Abstract
Mice deficient in Latent TGFβ Binding Protein 4 (Ltbp4) display a defect in lung septation and elastogenesis. The lung septation defect is normalized by genetically decreasing TGFβ2 levels. However, the elastic fiber assembly is not improved in Tgfb2−/−;Ltbp4S−/− compared to Ltbp4S−/− lungs. We found that decreased levels of TGFβ1 or TGFβ3 did not improve lung septation indicating that the TGFβ isoform elevated in Ltbp4S−/− lungs is TGFβ2. Expression of a form of Ltbp4 that could not bind latent TGFβ did not affect lung phenotype indicating that normal lung development does not require the formation of LTBP4-latent TGFβ complexes. Therefore, the change in TGFβ-level in the lungs is not directly related to Ltbp4 deficiency but probably is a consequence of changes in the extracellular matrix. Interestingly, combination of the Ltbp4S−/− mutation with a fibulin-5 null mutant in Fbln5−/−;Ltbp4S−/− mice improves the lung septation compared to Ltbp4S−/− lungs. Large globular elastin aggregates characteristic for Ltbp4S−/− lungs do not form in Fbln5−/−;Ltbp4S−/− lungs and EM studies showed that elastic fibers in Fbln5−/−;Ltbp4S−/− lungs resemble those found in Fbln5−/− mice. These results are consistent with a role for TGFβ2 in lung septation and for Ltbp4 in regulating fibulin-5 dependent elastic fiber assembly.
Keywords: TGFβ, LTBP-4 function, fibulin-5, lung development, elastogenesis
Introduction
Latent TGFβ binding protein 4 (LTBP-4) belongs to a family of four extracellular matrix (ECM) proteins (LTBP-1 to −4) that are structurally similar to the fibrillins (Todorovic and Rifkin, 2012). Both LTBPs and fibrillins are multidomain proteins consisting of epidermal growth factor (EGF)-like domains and signature 8-Cys domains. Although both the LTBPs and fibrillins each have multiple 8-Cys domains, only the 3rd 8-Cys domains of LTBP-1, −3 and −4 bind TGFβ, which is secreted as part of a latent complex consisting of the growth factor plus its cleaved propeptide (LAP) held together by non-covalent bonds. The 3rd 8-Cys LTBP domains form disulfide bonds with the latent TGFβ propeptide (Gleizes et al., 1996; Saharinen et al., 1996). In this large tripartite latent complex, LTBPs may modulate TGFβ activity by facilitating its secretion, specifying its location in the ECM, and by directly participating in latent TGFβ activation (Annes et al., 2004; Miyazono et al., 1991). LTBPs interact with fibrillins, fibronectin and fibulin-5 (Isogai et al., 2003; Noda et al., 2013; Taipale et al., 1996) and in this manner sequester latent TGFβ within the ECM. This may increase the local concentration of the latent cytokine and target it for subsequent activation.
The four LTBP isoforms are structurally similar, however, they share only about 30% identity and 40% similarity of amino acid sequences (Koli et al., 2001; Saharinen et al., 1999; Saharinen et al., 1998; Todorovic et al., 2005). In addition, each LTBP isoform is evolutionally conserved, which might indicate unique biological requirements. Indeed, in vivo studies have demonstrated specific functions for individual LTBP isoforms; Ltbp1L−/− mice have an abnormal cardiac outflow tract with persistent truncus arteriosis (Todorovic et al., 2007), Ltbp3−/− mice have skeletal and lung abnormalities (Colarossi et al., 2005; Dabovic et al., 2002), and Ltbp4S−/− mice have a severe defect in terminal air sac septation and rectal prolapse (Sterner-Kock et al., 2002). In addition, neither Ltbp1L−/−, nor Ltbp3−/− mice show defective elastic fibers, whereas LTBP-4 is essential for elastic fiber assembly (Dabovic et al., 2009).
Mutations in the fibulin-5 gene (Fbln5) also result in lung abnormalities similar to those observed in Ltbp4S−/− mice. Fibulin-5 belongs to a family of seven ECM glycoproteins (Yanagisawa and Davis, 2010). Fibulin-3, −4 and −5 form a subfamily of short fibulins with a similar structure consisting of six EGF-like domains followed by a fibulin-type C terminal domain. All three short fibulins are important in elastin fiber assembly and homeostasis, and mice deficient for either fibulin-3, −4 or −5 display phenotypes attributed to elastic fiber abnormalities. Fibulin-3 mice show reduced bone density, premature aging, inguinal hernias and both uterine and rectal prolapse (McLaughlin et al., 2007). The herniation and prolapse may be explained by reduced elastic fibers in the fascia of the fibulin-3-deficient mice. Fibulin-4 mice die shortly after birth from aortic rupture and display developmental emphysema and aortic tortuosity (McLaughlin et al., 2006). Elastic fiber formation is severely affected in fibulin-4 mice and only a small amount of amorphous elastin is present in the skin, lungs and blood vessels. Fibulin-5 deficient mice have abnormal elastic fibers in lungs, skin and blood vessels and display emphysematous lungs, loose and inelastic skin, aortic tortuosity and pelvic organ prolapse (Choi et al., 2009; Nakamura et al., 2002; Yanagisawa et al., 2002). Thus, both LTBP-4 and fibulin-5 participate in elastic fiber assembly.
Elastogenesis is a complex multi-step process in which elastin is incorporated into microfibril bundles to generate elastic fibers (Wagenseil and Mecham, 2007). The process of elastic fiber assembly includes synthesis and secretion of tropoelastin, initial formation of elastin microaggregates by elastin crosslinking catalyzed by enzymes lysyl oxidase (LOX) and lysyl oxidase-like 1 (LOXL1), incorporation of elastin microaggregates into the fibrillin-rich microfibril bundles and further crosslinking of the elastin. In addition to elastin and fibrillin, elastic fibers contain multiple associated proteins including microfibril-associated glycoproteins (MAGPs), emilin, fibulins and LTBPs. The function of many of the associated molecules in elastic fiber assembly remains largely unknown. However, as mentioned above, fibulin-4 and −5 deficiencies result in abnormal elastogenesis affecting multiple organ systems in both mice and humans (McLaughlin et al., 2006; Nakamura et al., 2002; Urban, 2012; Yanagisawa et al., 2002), therefore, these proteins must play critical roles in the assembly of elastic fibers.
In the absence of LTBP-4, elastin forms large aggregates adjacent to microfibers, with little elastin incorporated into the microfibril bundles (Dabovic et al., 2009). The developmental emphysema and rectal prolapse seen in the Ltbp4S−/− mice, and the disrupted pulmonary, gastrointestinal, urinary, musculoskeletal, craniofacial and dermal development in humans with LTBP-4 deficiency are all consistent with disrupted elastin-microfibril assembly (Sterner-Kock et al., 2002; Urban et al., 2009).
The molecular mechanism(s) by which LTBP-4 regulates elastic fiber assembly is largely unknown. LTBP-4 interacts with fibrillin and fibulin-5 (Isogai et al., 2003; Noda et al., 2013), and therefore, LTBP-4 might target elastin-fibulin-5 complexes to the microfibrils and hence promote elastic fiber assembly. In addition, the elastin aggregates that accumulate in the absence of LTBP-4 may impede proper elastic fiber formation.
Interestingly, the septation defect observed in the Ltbp4S−/− embryonic lungs is normalized by lowering the level of TGFβ2 suggesting that elevated TGFβ contributes to the developmental emphysema in Ltbp4S−/−mice. However, earlier studies indicated that LTBP-4 binds only TGFβ1, therefore, the relationship between TGFβ2 and LTBP-4 may be indirect. Although TGFβ2 deficiency normalized alveolar wall septation, Tgfb2−/−;Ltbp4S−/− mice continued to have abnormal elastic fiber assembly similar to that observed in Ltbp4S−/− lungs, indicating that abnormal elastic fiber formation is not a direct result of perturbations in TGFβ2 levels.
To gain insight into the function of LTBP-4 in lung development, the present study examines the role of Ltbp-4 in regulating lung TGFβ levels and the relevance of LTBP-4 and fibulin-5 interaction in lung septation. Our results indicate that TGFβ2 is the only TGFβ isoform involved with the observed septation abnormality, as genetic ablation of TGFβ1 or 3 had no effect on the Ltbp4S−/− lung phenotype. The interference of TGFβ binding to LTBP-4 demonstrated no requirement for a covalent association of cytokine and LTBP-4 for terminal air sac septation and elastin maturation, indicating a structural role for LTBP-4. Fbln5−/−;Ltbp4S−/− mice had no additional phenotypic defects compared to Ltbp4S−/− or Fbln5−/− mice and Fbln5−/−;Ltbp4S−/− mice displayed elastogenesis defects similar to Fbln5−/− mice. We hypothesize that LTBP-4 regulates the rate of incorporation of elastin-fibulin-5 complexes into the microfibril bundles and that elastin-fibulin-5 aggregates accumulate in the absence of LTBP-4. We suggest that in Fbln5;Ltbp4S−/−lungs, as in Fbln5−/− lungs, elastogenesis occurs through an alternative pathway involving fibulin-4 and interacting proteins that promote incorporation of fibulin-4-elastin complexes into the microfibril bundles.
Materials and Methods
Mice
Ltbp4S−/− mice were a generous gift of H. von Melchner and were previously described (Sterner-Kock et al., 2002). Fbln5−/− mice were a generous gift from Hiromi Yanagisawa (UT Southwestern) (Yanagisawa et al., 2002). Mice with two point mutations in the Ltbp4 gene leading to substitution of cysteines 1235 and 1260 with serines were generated by InGenious Targeting Laboratory (Supplemental Figure 1). The neomycin resistance cassette was deleted using Rosa26-Cre mice (C57BL/6NTac-Gt(ROSA)26Sortm16(cre)Arte) purchased from Taconic. Mice heterozygous for the mutated locus were bred to generate homozygous mutants. Mice carrying null mutations for Tgfb1 (Tgfb1tm1Doe/J), and Tgfb3 (B6.129-Tgfb3tm1Doe/J) were purchased from The Jackson Laboratory. Tgfb1+/− and Tgfb3+/− mice were crossed with Ltbp4S+/−mice to generate Ltbp4S+/−;Tgfb1+/− and Ltbp4S+/−;Tgfb3+/−. Ltbp4S+/−;Tgfb3+/−mice were crossed to generate Ltbp4S−/−;Tgfb3−/− mice. As Ltbp4 and Tgfb1 reside on the same chromosome (chromosome 7), we first generated mice that had both mutated alleles on the same chromosome by crossing Ltbp4S+/−;Tgfb1+/− mice with WT mice and looking for Ltbp4S+/−;Tgfb1+/− offspring. These mice were crossed to generate Ltbp4S−/−;Tgfb1−/− mice. Tgfb1+/− mice were crossed to generate Tgfb1−/− mice. Ltbp4S+/− mice were crossed to generate Ltbp4S−/− mice. All mice were maintained on normal lab diet. All procedures were performed according to the regulations of the NYU Langone Medical Center IACUC.
Genotyping
All mice were genotyped by PCR. Ltbp4 alleles were amplified using reverse primers 3C7Wt: GGCTCATGCTTGAATGTTCAG and 3C7Tg: ATCATGCAAGCTGGTGGCTG specific for the WT and the mutated allele, respectively, and a common forward primer P3: CCAATCTTGCTTCTTTGCTG AGC. Fbln5 alleles were amplified using reverse primers Ex1arev: ACAGCTGGTTCAGATTACAGCGC and Inton1brev: AGCATCTATCCAAGCAACTA specific for the WT and the mutated allele, respectively, and a common forward primer Safwd: GCTAAGGATAACGAGGTGAG. Mice carrying Tgfb1 null allele were genotyped using forward primers B16F: GAGAAGAACTGCTGTGTGCG and Neo1L: CGACCACCAAGCGAAACATCGC specific for WT and mutant alleles, respectively, and a common reverse primer B16R: GTGTCCAGGCTCCAAATATAGG. Mice carrying Tgfb3 null allele were genotyped using reverse primers B3wtR: AGCAGTTCTCCTCCAGGTTG and B3muR: AATTCGCCAATGACAAGACG specific for WT and mutant alleles, respectively, and a common forward primer B3wtF: AATCAAGTGTCGTTGCCAGA.
Genotyping of LtbpCC>SS mice. Both WT and mutated alleles were amplified using primers: L4GF2: GTCTACAGAGTGGGTTGCAGG and L4GR2: GCACCACTAACCCAATCCTTAG. The PCR product was digested with BstAPI restriction enzyme. The absence of BstAPI recognition site demonstrates presence of the mutated allele. The presence of the neo cassette was determined using primers: Neo2F: GGAGAGGCTATTCGGCTATGACTG and Neo2R: CTCTTCGTCCAGATCATCCTGATC. The presence of Rosa26:Cre transgene was determined using primers: Cre800S: GCTGCCACGACCAAGTGACAGCAATG and Cre1200As: GTAGTTATTCGGATCATCAGCTACAC.
Quantitative real time RT-PCR
To prepare cDNA from mouse lungs, newborn pups were euthanized and dissected. Lungs were stored in RNA later stabilization buffer (Invitrogen) prior to RNA extraction. In order to extract RNA, lungs were frozen in liquid nitrogen and homogenised with the aid of a pellet pestle motor (Kontes) in RNA extraction buffer (Qiagen). RNA extraction was carried out using an RNAeasy RNA purification kit (Qiagen) following the manufacturer’s instructions (including DNAaseI digestion of genomic DNA on the purification column). RNA concentrations were measured by Nanodrop 8000 (Thermo Scientific). cDNA was generated in an retro transcription (RT) reaction using the Superscript III enzyme buffer system (Invitrogen). 300ng RNA was used per reaction, and after the RT reaction, the cDNA solution was diluted with water to a total volume of 300 ul.
Two approaches, both of which were conducted using a one step RT-PCR and the primers as described in Supplemental Table 1, were employed to investigate expression of Tgfb, Fbln, and Ltbp isoforms by WT and LtbpS−/− mouse lungs. For relative comparison of specific mRNA expression between WT and Ltbp4S−/− samples, CT (threshold cycle) measurements were normalized using glycerol 3-phosphate dehydrogenase (G3pdh) measurement as a control for RNA levels. The relative differences between three WT and three Ltbp4S−/− lungs were plotted using the Applied Biosystems StepOne software (v2.2.2). Error bars demonstrate maximum and minimum relative expression values with 95% confidence as determined by the Singleplex algorithm within the StepOne software.
To compare expression levels of different Ltbp, Tgfb and Fbln isoforms in WT lungs, we generated DNA standards of known concentration in order to estimate RNA copy numbers. To make these standards, we designed primers to amplify cDNA regions encompassing the Q-PCR primer binding sites plus 100 or more base pairs on either side. The sequences of these primers are given in Supplemental Table 2. Once synthesized from WT mouse lung, cDNA (produced as described above) was amplified by PCR and obtained products were cloned using the pGEM-T Easy vector system (Promega). The plasmids were purified using the Miniprep kit (Qiagen) and sequenced to confirm the absence of mutations that could affect the Q-PCR reaction. Duplicate standard curves were generated using six, four fold serial dilutions from a starting concentration of 100,000 plasmids per µl. These standard curves were used to estimate the number of RNA copies in cDNA samples from each individual lung tested.
Histology and Elastin Staining
Mouse lungs were inflated with 10% buffered formalin (Sigma-Aldrich) through the cannulated trachea under water pressure of 25 cm for day 7 and older lungs and 15 cm for newborn lungs. The inflated lungs were fixed in 10% buffered formalin, processed and embedded in paraffin. Five-micrometer sections were used in all studies. The sections were stained with hematoxylin and eosin (H&E) (Sigma) for histological and histomorphometric studies. Elastin was stained using orcinol – new fuchsin technique (Sheehan and Hrapchak, 1980).
Immunofluorescence
Antibodies against fibrillin-1 and fibulin-5 were generous gifts from Lynn Sakai (Shriners Research Center, Portland, OR) and Takako Sasaki (Oita University, Japan), respectively. Mouse monoclonal antibody against elastin was purchased from Abcam (Cambridge, MA). All secondary antibodies used for immunofluorescence studies were purchased from Molecular Probes, Life Technologies. Antigen retrieval for ECM proteins was done by digestion with 20 µg/ml of Proteinase K (Roche) for 5 min at 37°C.
Histomorphometric analysis
For histomorphometric studies, five lung sections were stained with H&E and images were digitally captured using whole slide digital scanner Leica SCN 400 and Leica SlidePath software. The mean terminal sac diameters were assessed using Image J and the method described by Mitzner W et al. (Mitzner et al., 2008).
Western Blot Analysis
Western blot analysis on lung extracts from P7 mice. Tissues were snap-frozen in liquid nitrogen and homogenized in Pierce RIPA buffer (Thermo Scientific) containing protease and phosphatase inhibitor cocktail (Thermo Scientific). After constant agitation for 30 min at 4°C homogenates were centrifuged for 10 min at full speed in a microcentrifuge at 4°C and the supernatants were collected. Protein concentrations were determined using Pierce BCA kit (Thermo Scientific). Equivalent amounts of protein from each sample were used for further analysis. Proteins were separated by SDS–PAGE using NuPAGE 4–20% gradient polyacrylamide mini gels (Novex, Life Technologies) under reducing conditions and transferred to a nitrocellulose membrane (Whatman) by electroblotting. Western blotting with P-Smad3 (Abcam) and β actin (Sigma) antibodies was performed according to the manufacturers’ instructions. Membranes were blocked in TBS / 0.1% Tween-20 containing 5% nonfat milk for 1 h at room temperature and incubated with primary antibodies overnight at 4°C. The membranes were washed five times in TBS / 0.1% Tween-20 and incubated with horseradish peroxidase-linked secondary antibodies (GE Healthcare Life Sciences) for 1 h at room temperature.
For analysis of conditioned medium from LtbpCC>SS and WT mouse cells, primary lung fibroblasts from WT and LtbpCC>SS mice were put in culture as described (Zilberberg et al., 2012). Serum-free conditioned media were collected after 20 h of culture and were used for western blot analysis with an antibody against Ltbp-4 (R&D).
Immunoreactive bands were revealed using Amersham ECL Prime Western Blotting Detection Reagent and images acquired with ImageQuant LAS 4000 digital imaging system (GE Healthcare Life Sciences). Relative intensity of the bands was evaluated using ImageJ software.
Transmission electron microscopy
For transmission electron microscopy of elastic fibers, lungs were inflated with ice-cold 3% gluteraldehyde in 0.1 M cacodylate buffer (pH 7.4) and the left lobe was removed and placed in fresh fixative overnight. Samples were cut into 1.5 mm3 pieces that included small airways, and sequentially stained en bloc with 1% osmium tetroxide, 2% tannic acid and 2% uranyl acetate as previously described (Davis, 1993). Stained samples were dehydrated, Epon embedded and cut. Thin sections (60 nm) were placed on formvar-coated grids and counterstained with 7% methanolic uranyl acetate followed by lead citrate. Sections were viewed using a Tecnai 12 transmission electron microscope at 120 kV. Images were digitally captured.
Results
TGFβ levels in Ltbp-4 null lungs
We previously reported that the elimination of TGFβ2 in the developing lungs of Ltbp4S−/− mice normalized septation but not elastogenesis. This observation raised two questions. First, would the elimination of other TGFβ isoforms also result in improved air sac septation, and second, was binding of TGFβ to LTBP-4 essential for LTBP-4 action? We addressed these two points by genetic approaches.
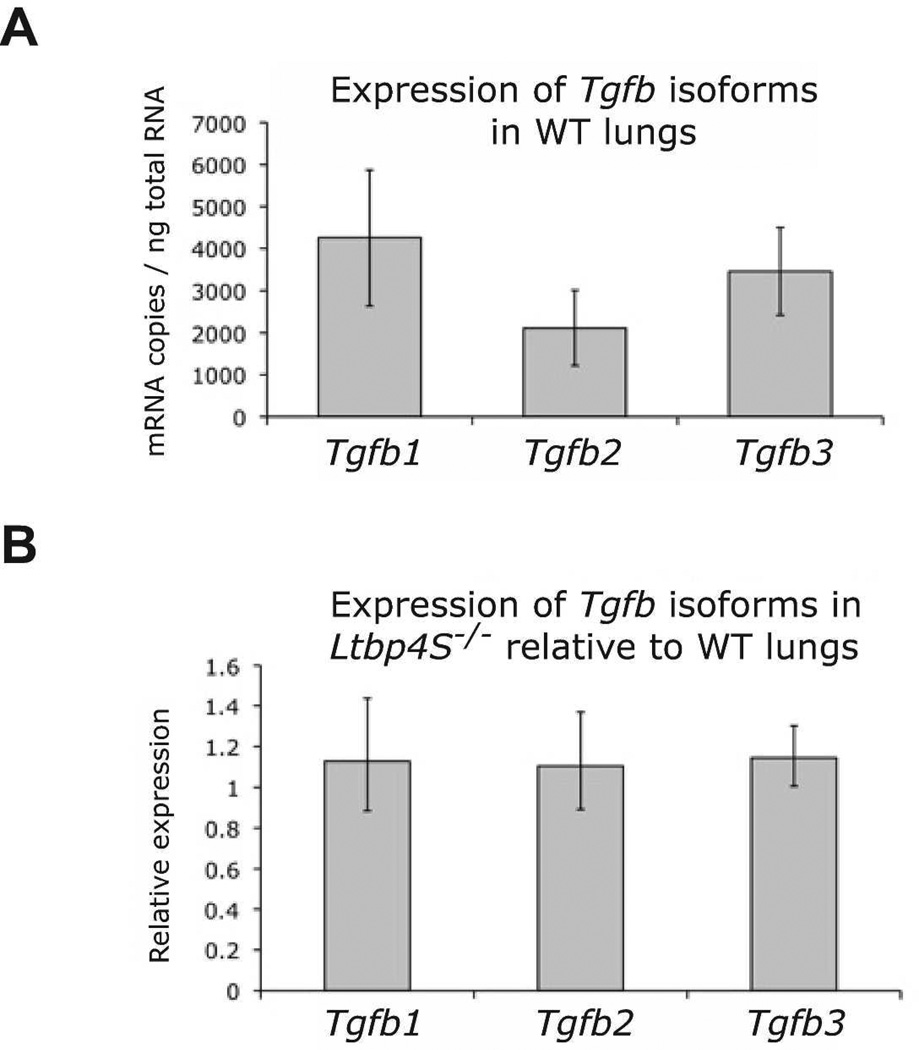
In the first approach, we initially examined expression of Tgfb genes in mouse lungs by quantitative RT-PCR to determine if there was preferential expression of a specific TGFβ isoform. Our data indicated that all three Tgfb genes are expressed at similar levels in both WT and Ltbp4S−/− P0.5 mouse lungs (Fig 1A) and that the lack of Ltbp-4 did not alter Tgfb isoform expression levels (Fig 1B). In order to test whether the improved terminal septation observed in Tgfb2−/− mice simply represented a decrease in the total TGFβ pool, we generated Tgfb1−/−;Ltbp4S−/− and Tgfb3−/−;Ltbp4S−/− animals and examined the development of their lungs. In both Tgfb1−/−;Ltbp4S−/− and Tgfb3−/−;Ltbp4S−/− mice, there was no improvement either lung architecture or elastogenesis (Supplemental Figures 2 and 3). Therefore, the pathological inhibition of lung septation in Ltbp4S−/− mice is caused by elevated TGFβ2 levels.
Figure 1.
Expression of Tgfb isoforms in WT and Ltbp4−/− lungs. A. Quantitative RT-PCR for Tgfb isoforms indicated that all three Tgfb isoforms are expressed at similar levels in P0.5 WT lungs. B. The expression levels of Tgfb1, 2 and 3 are similar in both WT and Ltbp4S−/− P0.5 lungs indicating that there is no alteration in Tgfb isoform expression due to the Ltbp-4 loss. Expression levels in B were normalized to levels of G3pdh. Three samples were analyzed per group.
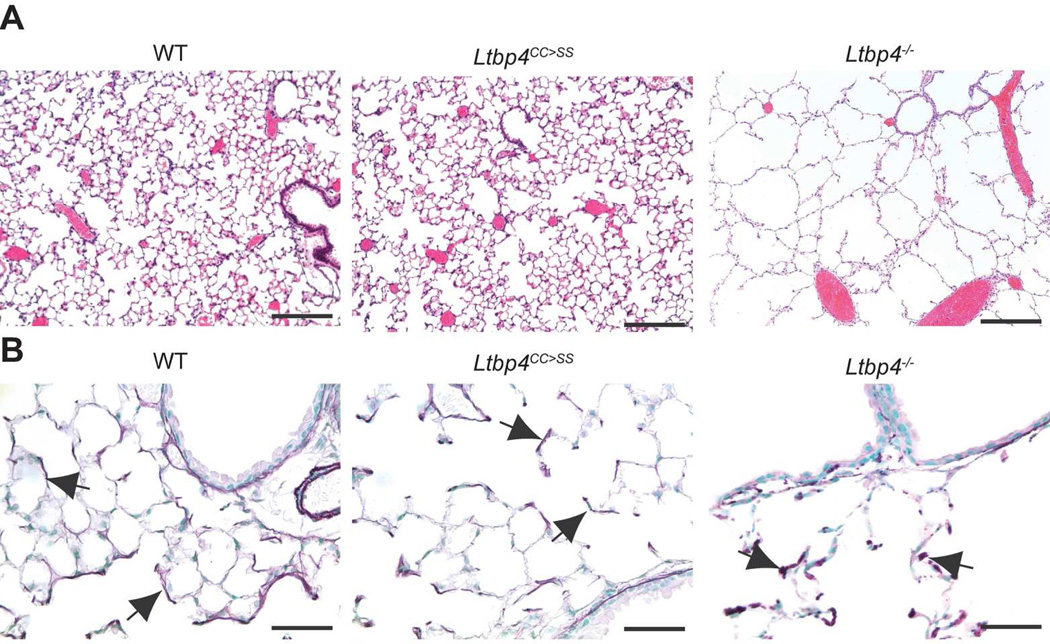
In our second approach, we asked whether binding of latent TGFβ is crucial for LTBP-4 function in lung development because LTBP-4 was reported to bind only TGFβ1, yet only a decrease in TGFβ2 modifies Ltbp4S−/− lung septation. To address this question, we generated Ltbp4CC>SS mice in which the Ltbp4 gene was mutated to produce a form of LTBP-4 where the two cysteines (Cys 1235 and 1260) in the 8-Cys 3 domain that bind to latent TGFβ were substituted by serines (Supplemental Figure 1). Previously, we showed that inhibition of the covalent association of TGFβ1 with LTBPs by mutation of the Tgfb1 gene yielded animals with a phenotype resembling TGFβ1 null mice (Shibahara et al., 2013), thus indicating the importance of TGFβ-LTBP complex formation for TGFβ1 function. Therefore, we reasoned that the specific inhibition of LTBP-4 binding to TGFβ should produce a similar TGFβ-mediated phenotype, if LTBP-4 binding to latent TGFβ1 was critical for regulating tissue levels of TGFβ. Our analysis of cells expressing the mutant Ltbp-4 showed that the mutant protein is secreted in normal amounts into the culture medium indicating that the mutation had no adverse effects on protein trafficking (Supplemental Figure 4). The migration of the mutant LTBP-4 as visualized after SDS-PAGE was equivalent to that of the WT protein that has no bound LAP (Saharinen et al., 1998). Mice with the homozygous LTBP-4 double cysteine to serine mutation were born at the expected Mendelian ratio (data not shown), had normal lung architecture (Fig. 2), and had no inflammation, which is characteristic for Tgfb1−/− mice. These results imply that LTBP-4 has a TGFβ-independent function in the lung.
Figure 2.
Lung septation and elastic fiber assembly are normal in Ltbp4CC>SS lungs. A. H&E staining of lungs from 6-week-old mice revealed normal architecture in Ltbp4CC>SS lungs, whereas there is decreased septation in Ltbp4−/− lungs. B. Elastin structure is normal in Ltbp4CC>SS lungs with a pattern in the lung parenchyma. In Ltbp4−/− lungs, the elastin appears globular. Arrows point to elastin. Bars: 200 µm in A and 50 µm in B.
Expression of Ltbps and fibulins in WT and Ltbp4−/− lungs
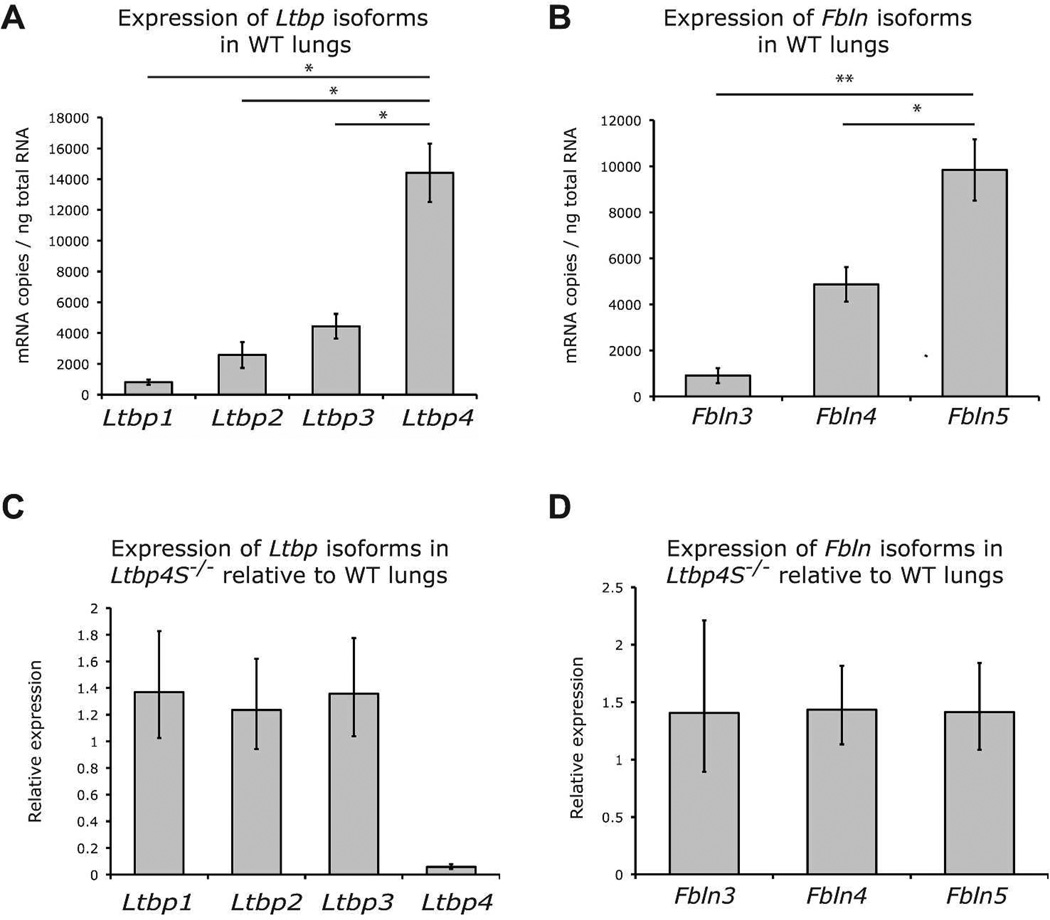
We next asked, if LTBP-4 does not bind to TGFβ, with what molecule(s) does it associate. We focused our attention on fibulin-5 because mice lacking either Ltbp-4 or fibulin-5 develop emphysema even though other isoforms of Ltbps and fibulins are expressed in the lung. These data suggest specific and non-redundant functions of Ltbp-4 and fibulin-5 in mouse lung development. Therefore, we investigated whether the expression levels of Ltbp4 and Fbln5 in the lung might explain that specificity. Quantitative RT-PCR analyses of Ltbps and Fblns from P0.5 WT lungs revealed that Ltbp4 is expressed at a significantly higher level than Ltbp1, 2 and 3 (Fig. 3A) and that Fbln5 is expressed at a significantly higher level than Fbln3 and 4 (Fig. 3B). These data suggest that Ltbp4 and Fbln5 are the major isoforms of their respective families expressed in mouse lungs. In addition, we performed semi-quantitative RT-PCR comparing Ltbp and Fbln levels in WT and Ltbp4S−/− lungs. The results showed that the expression levels of Ltbps and fibulins are similar in WT and Ltbp4S−/− lungs (Fig. 3 C and D). These data indicate that Ltbp4 and Fbln5 are the major isoforms of each of these two protein families expressed in the WT lungs and that expression of Ltbp1, 2 and 3 and Fbln3, 4 and 5 is not affected by lack of Ltbp-4 in Ltbp4S−/−lungs.
Figure 3.
Expression of Ltbps and fibulins in WT and Ltbp4−/− lungs. A and B. Quantitative RT-PCR of Ltbps and fibulins in WT mouse lungs. A. Ltbp expression levels in P0.5 lungs. Ltbp4 is expressed at higher levels than Ltbp1, 2 and 3 in WT lungs. B. Fbln5 is expressed at higher levels than Fbln3 and 4 in WT lungs. C and D Semi-quantitative RT-PCR comparison of Ltbps and Fblns expression in WT and Ltbp4S−/− P0.5 mouse lungs. C. Ltbp1, 2 and 3 expression levels are equivalent in Ltbp4S−/− and WT lungs. D. Fbln3, 4 and 5 expression levels are similar in WT and Ltbp4S−/− lungs. Expression levels in C and D were standardized to expression levels of G3pdh. Three samples were analyzed per group. * P<0.05, ** P<0.01, determined by Student t-test.
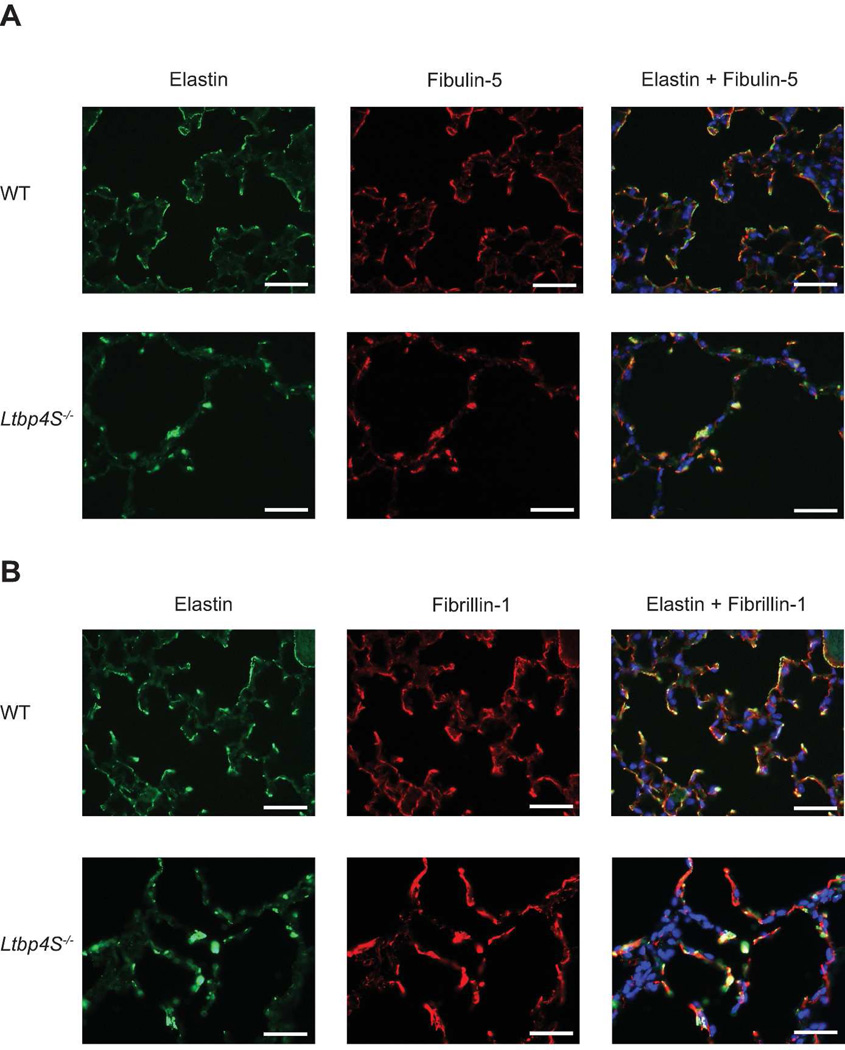
Elastin colocalizes with fibulin-5 but does not colocalize with fibrillin-1 in Ltbp4S−/− lungs
Previously published data showed that fibulin-5 interacts with both elastin and fibrillin-1 in vitro (El-Hallous et al., 2007; Freeman et al., 2005; Kobayashi et al., 2007; McLaughlin et al., 2006). We have shown that the distribution of fibulin-5 in Ltbp4S−/− lungs is abnormal and that its distribution resembles the distribution of elastin (Dabovic et al., 2009). However, the relationship of fibulin-5 with fibrillin-1 was not determined under these conditions. Therefore, we performed immunofluorescence analyses to investigate the localization of elastin, fibulin-5 and fibrillin-1 in WT and Ltbp4S−/− lungs. Staining with an anti-fibulin-5 antibody and an anti-elastin antibody revealed a fibrillar pattern and overlapping localization in WT lungs (Fig. 4A). In Ltbp4S−/−lungs, both antibodies detected a globular staining pattern indicating colocalization of fibulin-5 and elastin. Immunostaining with an anti-fibrillin-1 antibody and an anti-elastin antibody detected fibrillar staining patterns in WT lungs (Fig. 4B). There was a complete overlap of the staining patterns, indicating colocalization of the two proteins in WT mouse lungs. In Ltbp4S−/− lungs, staining with the anti-elastin antibody revealed a globular pattern, whereas staining with the fibrillin-1 antibody revealed fibrillar structures. These data suggest that in Ltbp4S−/− lungs, fibulin-5 colocalizes with elastin but colocalization with fibrillin-1 is impaired.
Figure 4.
Elastin colocalizes with fibulin-5 but does not colocalize with fibrillin-1 in Ltbp4S−/− lungs. A. Immunofluorescent staining of 10w old WT and Ltbp4S−/− lungs. Staining with an anti fibulin-5 antibody and an anti elastin antibody revealed overlapping signals in a fibrillar pattern in WT lungs. In Ltbp4S−/− lungs both antibodies detected a globular staining pattern. Thus, the signals for both fibulin-5 and elastin colocalize in Ltbp4S−/− lungs. B. Immunostaining with anti fibrillin-1 and elastin antibodies. Both anti fibrillin-1 and elastin antibodies detect fibrillar staining pattern in WT lungs with a complete overlap of the signals, indicating colocalization of the two proteins in WT mouse lungs. In Ltbp4S−/− lungs staining with the elastin antibody revealed a globular pattern, whereas staining with the fibrillin antibody revealed fibrillar structures. There was no overlap between fibrillin-1 and elastin staining in Ltbp4S−/− lungs, indicating a lack of association of the two proteins. Bars: 50 µm.
Improved lung septation in Fbln5−/−;Ltbp4S−/− lungs
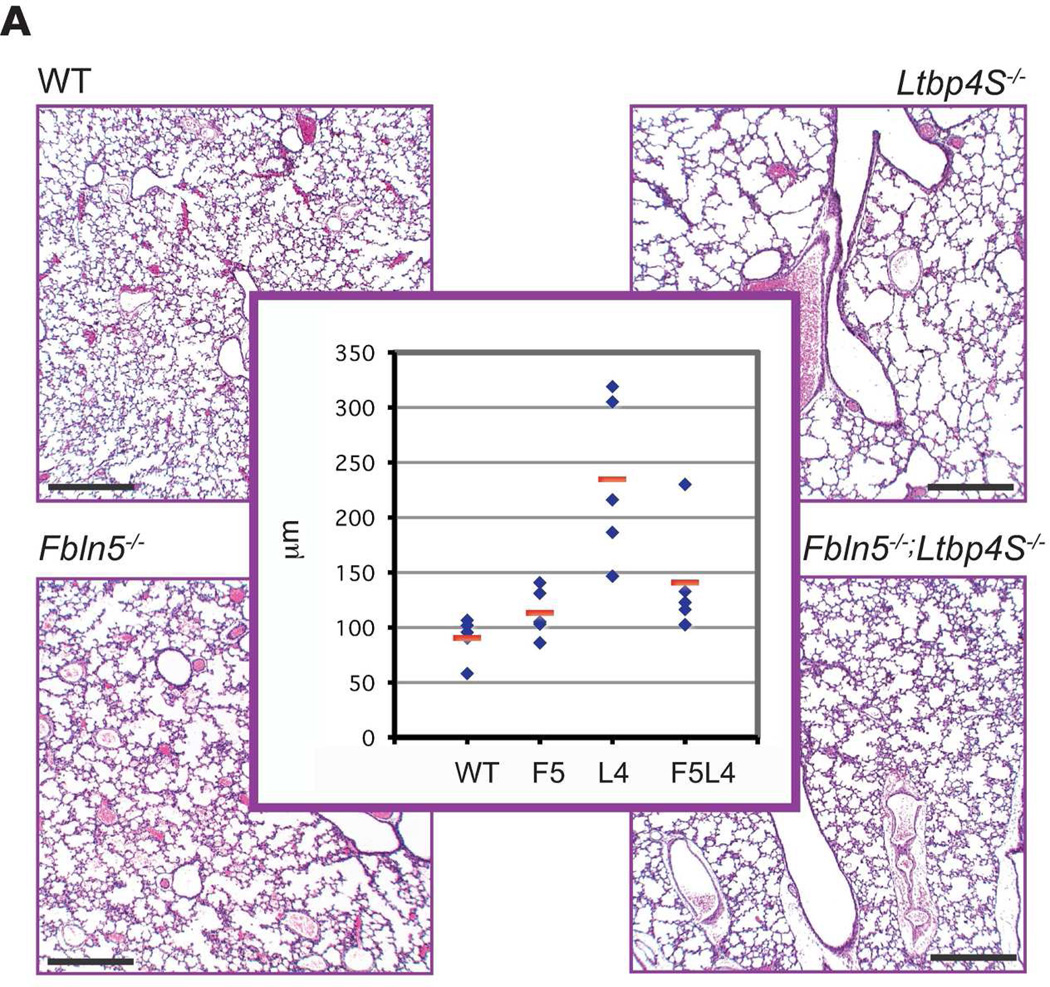
Both Ltbp4S−/− and Fbln5−/− mice have developmental emphysema. Defective lung septation in Ltbp4S−/− animals is evident at E18.5, whereas Fbln5−/− mice develop emphysema after birth (Dabovic et al., 2009; Yanagisawa et al., 2009). To investigate the relevance of the LTBP-4–fibulin-5 interaction in lung development, we generated Fbln5−/−;Ltbp4S−/− mice and analyzed lung septation. Histomorphometric analysis of P7 WT, Ltbp4S−/−, Fbln5−/− and Fbln5−/−;Ltbp4S−/− lungs revealed a decrease in terminal air-sac diameter in Fbln5−/−;Ltbp4S−/− lungs compared to Ltbp4S−/− lungs (Fig. 5A). There was no enlargement of terminal air-sacs in Fbln5−/− lungs at this time point. These data indicate improved terminal air-sac septation in Fbln5−/−;Ltbp4S−/− compared to Ltbp4S−/− lungs.
Figure 5.
Improved lung septation and elastogenesis in Fbln5−/−;Ltbp4S−/− compared to Ltbp4S−/− lungs. A. Terminal air-sac septation. Histological analysis of P7 WT, Ltbp4S−/−, Fbln5−/− and Fbln5−/−;Ltbp4S−/− lungs indicated improved terminal air-sac septation in Ltbp4S−/−;Fbln5−/− compared to Ltbp4S−/− lungs. Histomorphometric studies (center panel) illustrated a rescue of lung development in Fbln5−/−;Ltbp4S−/− lungs compared to Ltbp4S−/− lungs, as measured by average terminal air-sac diameter. Five animals of each genotype were analyzed in this study. P = 0.05. B. Elastin distribution in P0.5 lungs. In P0.5 mouse lungs elastin in the WT, Fbln5−/− and Fbln5−/−;Ltbp4S−/− alveolar walls appears fibrillar, whereas in the Ltbp4S−/− alveolar walls only globules of elastin were observed. Note the difference in thickness of the elastic fibers in WT and Fbln5−/−;Ltbp4S−/− lungs. C. Elastin distribution in 6w lungs. The differences in elastin organization between WT and mutant lungs at this time point are similar to those illustrated in B except that the thickness of the fibers is greater. Arrows point to elastin. Bars: 200 µm in A, 20 µm in B and 50 µm in C.
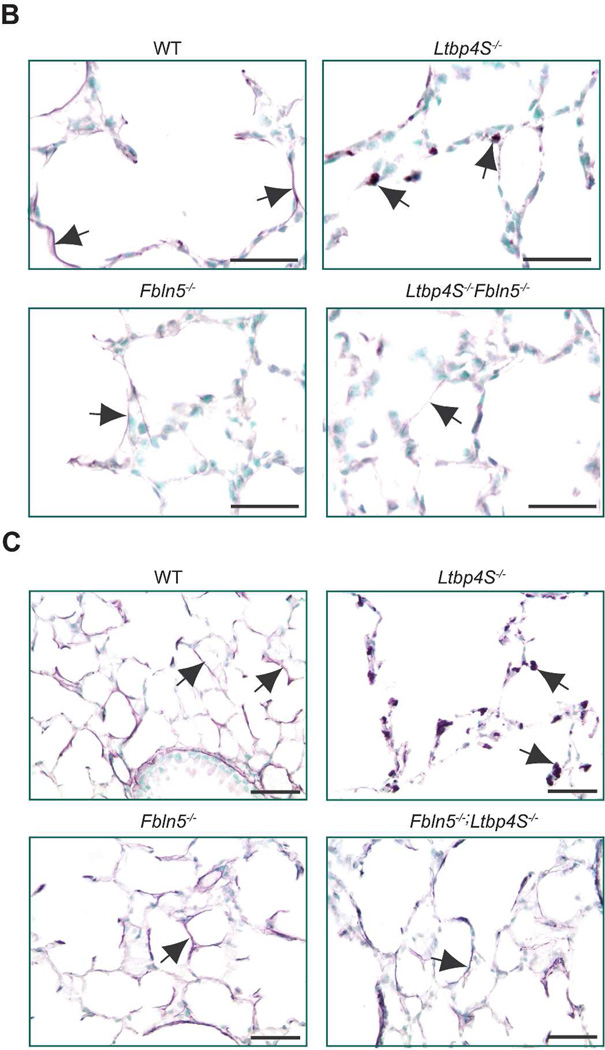
Elastogenesis in Fbln5−/−;Ltbp4S−/− lungs
Ltbp4S−/− mice show severely impaired elastogenesis, with large aggregates of elastin localized adjacent to the microfibrils with very little elastin incorporated into the microfibrillar bundles (Dabovic et al., 2009). Electron microscopy studies have revealed similar elastogenic defects in tissues of Fbln5−/− mice and humans (Choi et al., 2009; Hu et al., 2006; Yanagisawa et al., 2002). To study the fibulin-5-LTBP-4 interaction in the regulation of elastogenesis in vivo, we examined elastic fiber assembly in Fbln5+/−;Ltbp4S+/− and Fbln5−/−;Ltbp4S−/− mice using light microscopy. The Fbln5+/−;Ltbp4S+/− mice showed no abnormalities in elastic fiber assembly in the lungs (data not shown). In the Ltbp4S−/− lungs at P0.5, only globules of elastin were detected in alveolar walls. In contrast, the alveolar walls of Fbln5−/−;Ltbp4S−/− lungs showed elastin in a fibrillar network, which was also observed in WT and Fbln5−/− lungs (Fig. 5B). We noted, however, that the elastic fibers in Fbln5−/− and Fbln5−/−;Ltbp4S−/− lungs appeared thinner than the fibers in WT lungs. At 6 weeks, the difference in elastin organization between WT and mutant lungs remained similar to that observed at birth (Fig. 5C). Our data indicate that the defect in elastic fiber assembly in Fbln5−/−;Ltbp4S−/− does not resemble the defect observed in Ltbp4S−/− lungs.
EM studies of elastin of Fbln5−/−;Ltbp4S−/− lungs
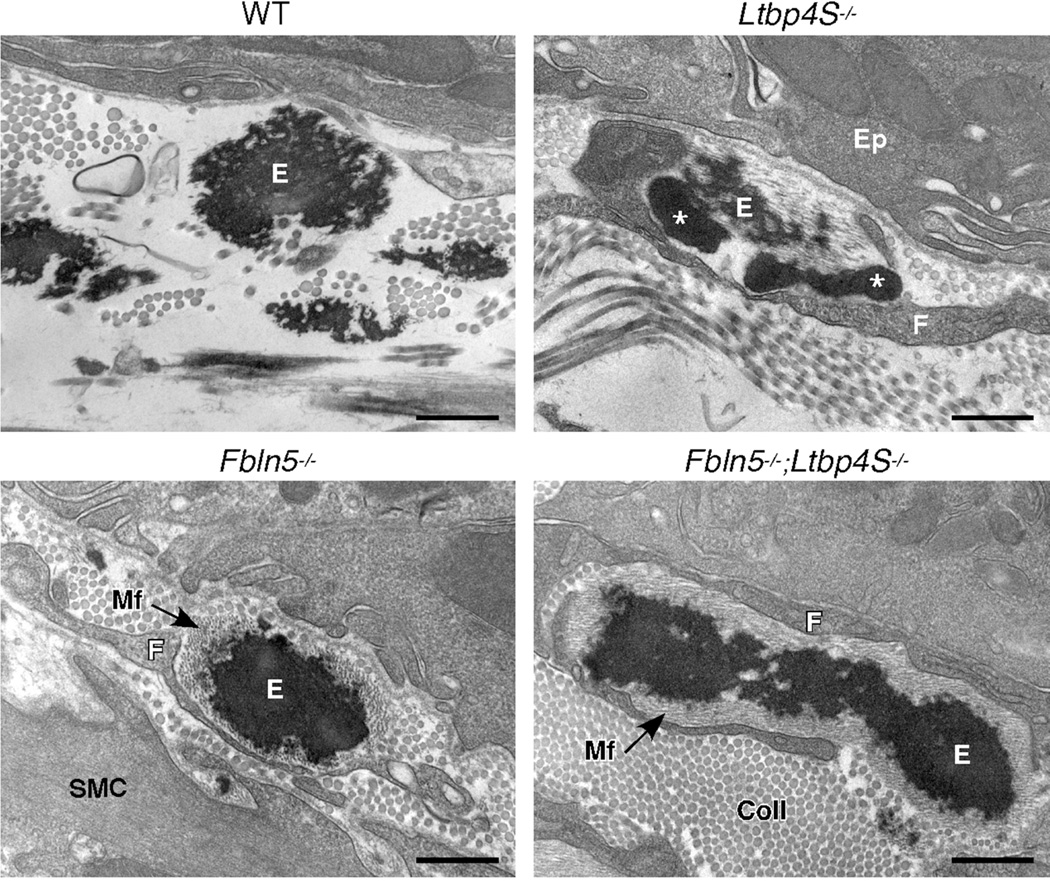
In previous work, we have found that the investigation of elastic fiber ultrastructure at the EM level to be very useful with respect to the study of elastic fiber assembly (Dabovic et al., 2011; Dabovic et al., 2009). The elastic fibers in the subepithelial matrix of lung airways are especially informative as they clearly show the degree of the incorporation of elastin into the microfibril bundles. As seen in figure 6, the microfibrils and elastin are completely integrated in the lungs of normal (WT) mice. In contrast, and consistent with previous findings (Dabovic et al., 2009), elastic fibers in the lungs of Ltbp4S−/− show the interesting feature of large globules of elastin that are associated with, but devoid of microfibrils. Although we have studied the appearance of the elastic laminae in the aorta and elastic fibers in the dermis of Fbln5−/− mice (Choi et al., 2009; Yanagisawa et al., 2002), this is the first report of the assembly of elastic fibers in the lung of this mouse model. Here, we see little incorporation of elastin into the microfibrils and instead, the fibers show a solid core of elastin surrounded by a peripheral mantle of microfibrils. Interestingly, the elastic fibers in the airways of the Fbln5−/−;Ltbp4S−/− show the same phenotype. Finally, two additional observations were made for the Fbln5−/− and Fbln5−/−;Ltbp4S−/−lungs; an excess of type I collagen fibers in the subepithelial matrix and an intimate association of interstitial fibroblasts with the elastic fibers. Both of these features were not seen in WT or Ltbp4S−/− lungs.
Figure 6.
Electron micrographs of WT, Ltbp4S−/−, Fbln5−/− and Fbln5−/−;Ltbp4S−/− elastic fibers in the subepithelial region of lung airways. Elastic fibers in normal lung airways (WT) show good incorporation of the elastin into the microfibril bundles, such that the microfibrils can only be seen in small holes around the periphery of the fiber. In Ltbp4S−/− airways, small, normal appearing elastic fibers (E) can be seen, however, these fibers are always closely associated with large, irregular elastin globules that are devoid of microfibrils (asterisks). In contrast, elastic fibers in the airways of Fbln5−/− mice show a solid core of elastin (E) surrounded by an abundance of microfibrils (Mf). Elastic fibers in the Fbln5−/−;Ltbp4S−/− airways showed a similar appearance. In both the Fbln5−/− and Fbln5−/−;Ltbp4S−/− airways, the subepithelial region was also filled with type I collagen fibrils (Coll). Additionally, in contrast to the normal elastic fibers, the abnormal elastic fibers in the airways of all three knockout mouse models were closely surrounded by interstitial fibroblasts (F). Ep, airway epithelial cell; SMC, smooth muscle cell. Bars = 500 µm.
TGFβ signaling is elevated in Ltbp4S−/− and Fbln5−/−;Ltbp4S−/− lungs
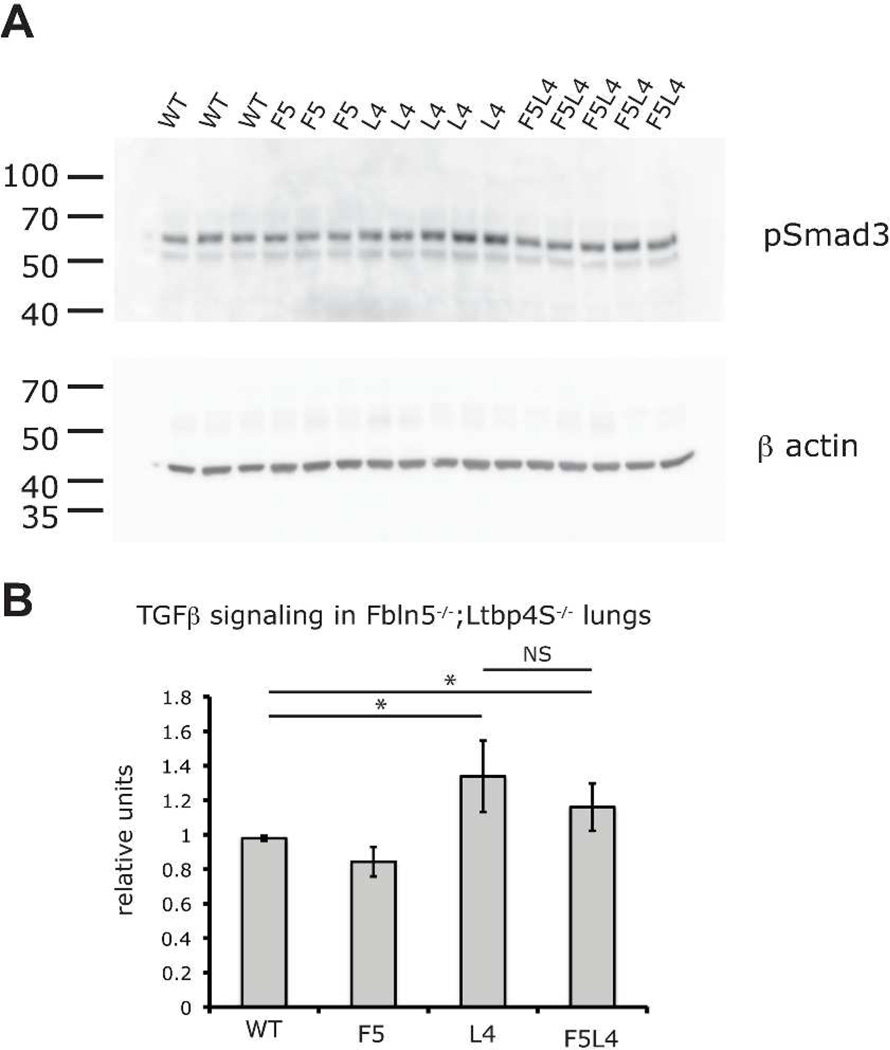
We have previously reported that TGFβ signaling is increased in Ltbp4S−/− compared to WT lungs (Dabovic et al., 2009). To measure levels of TGFβ signaling in WT, Ltbp4S−/−, Fbln5−/− and Fbln5−/−;Ltbp4S−/− lungs, we performed quantitative Western Blot analysis of P-Smad3 in the lungs from P7 mice (Fig. 7). Our data indicate modest increases in TGFβ signaling in both Ltbp4S−/− and Fbln5−/−;Ltbp4S−/− lungs. There was no statistically significant decrease in Smad3 levels in Fbln5−/−;Ltbp4S−/−compared to Ltbp4S−/− lungs. We suggest that the improved lung septation in Fbln5−/−;Ltbp4S−/− compared to Ltbp4S−/− lungs is not due to the normalization of TGFβ signaling but to the improved elastogenesis.
Figure 7.
TGFβ signaling is elevated in both Ltbp4S−/− and Fbln5−/−;Ltbp4S−/− P7 lungs. A. Western Blot analysis of P-Smad3 in lungs from WT, Ltbp4S−/− (L4), Fbln5−/− (F5) and Fbln5−/−;Ltbp4S−/− (F5L4) P7 lungs. Horizontal bars indicate position of molecular weight marker bands in kDa. B. Graphic presentation of the quantification of the data obtained in A. There is a modest increase in P-Smad3 in Ltbp4S−/− and Fbln5−/− ;Ltbp4S−/− lungs compared to WT and Fbln5−/− mice. There is no statistically significant difference between Ltbp4S−/− and Fbln5−/−;Ltbp4S−/−lungs. Abbreviations: L4, Ltbp4S−/−; F5, Fbln5−/−; F5L4, Fbln5−/−;Ltbp4S−/−.* P<0.05, determined by Student t-test; NS, statistically not significant.
Discussion
LTBP-4 and TGFβ interactions
We previously reported that Ltbp-4 deficiency causes abnormal elastogenesis and elevated TGFβ levels in the lung (Dabovic et al., 2009). Decreasing TGFβ levels in Tgfb2−/−;Ltbp4S−/− mice normalized lung septation but did not improve elastic fiber assembly (Dabovic et al., 2009). Here, we show that ablation of TGFβ1 or TGFβ3 did not rescue lung development or elastic fiber assembly in Ltbp4S−/− mice indicating that TGFβ2 is the unique TGFβ isoform that affects septation in Ltbp4S−/−lungs and that elastogenesis in lung tissue is not TGFβ dependent. Ltbp-4 binds only TGFβ1, however, TGFβ2 is increased in Ltbp4S−/− lungs. Therefore, we speculate that the increase of TGFβ2 levels in Ltbp4S−/− is not a consequence of the lack of binding of TGFβ to Ltbp4, but rather a result of perturbed ECM in Ltbp4S−/− lung tissue. This contention is supported by the fact that Ltbp4CC>SS mice, with a pair of mutations that prevent the covalent association of Ltbp-4 to latent TGFβ, yielded mice with normal lung development and elastic fiber assembly. We conclude that covalent binding of Ltbp-4 to latent TGFβ does not have an important function in lung development. It must be noted that a recent report described the binding of TGFβ1 to an extended form of LTBP-4 (Kantola et al., 2010). However, the position of binding was not described and there was no evidence that the bound TGFβ1 could be activated.
LTBP-4 and fibulin-5 interactions
Both LTBP-4 and fibulin-5 have important functions in lung biology, and in their absence, lung septation is defective and elastic fibers are abnormal. We have shown here that Ltbp4 and Fbln5 are the major isoforms expressed in mouse lungs and that expression of other isoforms is not affected by the absence of Ltbp-4 or fibulin-5. Therefore, other isoforms could not compensate for the lack of Ltbp-4 or fibulin-5 in Ltbp4S−/− lungs, assuming there is functional redundancy. We conclude that Ltbp-4 and fibulin-5 are essential for elastic fiber assembly.
Ltbp-4 and fibulin-5 interact and in vitro LTBP-4 enhances elastogenesis through its interaction with fibulin-5 (Noda et al., 2013). These observations suggested that, if the major function of LTBP-4 is to regulate the rate of incorporation of elastin-fibulin-5 complexes into the microfibril bundles, in the absence of both Ltbp-4 and fibulin-5, the lung phenotype would be similar to the phenotype observed in Fbln5−/− mice. Indeed, the elastic fiber assembly defect in Fbln5−/−;Ltbp4S−/− mice is similar to the defect observed in Fbln5−/− lungs (Fig. 5 and 6). We did not observe large elastin aggregates in Fbln5−/−;Ltbp4S−/− lungs either at P0.5 or at later time points as visualized by both light and electron microscopy (Fig. 5 and 6). However, the elastic fibers in Fbln5−/− and Fbln5−/−;Ltbp4S−/− are not normal and there are numerous microfibrils without incorporated elastin at the periphery of elastic fibers. (Fig. 6). Our data suggest that LTBP-4 is essential for fibulin-5 function in elastin-microfibril assembly.
Lung septation after birth is affected by TGFβ levels, as well as elastic fiber assembly (Hu et al., 2010; Vicencio et al., 2004). Interestingly, although Ltbp-4 deficiency results in increased TGFβ signaling (Dabovic et al., 2009) and decreasing TGFβ levels rescued lung septation in E18.5 Ltbp4S−/−;Tgfb2−/− lungs, TGFβ signaling is not normalized in Fbln5−/−;Ltbp4S−/− lungs (Fig. 7). Our apparently conflicting results, normalization of lung architecture by Tgfb2 deletion, but no change in TGFβ levels in Fbln5−/−;Ltbp4S−/− lungs where lung architecture is improved, may represent the state of TGFβ at two different times in the lung. Our earlier studies concerning Tgfb2 deletion were performed with 18.5 day old embryos, whereas our studies with Fbln5−/−;Ltbp4S−/− lungs were done at day 7 after birth. Therefore, we suggest that the improved terminal air sac septation in Fbln5−/−;Ltbp4S−/− lungs is a consequence of improved elastogenesis rather than normalized TGFβ tissue levels.
Model of elastic fiber assembly in Ltbp4S−/− and Fbln5−/−;Ltbp4S−/− lungs
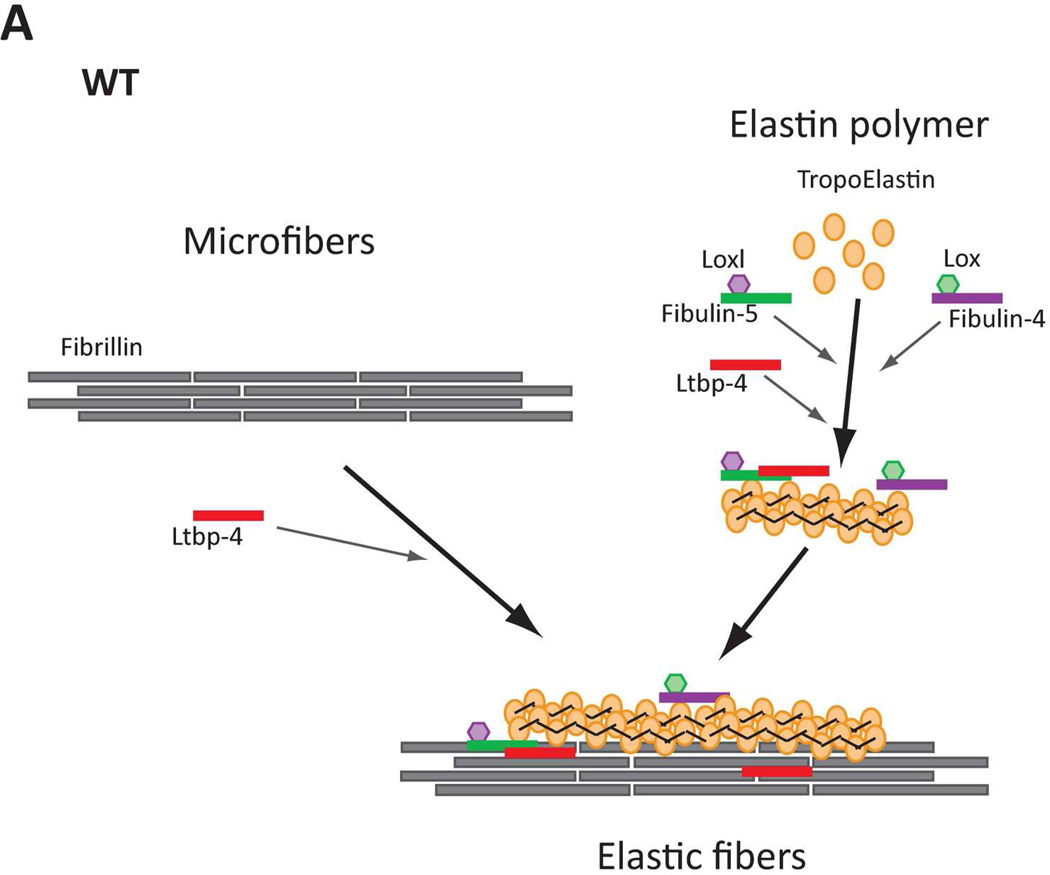
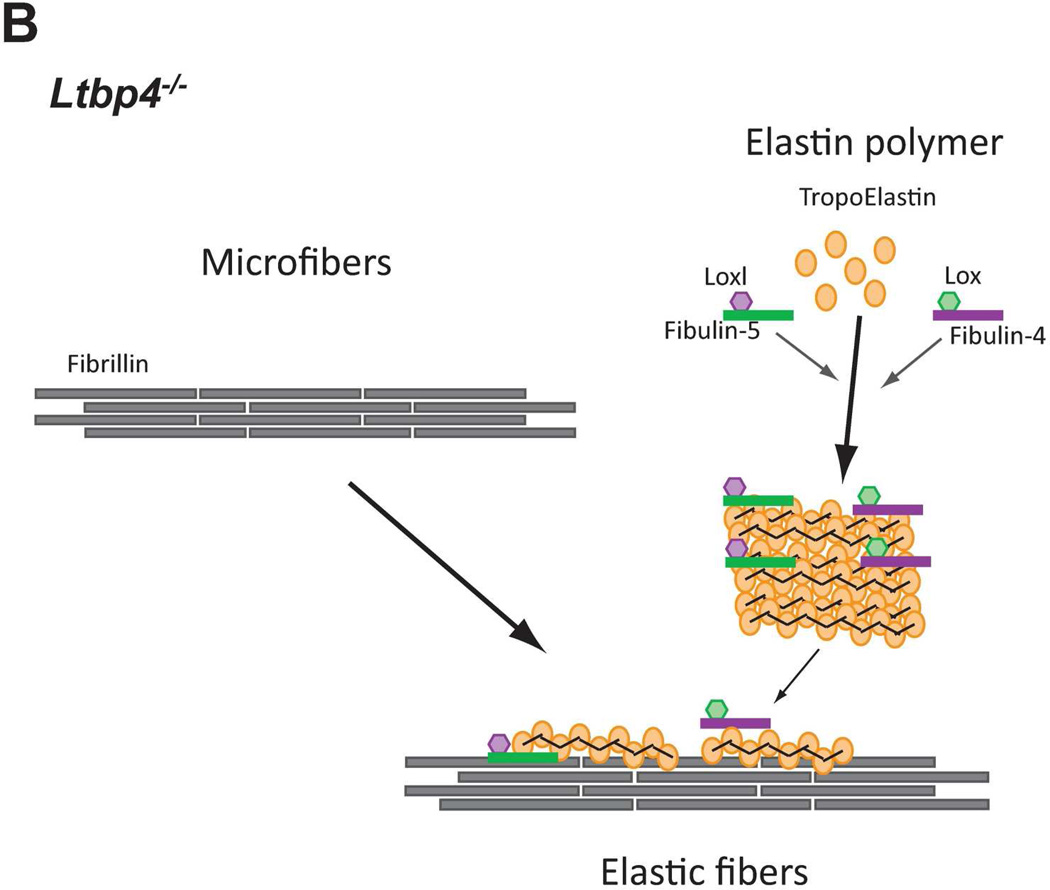
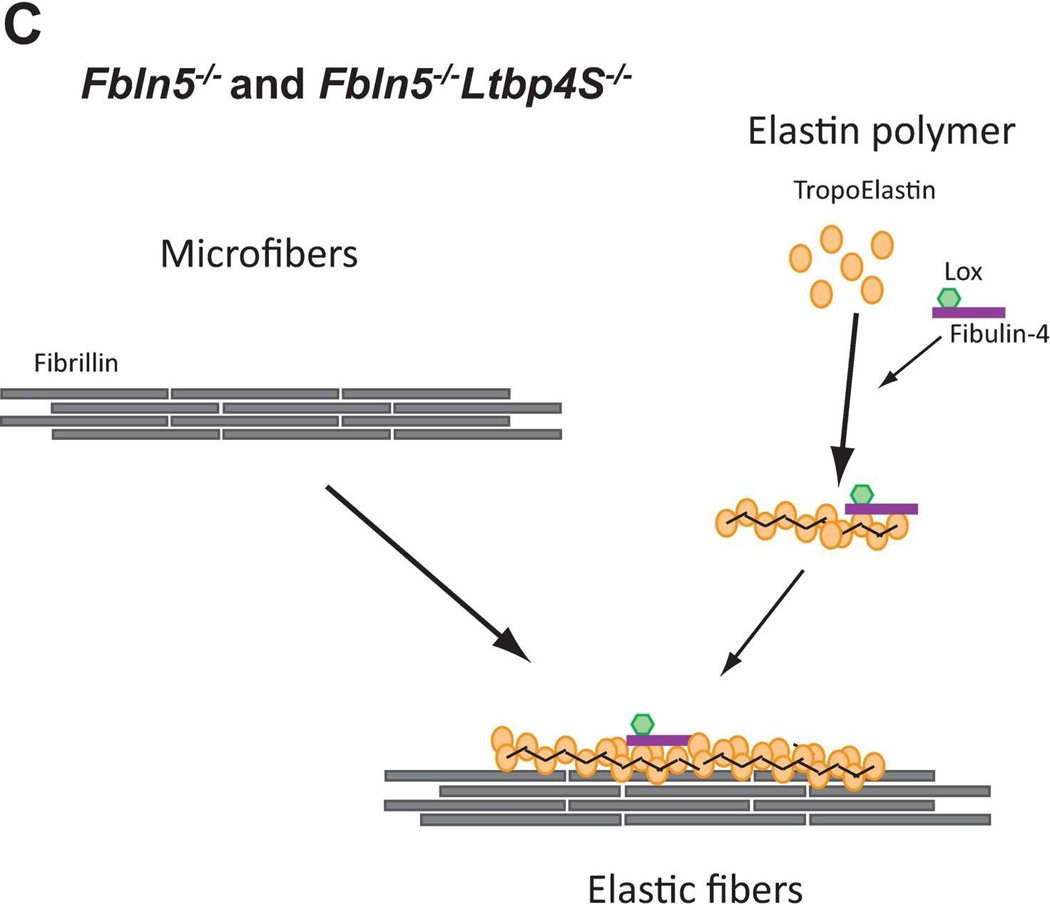
In normal (WT) lungs, elastic fiber assembly requires self-assembly of fibrillin molecules into microfibrils and secretion and micropolymerization of tropoelastin molecules (Fig. 8A). Elastin polymerization is catalyzed by lysyl oxidase (LOX) or lysyl oxidase like-1 (LOXL-1) enzymes. LOX interacts with fibullin-4, whereas LOXL-1 interacts with fibulin-5. Both fibulin-4 and fibulin-5 interact with elastin (El-Hallous et al., 2007). We propose that in the mouse lung, the interaction of elastin - fibulin-5 - LOXL-1 is rapid and rate limiting. LTBP- 4 interacts both with fibrillin and with fibulin-5 and promotes elastogenesis. In Ltbp4S−/− lungs, self-assembly of fibrillin is not affected and there is normal formation of microfibrils (Fig. 8B). However, large elastin-fibulin-5 aggregates (possibly cross-linked) form, as there is no carrier protein to chaperone the fibulin-5-elastin complexes to the microfibrils. A small fraction of elastin is integrated into microfibril bundles presumably through direct interaction of fibulin-4 and/or fibulin-5 with fibrillin-1 and −2. In Fbln5−/−;Ltbp4S−/− lungs, the large elastin aggregates do not form, as there is no fibulin-5-elastin interaction (Fig. 8C) and elastic fibers resemble those observed in Fbln5−/− lungs. In both Fbln5−/−;Ltbp4S−/− and Fbln5−/− lungs elastogenesis occurs through an alternative pathway presumably involving integration of fibulin-4-elastin complexes into the microfibrillar scaffold rather than the more efficient LTBP-4-fibulin-5 pathway. Fibulin-4 can interact with fibrillin-1 and therefore might itself promote elastin incorporation into the microfibrils (Sheehan and Hrapchak, 1980). However, our results suggest that this process is less efficient than Ltbp-4-guided incorporation of elastin-fibulin-5 complexes into microfibrils.
Figure 8.
Models of elastic fiber assembly in Ltbp4S−/− and Fbln5−/−;Ltbp4S−/− lungs. A. Normal Lungs. In normal (WT) lungs elastic fiber assembly requires self-assembly of fibrillin molecules into microfibers as well as secretion and micropolymerization of tropoelastin molecules. Elastin polymerization is catalyzed by lysyl oxidase (LOX) or lysyl oxidase like-1 (LOXL-1) enzymes. LOX interacts with fibullin-4, whereas LOXL-1 interacts with fibulin-5. LTBP-4 interacts with both fibrillin and fibulin-5 and promotes elastogenesis. B. Ltbp4S−/− Lungs. In Ltbp4S−/− lungs self-assembly of fibrillin is not affected and there is normal formation of microfibers. However, large elastin aggregates form, as there is no carrier protein to target fibulin-5-elastin complexes to or along microfibers. Only a small fraction of elastin is integrated into the microfibers, presumably through the direct interaction of fibulin-4 and fibulin-5 with fibrillin-1. C. Fbln5−/−;Ltbp4S−/− and Fbln5−/− lungs. In Fbln5−/−;Ltbp4S−/− as in Fbln5−/− lungs, the large elastin aggregates do not form, because there is no initial accumulation of fibulin-5-elastin complexes. Elastogenesis occur through an alternative pathway presumably involving the integration of fibulin-4-elastin complexes into microfibrillar scaffolds rather than the integration of LTBP-4-fibulin-5-elastin complexes into microfibers.
Supplementary Material
Acknowledgement
The authors thank Lea-Jeanne Ringuette for technical assistance for the EM studies.
Contract grant sponsor: NIH; Contract grant numbers: CA034282 AR49698 to DBR
Contract grant sponsor: NSERC RGPIN 355710 grant to ECD
References
- Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165:723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Bergdahl A, Zheng Q, Starcher B, Yanagisawa H, Davis EC. Analysis of dermal elastic fibers in the absence of fibulin-5 reveals potential roles for fibulin-5 in elastic fiber assembly. Matrix Biol. 2009;28:211–220. doi: 10.1016/j.matbio.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colarossi C, Chen Y, Obata H, Jurukovski V, Fontana L, Dabovic B, Rifkin DB. Lung alveolar septation defects in Ltbp-3-null mice. Am J Pathol. 2005;167:419–428. doi: 10.1016/S0002-9440(10)62986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabovic B, Chen Y, Choi J, Davis EC, Sakai LY, Todorovic V, Vassallo M, Zilberberg L, Singh A, Rifkin DB. Control of lung development by latent TGF-beta binding proteins. J Cell Physiol. 2011;226:1499–1509. doi: 10.1002/jcp.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabovic B, Chen Y, Choi J, Vassallo M, Dietz HC, Ramirez F, von Melchner H, Davis EC, Rifkin DB. Dual functions for LTBP in lung development: LTBP-4 independently modulates elastogenesis and TGF-beta activity. J Cell Physiol. 2009;219:14–22. doi: 10.1002/jcp.21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabovic B, Chen Y, Colarossi C, Obata H, Zambuto L, Perle MA, Rifkin DB. Bone abnormalities in latent TGF-[beta] binding protein (Ltbp)-3-null mice indicate a role for Ltbp-3 in modulating TGF-[beta] bioavailability. J Cell Biol. 2002;156:227–232. doi: 10.1083/jcb.200111080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EC. Smooth muscle cell to elastic lamina connections in developing mouse aorta. Role in aortic medial organization. Lab Invest. 1993;68:89–99. [PubMed] [Google Scholar]
- El-Hallous E, Sasaki T, Hubmacher D, Getie M, Tiedemann K, Brinckmann J, Batge B, Davis EC, Reinhardt DP. Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adaptor function to tropoelastin. J Biol Chem. 2007;282:8935–8946. doi: 10.1074/jbc.M608204200. [DOI] [PubMed] [Google Scholar]
- Freeman LJ, Lomas A, Hodson N, Sherratt MJ, Mellody KT, Weiss AS, Shuttleworth A, Kielty CM. Fibulin-5 interacts with fibrillin-1 molecules and microfibrils. The Biochemical journal. 2005;388:1–5. doi: 10.1042/BJ20050368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes PE, Beavis RC, Mazzieri R, Shen B, Rifkin DB. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-beta binding protein-1 that mediates bonding to the latent transforming growth factor-beta 1. J Biol Chem. 1996;271:29891–29896. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- Hu Q, Loeys BL, Coucke PJ, De Paepe A, Mecham RP, Choi J, Davis EC, Urban Z. Fibulin-5 mutations: mechanisms of impaired elastic fiber formation in recessive cutis laxa. Human molecular genetics. 2006;15:3379–3386. doi: 10.1093/hmg/ddl414. [DOI] [PubMed] [Google Scholar]
- Hu Q, Shifren A, Sens C, Choi J, Szabo Z, Starcher BC, Knutsen RH, Shipley JM, Davis EC, Mecham RP, Urban Z. Mechanisms of emphysema in autosomal dominant cutis laxa. Matrix Biol. 2010;29:621–628. doi: 10.1016/j.matbio.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Z, Ono RN, Ushiro S, Keene DR, Chen Y, Mazzieri R, Charbonneau NL, Reinhardt DP, Rifkin DB, Sakai LY. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J Biol Chem. 2003;278:2750–2757. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- Kantola AK, Ryynanen MJ, Lhota F, Keski-Oja J, Koli K. Independent regulation of short and long forms of latent TGF-beta binding protein (LTBP)-4 in cultured fibroblasts and human tissues. J Cell Physiol. 2010;223:727–736. doi: 10.1002/jcp.22082. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Kostka G, Garbe JH, Keene DR, Bachinger HP, Hanisch FG, Markova D, Tsuda T, Timpl R, Chu ML, Sasaki T. A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J Biol Chem. 2007;282:11805–11816. doi: 10.1074/jbc.M611029200. [DOI] [PubMed] [Google Scholar]
- Koli K, Saharinen J, Hyytiainen M, Penttinen C, Keski-Oja J. Latency, activation, and binding proteins of TGF-beta. Microsc Res Tech. 2001;52:354–362. doi: 10.1002/1097-0029(20010215)52:4<354::AID-JEMT1020>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- McLaughlin PJ, Bakall B, Choi J, Liu Z, Sasaki T, Davis EC, Marmorstein AD, Marmorstein LY. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Human molecular genetics. 2007;16:3059–3070. doi: 10.1093/hmg/ddm264. [DOI] [PubMed] [Google Scholar]
- McLaughlin PJ, Chen Q, Horiguchi M, Starcher BC, Stanton JB, Broekelmann TJ, Marmorstein AD, McKay B, Mecham R, Nakamura T, Marmorstein LY. Targeted disruption of fibulin-4 abolishes elastogenesis and causes perinatal lethality in mice. Mol Cell Biol. 2006;26:1700–1709. doi: 10.1128/MCB.26.5.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitzner W, Fallica J, Bishai J. Anisotropic nature of mouse lung parenchyma. Annals of biomedical engineering. 2008;36:2111–2120. doi: 10.1007/s10439-008-9538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Lozano PR, Ikeda Y, Iwanaga Y, Hinek A, Minamisawa S, Cheng CF, Kobuke K, Dalton N, Takada Y, Tashiro K, Ross J, Jr, Honjo T, Chien KR. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature. 2002;415:171–175. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- Noda K, Dabovic B, Takagi K, Inoue T, Horiguchi M, Hirai M, Fujikawa Y, Akama TO, Kusumoto K, Zilberberg L, Sakai LY, Koli K, Naitoh M, von Melchner H, Suzuki S, Rifkin DB, Nakamura T. Latent TGF-beta binding protein 4 promotes elastic fiber assembly by interacting with fibulin-5. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2852–2857. doi: 10.1073/pnas.1215779110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen J, Hyytiainen M, Taipale J, Keski-Oja J. Latent transforming growth factor-beta binding proteins (LTBPs) structural extracellular matrix proteins for targeting TGF-beta action. Cytokine Growth Factor Rev. 1999;10:99–117. doi: 10.1016/s1359-6101(99)00010-6. [DOI] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-beta with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15:245–253. [PMC free article] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Monni O, Keski-Oja J. Identification and characterization of a new latent transforming growth factor-beta-binding protein, LTBP-4. J Biol Chem. 1998;273:18459–18469. doi: 10.1074/jbc.273.29.18459. [DOI] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Theory and practice of histotechnology. Mosby: St. Louis; 1980. [Google Scholar]
- Shibahara K, Ota M, Horiguchi M, Yoshinaga K, Melamed J, Rifkin DB. Production of gastrointestinal tumors in mice by modulating latent TGF-beta1 activation. Cancer research. 2013;73:459–468. doi: 10.1158/0008-5472.CAN-12-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner-Kock A, Thorey IS, Koli K, Wempe F, Otte J, Bangsow T, Kuhlmeier K, Kirchner T, Jin S, Keski-Oja J, von Melchner H. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev. 2002;16:2264–2273. doi: 10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Saharinen J, Hedman K, Keski-Oja J. Latent transforming growth factor-β1 and its binding protein are components of extracellular matrix microfibrils. J Histochem Cytochem. 1996;44:875–889. doi: 10.1177/44.8.8756760. [DOI] [PubMed] [Google Scholar]
- Todorovic V, Frendewey D, Gutstein DE, Chen Y, Freyer L, Finnegan E, Liu F, Murphy A, Valenzuela D, Yancopoulos G, Rifkin DB. Long form of latent TGF-beta binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development. 2007;134:3723–3732. doi: 10.1242/dev.008599. [DOI] [PubMed] [Google Scholar]
- Todorovic V, Jurukovski V, Chen Y, Fontana L, Dabovic B, Rifkin DB. Latent TGF-beta binding proteins. Int J Biochem Cell Biol. 2005;37:38–41. doi: 10.1016/j.biocel.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Todorovic V, Rifkin DB. LTBPs, more than just an escort service. J Cell Biochem. 2012;113:410–418. doi: 10.1002/jcb.23385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban Z. The complexity of elastic fiber biogenesis: the paradigm of cutis laxa. The Journal of investigative dermatology. 2012;132:E12–E14. doi: 10.1038/skinbio.2012.4. [DOI] [PubMed] [Google Scholar]
- Urban Z, Hucthagowder V, Schurmann N, Todorovic V, Zilberberg L, Choi J, Sens C, Brown CW, Clark RD, Holland KE, Marble M, Sakai LY, Dabovic B, Rifkin DB, Davis EC. Mutations in LTBP4 cause a syndrome of impaired pulmonary, gastrointestinal, genitourinary, musculoskeletal, and dermal development. Am J Hum Genet. 2009;85:593–605. doi: 10.1016/j.ajhg.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicencio AG, Lee CG, Cho SJ, Eickelberg O, Chuu Y, Haddad GG, Elias JA. Conditional overexpression of bioactive transforming growth factor-beta1 in neonatal mouse lung: a new model for bronchopulmonary dysplasia? American journal of respiratory cell and molecular biology. 2004;31:650–656. doi: 10.1165/rcmb.2004-0092OC. [DOI] [PubMed] [Google Scholar]
- Wagenseil JE, Mecham RP. New insights into elastic fiber assembly. Birth Defects Res C Embryo Today. 2007;81:229–240. doi: 10.1002/bdrc.20111. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Davis EC. Unraveling the mechanism of elastic fiber assembly: The roles of short fibulins. Int J Biochem Cell Biol. 2010;42:1084–1093. doi: 10.1016/j.biocel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Davis EC, Starcher BC, Ouchi T, Yanagisawa M, Richardson JA, Olson EN. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature. 2002;415:168–171. doi: 10.1038/415168a. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Schluterman MK, Brekken RA. Fibulin-5, an integrin-binding matricellular protein: its function in development and disease. Journal of cell communication and signaling. 2009;3:337–347. doi: 10.1007/s12079-009-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberberg L, Todorovic V, Dabovic B, Horiguchi M, Courousse T, Sakai LY, Rifkin DB. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J Cell Physiol. 2012;227:3828–3836. doi: 10.1002/jcp.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.