Abstract
Basic/helix-loop-helix (bHLH) proteins comprise one of the largest transcription factor families and play important roles in diverse cellular and molecular processes. Comprehensive analyses of the composition and evolution of the bHLH family in cotton are essential to elucidate their functions and the molecular basis of cotton development. By searching bHLH homologous genes in sequenced diploid cotton genomes (Gossypium raimondii and G. arboreum), a set of cotton bHLH reference genes containing 289 paralogs were identified and named as GobHLH001-289. Based on their phylogenetic relationships, these cotton bHLH proteins were clustered into 27 subfamilies. Compared to those in Arabidopsis and cacao, cotton bHLH proteins generally increased in number, but unevenly in different subfamilies. To further uncover evolutionary changes of bHLH genes during tetraploidization of cotton, all genes of S5a and S5b subfamilies in upland cotton and its diploid progenitors were cloned and compared, and their transcript profiles were determined in upland cotton. A total of 10 genes of S5a and S5b subfamilies (doubled from A- and D-genome progenitors) maintained in tetraploid cottons. The major sequence changes in upland cotton included a 15-bp in-frame deletion in GhbHLH130D and a long terminal repeat retrotransposon inserted in GhbHLH062A, which eliminated GhbHLH062A expression in various tissues. The S5a and S5b bHLH genes of A and D genomes (except GobHLH062) showed similar transcription patterns in various tissues including roots, stems, leaves, petals, ovules, and fibers, while the A- and D-genome genes of GobHLH110 and GobHLH130 displayed clearly different transcript profiles during fiber development. In total, this study represented a genome-wide analysis of cotton bHLH family, and revealed significant changes in sequence and expression of these genes in tetraploid cottons, which paved the way for further functional analyses of bHLH genes in the cotton genus.
Introduction
Basic/helix-loop-helix (bHLH) transcription factors, named from their signature bHLH domains, are ubiquitously distributed in major eukaryotes and involved in diverse cellular and molecular processes [1–3]. A bHLH domain generally comprises around 60 amino acids and two functionally distinct segments, i.e., the basic and helix-loop-helix regions. Structural analyses have indicated that the basic region forms the major interface contacting DNA, whereas the helix-loop-helix region mediates protein-protein interactions regulating DNA binding activity [3]. By interacting with DNA and different proteins simultaneously, bHLH proteins frequently act as central integrators in gene regulation networks [1,2,4–7]. For example, phytochrome-interacting factors (PIFs), the major regulators of plant photomorphogenic development, interact with multiple regulatory proteins (such as DELLA, HY5, phy, BZR1) from different pathways and integrate diverse signals to control plant growth [1].
With more and more genomes sequenced, increasing number of bHLH proteins have been identified and employed in classification and evolutionary comparison across a wide range of organisms [3,8–16]. Compared to fungi and metazoans, the bHLH family expands significantly in higher plants, harboring 88 to 289 bHLH genes in a single genome [9,12,13]. Based on their evolutionary relationships, bHLH domains identified from representative species (Arabidopsis, poplar, rice, moss and algae) were classified into 32 subfamilies with 2 moss-specific (S6 and S29), and 1 algae-specific subfamily (S32) [13].
Cotton is the leading fiber crop and provides the majority of natural fibers in the worldwide textile market. Among around 50 species in cotton (Gossypium) genus, two diploids (G. arboreum and G. herbaceum, 2n = 2X = AA = 26) and two allotetraploids (upland cotton, G. hirsutum, and sea island cotton, G. barbadense, 2n = 4X = AADD = 52), have been cultivated to produce economically valuable fibers [17]. It is believed that allotetraploid cottons all derive from an interspecific hybrid formed during the Pleistocene (1–2 millions of years ago). G. raimondii is the closest living relative of the D-genome donor of allotetraploid cottons, but it does not produce significantly elongated fibers as the A-genome donors (G. arboreum and G. herbaceum) [17]. Upland cotton is most widely used in the modern cotton industry, and accounts for most of the world’s cotton yield. Compared to their diploid progenitors, upland cotton and sea island cotton have significantly higher yield and fiber quality. Agronomists and biologists have long been interested in genomic evolution of tetraploidization and domestication of cottons [17–22]. Transcriptomic analyses indicated that several pathways were up-regulated in developing fibers and homoeologous genes might be differentially regulated in tetraploid cotton tissues, implying that genes from different genomes may act synergistically to enhance fiber production in tetraploid cottons [19–26]. However, the importance of these up-regulated pathways and the mechanisms to control these pathways still need to be elucidated. As one of the largest transcription factor families in plants, bHLH proteins may play important roles in regulating various pathways and cotton development. Comprehensive analysis of cotton bHLH proteins and their evolutionary changes during tetraploidization may help to reveal molecular mechanisms underlying the varied pathways and super quality and yield in modern tetraploid cottons. On the other hand, evolutionary effect of alloploidy has long been an attracting theme in plant biology [18,27–32], since an estimate of 30–70% plant species, including many important crops such as cotton, wheat, oilseed rape, and tobacco, are of alloploid origin. Analyses of genetic and transcriptional alterations of bHLH homoeologous genes in allotetraploid cotton may provide useful clues to elucidate the influence of alloploidy in plant evolution and the molecular basis of speciation and domestication of alloploid crops.
In this study, we identified bHLH genes comprehensively from the known cotton sequences, including the genomes of G. raimondii and G. arboreum which were recently sequenced [18,33,34]. A set of Gossypium bHLH reference genes were constructed and employed to analyze their evolutionary relationships with homologs from the model plant Arabidopsis, and cacao (Theobroma cacao), a sequenced species most closely related to the cotton genus. To explore evolutionary changes of bHLH genes during tetraploidization, all genes of two subfamilies (S5a and S5b) in upland cotton and its diploid progenitors were cloned and compared, and transcription profiles of the genome-specific orthologous genes were further determined in upland cotton.
Materials and Methods
Sequence sources
All sequence data were obtained from the internet (S1 Table). Arabidopsis bHLH proteins and reference bHLH sequences of Oryza sativa, Physcomitrella patens, and Chlamydomonas reinhardtii (S2 Table) were retrieved from Phytozome (http://www.phytozome.net/search.php) [35] according to Carretero-Paulet et al [13]. The annotated genome sequences of T. cacao and G. raimondii (Gr-JGI) were downloaded from Phytozome [18,35,36]. The annotations of G. raimondii and G. arboreum genomes (Gr-CGP and Ga-CGP) were from the Cotton Genome Project in the Institute of Cotton Research of Chinese Academy of Agricultural Sciences (http://cgp.genomics.org.cn/page/species/download.jsp?category=raimondii and = arboreum, respectively) [33,34]. Upland cotton unigenes (Gh-Uni) were obtained from Plant Transcription Factor Database (PlantTFDB, http://planttfdb.cbi.pku.edu.cn/family.php?sp=Ghi&fam=bHLH) [37]. Gossypium bHLH contigs (Go-con) and mRNA sequences were retrieved from Cottongen (http://www.cottongen.org/retrieve/sequences) by searching sequences containing bHLH domain (IPR011598) [38].
Identification of bHLH proteins and corresponding bHLH domains
According to classification of plant bHLH proteins reported previously [13], 32 representative bHLH domains (one per subfamily) and three Arabidopsis orphans were selected to constitute a set of probe sequences (S2 Table). To identify bHLH proteins from annotated genomes (G. raimondii, G. arboreum and T. cacao), all probe bHLHs were employed to query primary-transcript-only proteins of the sequenced genomes by a standalone BLAST program [39]. For each genome, repeated entries were eliminated by Microsoft Excel program, and putative bHLH domains were retrieved according to two-sequence alignments in BLAST. The resulting bHLH sequences were further aligned with all probe sequences and Arabidopsis bHLH domains. The sequences conforming to the following rules were validated as bHLH domains. A bHLH domain should contain 1) at least two continuous sub-regions of basic, helix1, and helix2 and 2) over 60% consensus amino acid residuals identified in plant bHLHs by Carretero-Paulet et al [13].
To determine Gossypium reference bHLH genes, cotton bHLH proteins identified from various sources (Gr-JGI, Gr-CGP, Ga-CGP, Gh-Uni, Go-con, and mRNA) were aligned using AlignX program in Vector II software (Invitrogen). The branch lengths (BL) in the alignment guide tree reflecting the genetic divergences between sequences were employed as an arbitrary standard to group corresponding sequences. The proteins with BL<0.03, 0.03 to 0.15, and >0.15 were regarded as originating from an allele gene, orthologous genes of different genomes and different paralogous genes, respectively. For each orthologous group, a single representative member (mainly from Gr-JGI) was selected as reference gene, and the Gossypium reference bHLH genes included all non-overlapping paralogous genes (S3 Table).
Phylogenetic analysis and classification of Gossypium bHLHs
Phylogenetic analysis was performed using MEGA6.0 [40]. All cotton reference bHLH domains were aligned with the bHLH domains from Arabidopsis and T. cacao. Since members of several plant bHLH subfamilies (S6, S8, S29 and S32) were not identified in Arabidopsis [13], each two representative sequences of these subfamilies from other plants (S4 Table) were also included in the multiple sequence alignment. The alignment was performed using clustalW with default settings. Phylogenetic trees were constructed and tested by neighbor joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) methods, and bootstrap test was set as 1000 replicates. Classification of bHLH proteins was performed according to evolutionary relationships of bHLH domains. The bHLHs on a branch supported by at least two methods and with high bootstrap (>88%) in NJ test were clustered into a subfamily. The resultant bHLH subfamilies were compared to those determined by Carretero-Paulet et al [13], and named accordingly.
Cloning and sequence analysis of Gossypium S5a and S5b bHLH genes
Cotton DNAs were extracted from fresh leaves using a plant DNA extraction kit (Aidlab, Beijing, China). The S5a and S5b bHLH genes (except for GhbHLH062A) from G. hirsutum, G. arboreum, and G. raimondii were amplified with primers encompassing the coding regions (S5 Table). The 25-μl PCR reactions included 100 ng cotton genomic DNA, 1×PrimerSTAR mix (TaKaRa), 200 nM upstream and downstream primers. The PCR thermocycling parameters were as follows: 98°C for 1 min, followed by 35 cycles of 98°C for 10s, 55°C for 15 s and 72°C for 1 min, and a final extension of 3 min at 72°C. After A-tailing, all PCR products were cloned into pGEM-T (Promega) and sequenced by Invitrogen (Shanghai, China).
For GhbHLH062A, we first cloned the 3’-end of coding region using the A-specific primer and downstream coding-region primer. The upstream sequences were then amplified by two rounds of Y-shaped adaptor dependent extension (YADE) [41] until a long terminal repeat (LTR) retrotransposon insertion was found. Finally, the 5’-end of coding region were amplified using the upstream coding-region primer and a LTR primer.
The exon sequences and ORFs were determined by comparing genomic sequences to EST, mRNA or homologous proteins using BLAST program in NCBI. The deduced protein sequences were aligned and subjected to construction and test of phylogenetic tree using NJ method in MEGA6.0 [40]. The genome origin (A or D) of a certain gene from tetraploid cotton was determined according to its evolutionary relatedness to the orthologous genes from progenitor diploids (G. arboreum and G. raimondii).
To detect a certain gene in tetraploid and diploid cottons, genome-specific primers were designed to amplify fragments of 100 to 250 bp from genomic DNAs using 2×Taq PCR mixture (Tiangen, China). The PCRs contained around 100 ng genomic DNAs and 200 nM upstream and downstream primers, and amplified for 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s. The PCR products were detected by EtBr staining in agarose gel.
Sequences of all PCR primers employed in this study are shown in S5 Table.
RNA extraction and real-time RT-PCR analysis
Upland cotton RNAs were extracted from roots, stems, leaves, petals, ovules, and fibers of different developmental stages using a rapid plant RNA extraction kit (Aidlab, Beijing, China). The cDNAs were synthesized from total RNA using a first-strand cDNA synthesis kit (TaKaRa, Dalian, China), and then subjected to real-time PCR analyses. Real-time PCRs were performed on a CFX96 real-time PCR detection system using SYBR Green Supermix (Bio-Rad, CA, USA) according to the manufacturer’s introductions. The thermocycling parameters were as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 10 s, 57°C for 20 s, followed by a standard melting curve to monitor PCR specificity. Cotton histone3 (AF024716) [42] and GhUBQ14 [43] genes were amplified as internal standards. Reactions were performed for three replicates. Data were analyzed using the software Bio-Rad CFX Manager 2.0 provided by the manufacturer.
Results
Identification of cotton bHLH genes in Gossypium genomes
The D and A genomes of diploid cottons were recently sequenced by different projects (S1 Table) [18,33,34]. To perform a comprehensive analysis of cotton bHLH family, a set of plant bHLH probe sequences were employed to identify bHLH genes from annotated genomes of G. raimondii and G. arboreum. Consequently, 272, 255, and 256 distinct bHLH proteins were identified in the genomic sequences from Gr-JGI, Gr-CGP, and Ga-CGP, respectively. These sequences were further aligned with bHLH proteins annotated in Gossypium EST contigs (Go-con), G. hirsutum unigenes (Gh-Uni), and cotton mRNAs. Finally, 919 Gossypium bHLH proteins were clustered into 289 orthologous groups (S3 Table). Each orthologous group in the alignment might represent a distinct bHLH gene in Gossypium reference genome. Therefore, we selected a single representative from each orthologous group to constitute Gossypium reference bHLH genes, which were coded alphabetically as GobHLH001-289 (S3 Table).
As shown in Table 1 and S3 Table, most bHLH genes (>89%) had orthologous or overlapped genes from other source(s). All proteins from Go-con, Gh-Uni, and mRNA could be assigned to a certain gene in sequenced genomes, suggesting that most bHLH-coding genes in Gossypium might have been revealed by genome sequencing. On the other hand, none of the three genomic sequencing projects had annotated all the bHLH reference genes, indicating that the cotton genome sequences were still relatively fragmented and might have missed a portion of information.
Table 1. Numbers and overlapping of cotton bHLH genes from different sources.
| Gr-JGI | Gr-CGP | Ga-CGP | Gh-Uni | Go-con | mRNA | |
|---|---|---|---|---|---|---|
| Gr-JGI | 272 | 249 | 243 | 79 | 28 | 4 |
| Gr-CGP | 255(257)* | 236 | 78 | 27 | 4 | |
| Ga-CGP | 256(259)* | 79 | 28 | 4 | ||
| Gh-Uni | 79(92) | 27 | 4 | |||
| Go-con | 28(29) | 2 | ||||
| mRNA | 4(10) |
Numbers of non-overlapping paralogous genes from a certain source are shown on the diagonal line with the initial sequence number in brackets. Other numbers indicate overlapped or orthologous genes between two sources.
* The tandem repeat genes encoding identical proteins and orthologous to a single protein from other sources are regarded as a single gene.
Phylogenetic analysis and classification of Gossypium bHLH family
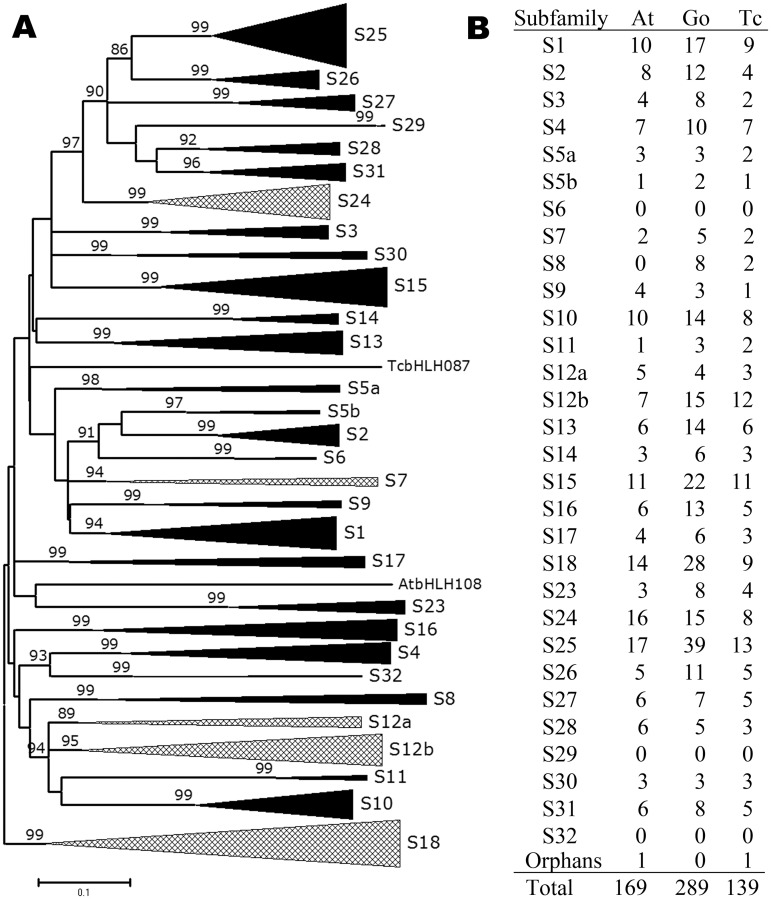
To analyze evolutionary relationship of Gossypium bHLH proteins, 289 Gossypium reference bHLH domains were aligned with bHLHs from Arabidopsis and cacao, and representative sequences of S6, S8, S29, and S32 bHLH subfamilies from P. patens, C. reinhardtii, and O. sativa (S1 Fig). Based on the resultant phylogenetic tree (Fig 1A; S2 Fig), 605 bHLH sequences were grouped into 30 subfamilies (Fig 1A and 1B; S6 Table). The majority of plant bHLH subfamilies determined by Carretero-Paulet et al [13] remained in our classification, except that S5 and S12 were divided into two subfamilies (S5a and S5b, S12a and S12b, respectively), and that S18, S19, S20, S21 and S22 were merged into a single subfamily S18.
Fig 1. Phylogenetic relationships and classification of Arabidopsis, Gossypium and Theobroma bHLH domains.
A, NJ tree of 605 bHLH domains (169 from A. thaliana, 289 from Gossypium, 139 from T. cacao, 4 from P. patens, 2 from C. reinhardtii, and 2 from O. sativa). Subfamilies are collapsed and represented as triangles with both depth and width proportional to sequence divergence and size, respectively. Orphans are represented as single lines. The scale bar indicates the estimated number of amino acid replacements per site. For subfamilies S6, S29 and S32, only 2 reference sequences from P. patens or C. reinhardtii are used in the alignment. The alignment used for tree construction and the full representation of the tree are shown in S1 and S2 Figs, respectively. B, Member numbers of different bHLH subfamilies in A. thaliana (At), Gossypium (Go) and T. cacao (Tc).
In the phylogenetic tree, Gossypium bHLHs are assigned to 27 subfamilies along with those from Theobroma and Arabidopsis (Fig 1A; S2 Fig). Although Gossypium contains many more bHLH members, the subfamily numbers in Gossypium and Theobroma are the same (27 subfamilies), and only one (S8) more than that in Arabidopsis (S6 Table). Consistent with the recent genome polyploidization in Gossypium [18,33,34], Gossypium bHLH subfamily members generally increase in number, compared to Theobroma. However, this expansion (1- to 4-fold in gene number) is uneven, suggesting that the extent of gene deletion after polyploidization varies among subfamilies (Fig 1B).
Origin and variations of S5a and S5b bHLH genes in tetraploid cottons
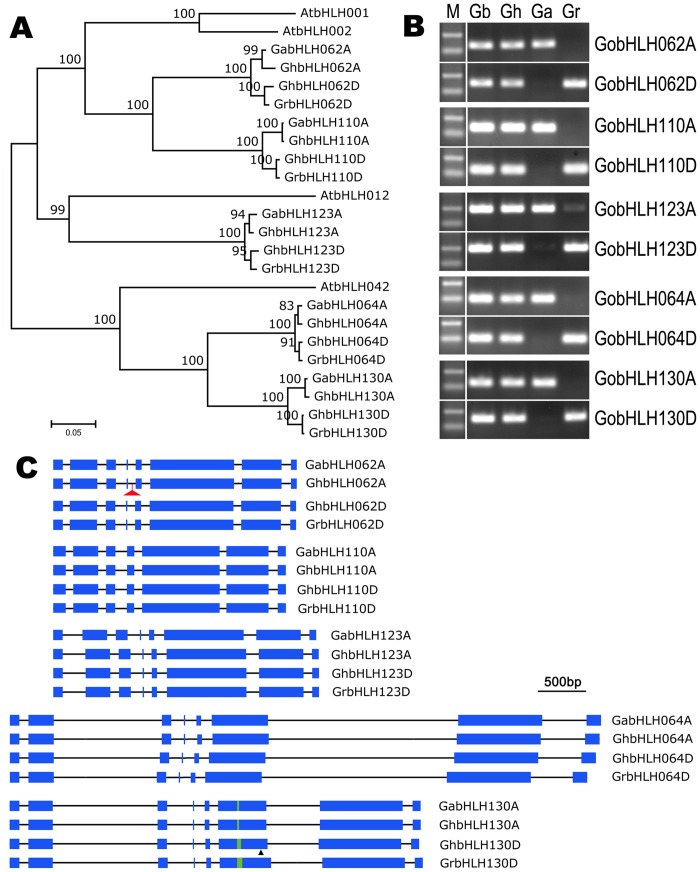
Cotton fibers are ovule epidermal trichomes and flavonoids may be involved in the regulation of fiber development [44,45]. Thus, we selected S5a and S5b bHLHs, acting as important regulators of trichome differentiation and flavonoid biosynthesis in Arabidopsis (S6 Table) [4,46,47], to explore evolutionary changes of bHLH genes in tetraploid cottons. All bHLH genes of S5a and S5b subfamilies (GobHLH062, GobHLH064, GobHLH110, GobHLH123 and GobHLH130) were cloned from upland cotton and its diploid progenitors (G. arboreum and G. raimondii). These sequences were deposited in GenBank under the accession nos KP698854-KP698873. Consistent with the number of reference genes, both diploid cottons contained five genes, while upland cotton harbored 10 genes of S5a and S5b bHLH subfamilies. Based on deduced protein sequences, these cotton bHLH genes could be assigned to five orthologous groups, and each group included four genes, two from each genome (A or D; Fig 2A; S3 Fig). All of these genes could be also detected in another tetraploid species (G. barbadense) using genome-specific primers (Fig 2B). These data suggest that bHLH genes doubled by tetraploidization are generally maintained in tetraploid cottons. For the convenience of discrimination, we name a certain gene like GhbHLH062A, i.e., the abbreviation of species (the first two letters) plus reference gene code plus genome name (the last letter).
Fig 2. S5a and S5b bHLH genes in Gossypium.
A, NJ tree of S5a and S5b bHLH proteins from A. thaliana and Gossypium. The scale bar indicates the estimated number of amino acid replacements per site. The tree was constructed and tested using the alignment shown in S3 Fig. Percentage of supported bootstrap for every branch in a test of 1000 replicates is indicated. B, PCR confirmation of the genome-specific S5a and S5b bHLH genes in tetraploid cottons G. barbadense (Gb) and G. hirsutum (Gh), and their diploid progenitors G. arboreum (Ga) and G. raimondii (Gr). M, DNA marker with two bands of 250 bp (upper) and 100 bp (lower) indicated. C, Gene structures of cotton S5a and S5b bHLH genes. The sequences corresponding to the ORFs are depicted proportionally. Exons and introns are represented by blue bars and black lines, respectively. Red triangle indicates the long terminal repeat (LTR) retrotransposon inserted in GhbHLH062A (Details are shown in S4 Fig). For GobHLH130s, the simple sequence repeats (GAA)n of various lengths in the 6th exon are presented in green and a black triangle directs the site of a 15-bp deletion in GhbHLH130D (see details in S5 Fig).
The structures of S5a and S5b bHLH genes from different cotton species are shown in Fig 2C. The exon-intron patterns of these genes are highly conserved, except that the 4th intron is lost in GobHLH110s. Comparing to their orthologs from diploid cottons, there exist three major sequence variations, and several single-nucleotide changes in the upland cotton genes. Firstly, a LTR retrotransposon (~5 kb in length) is inserted in the 4th intron of G. hirsutum bHLH062A (Fig 2C). As shown in S4 Fig, this LTR retrotransposon insertion exists in different G. hirsutum lines and also in G. barbadense, suggesting that this LTR retrotransposon duplication is a common evolutionary event during tetraploidization. Secondly, a 15-bp fragment is deleted in the 6th exon of GhbHLH130D (Fig 2C). This deletion may be specifically in the G. hirsutum lineage, as it is detected in two G. hirsutum lines, but not in G. barbadense and G. raimondii (S5 Fig). Finally, the length of a simple sequence repeats (GAA)n in the 6th exon of GhbHLH130D is 12-bp shorter than that of GrbHLH130D (Fig 2C).
Transcriptional profiling of S5a and S5b bHLH genes in upland cotton
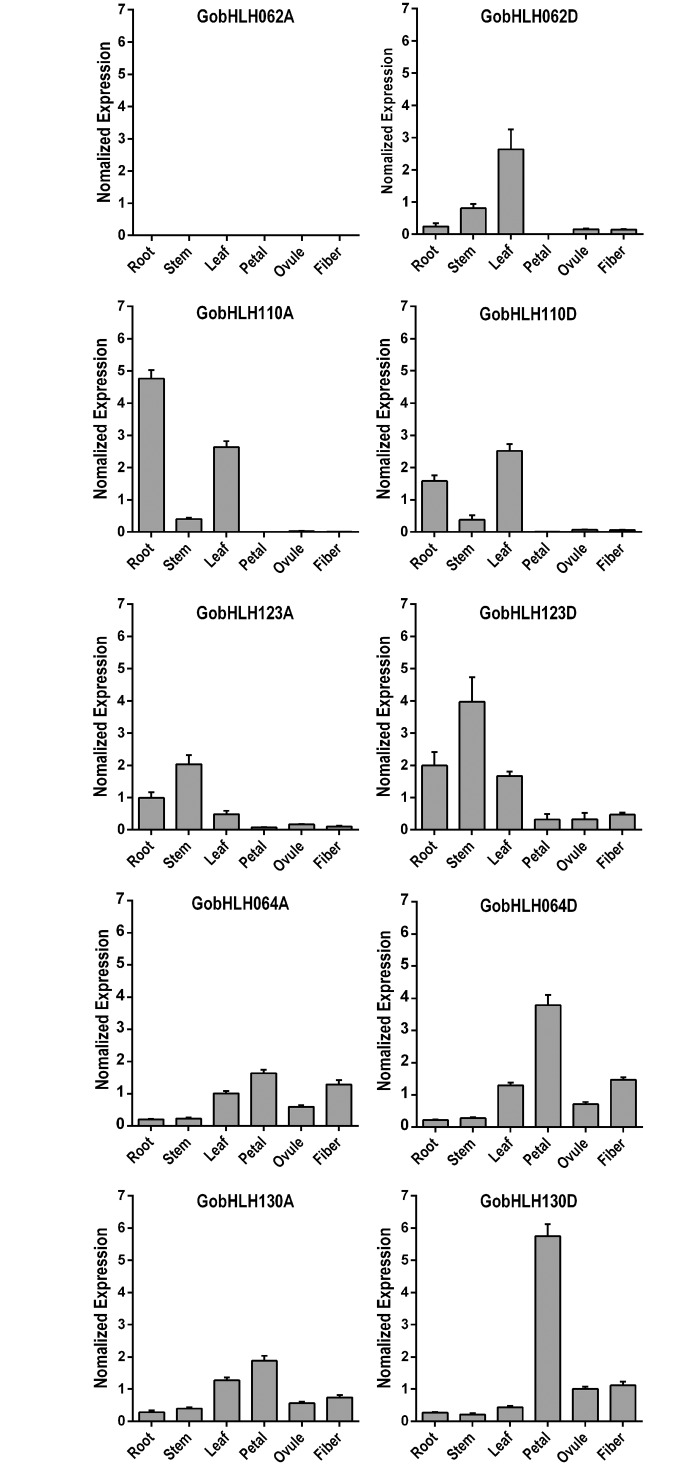
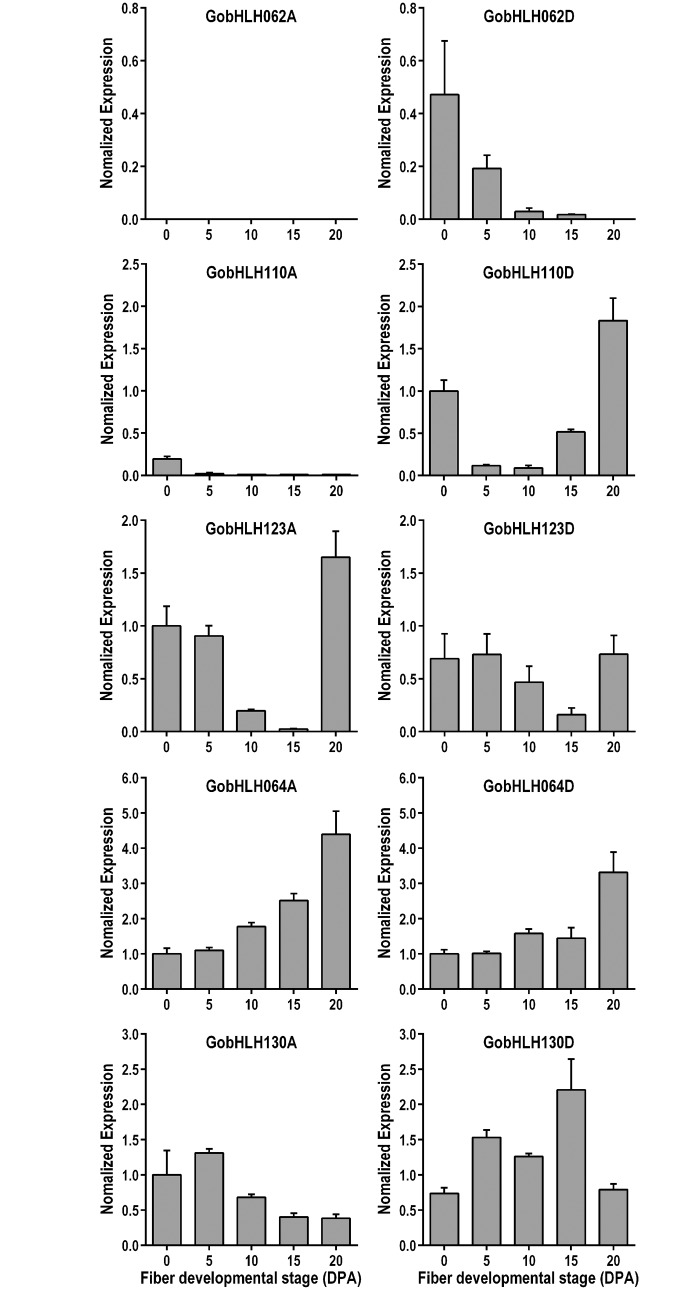
To elucidate whether the doubled bHLH genes were differentially regulated at transcriptional level in tetraploid cotton, real-time RT-PCR was employed to detect the transcript levels of 10 S5a and S5b bHLH genes in various tissues and at different fiber developmental stages (Figs 3 and 4). The expression of GhbHLH062A was totally undetectable in all investigated tissues, suggesting that the LTR retrotransposon insertion in this gene might lead to gene disruption and loss of function. The rest nine bHLH genes all showed significant expression in certain tissues and the expression levels varied in a developmentally regulated pattern. Four pairs of homoeologous bHLH genes from different genomes (GobHLH064A and D, GobHLH110A and D, GobHLH123A and D, and GobHLH130A and D) exhibited similar expression profiles among various tissues (Fig 3). During fiber development, similar expression profiles of homoeologous genes occurred for GobHLH064s and GobHLH123s, but not for GobHLH110s and GobHLH130s (Fig 4). GobHLH110A only showed significant expression in 0-DPA ovules, while GobHLH110D was also highly expressed in late-stage fibers (20DPA). GobHLH130A had relatively high expression levels at early stage (0 and 5DPA), while high expression levels of GobHLH130D was maintained from 5 to 15 DPA. These data show that the homoeologous genes of different genomes may express differentially in tetraploid cottons, especially in developing fibers.
Fig 3. Expression profiles of S5a and S5b bHLH genes in various upland cotton tissues.
Genome-specific primers were employed to detect relative transcript levels of 10 S5a and S5b bHLH genes in various tissues of upland cotton. Both cotton histone3 (AF024716) [42] and GhUBQ14 [43] genes were amplified and set as references. Error bars indicate SEM of three technical replicates.
Fig 4. Expression profiles of S5a and S5b bHLH genes at different fiber developmental stages.
Relative transcript levels of 10 S5a and S5b bHLH genes at different fiber developmental stages in upland cotton were detected by qRT-PCR. DPA, day(s) post anthesis. Total RNAs were extracted from ovules with fiber initials at anthesis (0 DPA) or fibers of different developmental stages (5~20 DPA). Data were analyzed and presented as in Fig 3.
Discussion
It is rather difficult to thoroughly identify bHLH proteins or coding genes in a certain genome. Traditionally, one or several bHLH sequences were employed as probe to search homologs in sequenced genomes. Due to the complexity and diversity of bHLH families, this method generally resulted in missing of some members. For example, different probe sequences used in homolog searching detected partially-overlapped sets of bHLH proteins in Arabidopsis [13–16,48]. In this study, a total of 35 probe sequences, including representative sequences of 32 plant bHLH subfamilies and three Arabidopsis orphans determined by Carretero-Paulet et al [13], were used to search homologs in sequenced cotton and cacao genomes. These probe sequences represented a much broader set of bHLH domains, and might cover most diversity in plant bHLH domains. Finally, we identified 272, 255, and 256 distinct bHLH genes from Gr-JGI, Gr-CGP, and Ga-CGP, respectively. In contrast, Cottongen detected less bHLH genes (258 and 243 in Gr-JGI and Gr-CGP, respectively) by using a bHLH consensus (IPR011598) as searching probe [38]. On the other hand, the bHLH domains determined in this study (except TcbHLH087) were all clustered along with plant reference bHLHs, indicating that multiple sequence alignment excluded false positives efficiently. These data suggest that searching with diverse probes and re-confirming by multiple sequence alignment may facilitate comprehensive detection of homologs in a certain genome.
Genome sequences are the major sources to systematically detect protein family members, and most of cotton bHLH genes are identified from sequenced genomes (Table 1; S3 Table). However, the genome sequencing projects in cottons are still in infancy compared to those in model plants, and currently available cotton genome sequences are relatively fragmented [18,33,34]. Consequently, only a part of the 'complete' set of cotton bHLH genes could be detected in a single sequenced genome (Table 1; S3 Table). For example, GobHLH130 was first identified only in Gr-JGI (D-genome). But cloning and PCR detection showed that a full-length GobHLH130A gene did exist in tetraploid cottons and A-genome donor G. arboreum, indicating that the corresponding sequences were not correctly assembled or annotated in Gr-CGP and Ga-CGP. Therefore, it is reasonable currently to integrate all the bHLHs detected in different sources to constitute the cotton bHLH reference genes.
Classification of plant bHLH proteins varied in different studies, probably due to different methods and sequences adopted [12–16,48]. A genome-wide classification of bHLH family in Arabidopsis, poplar, rice, moss, and algae assigned 638 bHLHs into 32 subfamilies [13]. In this study, the bHLHs from Arabidopsis, cotton, cacao, and some representatives from other plants were clustered into 30 subfamilies, a little less than those determined by Carretero-Paulet et al [13]. In our classification, S5 and S12 were divided into two subfamilies (S5a and S5b, S12a and S12b, respectively; Fig 1; S2 Fig), and S18, S19, S20, S21, and S22 were merged into a single group (S18). This difference may be attributed to different methods and less sequences used in our study. Nevertheless, most subfamilies in our classification are consistent with the previous system [13]. S5 and S12 subfamilies in Carretero-Paulet’s system are also divided into two subgroups [13], corresponding to the separate subfamilies (S5a and S5b, S12a and S12b, respectively) in our classification. Meanwhile, the bHLH proteins of the present S18 subfamily (including S18-S22 in Carretero-Paulet’s system) share similar structures, i.e., they are short in length, and contain little conserved domains other than bHLH domain (S6 Table) [13,46]. Taken together, the classification of bHLH proteins in our study is comparable to the previous system [13], and lays a good foundation for exploring the evolutionary characteristics of cotton bHLH family.
The bHLH protein family expanded significantly in higher plants and formed one of the largest transcription factor families [9,12,13]. In this study, we identified a total of 289 cotton reference bHLH genes from three independent cotton genome sequencing projects. This may be very close to the number of complete bHLH genes in a single cotton genome, although small changes in this data can’t be excluded. Considered that the coding genes are doubled and basically maintained after tetraploidization (Fig 2), bHLH family in tetraploid cottons may harbor around 580 genes. Expression analyses suggest that most of bHLH genes in tetraploids may express in certain tissues (Figs 3 and 4). Although cotton bHLHs are clustered into the similar subfamilies as Arabidopsis and cacao, multiple copies of bHLH genes may increase the complexity in regulation and also the possibility of mutation, neofunctionalization, and subfunctionalization in cotton species [1,49].
Retrotransposons comprise a large part of genomes in higher plants [18,33,34,50,51]. However, little is known about their biological function and evolutionary effects in cotton genomes. Recently, Woodhouse et al indicated that transponson-derived small RNA might induce differential silencing of homoeologous genes from different subgenomes and lead to genome dominance in hexaploid Brassica rapa [27]. In this study, we revealed that a LTR retrotransposon inserted in GhbHLH062A gene eliminated the transcription of this gene in tetraploid cotton. Interestingly, this retrotransposon may originate from D-genome progenitor, for it shares high sequence similarity with some LTR retrotransposons from G. ramondii rather those from G. arboreum (data not shown). Furthermore, this retrotransposon insertion can be detected both in G. hirsutum and G. barbadense (S4 Fig), suggesting that retrotransposon duplication and insertion may be a common event during tetraploidization in cotton. It may be valuable to elucidate the evolutionary and genetic effects of retrotransposon duplication in tetraploid cottons.
New allopolyploids harbor divergent genomes of their progenitors, and thus entail extensive genetic and epigenetic changes, including gene deletion, recombination, gene conversion, and varied expression, to conciliate different sets of genetic materials [18,24,28,29,31,52]. Nevertheless, recent genome sequencing researches indicated that, in oilseed and cotton, most of orthologous genes from progenitors maintained as homoeologs in allotetraploid, and expressed in certain tissues [18,28]. Consistent with previous reports [18,24,30], all the S5a and S5b bHLH genes remained in upland cotton. Except GhbHLH062A which was disrupted by retrotransposon insertion, all the rest genes expressed significantly in upland cotton. Interestingly, two pairs of homoeologous bHLH genes (GobHLH064A and D, GobHLH130A and D) had different, but complementary expression profiles during fiber development. The A-genome genes expressed predominantly at the early stages (<10DPA), while D-genome ones mainly at the late stages (>10DPA; Fig 4). S5 bHLHs in Arabidopsis function as important regulator of trichome differentiation and flavonoid biosynthesis [4,46,47]. We envision that the complementary expression of S5a and S5b bHLH homoeologs may play a role in promoting fiber development in allotetraploid cottons.
Supporting Information
A total of 605 bHLH domains (169 from A. thaliana, 289 from Gossypium, 139 from T. cacao, four from P. patens, two from C. reinhardtii and two from O. sativa) were aligned using AlignX program in software Vector NTI (Invitrogen) with default parameters. The consensus sequence is indicated at the bottom. The amino acid residuals conserved in over 80% and 60% sequences are shaded in light blue and green, respectively.
(TIF)
The phylogenetic trees were constructed and tested using NJ, ML, and MP methods on the basis of the alignment shown in S1 Fig. The NJ tree is presented, and subfamilies are indicated by square brackets with bootstraps (%) supported in NJ, ML and MP test (-, not supported). The scale bar indicates the estimated number of amino acid replacement per site. The sequences from A. thaliana (At), Gossypium (Go) and T. cacao (Tc) are marked with solid dots, open circles and squares, respectively.
(PDF)
The S5a and S5b bHLH proteins from G. hirsutum (Gh) and the diploid progenitors G. arboreum (Ga) and G. raimondii (Gr) are aligned with Arabidopsis homologous proteins. The amino acid residuals conserved in 100%, over 80% and 60% sequences are shaded in black, dark grey and light grey, respectively. The conserved domains identified by Carretero-Paulet et al (2010) in S5 bHLH subfamily are marked by black bars, and red bar indicates the bHLH domains.
(TIF)
The sequences are from GobHLH062D of G. hirsutum and G. raimondii, GobHLH062A of G. arboreum, G. hirsutum Yumian No.1 (-Y) and T586 (-T), and G. barbadense. Intron, exon, and LTR retrotransposon sequences are marked by black lines, blue and red bars, respectively. Identical sequences are shaded in grey. Dashes indicate gaps in the alignment, while dots represent the omitted LTR sequences.
(TIF)
The sequences are from GobHLH130A of G. hirsutum and G. arboreum, GobHLH130D of G. raimondii, G. hirsutum Yumian No.1 (-Y) and T586 (-T), and G. barbadense. Identical and conserved (>60%) sequences are shaded in light blue and pink, respectively. Dashes indicate the deleted sequences in GhbHLH130D.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOC)
Data Availability
The sequences were deposited in GenBank under the accession nos KP698854-KP698873. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was partially supported by the National Natural Science Foundation of China (31130039 to Y.P., 30971713 and 31271769 to Y.H.X.) and by the Genetically Modified Organisms Breeding Major Project of China (2014ZX08005005-001 to Y.H.X.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78. 10.1105/tpc.113.120857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6: 686–703. 10.1093/mp/sss128 [DOI] [PubMed] [Google Scholar]
- 3. Jones S (2004) An overview of the basic helix-loop-helix proteins. Genome Biol 5: 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu W, Grain D, Bobet S, Le Gourrierec J, Thevenin J, Kelemen Z, et al. (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol 202: 132–144. 10.1111/nph.12620 [DOI] [PubMed] [Google Scholar]
- 5. Sasaki-Sekimoto Y, Saito H, Masuda S, Shirasu K, Ohta H (2014) Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal Behav 9: e27639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lockhart J (2013) Frenemies: antagonistic bHLH/bZIP transcription factors integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1483 10.1105/tpc.113.250510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bai M-Y, Fan M, Oh E, Wang Z-Y (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929. 10.1105/tpc.112.105163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gyoja F (2014) A genome-wide survey of bHLH transcription factors in the Placozoan Trichoplax adhaerens reveals the ancient repertoire of this gene family in metazoan. Gene 542: 29–37. 10.1016/j.gene.2014.03.024 [DOI] [PubMed] [Google Scholar]
- 9. Song XM, Huang ZN, Duan WK, Ren J, Liu TK, Li Y, et al. (2014) Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol Genet Genomics 289: 77–91. 10.1007/s00438-013-0791-3 [DOI] [PubMed] [Google Scholar]
- 10. Liu A, Wang Y, Zhang D, Wang X, Song H, Dang C, et al. (2013) Classification and evolutionary analysis of the basic helix-loop-helix gene family in the green anole lizard, Anolis carolinensis. Mol Genet Genomics 288: 365–380. 10.1007/s00438-013-0755-7 [DOI] [PubMed] [Google Scholar]
- 11. Sailsbery JK, Atchley WR, Dean RA (2012) Phylogenetic analysis and classification of the fungal bHLH domain. Mol Biol Evol 29: 1301–1318. 10.1093/molbev/msr288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pires N, Dolan L (2010) Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol 27: 862–874. 10.1093/molbev/msp288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153: 1398–1412. 10.1104/pp.110.153593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, et al. (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, et al. (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wendel J, Cronn R (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186. [Google Scholar]
- 18. Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, et al. (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- 19. Bao Y, Hu G, Flagel LE, Salmon A, Bezanilla M, Paterson AH, et al. (2011) Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proc Natl Acad Sci U S A 108: 21152–21157. 10.1073/pnas.1115926109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaudhary B, Hovav R, Flagel L, Mittler R, Wendel J (2009) Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium). BMC Genomics 10: 378 10.1186/1471-2164-10-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hovav R, Udall JA, Chaudhary B, Hovav E, Flagel L, Hu G, et al. (2008) The evolution of spinnable cotton fiber entailed prolonged development and a novel metabolism. PLoS Genet 4: e25 10.1371/journal.pgen.0040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams KL, Percifield R, Wendel JF (2004) Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168: 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen X, Guo W, Liu B, Zhang Y, Song X, Cheng Y, et al. (2012) Molecular mechanisms of fiber differential development between G. barbadense and G. hirsutum revealed by genetical genomics. PLoS One 7: e30056 10.1371/journal.pone.0030056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu H, Han X, Lv J, Zhao L, Xu X, Zhang T, et al. (2011) Structure, expression differentiation and evolution of duplicated fiber developmental genes in Gossypium barbadense and G. hirsutum. BMC Plant Biol 11: 40 10.1186/1471-2229-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yuan D, Tu L, Zhang X (2011) Generation, annotation and analysis of first large-scale expressed sequence tags from developing fiber of Gossypium barbadense L. PLoS One 6: e22758 10.1371/journal.pone.0022758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi Y-H, Zhu S-W, Mao X-Z, Feng J-X, Qin Y-M, Zhang L, et al. (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X (2014) Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci U S A 111: 5283–5288. 10.1073/pnas.1402475111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, et al. (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953. 10.1126/science.1253435 [DOI] [PubMed] [Google Scholar]
- 29. Zhou R, Moshgabadi N, Adams KL (2011) Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc Natl Acad Sci U S A 108: 16122–16127. 10.1073/pnas.1109551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flagel L, Udall J, Nettleton D, Wendel J (2008) Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 6: 16–16. 10.1186/1741-7007-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feldman M, Levy AA (2012) Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774. 10.1534/genetics.112.146316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, et al. (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46: 567–572. 10.1038/ng.2987 [DOI] [PubMed] [Google Scholar]
- 34. Wang K, Wang Z, Li F, Ye W, Wang J, Song G, et al. (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44: 1098–1103. 10.1038/ng.2371 [DOI] [PubMed] [Google Scholar]
- 35. Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Motamayor J, Mockaitis K, Schmutz J, Haiminen N, Livingstone D, Cornejo O, et al. (2013) The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol 14: r53 10.1186/gb-2013-14-6-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42: D1182–D1187. 10.1093/nar/gkt1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu J, Jung S, Cheng C-H, Ficklin SP, Lee T, Zheng P, et al. (2014) CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res 42: D1229–D1236. 10.1093/nar/gkt1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Camacho C, Madden T, Ma N, Tao T, Agarwala R, Morgulis A (2008) BLAST command line applications user manual In: Bethesda M, editor. BLAST Help [Internet] National Center for Biotechnology Information (US). [Google Scholar]
- 40. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiao YH, Luo M, Fang WG, Luo KM, Hou L, Luo XY, et al. (2002) PCR walking in cotton genome using YADE method. Yi Chuan Xue Bao 29: 62–66. [PubMed] [Google Scholar]
- 42. Zhu Y-Q, Xu K-X, Luo B, Wang J-W, Chen X-Y (2003) An ATP-binding cassette transporter GhWBC1 from elongating cotton fibers. Plant Physiol 133: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10: 49 10.1186/1471-2229-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao Y-H, Yan Q, Ding H, Luo M, Hou L, Zhang M, et al. (2014) Transcriptome and biochemical analyses revealed a detailed proanthocyanidin biosynthesis pathway in brown cotton fiber. PLoS One 9: e86344 10.1371/journal.pone.0086344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan J, Tu L, Deng F, Hu H, Nie Y, Zhang X (2013) A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol 162: 86–95. 10.1104/pp.112.212142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, et al. (2013) The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genomics 13: 75–98. 10.1007/s10142-012-0301-4 [DOI] [PubMed] [Google Scholar]
- 47. Zhao H, Li X, Ma L (2012) Basic helix-loop-helix transcription factors and epidermal cell fate determination in Arabidopsis. Plant Signal Behav 7: 1556–1560. 10.4161/psb.22404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747. [DOI] [PubMed] [Google Scholar]
- 49. Ran JH, Shen TT, Liu WJ, Wang XQ (2013) Evolution of the bHLH genes involved in stomatal development: implications for the expansion of developmental complexity of stomata in land plants. PLoS One 8: e78997 10.1371/journal.pone.0078997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo W, Cai C, Wang C, Zhao L, Wang L, Zhang T (2008) A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genomics 9: 314 10.1186/1471-2164-9-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hawkins J, Kim H, Nason J, Wing R, Wendel J (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16: 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoo MJ, Szadkowski E, Wendel JF (2013) Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110: 171–180. 10.1038/hdy.2012.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A total of 605 bHLH domains (169 from A. thaliana, 289 from Gossypium, 139 from T. cacao, four from P. patens, two from C. reinhardtii and two from O. sativa) were aligned using AlignX program in software Vector NTI (Invitrogen) with default parameters. The consensus sequence is indicated at the bottom. The amino acid residuals conserved in over 80% and 60% sequences are shaded in light blue and green, respectively.
(TIF)
The phylogenetic trees were constructed and tested using NJ, ML, and MP methods on the basis of the alignment shown in S1 Fig. The NJ tree is presented, and subfamilies are indicated by square brackets with bootstraps (%) supported in NJ, ML and MP test (-, not supported). The scale bar indicates the estimated number of amino acid replacement per site. The sequences from A. thaliana (At), Gossypium (Go) and T. cacao (Tc) are marked with solid dots, open circles and squares, respectively.
(PDF)
The S5a and S5b bHLH proteins from G. hirsutum (Gh) and the diploid progenitors G. arboreum (Ga) and G. raimondii (Gr) are aligned with Arabidopsis homologous proteins. The amino acid residuals conserved in 100%, over 80% and 60% sequences are shaded in black, dark grey and light grey, respectively. The conserved domains identified by Carretero-Paulet et al (2010) in S5 bHLH subfamily are marked by black bars, and red bar indicates the bHLH domains.
(TIF)
The sequences are from GobHLH062D of G. hirsutum and G. raimondii, GobHLH062A of G. arboreum, G. hirsutum Yumian No.1 (-Y) and T586 (-T), and G. barbadense. Intron, exon, and LTR retrotransposon sequences are marked by black lines, blue and red bars, respectively. Identical sequences are shaded in grey. Dashes indicate gaps in the alignment, while dots represent the omitted LTR sequences.
(TIF)
The sequences are from GobHLH130A of G. hirsutum and G. arboreum, GobHLH130D of G. raimondii, G. hirsutum Yumian No.1 (-Y) and T586 (-T), and G. barbadense. Identical and conserved (>60%) sequences are shaded in light blue and pink, respectively. Dashes indicate the deleted sequences in GhbHLH130D.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOC)
Data Availability Statement
The sequences were deposited in GenBank under the accession nos KP698854-KP698873. All relevant data are within the paper and its Supporting Information files.