Abstract
Purpose
Interleukin-6 (IL-6) encodes a cytokine protein, which causes inflammation, maintains immune homeostasis and plays an essential role in oral pathogenesis. The aim of this study was to evaluate the association between IL-6 (− 174 and − 572) G/C promoter gene polymorphisms and risk of OSCC among Indians.
Methods
Single nucleotide polymorphism in IL-6 genes was genotyped in OSCC patients and healthy controls by PCR-RFLP method. Genotype and allele frequencies were analyzed by chi-square test and strength of associations by odds ratio with 95% confidence intervals.
Results
Frequency distribution of IL-6 (− 174) G/C gene polymorphism was significantly associated with OSCC patients in comparison to healthy controls (OR: 0.541, CI: 0.356–0.822; p: 0.004. However, frequency of IL-6 (− 572) G/C gene polymorphism was not significantly associated with OSCC patients (p > 0.05).
Conclusion
The genotype GC and allele C of IL-6 (− 174) G/C gene polymorphism play a significant role in OSCC susceptibility.
Keywords: OSCC, Inflammation, Immune response, Genotype, PCR-RFLP
Highlights
-
•
We first demonstrate the IL-6 polymorphism in OSCC patients in Indian population.
-
•
We obtained the SNP of IL-6 (-174) is increase the risk of OSCC.
-
•
We also obtained the SNP of IL-6 (-572) and risk of OSCC
-
•
We evaluate the correlation of these IL-6 polymorphisms and progression of OSCC.
-
•
We identified the environmental factors and gene interactions with pathogenesis of OSCC.
Introduction
Oral squamous cell carcinoma (OSCC) is a common malignant tumor of the oral cavity and has a high incidence with poor prognosis (Alvarez Marcos et al., 2006, Argiris et al., 2008). In India, OSCC is one of the major causes of cancer-related deaths. It is predominant in Indian males (Dikshit et al., 2012, Kuruvilla, 2008). OSCC is a multistep progression which is influenced by several environmental factors, such as tobacco chewing, smoking and alcohol consumption and alteration in genes, such as genetic tumor suppressor genes and oncogenes (Ko et al., 1995, Williams, 2000). Several studies have observed that the common polymorphisms in angiogenesis, inflammation and thrombosis-related genes, are associated with increased risk for oral cancer (Dikshit et al., 2012, Ko et al., 1995, Kuruvilla, 2008, Sansone and Bromberg, 2012, Tan et al., 2005). One such factor, related with both thrombosis and malignancies, is interleukin-6 (IL-6) (Guo et al., 2012, Seike et al., 2011, Terry et al., 2000, Wojcik et al., 2010). IL-6, also known as B cell differentiation factor, is an immune regulatory cytokine, involved in the regulation of various cellular functions (angiogenesis, apoptosis, proliferation, differentiation and regulation of immune response) (Culig et al., 2005). IL-6 may also have an essential role in the growth and differentiation of malignant tumors (Mandal et al., 2014, Zarogoulidis et al., 2013).
IL-6 is a multifunctional cytokine of Th2 type which has both, pro-inflammatory and anti-inflammatory cytokine activities (Sansone and Bromberg, 2012). It is a glycoprotein which consists of 184 amino acids and has a molecular weight of 26 kDa (Guo et al., 2012). It is involved in tumor growth, differentiation of malignant cancer cells and microenvironment immune-modulation (Lagmay et al., 2009, Seike et al., 2011). These properties are the result of enhanced neo-angiogenesis, inhibition of cancer cell apoptosis and acquired cell resistance (Wojcik et al., 2010). IL-6 is secreted by T cells, which is capable of stimulating immune responses, including responses to tumors. The IL-6 gene is located on chromosome 7p21 and the SNPs at the 5′ flanking region of the IL-6 promoter, to be identified as IL-6 − 174 and − 572 (Terry et al., 2000). IL-6 (− 174) G/C genepolymorphism is one of the most frequently studied polymorphisms. IL-6 (− 174) G/C gene polymorphism alters the expression of IL-6 gene. The genotypes GC, CC and allele C of IL-6 (− 174) G/C gene polymorphism are associated with increased risk of oral cancer (Vairaktaris et al., 2006). Some studies observed that the IL-6 (− 174) G/C gene polymorphism was significantly associated with some cancers (breast, ovarian, prostatic, gastric, cervical and colorectal cancer) (Berek et al., 1991, DeMichele et al., 2003, Landi et al., 2003, Slattery et al., 2007, Yeh et al., 2010). The IL-6 (− 572) G allele is also associated with increased level of IL-6 protein in serum than C/C allele and is significantly associated with the risk of lung cancer (Jerrard-Dunne et al., 2004, Seow et al., 2006). On the contrary IL-6 (− 174 and − 572) C/G gene polymorphisms are not associated with the risk of gastric cancer, moreover they lower the risk of bladder cancer (Xu et al., 2011, Yin et al., 2012).
Since the association between IL-6 (− 174G/C and − 572G/C) gene polymorphism with OSCC in the Indian population has yet not been studied, we investigated the role of IL-6 promoter (− 174) G/C and (− 572) G/C gene polymorphisms in pathogenesis of OSCC in Indian population.
Materials and methods
The current study was undertaken on a total of 272 OSCC patients and 185 healthy controls. Histopathologically confirmed OSCC patients from the Department of Surgical Oncology, King George's Medical University, UP, Lucknow were recruited for this study. Clinical information, including age, sex, tobacco chewing, smoking, alcohol consumption and TNM stages, was obtained from patients' medical charts. This study was approved by the Institutional Ethics Committee of the King George's Medical University, Lucknow. An informed written consent was obtained from all the subjects.
Blood collection and molecular analyses
Venous blood samples were collected in EDTA tubes and stored at − 80 °C, till DNA extraction. Genomic DNA extraction from blood samples was carried out by salting out method.
Genotyping of IL-6 (− 174) G/C gene polymorphism
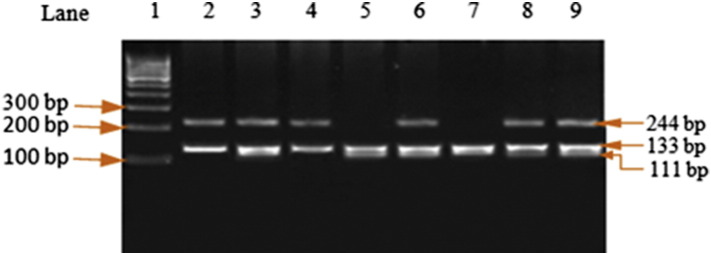
The PCR of IL-6 (− 174) G/C, the final volume of PCR reaction mixture, was 25 μl containing 40 ng genomic DNA, 10 picomole each of forward and reverse primers (5′-TTGTCAAGACATGCCAAGTGCT-3′ and 5′-GCCTCAGAGACATCTCCAGTCC-3′) at concentration of 1X, 1X PCR master mixture. Amplification was carried out at 94 °C for 5 min followed by 35 cycles of 30 s at 94 °C for denaturation, 45 s at 57 °C for annealing, 45 s at 72 °C for extension, with a final extension at 72 °C for 7 min (Table 1). An overnight digestion with 3 units NlaIII at 37 °C yielded G allele (244 + 133 + 11) bp and C allele (133 + 111 + 56) bp products. The obtained fragments' sizes were analyzed on a 2% agarose gel. After restriction digestion, the obtained predicted sizes of the fragments are shown in Table 1 and Fig. 1.
Table 1.
Primer sequences and restriction enzyme for IL-6 gene polymorphism.
| SNP locus | Primer sequence | Restriction enzyme | Product size (bp) |
|---|---|---|---|
| IL6 (− 174G/C) | F-5′-ttgtcaagacatgccaagtgct-3′ R-5′-gcctcagagacatctccagtcc-3′ |
NlaIII | G: 244 + 133 + 11 C: 133 + 111 + 56 |
| IL6 (− 572G/C) | F-5′-ggagacgccttgaagtaactgc-3′ R-5′-gagtttcctctgactccatcgcag-3′ |
MbiI | G: 102, 61 C:163 |
SNP, single nucleotide polymorphisms.
Fig. 1.
PCR-RFLP analysis for IL-6 − 174G/C genotyping. The PCR-amplified product was 300 bp. The fragments after digestion with NlaIII (lanes 1: 100 bp ladder, lanes 2, 4: GG, lanes 3, 6, 8, 9: GC and lanes 5, 7: CC genotype) were obtained.
Genotyping of IL-6 (− 572) G/C gene polymorphism
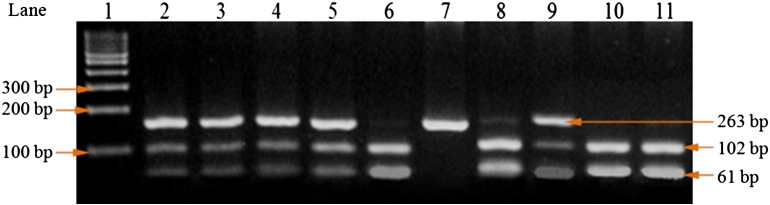
The PCR of IL-6 (− 572G/C), the final volume of PCR reaction mixture, was 25 μl containing 40 ng genomic DNA, 10 picomole each of forward and reverse primers (5′-GGAGACGCCTTGAAGTAACTGC-3′ and 5′-GAGTTTCCTCTGACTCCATCGCAG-3′) at concentration of 1X, 1X PCR master mixture. Amplification was carried out at 94 °C for 5 min followed by 35 cycle of 30 s at 94 °C for denaturation, 40 s at 55 °C for annealing, 30 s at 72 °C for extension, with a final extension at 72 °C for 7 min (Table 1). An overnight digestion with 3 units MbiI at 37 °C yielded G allele (102, 61) bp and C allele (163) bp products. The obtained fragments' sizes were analyzed on a 2% agarose gel. After restriction digestion, the obtained predicted sizes of the fragments are shown in Table 1 and Fig. 2.
Fig. 2.
PCR-RFLP analysis for IL-6 − 572G/C genotyping. The PCR-amplified product was 163 bp. The fragments after digestion with MbiI (lanes 1: 100 bp ladder, lanes 2, 3, 4, 5, 8, 9: GC, lanes 6, 10, 11: GG and lane 7: CC genotype) were obtained.
Statistical analysis
The obtained demographics and clinical data were analyzed using Chi-square test. Genotype and allele frequencies of IL-6 were compared between OSCC cases and controls, using the Chi-square test. Odds ratios (ORs) were calculated to estimate the strength of association between the two IL-6 promoter (− 174 and − 572) gene polymorphisms and risk of OSCC. ORs are given with 95% confidence interval (CI). A ‘p-value’ < 0.05 was considered to be statistically significant.
Results
Demographic characteristics, lifestyle and disease status of OSCC patients are previously published (Singh et al., 2014a, Singh et al., 2014b), which included age (47.67 ± 12.67 and 43.1 ± 8.31), sexes [(80.51% male and 76.76% male), (19.49% female and 23.24% female)], height (162.33 ± 6.0 and 163.10 ± 5.65) and weight (55.86 ± 8.50 and 57.78 ± 7.58) in OSCC patients and controls respectively. In this study we also observed tobacco chewing (61.40%, 47.57%), smoking (30.51%, 21.62%) and alcohol consumption (13.97%, 11.35%) in OSCC patients and controls respectively. In this study we observed that smoking and tobacco chewing are significantly (p < 0.05) associated, while alcohol consumption is not significantly associated with OSCC. The occurrence of tumor stages [I (8.09%) II (56.25), III (32.72) and (1.47) IV], tumor T status (≤ T2 71%, > T2 29%), and lymph node status (N0 76.47% and N1 + N2 23.53%) was presented in this study.
The genotype and allele frequencies of IL-6 (− 174) G/C and (− 572) G/C
The genotype and allele frequencies of the IL-6 (− 174) G/C and IL-6 (− 572) G/C gene polymorphisms among the controls and OSCC patients are shown in Table 2. The frequencies of the GG, GC and CC genotypes of IL-6 (− 174G/C) were 55.15%, 37.13% and 7.72% in the OSCC patients and 69.73%, 25.41% and 4.86% in controls, respectively. On the other hand the allele frequencies of the G and C were 73.71% and 26.29% in the OSCC patients, and 82.43% and 17.57% in controls, respectively (Table 2).
Table 2.
The genotype and allele frequencies of IL-6 (- 174 G/C and - 572 G/C) gene polymorphisms.
| Controls | OSCC n = 272 (%) | p- Value | OR (95% CI) | OSCC with cancer stages I & II | p- Value | OR (95% CI) | OSCC with cancer stages III & IV | p- Value | OR (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| IL-6 (174) | ||||||||||
| GG | 129 (69.73) | 150 (55.15) | - | - | 96 (54.86) | - | - | 54 (55.67) | - | - |
| GC | 47 (25.41) | 101 (37.13) | 0.005⁎ | 1.848 (1.216-2.809) |
66 (37.72) | 0.009⁎ | 1.887 (1.194-2.983) |
35 (36.08) | 0.05⁎ | 1.779 (1.036-3.055) |
| CC | 9 (4.86) | 21 (7.72) | 0.132 | 2.007 (0.888-4.537) |
13 (7.42) | 0.2092 | 1.941 (0.797-4.728) |
8 (8.25) | 0.222 | 2.123 (0.778-5.796) |
| Allele | ||||||||||
| G | 305 (82.43) | 401 (73.71) | - | - | 258 (73.72) | - | - | 143 (73.72) | - | - |
| C | 65 (17.57) | 143 (26.29) | 0.003⁎ | 1.673 (1.204-2.325) |
92 (26.28) | 0.006⁎ | 1.673 (1.169-2.394) |
51 (26.28) | 0.0201⁎ | 1.673 (1.103-2.540) |
| IL-6 (-572) | ||||||||||
| GG | 57 (30.81) | 75 (27.57) | - | - | 44 (25.14) | - | - | 31 (31.95) | - | - |
| GC | 115 (62.16) | 175 (64.34) | 0.564 | 1.157 (0.762-1.755) |
115 (65.72) | 0.337 | 1.295 (0.809-2.074) |
60 (61.87) | 0.988 | 0.959 (0.561-1.642) |
| CC | 13 (7.03) | 22 (8.09) | 0.6519 | 1.286 (0.597-2.771) |
16 (9.14) | 0.371 | 1.594 (0.695-3.660) |
6 (6.18) | 0.970 | 0.849 (0.294-2.454) |
| Allele | ||||||||||
| G | 219 (59.29) | 325 (59.74) | - | - | 203 (58.00) | - | - | 121 (62.37) | - | - |
| C | 141 (40.71) | 219 (40.26) | 0.796 | 1.047 (0.797-1.374) |
147 (42.00) | 0.489 | 1.125 (0.833-1.518) |
72 (37.63) | 0.736 | 0.924 (0.645-1.325) |
= Significant value
There were significant differences in the GC genotype and C allele frequencies of the IL-6 (− 174) G/C gene polymorphism between OSCC patients and controls. The difference between OSCC patients and controls was statistically significant in homozygous GG and heterozygous GC genotype (OR: 1.848, CI: 1.216–2.809; p: 0.005) and homozygous GG and CC genotype (OR: 2.007, CI: 0.888–4.537; p: 0.131). C allele was also significantly increased, as compared with the G allele [OR: 1.673, CI: 1.204 − 2.325; p: 0.003] (Table 2). The genotype (GG vs GC) and allele frequencies of the IL-6 (− 174) G/C gene polymorphism between control group and OSCC patients, with cancer stages (I/II and III/IV), were significantly different [p = 0.0089 and 0.05 respectively] (Table 2).
The frequencies of the GG, GC and CC genotypes of IL-6 (− 572) G/C were 27.57%, 64.34% and 8.09% in the OSCC patients and 30.81%, 62.16% and 7.03% in controls, respectively. On the other hand the allele frequencies of the G and C were 59.74% and 40.26% in the OSCC patients and 59.29% and 40.71% in controls, respectively (Table 3). The genotypes and allele frequencies of the IL-6 (− 572) G/C were not significantly different between controls and OSCC patients. The genotype (GG vs GC) and allele frequencies of the IL-6 (− 174) G/C gene polymorphism between controls and OSCC patients with cancer stages (I/II and III/IV), were not significantly different (p > 0.05) (Table 2).
Table 3.
Genotype distribution of IL-6 (− 174G/C) in tobacco chewing, smoking and alcohol consumption.
| OSCC patients | Controls | OR (95% CI) | p-Value | |
|---|---|---|---|---|
| Tobacco chewing | ||||
| Genotypes | n = 167 (%) | n = 88 (%) | ||
| GG | 87 (52.10) | 58 (65.91) | – | |
| GC | 67 (40.12) | 25 (28.41) | 0.560 (0.318–0.987) | 0.051 |
| CC | 13 (7.78) | 5 (5.68) | 0.578 (0.195–1.705) | 0.455 |
| Smoking | ||||
| Genotypes | n = 83 (%) | n = 40 (%) | ||
| GG | 47 (56.63) | 30 (75.00) | – | |
| GC | 31 (37.34) | 7 (17.5) | 0.354 (0.138–0.905) | 0.045⁎ |
| CC | 5 (6.03) | 3 (7.50) | 0.940 (0.209–4.23) | 0.936 |
| Alcohol consumption | ||||
| Genotypes | n = 38 (%) | n = 21 (%) | ||
| GG | 19 (50.00) | 16 (76.19) | – | |
| GC | 15 (39.47) | 3 (14.29) | 0.238 (0.058–0.970) | 0.074 |
| CC | 4 (10.53) | 2 (9.52) | 0.594 (0.096–3.677) | 0.905 |
Significant value.
Correlation between genotype distribution of IL-6 (− 174) G/C and (− 572) G/C in tobacco chewing, smoking and alcohol consumption in OSCC patients and controls
The correlation between genotypes and allele frequencies of IL-6 (− 174) G/C and (− 572) G/C gene polymorphisms with tobacco chewing, smoking and alcohol consumption, on OSCC susceptibility, is shown in Table 3, Table 4. In tobacco chewing, the genotype frequencies of GG, GC and CC of the IL-6 (− 174) G/C and (− 572) G/C gene polymorphisms were not significantly (p > 0.05) associated with the development of OSCC as compared with controls (Table 3, Table 4). However the GC genotype of the IL-6 (− 174) G/C and GC, CC genotype of the IL-6 (− 572) G/C gene polymorphism were significantly (p < 0.05) associated with the development of OSCC (Table 3, Table 4).
Table 4.
Association between IL-6 − 572G/C genetic variations and genetic variations and tobacco chewing, smoking and alcohol consumption.
| OSCC patients | Controls | OR (95% CI) | p-Value | |
|---|---|---|---|---|
| Tobacco chewing | ||||
| Genotypes | n = 167 (%) | n = 88 (%) | ||
| GG | 41 (24.56) | 30 (34.09) | – | |
| GC | 113 (67.66) | 54 (61.36) | 0.653 (0.369–1.157) | 0.188 |
| CC | 13 (7.78) | 4 (4.55) | 0.421 (0.125–1.418) | 0.251 |
| Smoking | ||||
| Genotypes | n = 83 (%) | n = 40 | ||
| GG | 17 (20.48) | 17 (42.5) | – | |
| GC | 56 (67.47) | 22 (55.0) | 0.421 (0.125–1.418) | 0.251 |
| CC | 10 (12.05) | 1 (2.5) | 0.137 (0.017–1.127) | 0.076 |
| Alcohol consumption | ||||
| Genotypes | n = 38 (%) | n = 21 | ||
| GG | 8 (21.06) | 13 (61.90) | – | |
| GC | 25 (65.78) | 8 (38.10) | 0.197 (0.06–0.646 | 0.013⁎ |
| CC | 5 (13.16) | 0 (0.00) | 0.057 (0.003–1.173) | 0.047⁎ |
Significant value.
Association of IL-6 (− 174) G/C and (− 572) G/C genotypes with tumor status
We stratified the OSCC patients into two categories on the basis of disease status, low risk OSCC group (stage I + II, ≤ T2 and N0) and high risk OSCC group (stage III + IV, > T2 and N1 + N2). Low risk OSCC group was taken as a reference. There was no significant difference between the genotype distribution of the IL-6 (− 174) G/C and (− 572) G/C gene polymorphism and tumor status (stage III + IV, > T2 and N1 + N2), except lymph node status and GG and GC genotype of IL-6 (− 572) G/C gene polymorphism [OR: 0.395; CI: 0.200–0.789; p: 0.0109] (Table 5).
Table 5.
IL-6 (− 174) G/C and (− 572) G/C gene polymorphisms and tumor stage, tumor T status and lymph node in OSCC patients.
| IL-6 (− 174) G/C genotype | ||||
|---|---|---|---|---|
| I + II n = 175 (%) |
III + IV n = 97 (%) |
OR (95% CI) | p-Value | |
| Genotypes | ||||
| GG | 96 (54.86) | 54 (55.67) | – | |
| GC | 66 (37.72) | 35 (36.08) | 0.9428 (0.5558–1.599) | 0.9329 |
| CC | 13 (7.42) | 8 (8.25) | 1.094 (0.4265–2.806) | 0.8516 |
| Alleles | ||||
| G | 258 (73.72) | 143 (73.72) | – | |
| C | 92 (26.28) | 51 (26.28) | 1.000 (0.6713–1.490) | 0.9994 |
| Tumor T status | ≤ T2 n = 193 (%) |
> T2 n = 79 (%) |
OR (95% CI) | p-Value |
| Genotypes | ||||
| GG | 103 (53.37) | 47 (59.49) | – | |
| GC | 76 (39.37) | 25 (31.65) | 0.7209 (0.4082–1.273) | 0.3231 |
| CC | 14 (7.26) | 7 (8.86) | 1.096 (0.4150–2.893) | 0.8535 |
| Alleles | ||||
| G | 282 (73.06) | 119 (75.32) | ||
| C | 104 (26.94) | 39 (24.68) | 0.8887 (0.5804–1.361) | 0.6627 |
| Lymph node | N0 n = 205 (%) |
N1 + N2 n = 67 (%) |
OR (95% CI) | p-Value |
| Genotypes | ||||
| GG | 112 (54.64) | 38 (56.71) | – | |
| GC | 79 (38.54) | 22 (32.84) | 0.8208 (0.4509–1.494) | 0.6199 |
| CC | 14 (6.82) | 7 (10.45) | 1.474 (0.5535–3.924) | 0.6064 |
| Alleles | ||||
| G | 303 (73.90) | 98 (73.13) | – | |
| C | 107 (26.10) | 36 (26.87) | 1.040 (0.6692–1.617) | 0.9503 |
| Il-6 (− 572) G/C gene polymorphisms | ||||
| I + II n = 175 (%) |
III + IV n = 97 (%) |
OR (95% CI) | p-Value | |
| Genotypes | ||||
| GG | 44 (25.14) | 31 (31.95) | – | |
| GC | 115 (65.72) | 60 (61.87) | 0.7405 (0.4248–1.291) | 0.3587 |
| CC | 16 (9.14) | 6 (6.18) | 0.5323 (0.1872–1.513) | 0.3450 |
| Alleles | ||||
| G | 203 (58.00) | 121 (62.37) | – | |
| C | 147 (42.00) | 72 (37.63) | 0.8217 (0.5728–1.179) | 0.3291 |
| Tumor T status | ≤ T2 n = 193 (%) |
> T2 n = 79 (%) |
OR (95% CI) | p-Value |
| Genotypes | ||||
| GG | 50 (25.91) | 25 (31.65) | – | |
| GC | 126 (65.28) | 49 (62.03) | 0.7778 (0.4343–1.393) | 0.4868 |
| CC | 17 (8.81) | 5 (6.32) | 0.5882 (0.1944–1.779) | 0.4939 |
| Alleles | ||||
| G | 226 (58.55) | 99 (62.66) | – | |
| C | 160 (41.45) | 59 (37.34) | 0.8418 (0.5752–1.232) | 0.4290 |
| Lymph node status | N0 n = 205 (%) |
N1 + N2 n = 67 (%) |
OR (95% CI) | p-Value |
| Genotypes | ||||
| GG | 55 (26.83) | 20 (29.85) | – | |
| GC | 133 (64.88) | 42 (62.69) | 0.8684 (0.4679–1.612) | 0.7736 |
| CC | 17 (8.29) | 5 (7.46) | 0.8088 (0.2636–2.482) | 0.9249 |
| Alleles | ||||
| G | 243 (59.27) | 82 (61.19) | – | |
| C | 167 (40.73) | 52 (38.81) | 0.9227 (0.6188–1.376) | 0.7694 |
Discussions
To the best of our knowledge, this is the first study that correlates the genotype and allele frequencies of IL-6 (− 174 and − 572) gene polymorphism with OSCC, in Indian population. The immune response to inflammation frequently plays an important role in the pathogenesis of a tumor (Savage et al., 2004). Genetic alteration also plays a significant role in the inflammatory response, which may be associated with the risk of development of various cancers, including the oral cancer (Savage et al., 2004). IL-6 is an immune-regulatory cytokine with biological functions of pro-inflammation, anti-inflammation and angiogenesis. These responses are triggered by activation of the JAK tyrosine kinase family and then further stimulated by the MAPK, PI3K, or STAT signaling pathways (Aggarwal et al., 2006, Chen et al., 2012, Kishimoto, 1989, Schafer and Brugge, 2007). It has been suggested that polymorphisms in genes, encoding inflammatory mediators, may influence the plasma level and the biological activity of the corresponding proteins (Jibiki et al., 2001). IL-6 (− 174G/C) promoter SNP is a crucial mediator of the Th2 response, that has been associated with increased IL-6 production (Savage et al., 2004). During the inflammation, the C allele of IL-6 (− 174) G/C gene polymorphism elevates the level of IL-6 protein (Belluco et al., 2003, Berger, 2004, DeMichele et al., 2003). In addition, several studies have reported that the increased serum level of IL-6 was obtained during development and progression of tumors, including oral cancer (DeMichele et al., 2003, Vucicević Boras et al., 2005, Wang et al., 2002). In fact, an increased level of IL-6 protein was associated with poor prognosis and high mortality in various cancers (Chung and Chang, 2003, Michalaki et al., 2004, Srivani and Nagarajan, 2003). St John et al. (2004) reported that the increased level of IL-6 was present in the serum of patients with OSCC (St John et al., 2004). Chen et al. (2012) reported that the increased expression of IL-6 in tumor cells was significantly associated with aggressive clinical manifestations and might be an independent survival predictor, particularly in male OSCC patients (Chen et al., 2012).
In the present study, we found that the promoter (− 174) GC genotype is significantly associated with increased risk of OSCC. Moreover, the − 174 C allele was also associated with a high risk for development of OSCC. The genotype and allele frequencies of IL-6 (− 572) G/C were not significantly associated with the risk of OSCC. Moreover these (− 174 and − 572) promoter region genotypes were not associated with the progression of OSCC. The reported observations of various studies, about the association of IL-6 gene polymorphism with various cancers, are variable. A previous study by Vairaktaris et al. (2006) reported that the IL-6 gene polymorphism is strongly correlated with increased risk for oral cancer by affecting IL-6 gene expression. The GC heterozygotes and CC homozygotes were significantly associated with the development and progression of oral cancer (Vairaktaris et al., 2006). Our finding are also supported by DeMichele et al. (2003) and Landi et al. (2003), who reported that the genotypes (GC, CC) and allele C frequencies of IL-6 (− 174) G/C were significantly associated with increased risk for breast and colorectal cancers (DeMichele et al., 2003, Landi et al., 2003). Several previous studies showed that the elevated level of IL-6 protein in serum is present in many cancers (OSCC, breast, prostate, ovarian, gastric, colorectal and cervical cancers), in which the C allele of promoter (− 174G/C) gene polymorphism was found associated with increased production of IL-6 protein and risk of cancers (Bachelot et al., 2003, Berek et al., 1991, Hefler et al., 2005, Kishimoto, 1989, Slattery et al., 2007, Zhang and Adachi, 1999). On the contrary, several studies reported that the IL-6 (− 174) G/C gene polymorphism was not significant associated with some cancers (breast, prostate, colorectal, lung and gastric cancer) lymphoma, melanoma and multiple myeloma (Xu et al., 2011, Yu et al., 2010). Balasubramanian et al. (2006) also did not observe any significant association between the (− 174) G/C polymorphism of IL-6 and breast cancer (Balasubramanian et al., 2006). Individuals with the C allele of IL-6 (− 572) G/C gene polymorphism and the GG genotype for the IL-6 (− 174) G/C gene polymorphism were associated with decreased risk of colon cancer, but with an increased risk of rectal cancer (Slattery et al., 2007). Liang et al. (2013) reported that the C allele at (− 572) locus of IL-6 genepolymorphism was significantly associated with lung cancer (Liang et al., 2013). Yin et al. (2012) suggested that the IL-6 (− 174 C/G and − 572 C/G) gene polymorphisms were not associated with the risk of gastric cancer (Yin et al., 2012).
In our study the genotypes and allele frequencies of IL-6 (− 174 and − 572) G/C gene polymorphism were not associated with environmental factors (tobacco chewing, smoking and alcohol consumption) and tumor progression. These results are similar to that of Balasubramanian et al. (2006), who showed that IL-6 (− 174) G/C gene polymorphism was not associated with either breast cancer risk or severity and prognosis, as assessed by tumor grade and lymph node status (Balasubramanian et al., 2006). In contrast, Vairaktaris et al. (2006) demonstrated that the CC and GC genotypes of IL-6 (− 174) G/C polymorphism were significantly associated with the development and progression of cancer, in alcohol consuming patients. Furthermore, C allele carriers have twice as much greater relative risk for developing oral cancer in stages III & IV than in I & II (Vairaktaris et al., 2006). It can be concluded that the present study provides evidence of the correlation between IL-6 (− 174) G/C gene polymorphism with risk of OSCC. The GC genotype and C allele are significantly associated with the risk of OSCC, whereas IL-6 (− 572) G/C polymorphism is not associated with OSCC. Moreover related environmental factors and gene interactions are not associated with pathogenesis of OSCC. IL-6 (− 174 and − 572) promoter gene polymorphisms are not significantly associated with progression of OSCC. This finding suggests that IL-6 may be used as a diagnostic marker for effective management of OSCC in future, though further studies with larger sample sizes will be necessary to confirm this.
References
- Aggarwal B.B., Shishodia S., Sandur S.K., Pandey M.K., Sethi G. Inflammation and cancer: how hot is the link? Biochem. Pharmacol. 2006;72:1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Alvarez Marcos C.A., Llorente Pendás J.L., Franco Gutiérrez V., Hermsen M.A., Franco Albalad M.P., Fernández Espina H., Suárez Nieto C. Second primary tumors in head and neck cancer. Acta Otorrinolaringol. Esp. 2006;57:462–466. doi: 10.1016/s0001-6519(06)78749-0. [DOI] [PubMed] [Google Scholar]
- Argiris A., Karamouzis M.V., Raben D., Ferris R.L. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelot T., Ray-Coquard I., Menetrier-Caux C., Rastkha M., Duc A., Blay J.Y. Prognostic value of serum levels of interleukin 6 and of serum and plasma levels of vascular endothelial growth factor in hormone-refractory metastatic breast cancer patients. Br. J. Cancer. 2003;88:1721–1726. doi: 10.1038/sj.bjc.6600956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.P., Azmy I.A.F., Higham S.E., Wilson A.G., Cross S.S., Cox A., Brown N.J., Reed M.W. Interleukin gene polymorphisms and breast cancer: a case control study and systematic literature review. BMC Cancer. 2006;6:188. doi: 10.1186/1471-2407-6-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluco C., Olivieri F., Bonafe M., Giovagnetti S., Mammano E., Scalerta R., Ambrosi A., Franceschi C., Nitti D., Lise M. 174G > C polymorphism of interleukin 6 gene promoter affects interleukin 6 serum levels in patients with colorectal cancer. Clin. Cancer Res. 2003;9:2173–2176. [PubMed] [Google Scholar]
- Berek J.S., Chung C., Kaldi K., Watson J.M., Knox R.M., Martínez-Maza O. Serum interleukin-6 levels correlate with disease status in patients with epithelial ovarian cancer. Am. J. Obstet. Gynecol. 1991;164:1038–1042. doi: 10.1016/0002-9378(91)90582-c. [DOI] [PubMed] [Google Scholar]
- Berger F.G. The interleukin-6 gene: a susceptibility factor that may contribute to racial and ethnic disparities in breast cancer mortality. Breast Cancer Res. Treat. 2004;88:281–285. doi: 10.1007/s10549-004-0726-0. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Sung W.W., Lin Y.M., Chen M.K., Lee C.H., Lee H., Yeh K.T., Ko J.L. Gender difference in the prognostic role of interleukin 6 in oral squamous cell carcinoma. PLoS One. 2012;7:e50104. doi: 10.1371/journal.pone.0050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y.C., Chang Y.F. Serum interleukin-6 levels reflect the disease status of colorectal cancer. J. Surg. Oncol. 2003;83:222–226. doi: 10.1002/jso.10269. [DOI] [PubMed] [Google Scholar]
- Culig Z., Steiner H., Bartsch G., Hobisch A. Interleukin-6 regulation of prostate cancer cell growth. J. Cell. Biochem. 2005;95:497–505. doi: 10.1002/jcb.20477. [DOI] [PubMed] [Google Scholar]
- DeMichele A., Martin A.M., Mick R., Gor P., Wray L., Klein-Cabral M., Athanasiadis G., Colligan T., Stadtmauer E., Weber B. Interleukin-6–174G > C polymorphism is associated with improved outcome in high-risk breast cancer. Cancer Res. 2003;63:8051–8056. [PubMed] [Google Scholar]
- Dikshit R., Gupta P.C., Ramasundarahettige C., Gajalakshmi V., Aleksandrowicz L., Badwe R. Cancer mortality in India: a nationally representative survey. Lancet. 2012;379:1807–1816. doi: 10.1016/S0140-6736(12)60358-4. [DOI] [PubMed] [Google Scholar]
- Guo Y., Xu F., Lu T., Duan Z., Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat. Rev. 2012;38:904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Hefler L.A., Grimm C., Lantzsch T., Lampe D., Leodolter S., Koelbl H., Heinze G., Reinthaller A., Tong-Cacsire D., Tempfer C., Zeillinger R. Interleukin-1 and interleukin-6 gene polymorphisms and the risk of breast cancer in Caucasian women. Clin. Cancer Res. 2005;15(11):5718–5721. doi: 10.1158/1078-0432.CCR-05-0001. [DOI] [PubMed] [Google Scholar]
- Jerrard-Dunne P., Sitzer M., Risley P., Buehler A., von Kegler S., Markus H.S. Inflammatory gene load is associated with enhanced inflammation and early carotid atherosclerosis in smokers. Stroke. 2004;35:2438–2443. doi: 10.1161/01.STR.0000144681.46696.b3. [DOI] [PubMed] [Google Scholar]
- Jibiki T., Terai M., Shima M., Ogawa A., Hamada H., Kanazawa M., Yamamoto S., Oana S., Kohno Y. Monocyte chemoattractant protein 1 gene regulatory region polymorphism and serum levels of monocyte chemoattractant protein 1 in Japanese patients with Kawasaki disease. Arthritis Rheum. 2001;44:2211–2212. doi: 10.1002/1529-0131(200109)44:9<2211::aid-art375>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. The biology of interleukin-6. Blood. 1989;74:1–10. [PubMed] [Google Scholar]
- Ko Y.C., Huang Y.L., Lee C.H., Chen M.J., Lin L.M., Tsai C.C. Betel quid chewing, cigarette smoking and alcohol consumption related to oral cancer in Taiwan. J. Oral Pathol. Med. 1995;24:450–453. doi: 10.1111/j.1600-0714.1995.tb01132.x. [DOI] [PubMed] [Google Scholar]
- Kuruvilla J. Utilizing dental colleges for the eradication of oral cancer in India. Indian J. Dent. Res. 2008;19:349–353. doi: 10.4103/0970-9290.44541. [DOI] [PubMed] [Google Scholar]
- Lagmay J.P., London W.B., Gross T.G., Termuhlen A., Sullivan N., Axel A., Mundy B., Ranalli M., Canner J., McGrady P., Hall B. Prognostic significance of interleukin-6 single nucleotide polymorphism genotypes in neuroblastoma: rs1800795 (promoter) and rs8192284 (receptor) Clin. Cancer Res. 2009;15:5234–5239. doi: 10.1158/1078-0432.CCR-08-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi S., Moreno V., Gioia-Patricola L., Guino E., Navarro M., de Oca J., Capella G., Canzian F. Bellvitge Colorectal Cancer Study Group. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63:3560–3566. [PubMed] [Google Scholar]
- Liang J., Liu X., Bi Z., Yin B., Xiao J., Liu H., Li Y. Relationship between gene polymorphisms of two cytokine genes (TNF-a and IL-6) and occurring of lung cancers in the ethnic group Han of China. Mol. Biol. Rep. 2013;40:1541–1546. doi: 10.1007/s11033-012-2199-2. [DOI] [PubMed] [Google Scholar]
- Mandal S., Abebe F., Chaudhary J. 174G/C polymorphism in the interleukin-6 promoter is differently associated with prostate cancer incidence depending on race. Genet. Mol. Res. 2014;13:139–151. doi: 10.4238/2014.January.10.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalaki V., Syrigos K., Charles P., Waxman J. Serum levels of IL-6 and TNF-alpha correlate with clinicopathological features and patient survival in patients with prostate cancer. Br. J. Cancer. 2004;90:2312–2316. doi: 10.1038/sj.bjc.6601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P., Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J. Clin. Oncol. 2012;30:1005–1014. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage S.A., Abnet C.C., Haque K., Mark S.D., Qiao Y.L., Dong Z.W., Dawsey S.M., Taylor P.R., Chanock S.J. Polymorphisms in interleukin − 2, − 6, and − 10 are not associated with gastric cardia or esophageal cancer in a high-risk Chinese population. Cancer Epidemiol. Biomarkers Prev. 2004;13:1547–1549. [PubMed] [Google Scholar]
- Schafer Z.T., Brugge J.S. IL-6 involvement in epithelial cancers. J. Clin. Invest. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seike T., Fujita K., Yamakawa Y., Kido M.A., Takiguchi S. Interaction between lung cancer cells and astrocytes via specific inflammatory cytokines in the microenvironment of brain metastasis. Clin. Exp. Metastasis. 2011;28:13–25. doi: 10.1007/s10585-010-9354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow A., Ng D.P., Choo S., Eng P., Poh W.T., Ming T., Wang Y.T. Joint effect of asthma/atopy and an IL-6 gene polymorphism on lung cancer risk among lifetime non-smoking Chinese women. Carcinogenesis. 2006;27:1240–1244. doi: 10.1093/carcin/bgi309. [DOI] [PubMed] [Google Scholar]
- Singh P.K., Ahmad M.K., Kumar V., Hussain S.R., Gupta R., Jain A., Mahdi A.A., Bogra J., Chandra G. Effects of interleukin-18 promoter (C607A and G137C) gene polymorphisms and their association with oral squamous cell carcinoma (OSCC) in northern India. Tumour Biol. 2014;35:12275–12284. doi: 10.1007/s13277-014-2538-0. [DOI] [PubMed] [Google Scholar]
- Singh P.K., Bogra J., Chandra G., Ahmad M.K., Gupta R., Kumar V., Jain A., Ali Mahdi A. Association of TNF-α (− 238 and − 308) promoter polymorphisms with susceptibility of oral squamous cell carcinoma in North Indian population. Cancer Biomark. 2014 doi: 10.3233/CBM-140444. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Slattery M.L., Curtin K., Baumgartner R., Sweeney C., Byers T., Giuliano A.R., Baumgartner K.B., Wolff R.R. IL6, aspirin, nonsteroidal anti-inflammatory drugs, and breast cancer risk in women living in the southwestern United States. Cancer Epidemiol. Biomarkers Prev. 2007;16:747–755. doi: 10.1158/1055-9965.EPI-06-0667. [DOI] [PubMed] [Google Scholar]
- Srivani R., Nagarajan B. A prognostic insight on in vivo expression of interleukin-6 in uterine cervical cancer. Int. J. Gynecol. Cancer. 2003;13:331–339. doi: 10.1046/j.1525-1438.2003.13197.x. [DOI] [PubMed] [Google Scholar]
- St John M.A., Li Y., Zhou X., Denny P., Ho C.M., Montemagno C., Shi W., Qi F., Wu B., Sinha U., Jordan R., Wolinsky L., Park N.H., Liu H., Abemayor E., Wong D.T. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2004;130:929–935. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- Tan D., Wu X., Hou M., Lee S.O., Lou W., Wang J., Janarthan B., Nallapareddy S., Trump D.L., Gao A.C. Interleukin-6 polymorphism is associated with more aggressive prostate cancer. J. Urol. 2005;174:753–756. doi: 10.1097/01.ju.0000168723.42824.40. [DOI] [PubMed] [Google Scholar]
- Terry C.F., Loukaci V., Green F.R. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J. Biol. Chem. 2000;275:18138–18144. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- Vairaktaris E., Yiannopoulos A., Vylliotis A., Yapijakis C., Derka S., Vassiliou S., Nkenke E., Serefoglou Z., Ragos V., Tsigris C., Vorris E., Critselis E., Avgoustidis D., Neukam F.W., Patsouris E. Strong association of interleukin-6–174G > C promoter polymorphism with increased risk of oral cancer. Int. J. Biol. Markers. 2006;21:246–250. doi: 10.1177/172460080602100409. [DOI] [PubMed] [Google Scholar]
- Vucicević Boras V., Cikes N., Lukać J., Virag M., Cekić-Arambasin A. Salivary and serum interleukin 6 and basic fibroblast growth factor levels in patients with oral squamous cell carcinoma. Minerva Stomatol. 2005;54:569–573. [PubMed] [Google Scholar]
- Wang Y.F., Chang S.Y., Tai S.K., Li W.Y., Wang L.S. Clinical significance of interleukin-6 and interleukin-6 receptor expressions in oral squamous cell carcinoma. Head Neck. 2002;24:850–858. doi: 10.1002/hed.10145. [DOI] [PubMed] [Google Scholar]
- Williams H.K. Molecular pathogenesis of oral squamous carcinoma. Mol. Pathol. 2000;53:165–172. doi: 10.1136/mp.53.4.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E., Jakubowicz J., Skotnicki P., Sas-Korczyńska B., Kulpa J.K. IL-6 and VEGF in small cell lung cancer patients. Anticancer Res. 2010;30:1773–1778. [PubMed] [Google Scholar]
- Xu B., Niu X.B., Wang Z.D., Cheng W., Tong N., Mi Y.Y., Min Z.C., Tao J., Li P.C., Zhang W., Wu H.F., Zhang Z.D., Wang Z.J., Hua L.X., Feng N.H., Wang X.R. IL-6–174G/C polymorphism and cancer risk: a meta-analysis involving 29,377 cases and 37,739 controls. Mol. Biol. Rep. 2011;38:2589–2596. doi: 10.1007/s11033-010-0399-1. [DOI] [PubMed] [Google Scholar]
- Yeh K.Y., Li Y.Y., Hsieh L.L., Chen J.R., Tang R.P. The -174G/C polymorphism in interleukin-6 (IL-6) promoter region is associated with serum IL-6 and carcinoembryonic antigen levels in patients with colorectal cancers in Taiwan. J. Clin. Immunol. 2010;30:53–59. doi: 10.1007/s10875-009-9324-6. [DOI] [PubMed] [Google Scholar]
- Yin Y.W., Sun Q.Q., Hu A.M., Wang Q., Liu H.L., Hou Z.Z., Zeng Y.H., Xu R.J., Shi L.B., Ma J.B. Associations between interleukin-6 gene − 174 C/G and − 572 C/G polymorphisms and the risk of gastric cancer: A meta-analysis. J. Surg. Oncol. 2012;106:987–993. doi: 10.1002/jso.23199. [DOI] [PubMed] [Google Scholar]
- Yu K.D., Di G.H., Fan L., Chen A.X., Yang C., Shao Z.M. Lack of an association between a functional polymorphism in the interleukin-6 gene promoter and breast cancer risk: a meta-analysis involving 25,703 subjects. Breast Cancer Res. Treat. 2010;122:483–488. doi: 10.1007/s10549-009-0706-5. [DOI] [PubMed] [Google Scholar]
- Zarogoulidis P., Yarmus L., Darwiche K., Walter R., Huang H., Li Z., Zaric B., Tsakiridis K., Zarogoulidis K. Interleukin-6 cytokine: a multifunctional glycoprotein for cancer. Immunome Res. 2013;9:16535. doi: 10.1186/2090-5009-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.J., Adachi I. Serum interleukin-6 levels correlate to tumor progression and prognosis in metastatic breast carcinoma. Anticancer Res. 1999;19:1427–1432. [PubMed] [Google Scholar]