotra A, Sudol M, et al. Biophysical basis of the binding of WWOX tumor suppressor to WBP1 and WBP2 adaptors. J Mol Biol 2012; 422: 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aragon E, Goerner N, Xi Q, Gomes T, Gao S, Massague J, Macias MJ. Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-beta Pathways. Structure 2012; 20: 1726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez-Espain X, Ruiz L, Martin-Malpartida P, Oschkinat H, Macias MJ. Structural characterization of a new binding motif and a novel binding mode in group 2 WW domains. J Mol Biol 2007; 373: 1255–68. [DOI] [PubMed] [Google Scholar]
- 45.Wiesner S, Stier G, Sattler M, Macias MJ. Solution structure and ligand recognition of the WW domain pair of the yeast splicing factor Prp40. J Mol Biol 2002; 324: 807–22. [DOI] [PubMed] [Google Scholar]
- 46.Fedoroff OY, Townson SA, Golovanov AP, Baron M, Avis JM. The structure and dynamics of tandem WW domains in a negative regulator of notch signaling, Suppressor of deltex. J Biol Chem 2004; 279: 34991–35000. [DOI] [PubMed] [Google Scholar]
- 47.Qi S, O’Hayre M, Gutkind JS, Hurley JH. Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3). J Biol Chem 2014; 289: 4743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 2004; 23: 1972–84. [DOI] [PubMed] [Google Scholar]
- 49.Klippel S, Wieczorek M, Schumann M, Krause E, Marg B, Seidel T, Meyer T, Knapp EW, Freund C. Multivalent binding of formin-binding protein 21 (FBP21)-tandem-WW domains fosters protein recognition in the pre-spliceosome. J Biol Chem 2011; 286: 38478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohnishi S, Guntert P, Koshiba S, Tomizawa T, Akasaka R, Tochio N, Sato M, Inoue M, Harada T, Watanabe S, et al. Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form. FEBS Lett 2007; 581: 462–8. [DOI] [PubMed] [Google Scholar]
- 51.Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci 2010; 19: 1354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem 2003; 278: 33334–41. [DOI] [PubMed] [Google Scholar]
- 53.Sudol M, McDonald CB, Farooq A. Molecular insights into the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1. FEBS Lett 2012; 586: 2795–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res 2000; 28: 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]