Abstract
Background
Shigella sonnei is considered as a major cause of diarrheal disease in both developing and developed countries. Iran is one of the endemic areas of shigellosis. The present study was undertaken to investigate the antibiotic susceptibility and genetic relatedness of S. sonnei strains isolated from pediatric patients in Tehran, Iran.
Methods
The study included all S. sonnei strains isolated from pediatric patients with diarrhea admitted to several hospitals in Tehran, Iran, during 2008-2010. Shigella spp. strains were recovered from patients using standard microbiological methods. S. sonnei strains were further studied by antimicrobial susceptibility testing and Enterobacterial Repetitive Intergenic Consensus (ERIC) - PCR analysis.
Results
Eighty nine Shigella isolates were isolated. S. sonnei was themost prevalent Shigella species (60.7%) followed by, S. flexneri (31.5%). Eleven antimicrobial resistance patterns (R1-R11) were identified among S. sonnei isolates. The majority of the strains were resistant to trimethoprim-sulfamethoxazole, tetracycline and streptomycin. All isolates were susceptible to ciprofloxacin, ceftizoxime and chloramphenicol. All strains were typable by ERIC-PCR. Five ERIC-PCR patterns (E1-E5) were found among S. sonnei isolates; however the half of the isolates was clustered in E4 pattern.
Conclusion
The antibiotic resistance rates are increasing among S. sonnei strains. Moreover, a predominant clone or limited clones of S. sonnei were responsible for shigellosis caused by this Shigella species in pediatric patients in Tehran, Iran.
Keywords: Shigella sonnei, Antibiotic resistance, ERIC-PCR
Introduction
Shigellosis is among the most common causes of bacterial diarrheal disease in both developing and developed countries. Shigella consists of four serogroups including serogroup A (S. dysenteriae), serogroup B (S. flexneri), serogroup C (S. boydii) and serogroup D (S. sonnei). It is one of the major causes of morbidity in children with diarrhea in Iran (1, 2). S. sonnei is the major cause of shigellosis in industrialized countries (3). It has been recently reportedit as the prevalent Shigella serotype in Iran (4, 5).
Treatment using adequate antibiotics is effective for shigellosis particularly for the children and immunosuppressed patients because it may shortens the clinical course of the disease, reduce the risk of transmission and prevent potentially lethal complications. However, resistance to commonly used antibiotics is increasing among Shigella spp. worldwide (6, 7). The antibiotic resistance among Shigella spp. is increasing in Iran (8–10).
In the recent years, the conventional bacterial typing methods such as antimicrobial resistance pattern, bacteriophage typing, or serotyping have been replaced by molecular techniques such as ribotyping, pulsed-field gel electrophoresis (PFGE), and PCR-based methods (1, 11, 12). An Entero-bacterial Repetitive Intergenic Consensus (ERIC) sequence is an imperfect palindrome of 127 bp. ERIC sequences have been found only in intergenic regions, apparently only within transcribed regions. PCR- mediated genomic fingerprinting based on ERIC sequences has been found to be useful for subtyping Gram-negative enteric bacteria and differentiation of their strains (13).
The present study was conducted to determine the antimicrobial susceptibility and genetic relationship among S. sonnei strains isolated from pediatric patients in Tehran, Iran.
Material and Methods
Bacterial strains
The study included all S. sonnei strains isolated from pediatric patients with diarrhea who were admitted to several hospitals in Tehran, Iran, during 2008-2010. A single specimen was obtained from each patient, and rectal swabs were collected from patients on the day of admission at the hospital. When the isolates were identified as Shigella by the conventional methods (14), these were serotyped using slide agglutination with specific antisera (MAST Group LTD, Merseyside, UK).
All ethical issues were considered. The name and characters, personal information and even patients’ illnesses and their medical information remained secret and the life, health, dignity, integrity, rights to self-determination, privacy, and confi-dentiality of personal information of research subjects were protected in this study.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed according to Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) (15).
The following antimicrobial agents were tested: ampicillin (AM, 10µg), amoxycillin-clavulanic acid (AMC20+10µg), amikacin (AN 30µg), ceftazidime (CAZ 30µg), cephalothin (CF 30µg), ceftriaxone (CRO 30µg), cefotaxime (CTX 30µg), kanamycin (K 30µg), nalidixic acid (NA 30µg), streptomycin (S 10µg), trimethoprim-sulfamethoxazole (SXT 23.75+1.25µg), tetracycline (TE 30µg), ticarcillin (TIC 75µg), tobramycin (TOB 10µg), gentamicin (GM 10µg), ceftizoxime (CT 30µg), ciprofloxacin (CP 5µg), chloramphenicol (C 30µg). Escherichia coli ATCC 25922 were used as control strain.
DNA preparation
One colony from overnight culture was inoculated into LB-broth (tryptone 1%, NaCl 1% and yeast extract 0.5%) and was grown with shaking overnight at 37°C. Bacterial DNA was extracted using the phenol-chloroform method. Extracted genome was dissolved in 50-100µl of TE buffer.
ERIC-PCR primers and PCR condition
ERIC-PCR was performed using the primers ERIC 1R (5’-ATGTAAGCTCCTGGGGATTCAC-3’) and ERIC2 (5’-AAGTAAGTGACTGGGGTGAGCG-3’) with minor modifications (16, 17).
The PCR reaction mixture (20µl) consisted of 10x reaction buffer [750 mM Tris-HCl (pH 8.8), 200 mM (NH4)2SO4, 0.1% Tween20], a 250 µM concentration of each dNTP, 20 pmol of primer ERIC 1R, 20 pmol of primer ERIC 2, 3.75 mM MgCl2, 100 ng of template DNA and 2 U of Taq polymerase. The reaction mixture was denatured for 7 min at 95 °C and then subjected to 30 cycles of denaturation for 30s at 90 °C, annealing for 1 min at 52 °C, extension for 3 min at 65 °C, and a final extension for 16 min at 65 °C. Negative controls without template DNA were included in each run. Amplified products were resolved by electrophoresis of 5 µl samples on 1% agarose gels in 1x TBE buffer at 70 V for 3 h and were visualized by ethidium bromide staining. The PCR patterns were visually compared and were considered to be identical on the basis of similar numbers and matching positions of all major bands. Small differences in bonds were ignored. The type strain of S. sonnei, ATCC 9290, was also included in this study for the comparison.
Results
Of 950 patients with acute diarrhoea, 89 Shigella spp. strains were isolated which were distributed as following; S. sonnei, 54 (60.7%), S. flexneri 28 (31.5%), S. boydii 5 (5.6%) and S. dysenteriae 2 (2.2%). The season distribution of the isolated strains was winter, 10; spring, 17; summer, 38 and fall, 24. Of all patients, 57.4% were male and 42.7% were female. The ratio of males to females was 1.34. Shigella was isolated frequently from children under 5 years of age, who accounted for 62.9.7% of all isolates. About 35.1 % of all isolates came from persons aged 5-12 years, and 1.8% from persons aged over 12 years of age.
Results of further examination of S. sonnei strains showed that the majority of the isolates (≥90%) were resistant to trimethoprim-sulfamethoxazole, tetracycline and streptomycin, 37% to ampicillin, 27.8% to nalidixic acid, 9.3% to amoxycillin-clavulanic acid and 11.3% tobramycin. The most of the isolates were susceptible to cephalothin, ticarcillin, cefotaxime, ceftriaxone, amikacin, ceftazidime, kanamycin and gentamicin. All strains were fully susceptible to ciprofloxacin, ceftizoxime and chloramphenicol. As shown in table 1, the majority of the isolates demonstrated multiple drug resistance profile, among which, 1.8% were resistant to 2, 57.4% to 3, 3.7% to 4, 16.7% to 5, 7.4% to 6, 1.8% to 9, 5.5% to 11 and 1.8% were resistant to 12 tested antibiotics. Eleven antibiotic resistance patterns (R1-R11) were observed among the strains, R1 (tetracycline/streptomycin/trimethoprim-sulfamethoxazole) was however found as most prevalent (51.8%) resistance pattern.
All strains were typable by ERIC-PCR. The number of ERIC-PCR bands produced for a given primer ranged from 10 to 15, with molecular sizes ranging from 100 to 3500 bp. ERIC-PCR analysis of the isolates resulted in five different patterns (E1-E5) with 9-13 DNA bands (Fig. 1.) ; however the half of the isolates was clustered in E4 pattern. Table 1 shows the distribution of ERIC-PCR patterns. No similarity was observed in the ERIC-PCR patterns between clinical isolates and ATCC type strain 9290 (Fig. 1).
Fig. 1:
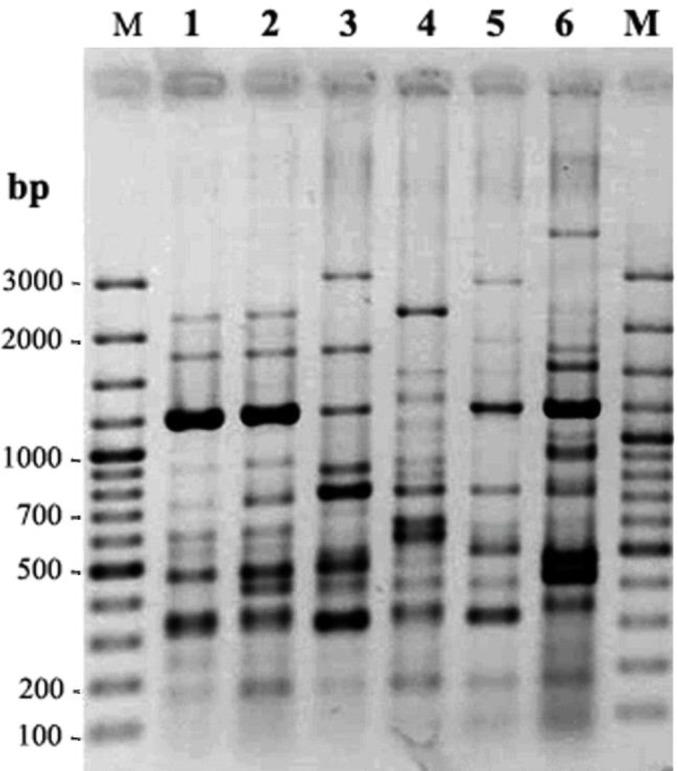
ERIC-PCR patterns of S. sonnei isolates. Lanes 1-5 are representative patterns E1, E2, E3, E4 and E5 respectively. Lane 6 is Es pattern from type strains ATCC 9290. Lane M is molecular size marker
Table 1:
Characteristics of S. sonnei isolates included in the study
| No. of isolates | Resistance pattern* (R1-R11) | ERIC-PCR pattern |
|---|---|---|
| Type strains (ATCC 9290) | - | Es |
| 1 | AMC, NA, CAZ, AM, TIC, CTX, CRO, TE, S, SXT, CF (R4) | E1 |
| 2 | AMC, NA, CAZ, AM, TIC, CTX, CRO, TE, S, SXT, CF (R4) | E1 |
| 3 | NA, AM, TE, S, SXT (R2) | E1 |
| 4 | NA, AM, TE, S, SXT (R2) | E1 |
| 5 | AMC, NA, CAZ, AM, TIC, CTX, CRO, TE, S, SXT, CF (R4) | E1 |
| 6 | - | E2 |
| 7 | NA, AM, TE, S, SXT (R2) | E4 |
| 8 | NA, AM, TE, S, SXT (R2) | E1 |
| 9 | NA, AM, TE, S, SXT (R2) | E2 |
| 10 | NA, AM, TE, S, SXT (R2) | E3 |
| 11 | NA, AM, TE, S, SXT (R2) | E1 |
| 12 | AMC, NA, CAZ, AM, TIC, CTX, CRO, TE, S, SXT, GM, CF (R7) | E1 |
| 13 | NA, AM, TE, S, SXT (R2) | E1 |
| 14 | AM, TE, S, SXT (R9) | E5 |
| 15 | NA, AM, TE, S, SXT (R2) | E4 |
| 16 | TOB, TE, SXT (R11) | E4 |
| 17 | AM, TOB, AN, TE, S, SXT (R3) | E4 |
| 18 | AM, TOB, AN, TE, S, SXT (R3) | E4 |
| 19 | TE, S, SXT (R1) | E4 |
| 20 | AM, TOB, AN, TE, S, SXT (R3) | E4 |
| 21 | TE, S, SXT (R1) | E4 |
| 22 | TE, S, SXT (R1) | E4 |
| 23 | AMC, AM, TOB, TIC, CTX, CRO, TE, S, SXT (R8) | E4 |
| 24 | AM, TOB, AN, TE, S, SXT (R3) | E4 |
| 25 | NA, S, SXT (R5) | E4 |
| 26 | TE, S, SXT (R1) | E4 |
| 27 | TE, S, SXT (R1) | E4 |
| 28 | NA, S, SXT (R5) | E4 |
| 29 | TE, S, SXT (R1) | E4 |
| 30 | - | E2 |
| 31 | TE, SXT (R10) | E2 |
| 32 | TE, S, SXT (R1) | E4 |
| 33 | TE, S, SXT (R1) | E2 |
| 34 | TE, S, SXT (R1) | E2 |
| 35 | TE, S, SXT (R1) | E1 |
| 36 | TE, S, SXT (R1) | E1 |
| 37 | TE, S, SXT (R1) | E4 |
| 38 | TE, S, SXT (R1) | E2 |
| 39 | TE, S, SXT (R1) | E2 |
| 40 | TE, S, SXT (R1) | E2 |
| 41 | TE, S, SXT (R1) | E2 |
| 42 | TE, S, SXT (R1) | E2 |
| 43 | TE, S, SXT (R1) | E4 |
| 44 | TE, S, SXT (R1) | E2 |
| 45 | TE, S, SXT (R1) | E4 |
| 46 | TE, S, SXT (R1) | E2 |
| 47 | TE, S, SXT (R1) | E4 |
| 48 | TE, S, SXT (R1) | E4 |
| 49 | TE, S, SXT (R1) | E4 |
| 50 | TE, S, SXT (R1) | E4 |
| 51 | TE, S, SXT (R1) | E4 |
| 52 | K, AM, TE, SXT (R6) | E4 |
| 53 | TE, S, SXT (R1) | E4 |
| 54 | TE, S, SXT (R1) | E4 |
*AM: ampicillin, AMC: amoxycillin-clavulanic acid, AN: amikacin, CAZ: ceftazidime, CF: cephalothin, CRO: ceftriaxone, CTX: cefotaxime, K: kanamycin, NA: nalidixic acid, S: streptomycin, SXT: trimethoprim-sulfamethoxazole, TE: tetracycline, TIC: ticarcillin, TOB: tobramycin, GM: gentamycin.
Discussion
The infections caused by Shigella spp. has been increasing continuously and has turned into a prominent public health concern worldwide. Shigella spp. is one of the major causes of diarrheal disease among children in Iran (18).
Shigella strains were isolated frequently from children under 5 years of age, who accounted for 62.9.7% of all isolates. Previous studies have also demonstrated the fact that age can be a risk factor in shigellosis where children are in the high risk zone for Shigella infections (19). The typical seasonal increase in shigellosis occurred during the summer, with peak incidence in August that confirmed the warm months of the year can intensify the frequency of Shigella associated infections.
Assessment of antibiotic resistance patterns among Shigella isolates revealed that the antibiotic resistance rates have increased in comparison with previous reports published from Iran. For examples, when compared to a previous study in Tehran, the percentage of resistance against ampicillin, and nalidixic acid has increased from 10% and 8.3% to 37% and 27.8% respectively in our study (20). Moreover, to our knowledge, resistance to ceftriaxone and cefotaxime has not been reported in clinical strains of S. sonnei in Iran to date. However we found here 5 strains were resistant to these antibiotics.
Only one strain was resistant to kanamycin where also more than half of isolates showed intermediate resistance to this antibiotic which can be result in the probable increase in Shigella resistance to this antibiotic in the future. Moreover, in a ten-year study conducted by Ashtiani et al, in Tehran, Iran, Shigella spp. were found to show noticeably increasing resistance to kanamycin between 2001 and 2005 (21).
Fortunately resistance to ciprofloxacin and gentamicin is still low among our Shigella isolates. These finding is consistent with those reported by Vrints et al. who showed that the all Shigella isolates were sensitive to ciprofloxacin as well as gentamicin in a 18-years study from 1990 to 2007 in Belgium (22).
Subtyping using phenotypic methods is hindered by the homogeneity of circulating strains causing infectious diseases. Molecular typing of microbial strains helps us to accurately interpret epidemiological evolution of infectious diseases in the communities. Several different molecular methods have been applied to study the molecular epidemiology of S. sonnei isolates. PCR-based typing methods have been applicable to many organisms including S. sonnei and can be completed easily within a single day (18, 23).
ERIC-PCR is less laborious and time-consuming than other DNAbased typing techniques. This method has been widely used for the molecular typing of different bacteria in epidemiological studies, and its advantages and disadvantages are well known (24). However a few limited studies have previously evaluated the ERIC-PCR for molecular typing of Shigella strains. However as reported by Liu et al., this technique has been established to be a reliable and rapid genotyping approach with high discriminatory power for the differentiation of Shigella strains (25).
Here we used ERIC-PCR to study the genetic relatedness among endemic S. sonnei strains isolated from pediatric patients in Tehran, Iran. This technique differentiated the isolates into five different clusters (E1-E5). Using one pair of ERIC-PCR primers in a study carried out by Penatti et al, three genetic patterns were reported from S. flexneri and S. sonnei strains isolated from bacillary dysentery cases in Southeast Brazil (26).
This method has been also used for subtyping of epidemic S. flexneri strains in Iran previously. In a recent outbreak of shigellosis occurred among prisoners in Isfahan, ERIC-PCR showed to be a powerful method for molecular typing of Shigella strains. This technique differentiated the epidemic causative agent of outbreak from endemic and type standard strains and showed that a single clone of S. flexneri serotype 3a was responsible for the outbreak since all tested isolates had a single pattern (27). Kosek et al. also reported that this method was highly reproducible and could provide highly similar and supplementary data compared with serotyping regarding the transmission dynamics of shigellosis in the community studied (28). Otherwise, in another study for investigation of shigellosis outbreaks occurred in school children in Lungtan and Bade in Taoyuan county in northern Taiwan, this technique showed poor discriminatory power where ERIC-PCR analysis could not discriminate an epidemiologically unrelated strain from some outbreak strains. Epidemic S. sonnei strains could not be differentiated from type strain ATCC 9290 (29).
Here we also used type strain ATCC 9290 for the comparison; however ERIC-PCR was capable to differentiate endemic S. sonnei strains from this standard strain and from each other clinical strains distributed in different clusters.
Navia et al. used another PCR-based technique, in which the amplification of the regions between repetitive extragenic palindromic (REP) sequences gave a fingerprinting pattern valid for epidemiological typing of different species of Shigella (30). Also in a study carried out on 60 S. sonnei strains isolated from children hospitalized at five hospitals in Tehran during 2003, a similar technique, known as Arbitrarily Primed PCR (AP-PCR), was evaluated for subtyping of endemic S. sonnei isoaltes. Only a single AP-PCR pattern was observed among all S. sonnei strains (23).
When reviewing the previous reports on the distribution of molecular types of endemic S. sonnei in Iran, our findings further confirm the involvement of our geographic area within an epidemiological global picture of dissemination of a limited number of well-defined clones of S. sonnei. The results obtained from our previous global project on 1,672 S. sonnei isolates obtained since 1943 from 50 countries including Iran revealed that three major S. sonnei groups were responsible for shigellosis caused by this serogroup in which two groups were globally spread (3).
Conclusion
Considering that more than half of isolates were clustered into E4, it is concluded that one predominant clone or limited clones of S. sonnei are responsible for shigellosis caused by this Shigella species in pediatric patients during our research period. We hope our finding could be helpful for further epidemiological surveillance of S. sonnei in our country in the future.
Continuous studies using more discriminating molecular methods is needed to be conducted in Tehran and other parts of Iran in order to determine molecular subtypes of S. sonnei and other Shigella species in the future.
Ethical Considerations
All ethical issues including plagiarism, Informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc. have been completely observed by the author.
Acknowledgment
This study was financially supported in part by a grant from Iranian Ministry of Health and Medical Education, Deputy of Research and Technology. The authors declare that there is no conflict of interest.
References
- 1.Ranjbar R, Mammina C, Pourshafie MR, Soltan-Dallal MM (2008). Characterization of endemic Shigella boydii strains isolated in Iran by serotyping, antimicrobial resistance, plasmid profile, ribotyping and pulsed-field gel electrophoresis. BMC Res Notes, 1: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranjbar R, Pourshafie MR, Soltan-Dallal MM, Rahbar M, Farshad S, Parvaneh N, et al. (2010). Fatality due to shigellosis with special reference to molecular analysis of Shigella sonnei strains isolated from the fatal cases. Iranian J Clin Infect Dis, 5: 36 – 39. [Google Scholar]
- 3.Filliol-Toutain I, Chiou CS, Mammina C, Gerner-Smidt P, Thong KL, Phung DC, et al. (2011). Global distribution of Shigella sonnei clones. Emerg Infect Dis, 17: 1910–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ranjbar R, Soltan Dallal MM, Talebi M, Pourshafie MR (2008). Increased isolation and characterization of Shigella sonnei obtained from hospitalized children in Tehran, Iran. J Health Popul Nutr, 26: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farshad S, Sheikhi R, Japoni A, Basiri E, Alborzi A (2006). Characterization of Shigella strains in Iran by plasmid profile analysis and PCR amplification of ipa genes. J Clin Microbiol, 44: 2879–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voogd CE, Schot CS, Van Leeuwen WJ, and Van Klingeren B (1992). Monitoring of antibiotic resistance in Shigella isolated in the Netherlands 1984–1989. Eur J Clin Microbiol Infect Dis, 11: 164–167. [DOI] [PubMed] [Google Scholar]
- 7.Salam M, Bennish ML (1991). Antimicrobial therapy for shigellosis. Res Infect Dis, 13 (Suppl 4): S332–S341. [DOI] [PubMed] [Google Scholar]
- 8.Soltan-Dallal MM, Ranjbar R, Pourshafie MR (2011). The study of antimicrobial resistance among Shigella flexneri strains isolated in Tehran, Iran. J Pediatr Infect Dis, 6 : 125–129. [Google Scholar]
- 9.Hosseini MJ, Ranjbar R, Ghasemi H, Jalalian HR (2007). The prevalence and antibiotic resistance of Shigella spp. recovered from patients admitted to Bouali Hospital, Tehran, Iran during 1999 – 2000. Pak J Biol Sci, 10: 2778 – 2780. [DOI] [PubMed] [Google Scholar]
- 10.Ranjbar R, Soltan-Dallal, MM, Pourshafie MR (2009). Antibiotic resistance among Shigella serogroups isolated in Tehran, Iran (2002–2004). J Infect Dev Ctries, 3: 647–648. [DOI] [PubMed] [Google Scholar]
- 11.Bentley CA, Frost JA, Rowe B (1996). Phage typing and drug resistance of Shigella sonnei isolated in England and Wales. Epidemiol Infect, 116: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendoza MC, Martin MC, Gonzalez-Hevia MA (1996). Usefulness of ribotyping in a molecular epidemiology study of shigellosis. Epidemiol Infect, 116: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson LA Sharp PM (2006). Enterobacterial Repetitive Intergenic Consensus (ERIC) Sequences in Escherichia coli: Evolution and Implications for ERIC-PCR. Mol Biol Evol, 23: 1156–1168. [DOI] [PubMed] [Google Scholar]
- 14.Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken FC editors (1995). Manual of clinical microbiology. 6th ed. Washington, DC: American Society for Microbiology, 1,482p. [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. Approved standard M100-S15. Wayne, PA: Clinical and Laboratory Standards Institute, 2005. [Google Scholar]
- 16.Versalovic1 J, Koeuth1 T, Lupski1 JR (1991). Distribution of repetitive DNA sequences in eubacteria andapplication to fingerprinting of bacterial genomes. Nucleic Acids Res, 19: 6831–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjbar R, Rahbar M, Naghoni A, Farshad S, Davari A, Shahcheraghi F (2011). A cholera outbreak associated with drinking contaminated well water. Arch Iran Med, 14 : 339–340. [PubMed] [Google Scholar]
- 18.Ranjbar R, Aleo A, Giammanco GM, Dionisi AM, Sadeghifard N, Mammina C (2007). Genetic relatedness among isolates of Shigella sonnei carrying class 2 integrons in Tehran, Iran, 2002 – 2003. BMC Infect Dis, 22: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herwana E, Surjawidjaja J, Salim OCH, Indriani N, Bukitwetan P, Lesmana M (2010). Shigella-associated diarrhea in children in south Jakarta, Indonesia. Southeast Asian J Trop Med Public Health, 41: 418–425. [PubMed] [Google Scholar]
- 20.Ranjbar R, Farshad S, Rahbar M, Safiri Z, Mammina C, Arjomanzadegan M (2010). Occurrence of class 2 integrons among multi-drug resistant Shigella sonnei isolated from Tehran, Iran in 2005. Iranian J Clin Infect Dis, 5: 156–160. [Google Scholar]
- 21.Ashtiani MT, Monajemzadeh M, Kashi L (2009). Trends in antimicrobial resistance of fecal Shigella and Salmonella isolates in Tehran, Iran. Indian J Pathol Microbiol, 52: 52–55. [DOI] [PubMed] [Google Scholar]
- 22.Vrints M, Mairiaux E, Meervenne V, Collard JM, Bertrand S (2009). Surveillance of antibiotic susceptibility pattern among Shigella sonnei strains isolated in Belgium during the 18 year period, 1990 to 2007. J Clin Microbiol, 47: 1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjbar R, Sadeghifard N, Soltan Dallal MM, Farshad S (2009). Evaluation of a PCR based approach to study the relatedness among S. sonnei strains isolated in Tehran. Iran J Clin Infect Dis, 4: 163–166. [Google Scholar]
- 24.Ranjbar R, Mirzaee A (2013). Determining of the variety of genotypes in Salmonella Typhi-murium by ERIC-PCR. J Babol Univ Med Sci, 15: 51–57. [Google Scholar]
- 25.Liu PY, Lau YJ, Hu BS, Shyr JM, Shi ZY, Tsai WS et al (1995). Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J Clin Microbiol, 33: 1779–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penatti MP, Hollanda LM, Nakazato G, Campos TA, Lancellotti M, Angellini M, et al. (2007). Epidemiology characterization of resistance and PCR typing of Shigella flexneri and Shigella sonnei strains isolated from bacillary dysentery cases in Southeast Brazil. Braz J Med Biol Res, 40: 294–258. [DOI] [PubMed] [Google Scholar]
- 27.Ranjbar R, Hosseini MJ, Kaffashian AR, Farshad S (2010). An outbreak of shigellosis due to Shigella flexneri serotype 3a in a prison in Iran. Arch Iran Med, 13: 413–416. [PubMed] [Google Scholar]
- 28.Kosek M, Yori PP, Gilman RH, Vela H, Olortegui MP, Chavez CB, et al. (2012). Facilitated molecular typing of Shigella isolates using ERIC-PCR. Am J Trop Med Hyg, 86: 1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TM, Chang CY, Chang LL, Chen WM, Wang TK, Chang SF (2003). One predominant type of genetically closely related Shigella sonnei prevalent in four sequential outbreaks in school children. Diagno Microbiol Infect Dis, 45: 173–181. [DOI] [PubMed] [Google Scholar]
- 30.Navia MM, Capitano L, Ruiz J, Vargas M, Urassa H, Schellemberg D (1999). Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol, 37: 3113–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]


