Abstract
Background
Xylose isomerase (XI) catalyzes the conversion of xylose to xylulose, which is the key step for anaerobic ethanolic fermentation of xylose. Very few bacterial XIs can function actively in Saccharomyces cerevisiae. Here, we illustrate a group of XIs that would function for xylose fermentation in S. cerevisiae through phylogenetic analysis, recombinant yeast strain construction, and xylose fermentation.
Results
Phylogenetic analysis of deposited XI sequences showed that XI evolutionary relationship was highly consistent with the bacterial taxonomic orders and quite a few functional XIs in S. cerevisiae were clustered with XIs from mammal gut Bacteroidetes group. An XI from Bacteroides valgutus in this cluster was actively expressed in S. cerevisiae with an activity comparable to the fungal XI from Piromyces sp. Two XI genes were isolated from the environmental metagenome and they were clustered with XIs from environmental Bacteroidetes group. These two XIs could not be expressed in yeast with activity. With the XI from B. valgutus expressed in S. cerevisiae, background yeast strains were optimized by pentose metabolizing pathway enhancement and adaptive evolution in xylose medium. Afterwards, more XIs from the mammal gut Bacteroidetes group, including those from B. vulgatus, Tannerella sp. 6_1_58FAA_CT1, Paraprevotella xylaniphila and Alistipes sp. HGB5, were individually transformed into S. cerevisiae. The known functional XI from Orpinomyces sp. ukk1, a mammal gut fungus, was used as the control. All the resulting recombinant yeast strains were able to ferment xylose. The respiration-deficient strains harboring B. vulgatus and Alistipes sp. HGB5 XI genes respectively obtained specific xylose consumption rate of 0.662 and 0.704 g xylose gcdw−1 h−1, and ethanol specific productivity of 0.277 and 0.283 g ethanol gcdw−1 h−1, much comparable to those obtained by the control strain carrying Orpinomyces sp. ukk1 XI gene.
Conclusions
This study demonstrated that XIs clustered in the mammal gut Bacteroidetes group were able to be expressed functionally in S. cerevisiae and background strain anaerobic adaptive evolution in xylose medium is essential for the screening of functional XIs. The methods outlined in this paper are instructive for the identification of novel XIs that are functional in S. cerevisiae.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0253-1) contains supplementary material, which is available to authorized users.
Keywords: Xylose isomerase, Bacteroidetes, Alistipes, Bacteroides, Mammal gut, Microbe, Adaptive evolution, Saccharomyces cerevisiae, Xylose fermentation, Cellulosic ethanol
Background
Bio-ethanol, currently blended in gasoline, is primarily produced from microbial fermentation of food-based sugars, such as corn starch and cane sugar. Bioethanol derived from food-based substrates is unsustainable as it competes with our food chain and may cause human starvation. On the other hand, ethanol production from the abundant lignocellulosic biomass, such as agricultural wastes and forest waste, promises a sustainable green fuel. However, effective conversion of xylose, the second abundant sugar after glucose in lignocellulosic biomass, is necessary to make this process more economically feasible and efficient [1]. Saccharomyces cerevisiae is an attractive host to produce biofuels and industrial chemicals from lignocellulosic biomass due to its robustness and fast fermentation rate [2]. However, it cannot naturally utilize xylose [3]. Xylose isomerase (XI) naturally catalyzes the bacterial conversion of xylose to xylulose, which is further metabolized to ethanol through central metabolic pathways. One of the essential metabolic engineering modifications for effective xylose fermentation by S. cerevisiae is expressing an XI with high activity. However, the species boundary makes most of eubacterial xylose isomerases fail to be functionally expressed in yeast.
Besides XI pathway, xylose reductase (XR) and xylitol dehydrogenase (XDH) pathway from Pichia stipitis (Scheffersomyces stipitis) was also engineered in S. cerevisiae for the construction of xylose-fermenting yeast [4–7]. In XI pathway, xylose is firstly isomerized into xylulose through one-step catalysis by XI and xylulose was then phosphorylated into xylulose 5-phosphate by xylulokinase. The latter is the intermediate metabolite of pentose phosphate pathway and can be further converted into ethanol through glycolysis [6]. In XR and XDH pathway, xylose is firstly reduced into xylitol by XR, and xylitol is then oxidized into xylulose by XDH [1]. This pathway has been successfully constructed in S. cerevisiae; however, xylitol accumulation is a problem. This is caused by the uncoupled cofactor consumption of NADPH-preferred XR and NAD+-preferred XDH [5]. Although anaerobic xylose-fermenting yeast was successfully constructed by the overexpression of NADH-preferred XR or NADP+-preferred XDH generated through protein engineering [1, 8], xylitol accumulation was still a problem and the specific xylose consumption was not as efficient as XI expressed strains [9–11]. Therefore, xylose isomerase pathway is still the method of choice for the construction of efficient xylose-fermenting yeast.
XI commonly exists in bacteria. However, because of unknown molecular barrier between yeast and bacteria, most Proteobacteria XI could not be expressed in S. cerevisiae functionally. It was found that XIs from Escherichia coli, Bacillus subtilis, and Streptomyces rubiginosus were expressed as the insoluble form in S. cerevisiae [12–14]. XI from Thermus thermophilus was firstly reported to be actively expressed in S. cerevisiae. However, its optimal reaction temperature was 85 °C, and at 30 °C it only demonstrated about 4 % of the maximum activity, which was insufficient for fast xylose consumption [4]. The first reported XI that guaranteed effective xylose fermentation in S. cerevisiae at 30 °C was from the anaerobic fungus Piromyces sp. E2, and the recombinants generated from XI gene transformation and adaptive evolution consumed xylose at the specific rate of 0.18 to 1.87 g−1 xylose g−1 biomass h−1 [6, 7, 15]. XIs from another anaerobic fungus Orpinomyces sp. ukk1 [16], several Bacteroid bacteria [17, 18], Clostridium phytofermentans [19], Ruminococcus flavefaciens [20] and Prevotella ruminicola [21] have also been successfully used to construct xylose-fermenting yeast. Information on the active XIs in yeast is scattered, and it would be instructive to rational selection of XIs to efficiently construct xylose-fermenting yeast if the evolutionary relationship among the active XIs can be elucidated.
In this study, phylogenetic analysis of the active XIs was conducted and it was found that majority of the reported active XIs were clustered in mammal gut Bacteroidetes group. This suggests that other XIs clustered in this group might also be able to function in S. cerevisiae. In order to confirm this, four different deposited XI genes from mammal gut Bacteroidetes group were tested for their functional expression in yeast. In addition, two XIs were cloned from snail manure metagenomic DNA by degenerate PCR and thermal asymmetric interlaced (TAIL) PCR, and they were used as the reference. The two cloned XIs were clustered with XIs from environmental Bacteroidetes bacteria. As expected, only XIs from the mammal gut Bacteroidetes group were expressed in S. cerevisiae with activity. Through combined optimization of xylose metabolic pathways and adaptive evolution in xylose medium, recombinant yeast strains containing mammal gut Bacteroidetes XIs were generated and all of them showed efficient xylose fermentation.
Results and discussion
Sequence analysis and cloning of xylose isomerase (XI) from environmental mega-genome
With the fast development of genome-sequencing technique, increasing numbers of XI sequences were deposited in public nucleotide/protein databases, such as Genbank, ENA and Uniprot. For the exploration of XIs that are potentially functional in yeast, XI sequences were retrieved and their phylogenetic relationship was analyzed. XIs were grouped into two classes (class I and class II) [22]. Class I XIs are distinguished from class II XIs by having a shorter N-terminal, and T. thermophilus XI, the first XI actively expressed in S. cerevisiae, belongs to class I. However, another class I enzyme from S. rubiginosus was misfolded when expressed in yeast. Class II represents a large number of XI families, and the relationship of XIs in phylogenetic tree is highly consistent with the taxonomic orders (Fig. 1). However, only a few of them are active in yeast. Currently, most of the active XIs in yeast belong to Class II. The phylogenetic tree shows that XI expressed actively at 30 °C in yeast are in three branches, Bacteroidetes, C. phytofermentans-R. flaverfaciens and higher eukaryotes (Ciona and Arabidopsis) (Fig. 1), and majority of the active XIs are from gut bacteria (or fungi) of mammals. This suggests that the active expression of microbial xylose isomerase in S. cerevisiae might be related to the diverse evolutionary events of mammalian gut commensals where xylan-degrading microorganisms are abundant [21].
Fig. 1.
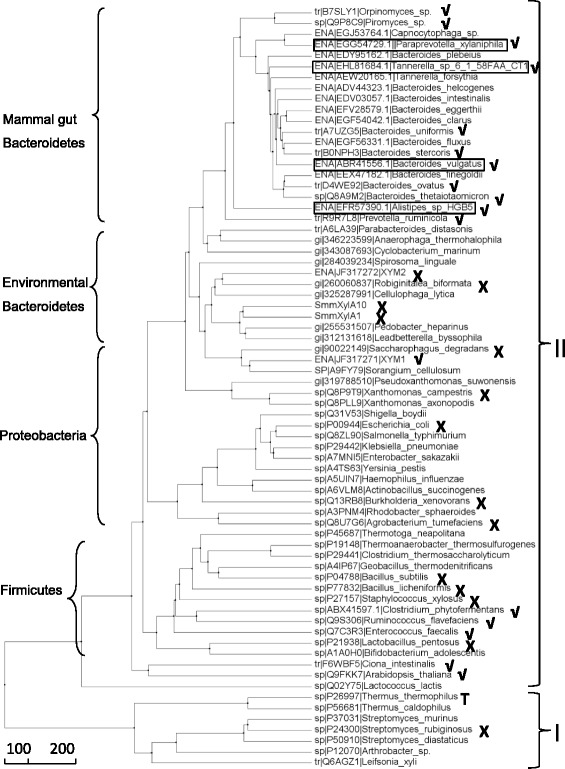
Phylogenetic tree analysis of amino acid sequences of Class I & II xylose isomerases: √, active in Saccharomyces cerevisiae at 30 °C; ×, not active at 30 °C for xylose fermentation when expressed in Saccharomyces cerevisiae [6, 12–14, 17–21, 23, 29–31]. The sequences were aligned by MAFFT algorithm with default setting and the tree was calculated by average distance using BLOSUM62 through software Jalview (http://www.jalview.org)
Among the active XIs in yeast, XIs originated from the phylum Bacteroidetes represent a significant number. XIs from gut Bacteroiedetes of mammals are categorized into a group different from environmental Bacteroidetes XIs, and XIs from fungi, Piromyces and Orpinomyces, are also clustered in this group. Therefore, it is very likely that other XIs clustered in this group might also be able to function in S. cerevisiae. Based on this hypothesis, in this study, the deposited gene sequence encoding XIs from Bacteroides vulgatus, Alistipes sp. HGB5, Paraprevotella xylaniphila, and Tannerella sp. 6_1_58FAA_CT1 from the mammal gut Bacteroidetes group were selected and tested for the construction of xylose-fermenting yeast.
In order to isolate novel functional XI genes in yeast, XI sequences from metagenomic DNA of snail manure and forest soil were directly amplified through degenerated PCR and TAIL-PCR. Interestingly, degenerated PCR products of snail manure sample and forest soil sample shared the same sequences, and the deducted amino acid sequences were related to environmental Bacteroidetes group (Fig. 1). Eleven unique sequences in total were amplified, but all were clustered in environmental Bacteroidetes group (data not shown). The upstream and downstream sequences of two degenerated products (SmmXylA1 and SmmXylA10) were amplified successfully through TAIL-PCR. Based on the sequencing results, the full-length genes of SmmXylA1 and SmmXylA10 (SEQ. A1 and SEQ. A2) were amplified directly from metagenomic DNA of snail manure sample and were also tested for their functional expression in yeast.
Expression of the xylose isomerase from two clades of Bacteroidetes xylose isomerases
B. vulgatus is a xylan-degrading bacterium habituating in human gastrointestinal tract, and its genome sequence has been released [23]. B. vulgatus XI shares 81 % identical amino acid sequence and 38 % identical nucleic sequence with Piromyces sp. E2 XI. The full length gene was directly synthesized according to the deposited sequence under EMBL-ENA accession No. ABR41556.1 without codon usage optimization so that the potential of the original XI in its functional expression in S. cerevisiae can be evaluated. B. vulgatus XI gene was fused with TDH3 promoter and PGK1 terminator. In addition, B. vulgatus XI gene, Piromyces sp. E2 XI (EMBL:AJ249909.1) gene and the two XI genes cloned from metagenomic DNA were cloned between TEF1 promoter and CYC1 terminator. Notably, only B. vulgatus and Piromyces sp. E2 XIs were expressed with activities, and the strains harboring the metagenomic XI genes and the reference strain carrying the empty vector almost showed no activity (Fig. 2).
Fig. 2.
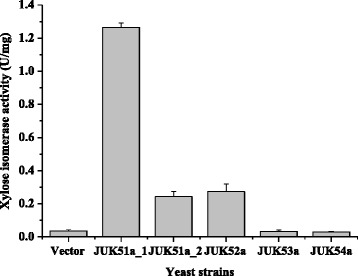
Xylose isomerase activities in the strains harboring empty vector (Vector), or xylose isomerases genes from Bacteroides vulgatus (JUK51a_1 and JUK51a_2), Piromyces sp. E2 (JUK52a) and snail manure/soil metagenomic DNA (JUK53a; JUK54a)
Active expression of B. vulgatus XI in S. cerevisiae is expected because its amino acid sequence is close to those of XIs from Piromyces sp. E2, Bacteroides stercoris [18] and P. ruminicola [21], which are all clustered with the XIs from mammal gut Bacteroidetes group. Interestingly, although the original B. vulgatus XI gene sequence had a low value (0.146) of codon adaptation index (CAI) to S. cerevisiae, the demonstrated activity of B. vulgatus XI was significantly high (Fig. 2), revealing the potential function of this XI in S. cerevisiae. While B. vulgatus XI under the promoter of TDH3 (JUK51a_1) displayed much higher activity than Piromyces sp. E2 XI under TEF1 promoter (JUK52a), under TEF1 promoter B. vulgatus XI (JUK51a_2) and Piromyces sp. E2 XI (JUK52a) exhibited comparable activities (Fig. 2). The fact that TDH3 promoter is a stronger promoter than TEF1 promoter [24] might contribute to the high XI activity in JUK51a_1. For the development of background strain, strain JUK51a_1 harboring B. vulgatus XI under TDH3 promoter (Table 1) was used. Consistent with the phylogenetic analysis (Fig. 1), the isolated metagenomics XI genes, SmmXylA1 and SmmXyl10, belonging to group of environmental Bacteroidetes, were not active in S. cerevisiae (JUK53a and JUK54a in Fig. 2).
Table 1.
Strains and plasmids
| Strain/Plasmid | Genotype/Property |
|---|---|
| Saccharomyces cerevisiae | |
| ATCC24860 | Obtained from American Type Culture Collection |
| MATα/MATa | |
| JUK36a | ATCC 24860 derivative; |
| MATa ura3::loxP TKL1(-268, -1):: T RKI1 - RKI1-P ADH1 -T RPE1 -RPE1-P TPI1 -LoxP-T XKS1 -XKS1-P PGK1 -P PDC1 -TAL1-T TAL1 -P FBA1 gre3::loxP | |
| JUK39a | JUK36a derivative; cyc3::loxP |
| JUK50a | JUK36a derivative; {pJFE11} |
| JUK51a_1 | JUK36a derivative; {pJFX11-TDH3p} |
| JUK51a_2 | JUK36a derivative; {pJFX11} |
| JUK52a | JUK36a derivative; {pJFX12} |
| JUK53a | JUK36a derivative; {pJFX13} |
| JUK54a | JUK36a derivative; {pJFX14} |
| JUK61a | JUK39a derivative; {pJFX11-TDH3p} |
| JUKx11a | Obtained from the adaptive cultivation of JUK61a on xylose |
| JUK36a1 | JUK51a_1 derivative with the loss of pJFX11-TDH3p |
| JUK39a1 | JUKx11a derivative with the loss of pJFX11-TDH3p |
| 36a(Bvu) | JUK36a1 derivative; {pPY1-Bvu} |
| 36a(XIq) | JUK36a1 derivative; {pPY1-XIq} |
| 36a(TAA) | JUK36a1 derivative; {pPY1-TAA} |
| 36a(HGB5) | JUK36a1 derivative; {pPY1-HGB5} |
| 36a(YIT) | JUK36a1 derivative; {pPY1-YIT} |
| 39a(Bvu) | JUK39a1 derivative; {pPY1-Bvu} |
| 39a(XIq) | JUK39a1 derivative; {pPY1-XIq} |
| 39a(TAA) | JUK39a1 derivative; {pPY1-TAA} |
| 39a(HGB5) | JUK39a1 derivative; {pPY1-HGB5} |
| 39a(YIT) | JUK39a1 derivative; {pPY1-YIT} |
| Plasmids | |
| pJPPP-XK | pUC19, genomic integrative plasmid used to overexpress Saccharomyces cerevisiae endogenous genes RPE1, RKI1, TAL1, TKL1 and XKS1 |
| pJFX11-TDH3p | YEp, PTDH3-TPGK1, Bacteroides vulgatus XI |
| pJFE11 | YEp, PTEF1-TCYC1 |
| pJFX11 | pJFE11, Bacteroides vulgatus XI |
| pJFX12 | pJFE11, Piromyces sp. E2 XI |
| pJFX13 | pJFE11, SmmXylA1 XI |
| pJFX14 | pJFE11, SmmXylA10 XI |
| pPY1 | pYES2, PPGK1-TCYC1 |
| pPY1-Bvu | pPY1, Bacteroides vulgatus XI |
| pPY1-XIq | pPY1, Orpinomyces sp. ukk1 XI |
| pPY1-TAA | pPY1, Tannerella sp. 6_1_58FAA_CT1 XI |
| pPY1-HGB5 | pPY1, Alistipes sp. HGB5 XI |
| pPY1-YIT | pPY1, Paraprevotella xylaniphila XI |
Improving xylose-fermenting background yeast strain using recombinant yeast strain harbouring B. vulgatus XI
Overexpressing of XI with high activities alone could not render the effective fermentation of xylose [6]. Therefore, xylose metabolic pathway in the above background S. cerevisiae strain was optimized by overexpressing xylulokinase (XKS1) and the four enzymes from non-oxidative pentose phosphate pathway, transaldolase (TAL1), transketolase (TKL1), ribose-5-phosphate isomerase (RKI1), and ribulose 5-phosphate 3-epimerase (RPE1), and disrupting the aldose reductase gene gre3. Host strain (JUK36a) was generated, and the recombinant yeast strains harboring B. vulgatus XI strains JUK51a_1 (XI controlled by TDH3 promoter) and JUK51a_2 (XI controlled by TEF1 promoter) exhibited aerobic growth on xylose. On the other hand, the control strain without XI genes (JUK50a) could not grow on xylose (data not shown). Adaptive laboratory evolution in xylose medium under the anaerobic conditions or using a respiration-deficient strain was shown effective to reinforce xylose catabolism to ethanol production [7, 10, 15, 21]. Therefore, before more XIs from the mammal gut Bacteroidetes cluster were tested, a respiration-deficient strain (JUK39a) was generated from JUK36a by disrupting cyc3, which encodes holocytochrome c synthase and attached heme to apo-cytochrome c [25], and it could not grow on glycerol (data not shown). Plasmid pJFX11-TDH3p was transformed to JUK39a to generate the respiration-deficient strain JUK61a. Strain JUK61a exhibited the delayed growth on xylose and the growth rate increased by adaptive evolution through continuous transferring cultivation in synthetic minimal medium (SMM) containing 6.7 g l−1 yeast nitrogen base (YNB) and 20 g l−1 xylose at pH 6.0 (Fig. 3a).
Fig. 3.
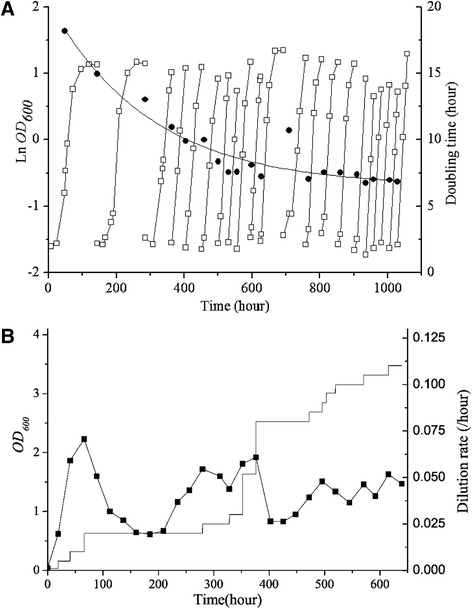
Continuous transferring cultivation of the respiration-deficient strain JUK61a (a) and anaerobic chemostat evolution of strain JUK51a_1 (b) in SMM with 20 g l−1 xylose at 30 °C and pH 6.0: empty square, LnOD600; filled circle, doubling time; filled square, OD600; solid line, dilution rate
Unlike strain JUK51a_1, for which xylose metabolism is dependent on respiration, strain JUKx11a, isolated from the continuous transferring cultivation of strain JUK61a on xylose, exhibited efficient xylose fermentation (Fig. 4). In batch aerobic cultivation, ~15 g l−1 xylose was consumed in 24 h, and ~6 g l−1 ethanol and ~1.4 g l−1 glycerol were produced. Ethanol yield reached 0.396 g g−1 consumed xylose. This strain was maintained in SMM with 20 g l−1 xylose and it was stable for xylose fermentation after more than 20 times of transferring. Similar results were obtained for anaerobic batch cultivation using strain JUKx11a indicating oxygen supply to this strain is not essential to this respiration-deficient strain (data not shown).
Fig. 4.
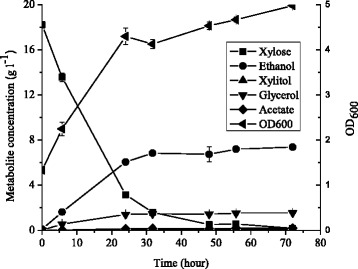
Ethanolic xylose fermentation by strain JUKx11a isolated from continuous transferring cultivation at SMM with 20 g l−1 xylose at 30 °C and pH. 6.0
Saccharomyces cerevisiae strain JUK51a_1 (Table 1) was also anaerobically evolved in a chemostat by continuous diluting cultivation until its dilution rate was increased from 0.01 to 0.11 h−1 (Fig. 3b). A fast growing strain was isolated and the plasmid of pJFX11-TDH3p (URA3) was lost by streaking on 5-fluoroorotic acid (5-FOA) plates [26] to obtain strain JUK36a1 (Table 1). Plasmid pJFX11-TDH3p was also lost from the evolved respiration-deficient strain JUKx11a similarly to obtain strain JUK39a1 (Table 1). Both strains JUK36a1 and JUK39a1 were used as hosts for further XI testing.
Parallel xylose utilization and cell growth testing of yeast strains expressing XI genes from mammal gut Bacteroidetes cluster
Based the background strains JUK36a1 and JUK39a1, more XIs from the mammal gut Bacteroidetes cluster (Fig. 1) were tested and compared. Beside B. vulgatus XI gene, XI genes from Tannerella sp. 6_1_58FAA_CT1, P. xylaniphila, Alistipes sp. HGB5 and Orpinomyces sp. ukk1 (used as a positive control) [16] were individually introduced into the evolved background strains under the control of PGK1 promoter. The five XIs respectively have an identity of 95 % (Orpinomyces sp.), 82 % (P. xylaniphila), 81 % (Tannerella sp.), 81 % (B. vulgatus) and 76 % (Alistipes sp.) to the amino acid sequence of Piromyces sp. XI.
SMM supplied with 40 g l−1 xylose was used in batch cultivation of 36a and 39a strains. Aerobic cultivation was performed in 24-well plates with 600 μl media at 218 rpm and 30 °C. Duplicate wells were used for each strain. Biomass was monitored by measuring the absorbance at the wavelength of 595 nm.
The concentrations of glucose, xylose, xylitol, glycerol, acetate, and ethanol were determined using HPLC and results are listed in Fig. 5. Notably, all 36a and 39a strains expressing the above five different xylose isomerase genes grew well in the SMM containing xylose. Interestingly, all 36a strains grew almost equally fast, indicated by very similar maximal specific growth rate of about 0.2 h−1 (Fig. 5), with strain 36a(HGB5) containing Alistipes sp. HGB5 XI gene presenting the highest specific growth rate of 0.2015 h−1, and strain 36a(Bvu) expressing B. vulgatus XI gene showed very similar specific growth rate of 0.2009 h−1, both were slightly higher than that (0.1977 h−1) obtained by the control strain 36a(XIq) harboring Orpinomyces sp. ukk1 XI gene. Strain 36a(TAA) carrying the Tannerella sp. 6_1_58FAA_CT1 XI gene and 36A(YIT) containing the P. xylaniphila XI gene presented equal specific rate of 0.1850 h−1, slightly lower than other three strains. The specific rate values were much comparable to that obtained for S. cerevisiae expressing codon-optimized P. ruminicola XI gene [21]. In the latter case, a specific rate of 0.23 h−1 was obtained after adaptive evolution of the recombinant strain in SMM containing xylose. Apparently, the relatively high specific rates obtained for the recombinant 36a strains is attributed to the background strain, JUK36a1, which has been anaerobically evolved in xylose medium. These results demonstrated that host strain anaerobic adaptive evolution in xylose medium followed by XI gene transformation might be an effective way for the screening of novel functional xylose isomerase genes in yeast. Orpinomyces sp. ukk1 is a mammal gut fungus and its XI gene is very well known for its functional expression in S. cerevisiae [16]. The comparable performance of xylose utilization between the recombinant strains harboring the four selected XI genes from the mammal gut Bacteroidetes cluster to the control strain points out the potential of such bacteria XI genes in functional expression in S. cerevisiae and xylose-fermenting yeast construction.
Fig. 5.
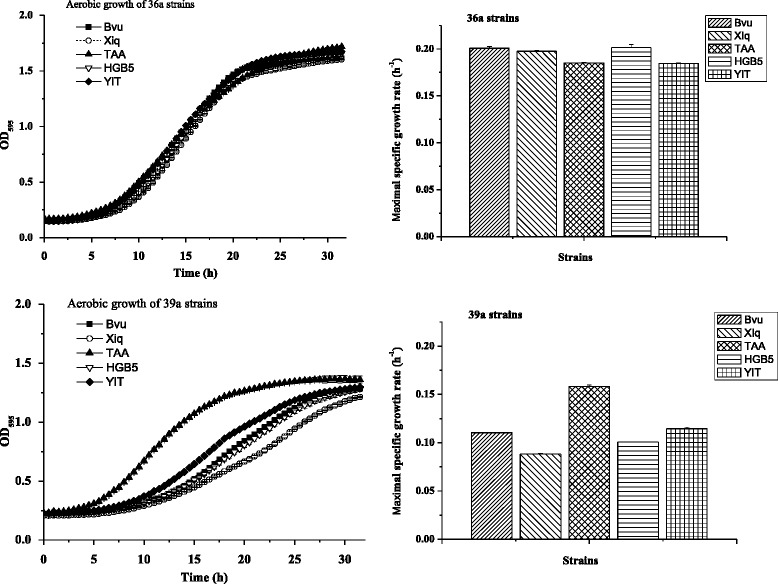
Aerobic growth and maximal specific growth rates of 36a and 39a strains in SMM with 40 g l−1 xylose at 30 °C and pH 6.0
On the other hand, cell growth rates varied among the 39a strains. Obviously, the knock-out of cyc3 gene, which encodes holocytochrome c synthase and attached heme to apo-cytochrome c, was able to differentiate the active expression of different xylose isomerase genes. Strain 39a(TAA) harboring the Tannerella sp. 6_1_58FAA_CT1 XI gene demonstrated the fastest growth with 0.1584 h−1 specific growth rate, followed by 39a(YIT) carrying the P. xylaniphila XI gene with about 0.1145 h−1 specific growth rate and strain 39a(Bvu) containing the B. vulgatus XI gene with specific growth rate of 0.1104 h−1. Notably, the control strain 39a(XIq) expressing the Orpinomyces sp. ukk1 XI gene in this case displayed the lowest specific growth rate of 0.0882 h−1 and strain 39a(HGB5) containing Alistipes sp. HGB5 XI gene displayed slightly higher specific growth rate of 0.1006 h−1. The above results suggest that Tannerella sp. XI is more favorable to cell growth on xylose using the respiration-deficient strain JUK39a1 as the host. In general, 36a strains grew faster than 39a strains. The specific growth rates of 36a strains were about 0.20 h−1 and those for 39a strains were about 0.10 h−1, except strain 39a(TAA) presenting a specific growth rate of about 0.16 h−1. The final OD595 values of 36a strains were above 1.6, whereas those for 39a strains were below 1.4.
Xylose fermentation by recombinant 36a and 39a strains
Better aerobic cell growth does not always correlate to better xylose fermentation. In order to identify the best xylose-fermenting recombinant yeast strains, in this part of the work, batch fermentation of recombinant 36a and 39a strains was performed in a 125 ml flask plugged with a rubber stopper with a syringe tip outlet at 30 °C and 200 rpm. SMM containing 40 g l−1 xylose was used in the cultivation. Interestingly, all 36a strains were able to utilize xylose as the sole carbon source and produced significant amount of ethanol (Fig. 6). This is different from the case of recombinant strains JUK51a_1 and JUK51a_2, whereby although both strains were able to grow in SMM containing xylose, no xylose-fermentation was observed. The ability of xylose-fermentation of the 36a strains was arisen from the anaerobic adaptive evolution of strain JUK51a_1 in xylose medium. Likewise, 39a strains were also able to grow in SMM containing xylose and produce ethanol, which otherwise was attributed to the anaerobic adaptive evolution of JUKx11a. For all the 36a strains tested, notably, strain 36a(HGB5) harboring Alistipes sp. HGB5 xylose isomerase gene was able to utilize xylose more rapidly than other strains and produced more ethanol (Fig. 6). On the other hand, for all 39a strains, strains harboring XI genes from B. vulgatus, Tannerella sp. 6_1_58FAA_CT1 and P. xylaniphila almost displayed almost identical rates of xylose consumption and ethanol production, with the control strain having the Orpinomyces sp. ukk1 XI gene showed the slowest xylose utilization and ethanol production rates (Fig. 6). Apparently, for strain 36a(HGB5) and 39a(TAA), the high rates of xylose consumption and ethanol production correlated well with the better cell growth and higher cell density. Correspondingly, the low xylose consumption and ethanol production by 39a(XIq) was due to its slow cell growth and lower cell density. However, the production of glycerol did not correlate with its cell growth. Strain 36a(HGB5) exhibited the lowest glycerol production among all the 36a strains, whereas strain 39a(TAA) displayed moderately high glycerol production among all the 39a strains.
Fig. 6.
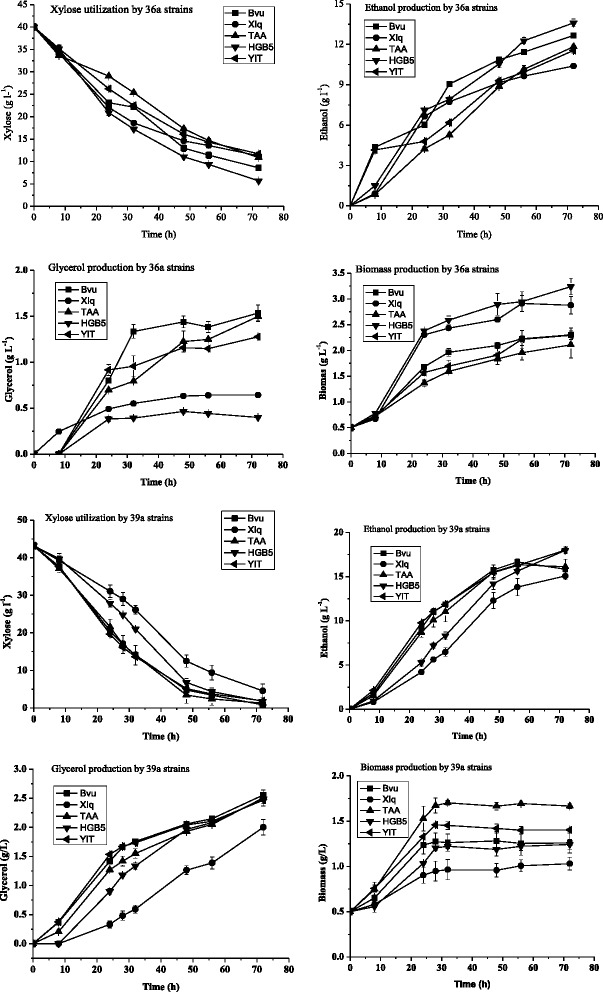
Xylose fermentation by 36a and 39a strains in SMM with 40 g l−1 xylose at 30 °C and pH 6.0
Fermentation parameters of 36a and 39a strains at 56 h are listed in Table 2 for detailed comparison, and the average specific xylose consumption, ethanol and glycerol production rates were calculated according to Peng and his colleagues [10]. Among all the 36a strains, notably, 36a(HGB5) presented the highest ethanol titer and xylose conversion, indicating its fastest xylose utilization rate. This might be associated with its highest cell growth rate revealed by its high final biomass concentration. Correspondingly, its specific ethanol productivity was lower than the rest of strains, except the control strain 36a(XIq). Interestingly, the control strain also displayed the lowest ethanol yield of 0.365 g g−1, corresponding to about 72 % of the theoretical value. This arose primarily from its lower ethanol titer than the rest strains (Table 2). The main by-product for 36a strain fermentation was glycerol, with its specific production rate of about 0.02 to 0.13 g gcdw−1 h−1. Very minimal amount of xylitol and acetic acid were produced with their specific production rates being lower than 0.01 g gcdw−1 h−1 (data not shown). For the 39a strains, the highest ethanol titer was obtained by 39a(Bvu), whereas the highest xylose conversion was obtained by strain 39a(TAA) with the 2nd highest ethanol titer. This is largely due to its high cell growth rate (Fig. 6) and cell density. Further analysis of the specific rates of xylose consumption and ethanol production suggest that strain 39a(HGB5) is the most potential by showing the highest rates, just below the control strain 39a(XIq). Strain 39a(Bvu) showed slightly lower specific rates of xylose consumption and ethanol production. The fact that 39a strains containing B. vulgatus and Alistipes sp. HGB5 XI genes exhibited moderately high cell growth rate and higher specific rates of xylose consumption and ethanol production recommends that these two XI genes are the most potential in xylose-fermenting yeast construction using JUK39a1 as the host. Similar to 36a strains, the main by-product for 39a strains was glycerol with specific rate of production of about 0.03 g gcdw−1 h−1. Production of xylitol and acetic acid was very minimal with both specific production rates being smaller than 0.01 g gcdw−1 h−1 (data not shown).
Table 2.
Xylose fermentation parameters obtained in SMM with 40 g l−1 xylose at 56 h
| Strains | Xylose conversion | Cell biomass | Ethanol | Ethanol yield | Average specific consumption/production rates | ||
|---|---|---|---|---|---|---|---|
| (%) | (g l−1) | (g l−1) | (g g−1) | g gcdw −1 h−1 | |||
| Xylose | Ethanol | Glycerol | |||||
| 36a(Bvu) | 71.42 ± 1.49 | 2.23 ± 0.02 | 11.43 ± 0.09 | 0.400 ± 0.001 | 0.324 ± 0.008 | 0.130 ± 0.001 | 0.012 ± 0.001 |
| 36a(XIq) | 66.09 ± 0.26 | 2.91 ± 0.03 | 9.64 ± 0.06 | 0.365 ± 001 | 0.241 ± 0.001 | 0.088 ± 0.001 | 0.004 ± 0.000 |
| 36a(TAA) | 63.30 ± 0.38 | 1.96 ± 0.02 | 10.15 ± 0.29 | 0.400 ± 0.009 | 0.333 ± 0.000 | 0.134 ± 0.004 | 0.013 ± 0.001 |
| 36a(HGB5) | 76.68 ± 0.64 | 2.95 ± 0.04 | 12.27 ± 0.23 | 0.400 ± 0.004 | 0.261 ± 0.002 | 0.104 ± 0.002 | 0.002 ± 0.000 |
| 36a(YIT) | 64.17 ± 0.05 | 2.22 ± 0.02 | 9.93 ± 0.16 | 0.387 ± 0.007 | 0.314 ± 0.000 | 0.121 ± 0.002 | 0.010 ± 0.000 |
| 39a(Bvu) | 92.13 ± 0.42 | 1.26 ± 0.11 | 16.67 ± 0.08 | 0.419 ± 0.003 | 0.662 ± 0.057 | 0.277 ± 0.022 | 0.036 ± 0.003 |
| 39a(XIq) | 78.26 ± 4.20 | 1.03 ± 0.07 | 13.82 ± 0.98 | 0.409 ± 0.014 | 0.721 ± 0.045 | 0.295 ± 0.008 | 0.030 ± 0.005 |
| 39a(TAA) | 94.47 ± 3.82 | 1.67 ± 0.03 | 16.35 ± 0.34 | 0.401 ± 0.006 | 0.532 ± 0.011 | 0.213 ± 0.001 | 0.027 ± 0.001 |
| 39a(HGB5) | 89.98 ± 2.08 | 1.24 ± 0.05 | 15.63 ± 0.30 | 0.402 ± 0.005 | 0.704 ± 0.005 | 0.283 ± 0.006 | 0.038 ± 0.001 |
| 39a(YIT) | 91.39 ± 0.35 | 1.40 ± 0.01 | 16.27 ± 0.01 | 0.412 ± 0.005 | 0.586 ± 0.002 | 0.242 ± 0.004 | 0.031 ± 0.001 |
Strain 39a(TAA) demonstrated the lowest specific ethanol productivity (0.213 g gcdw−1 h−1) with the highest being 0.295 g gcdw−1 h−1 obtained by the control strain 39a(XIq). Interestingly, such specific ethanol productivity values for 39a strains were much higher than those reported by Peng et al. [10] and Hector et al. [21], where they were below 0.100 g gcdw−1 h−1, and were comparable to those obtained by 36a strains ranged from 0.104 to 0.134. Our recombinant strains were constructed by strain adaptive evolution in xylose medium followed by XI gene transformation; whereas the recombinant strains by Peng et al [10] and Hector et al. [21] were constructed by XI gene transformation followed by adaptive evolution in xylose medium. Much comparable cell growth (Fig. 5) and fermentation performance (Table 2) were obtained, suggesting that the approach adopted in this work is effective in rapid screening of functional XI genes and efficient in construction of xylose-fermenting yeasts.
It is worthwhile noting that consistent ethanol yield of about 0.4 g g−1 was obtained for all the 39a strains tested, with the highest being 0.419 g g−1 for strain 39a(Bvu) and the lowest being 0.401 g g−1 for strain 39a(TAA), corresponding to 82.2 % and 78.6 %, respectively, of the theoretical yield. The control strain 39a(XIq) in this case obtained the highest ethanol yield of 0.409 g g−1, corresponding to 80.2 % of the theoretical yield. The significant difference of ethanol yield for strains 36a(XIq) and 39a(XIq) confirms that respiration-deficient background strain is more favorable for the expression of fungus Orpinomyces sp. XI gene. Moreover, all the 39a strains consumed xylose faster and produced more ethanol than 36a strains, further proving that respiration-deficient strain is more favorable for xylose fermentation although anaerobic adaptive evolution is able to enhance it for respiration-efficient 36a strains. The favorable xylose utilization and ethanol production by 39a strains is ascribed to the anaerobic nature of these mammal gut bacteria.
Conclusion
Xylose isomerase genes from two phylogenetic groups (mammal gut Bacteroidetes and environmental Bacteroidetes) were tested in S. cerevisiae strain with downstream xylose metabolic pathway optimization. Xylose isomerase from mammal gut microorganisms, B. vulgatus, Tannerella sp. 6_1_58FAA_CT1, Alistipes sp. HGB5 and P. xylaniphila, were actively expressed, and the two Bacteroidetes-related genes isolated from environmental metagenome did not exhibit activities. Expression of the above XI genes from the above listed mammal gut bacteria in a respiration-efficient host, S. cerevisiae JUK36a1, resulted in similar growth rate under aerobic conditions. On the other hand, the respiration-deficient host, JUK39a1, resulted in varied growth rates for strains harboring XI genes from different mammal gut bacteria. The expression of Tannerella sp. 6_1_58FAA_CT1 XI gene resulted in the fastest cell growth rate, maximal xylose conversion, and highest ethanol production under anaerobic cultivation conditions. Respiration-deficient S. cerevisiae strain carrying B. vulgatus and Alistipes sp. HGB5 XI genes presented moderate cell growth and relatively high specific ethanol productivity of 0.277 and 0.283 g gcdw−1 h−1, respectively. They are most potential in xylose-fermenting yeast construction. All respiration-deficient strains (39a strains) displayed higher rates of xylose consumption and ethanol production than 36a strains, confirming that cyc3 gene knock-out is favorable. In addition, this work also demonstrated that host strain adaptive evolution in xylose medium followed by XI gene transformation is effective in rapid functional XI gene screening in S. cerevisiae and is efficient in xylose-fermenting yeast construction.
Methods
Cloning of xylose isomerase genes from environmental metagenomic DNA
Primers used in this work are listed in Additional file 1. Metagenomic DNA from snail manure and soil from Singapore natural forest was extracted using FastDNA® Spin Kit for Soil (MP Biomedicals) and purified using E.Z.N.A. ® Cycle-Pure Kit (Omega Bio-tek). Degenerate primers were designed based on xylose isomerase conservative region (Additional file 1). Primers MetXIconss and MetXIconsa were used to amplify partial sequences of xylose isomerase from snail manure metagenomic DNA (Smm) and soil metagenomic DNA (Som). The PCR product of partial sequences of xylose isomerase was cloned into T-vector and sequenced. The 5′ and 3′ flanking regions of xylose isomerase from metagenomic DNA were obtained through thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) using LA Taq DNA polymerase (TAKARA), and arbitrary degenerated primers (AD1, AD2, AD3 or AD4) and nested gene-specific primers. TAIL-PCR was performed according to the previously reported protocol [27] and the TAIL-PCR products were sequenced to generate the whole gene sequences. Primers designed based on 5′ and 3′ regions of xylose isomerase were used to amplify the full-length genes directly from metagenomic DNA.
Plasmid construction
Strains and plasmids used in this work are listed in Table 1.
The gene fragments, FBA1p-TKL1(1318) (recombinant arm targeting to TKL1 open reading frame, ORF), TKL1(-501,-286) (recombinant arm targeting to TKL1 promoter), PDC1p-TAL1-TAL1t, ADH1p-RKI1-RKI1t, TPI1p-RPE1-RPE1t and PGK1p-XKS1-XKS1p were amplified using genomic DNA of S. cerevisiae ATCC 24860 and fused through overlapping extension polymerase chain reactions (PCR). G418 resistant marker loxP-KanMX4-loxP was amplified from plasmid pUG6. These DNA fragments were sequentially cloned into pUC19 to generate the plasmid pJPPP-XK (Fig. 7).
Fig. 7.
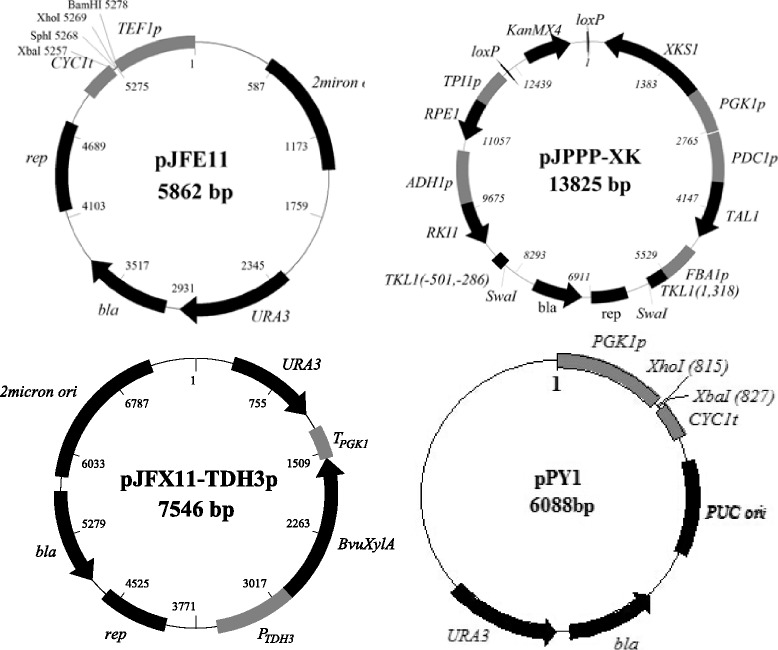
Plasmids used for the construction of xylose metabolic pathway
The coding sequence of B. vulgates xylose isomerase was synthesized according to the sequence under EMBL-ENA access No. ABR41556.1 and fused with TDH3 promoter and PGK1 terminator from S. cerevisiae through overlapping extension PCR to generate DNA fragment TDH3p-BvuXylA-PGK1t. Auxotrophic marker URA3 was cut out from pUG72 by XbaI and NdeI. DNA fragments of TDH3p-BvuXylA-PGK1 and URA3 were sequentially cloned into SalI/PstI sites and SpeI/NdeI sites of pRS327 to generate pJFX11-TDH3p (Fig. 7). The yeast expression vector pJFE11 was constructed by replacing GAL1 promoter of plasmid pYES2 by TEF1 promoter. The coding sequences of B. vulgates XI, Piromyces sp. XI, SmmXylA1 and SmmXylA10 were cloned into BamH/XhoI sites of pJFE11 (Fig. 7) between TEF1 promoter and CYC1 terminator to generate pJFX11, pJFX12, pJFX13 and pJFX14, respectively (Table 1).
Strain construction
S. cerevisiae haploid strain was isolated from the diploid strain S. cerevisiae ATCC 24860. The LiAc/ssDNA/PEG solution was used for yeast transformation [28]. Disruption cassettes of ura3, gre3 and cyc3 with G418 resistant marker loxP-KanMX4-loxP were amplified from pUG6 [26]. These disruption cassettes were transformed into yeast to generate the strains with relevant genotypes. Xylulokinase and non-oxidative pentose phosphate pathway were overexpressed by transforming SwaI-digested pJPPP-XK (Fig. 7). The KanMX4 markers for disruptions of ura3, gre3 and cyc3 and pJPPP-XK integration were separately removed by transforming a Cre plasmid pSH47 (URA3) and inducing Cre expression in YP galactose broth. Later on, pSH47 (URA3) was lost by cultivation on 5-FOA plates [26].
Strain cultivation
Synthetic minimal media (SMM, Yeast nitrogen base, YNB, 6.7 g l−1, pH 6.0) supplied with glucose and/or xylose was used in batch cultivation. Aerobic cultivation was performed in 24-well plate with 600 μl medium at 218 rpm and 30 °C and cell growth was monitored by measuring the absorbance at the wavelength of 595 nm. Oxygen-limited cultivation was performed in 125 ml Erlenmeyer flasks, plugged by rubber stoppers with syringe needle outlets, with 60 ml media incubated at 200 rpm and 30 °C. Cells in late exponential-phase in SMM with 20 g l−1 xylose were harvested by centrifuge and inoculated into the cultivation medium for fermentation evaluation. Cell biomass was monitored by measuring the absorbance at the wavelength of 600 nm.
Adaptive transferring cultivation was performed in a 125 ml Erlenmeyer flasks, plugged by rubber stoppers with syringe needle outlets, containing 20 ml SMM with 20 g l−1 xylose incubated at 200 rpm and 30 °C. In each batch of adaptation, the initial OD600 was set as 0.2, and when OD600 reached about 2.5, cells were inoculated into a new batch with an initial OD600 of 0.2 for further adaptation. Finally, single clones were separated by streaking on the SMM xylose plate, and the biggest clone was selected and named as JUKx11a after seven day incubation.
Continuous evolutionary cultivation was performed in a 200 ml feeding bottle with 150 ml SMM supplied with 12 g l−1 xylose as the carbon source and a stirring magnetic bar at 30 °C, under anaerobic conditions. The cultivation was initiated by inoculating the JUK51a_1 cells cultivated in the aerobic conditions. The fresh media was continuously added through a feeding pump. Biomass was monitored by measuring the absorbance at wavelength of 600 nm. According to cell growth, the dilution rate was increased gradually from 0.01 to 0.11 h−1.
The concentrations of glucose, xylose, xylitol, glycerol, acetate, and ethanol were determined using HPLC with an Aminex HPX-87H ion exchange column (Bio-Rad) at 60 °C, and a refractive index detector; 5 mmol l−1 H2SO4 was used as the mobile phase with a flow rate of 0.6 ml min−1 [6]. All aerobic cultivation and xylose fermentation experiments were conducted in duplicate, and the average value with standard deviation was reported.
Xylose isomerase activity assay
Cells were cultivated in SMM supplemented with 20 g l−1 xylose and were harvested at OD600 of 2.5. Cell-free extracts were prepared in 100 mM tris-HCl buffer (pH 7.5) using a glass bead beater and protease inhibitor cocktail set IV (Merck) was added. The xylose isomerase activity of the cell extracts was determined at ambient temperature (25 °C) using the UV-VIS Spectrophotometer 1240 (Shimadzu). The 1-ml reaction mixture contained 100 mmol l−1 Tris–HCl buffer (pH 7.5), 10 mmol l−1 MgCl2, 500 mmol l−1 xylose, 1 U of sorbitol dehydrogenase (Roche), 0.15 mmol l−1 NADH, and 0.05 ml of the cell extract [6]. Protein concentration was measured using the Coomassie protein assay kit (Thermo Scientific). One unit of enzyme activity was defined as the amount of enzyme required to oxidize 1 μmol of coenzyme/min, and the specific activity was expressed in units per milligram of protein.
Additional file
Supplementary material.
Acknowledgements
The authors are grateful for the financial support to this work provided by the Science and Engineering Research Council of the Agency for Science, Technology and Research (A*STAR) and National Research Foundation, Singapore.
Abbreviations
- ADH1
Alcohol dehydrogenase
- ATCC
American Type Culture Collection
- CAI
Codon adaptation index
- CYC1
Cytochrome c
- cyc3
Holocytochrome c synthase gene
- cdw
Cell dry weight
- EMBL-ENA
European Molecular Biology Laboratory – European Nucleotide Archive
- FBA1
1,6-bisphosphate aldolase
- FOA
Fluoroorotic acid
- gre3
Aldose reductase gene
- NADP+
Oxidized nicotinamide adenine dinucleotide phosphate
- NADPH
Reduced nicotinamide adenine dinucleotide phosphate
- NAD+
Oxidized nicotinamide adenine dinucleotide
- NADH
Reduced nicotinamide adenine dinucleotide
- PDC1
Pyruvate decarboxylase
- PGK1
3-phosphoglycerate kinase promoter
- RKI1
Ribose-5-phosphate isomerase
- RPE1
Ribulose 5-phosphate 3-epimerase
- SMM
Synthetic minimal medium
- TAIL-PCR
Thermal asymmetric interlaced polymerase chain reaction
- TDH3
Glyceraldehyde-3-phosphate dehydrogenase
- TAL1
Transaldolase
- TKL1
Transketolase
- TEF1
Transcription elongation factor
- XI
Xylose isomerase
- XKS1
Xylulokinase
- XR
Xylose reductase
- XDH
Xylitol dehydrogenase
- YEp
Yeast episomal plasmid
Footnotes
Competing interests
PCT specifications were filed on 16 December 2014. Patent Cooperation Treaty Application No. PCT/SG2014/000600.
Authors’ contributions
BP, SH, and TL carried out all the experiments, acquired the data and BP drafted the manuscript. AG designed the study, performed results interpretation and analysis, and critically revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Authors’ information
Mr. Bingyin Peng was a Research Scientist at School of Life Sciences and Chemical Technology, Ngee Ann Polytechnic, Singapore when he conducted this research. He was working on the metabolic engineering of yeast strains for cellulosic ethanol production.
Mr. Shuangcheng Huang is currently a Research Assistant at School of Life Sciences and Chemical Technology, Ngee Ann Polytechnic, Singapore. He is working on the metabolic engineering of yeast strains for cellulosic ethanol and biochemical production.
Miss Tingting Liu is currently a Research Scientist at School of Life Sciences and Chemical Technology, Ngee Ann Polytechnic, Singapore. She is working on the metabolic engineering of yeast strains for cellulosic ethanol and biochemical production.
Dr. Anli Geng is currently the Assistant Director (Technology) and Senior Lecturer at School of Life Sciences and Chemical Technology, Ngee Ann Polytechnic, Singapore. She is the Principal Investigator of the yeast metabolic engineering research.
Contributor Information
Bingyin Peng, Email: bingyin.peng@yahoo.com.
Shuangcheng Huang, Email: Higle945@gmail.com.
Tingting Liu, Email: lti3@np.edu.sg.
Anli Geng, Email: gan2@np.edu.sg.
References
- 1.Kim SR, Park YC, Jin YS, Seo JH. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv. 2013;31:851–61. doi: 10.1016/j.biotechadv.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Hong KK, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci. 2012;69:2671–90. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffries TW, Jin YS. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol. 2004;63:495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 4.Walfridsson M, Bao X, Anderlund M, Lilius G, Bulow L, Hahn-Hagerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol. 1996;62:4648–51. doi: 10.1128/aem.62.12.4648-4651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol. 2000;66:3381–6. doi: 10.1128/AEM.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MS, de Laat WT, et al. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae. FEMS Yeast Res. 2003;4:69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y, Chen X, Peng B, Chen L, Hou J, Bao X. An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl Microbiol Biotechnol. 2012;96:1079–91. doi: 10.1007/s00253-012-4418-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee S-H, Kodaki T, Park Y-C, Seo J-H. Effects of NADH-preferring xylose reductase expression on ethanol production from xylose in xylose-metabolizing recombinant Saccharomyces cerevisiae. J Biotechnol. 2012;158:184–91. doi: 10.1016/j.jbiotec.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Olofsson K, Runquist D, Hahn-Hagerdal B, Liden G. A mutated xylose reductase increases bioethanol production more than a glucose/xylose facilitator in simultaneous fermentation and co-fermentation of wheat straw. AMB Express. 2011;1:4. doi: 10.1186/2191-0855-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng B, Shen Y, Li X, Chen X, Hou J, Bao X. Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab Eng. 2012;14:9–18. doi: 10.1016/j.ymben.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Khattab SM, Saimura M, Kodaki T. Boost in bioethanol production using recombinant Saccharomyces cerevisiae with mutated strictly NADPH-dependent xylose reductase and NADP(+)-dependent xylitol dehydrogenase. J Biotechnol. 2013;165:153–6. doi: 10.1016/j.jbiotec.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Sarthy AV, McConaughy BL, Lobo Z, Sundstrom JA, Furlong CE, Hall BD. Expression of the Escherichia coli xylose isomerase gene in Saccharomyces cerevisiae. Appl Environ Microbiol. 1987;53:1996–2000. doi: 10.1128/aem.53.9.1996-2000.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amore R, Wilhelm M, Hollenberg CP. The fermentation of xylose - an analysis of the expression of Bacillus and Actinoplanes xylose isomerase genes in yeast. Appl Microbiol Biotechnol. 1989;30:351–7. doi: 10.1007/BF00296623. [DOI] [Google Scholar]
- 14.Gárdonyi M, Hahn-Hägerdal B. The Streptomyces rubiginosus xylose isomerase is misfolded when expressed in Saccharomyces cerevisiae. Enzym Microb Tech. 2003;32:252–9. doi: 10.1016/S0141-0229(02)00285-5. [DOI] [Google Scholar]
- 15.Zhou H, Cheng JS, Wang BL, Fink GR, Stephanopoulos G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng. 2012;14:611–22. doi: 10.1016/j.ymben.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Madhavan A, Tamalampudi S, Ushida K, Kanai D, Katahira S, Srivastava A, et al. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl Microbiol Biotechnol. 2009;82:1067–78. doi: 10.1007/s00253-008-1794-6. [DOI] [PubMed] [Google Scholar]
- 17.van Maris AJ, Winkler AA, Kuyper M, de Laat WT, van Dijken JP, Pronk JT. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv Biochem Eng Biotechnol. 2007;108:179–204. doi: 10.1007/10_2007_057. [DOI] [PubMed] [Google Scholar]
- 18.Ha S-J, Kim SR, Choi J-H, Park MS, Jin Y-S. Xylitol does not inhibit xylose fermentation by engineered Saccharomyces cerevisiae expressing xylA as severely as it inhibits xylose isomerase reaction in vitro. Appl Microbiol Biotechnol. 2011;92:77–84. doi: 10.1007/s00253-011-3345-9. [DOI] [PubMed] [Google Scholar]
- 19.Brat D, Boles E, Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:2304–11. doi: 10.1128/AEM.02522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aeling KA, Salmon KA, Laplaza JM, Li L, Headman JR, Hutagalung AH, et al. Co-fermentation of xylose and cellobiose by an engineered Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2012;39:1597–604. doi: 10.1007/s10295-012-1169-y. [DOI] [PubMed] [Google Scholar]
- 21.Hector RE, Dien BS, Cotta MA, Mertens JA. Growth and fermentation of D-xylose by Saccharomyces cerevisiae expressing a novel D-xylose isomerase originating from the bacterium Prevotella ruminicola TC2-24. Biotechnol Biofuels. 2013;6:84. doi: 10.1186/1754-6834-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng M, Bagdasarian M, Zeikus JG. The role of active-site aromatic and polar residues in catalysis and substrate discrimination by xylose isomerase. Proc Natl Acad Sci U S A. 1993;90:8459–63. doi: 10.1073/pnas.90.18.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu C, Jeffries T. Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol. 2007;73:6072–7. doi: 10.1128/AEM.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumont ME, Ernst JF, Hampsey DM, Sherman F. Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae. EMBO J. 1987;6:235–41. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–24. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–63. doi: 10.1046/j.1365-313X.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 28.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 29.Li GY, Wang Z, Lu M, Dun BQ. Function and application of D-xylose isomerase gene. Chinese Patent. 2012. [Google Scholar]
- 30.Parachin NS, Gorwa-Grauslund MF. Isolation of xylose isomerases by sequence- and function-based screening from a soil metagenomic library. Biotechnol Biofuels. 2011;4:9. doi: 10.1186/1754-6834-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teunissen AWRH, de Bont JAM. Xylose isomerase genes and their use in fermentation of pentose sugars. 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.


