Lipid homeostasis is modulated upon starvation at three different levels manifested in reversible 1) spatial confinement of lipid biosynthetic enzymes, 2) mitochondrial and endoplasmic reticular reorganization, and 3) loss of organelle contact sites, thus highlighting a novel mechanism regulating lipid biosynthesis by simply modulating flux through the pathway.
Abstract
Cells adapt to changing nutrient availability by modulating a variety of processes, including the spatial sequestration of enzymes, the physiological significance of which remains controversial. These enzyme deposits are claimed to represent aggregates of misfolded proteins, protein storage, or complexes with superior enzymatic activity. We monitored spatial distribution of lipid biosynthetic enzymes upon glucose depletion in Saccharomyces cerevisiae. Several different cytosolic-, endoplasmic reticulum–, and mitochondria-localized lipid biosynthetic enzymes sequester into distinct foci. Using the key enzyme fatty acid synthetase (FAS) as a model, we show that FAS foci represent active enzyme assemblies. Upon starvation, phospholipid synthesis remains active, although with some alterations, implying that other foci-forming lipid biosynthetic enzymes might retain activity as well. Thus sequestration may restrict enzymes' access to one another and their substrates, modulating metabolic flux. Enzyme sequestrations coincide with reversible drastic mitochondrial reorganization and concomitant loss of endoplasmic reticulum–mitochondria encounter structures and vacuole and mitochondria patch organelle contact sites that are reflected in qualitative and quantitative changes in phospholipid profiles. This highlights a novel mechanism that regulates lipid homeostasis without profoundly affecting the activity status of involved enzymes such that, upon entry into favorable growth conditions, cells can quickly alter lipid flux by relocalizing their enzymes.
INTRODUCTION
Cells respond to environmental stress such as nutrient depletion or heat shock by modulating a variety of processes such as transcription, translation, protein quality control, and activity status of individual cellular factors aiding in cellular adaptation and survival (Zaman et al., 2008; Galdieri et al., 2010; Anckar and Sistonen, 2011). Recent evidence suggests sequestration of proteins into deposits as yet another adaptive response of cells to nutrient limitation (Narayanaswamy et al., 2009; Noree et al., 2010; Petrovska et al., 2014). A global screen for yeast proteins forming insoluble deposits upon starvation, appearing as foci of respective green fluorescent protein (GFP) fusions, identified 180 proteins, many of which are metabolic enzymes (Narayanaswamy et al., 2009). Further analysis of some of the protein sequestrations formed during the stationary phase revealed that they are reversible and solubilized upon addition of specific metabolites (An et al., 2008; Narayanaswamy et al., 2009; Noree et al., 2010; Laporte et al., 2011), which prompted the proposal that they are storage assemblies. Cosequestration has been reported for some functionally related proteins, such as Ura7 and Ura8 involved in CTP biosynthesis in yeast and enzymes involved in purine biosynthesis in mammalian cells (An et al., 2008; Noree et al., 2010). It was postulated that coclustering of enzymes during metabolic stress aids in enhanced substrate channeling and more efficient biosynthetic activity.
In contrast to these reports, a recent study suggests that the majority of starvation-induced protein sequestrations represent aggregates of misfolded conformers (O'Connell et al., 2014). Moreover, molecular chaperones, including Hsp104 and Hsp42, that play key roles in aggregate formation and solubilization (Glover and Lindquist, 1998; Specht et al., 2011; Saibil, 2013), also accumulate into foci in stationary-phase yeast cells, suggesting increased protein misfolding and aggregation during starvation (Narayanaswamy et al., 2009). Given these diverse observations, the relationship of starvation-induced protein sequestrations to misfolded protein aggregates, their activity status, and their physiological relevance remain unclear.
Interestingly, similar to cytosolic proteins, organelle proteins also respond dynamically to a variety of stress treatments, including exposure to H2O2, dithiothreitol (DTT), and nitrogen starvation, leading to their spatial alterations (Breker et al., 2013). Extending the organelle protein remodeling, a nucleus–vacuole contact site factor, Nvj2, accumulates into cytosolic punctate structures upon treatment of cells with H2O2 (Breker et al., 2013). However, the physiological significance of these observations remains unexplored.
To obtain insights into the nature and functional significance of starvation-induced protein sequestrations, we performed a systematic analysis of starvation-induced changes of the enzymes constituting one major metabolic pathway—phospholipid biosynthesis. This pathway is central to cell physiology yet poorly understood in terms of adaptive changes during nutrient starvation. The starting point for this analysis was the reported finding that Fas2, one of the two subunits of the heterooligomeric key enzyme fatty acid synthetase (FAS), sequesters into foci under a variety of stress conditions, including nutrient starvation (Narayanaswamy et al., 2009; Tyedmers et al., 2010; Jacobson et al., 2012).
We show here that intact FAS and multiple other enzymes of lipid biosynthesis localized to different cellular compartments sequester into distinct foci upon glucose starvation. Using FAS as model for further analysis, we show FAS foci are active and dynamic and dissolve upon addition of glucose independent of chaperone disaggregase activity, thus separating them from misfolded protein aggregates. Sequestration of ER and mitochondria-localized enzymes coincides with the concomitant loss of endoplasmic reticulum–mitochondria encounter structure (ERMES) and vacuole and mitochondria patch (vCLAMP) organelle contact sites that are reflected in changes in phospholipid profiles.
RESULTS
Lipid biosynthetic enzymes are sequestered upon starvation
A recent study reported that the Fas2 subunit of fatty acid synthetase fused with GFP was sequestered into cytosolic foci in stationary-phase yeast cells (Narayanaswamy et al., 2009). We also determined whether Fas1, the second subunit of the heterooligomeric FAS enzyme, forms foci and how it relates to Fas2 foci. Fas1 and Fas2 were genomically C-terminally tagged with mcherry and GFP fluorescent markers, respectively. Tagging did not affect cellular growth rates, indicating that the fusions with mcherry/GFP do not affect the essential function of FAS.
Fluorescence microscopic analysis revealed that, during logarithmic growth, both subunits exhibit a diffuse cytosolic distribution, whereas they coaccumulated into one cytosolic focus in most (68%) cells when they entered the quiescence stage upon prolonged growth in stationary phase (Supplemental Figure S1A). The fraction of cells showing one or more FAS foci steadily increased during starvation, until 100% was reached after 10 d of starvation (Supplemental Figure S1B).
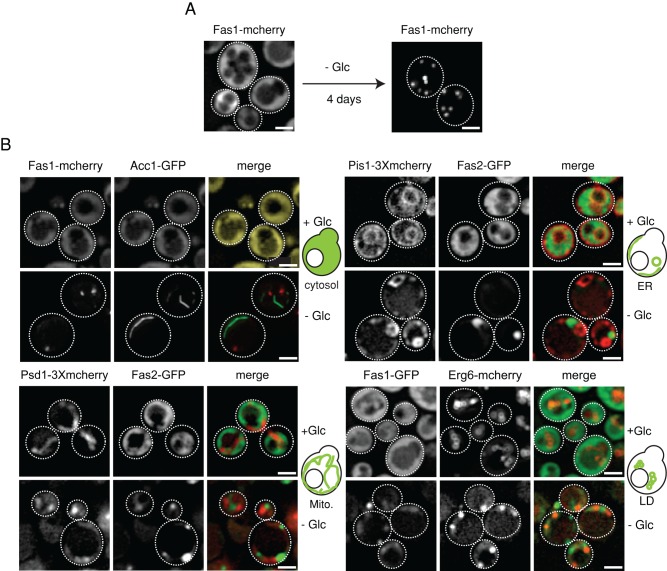
When an alternate form of metabolic stress such as glucose depletion was used, Fas1-mcherry also accumulated into foci. In this case, multiple Fas1-mcherry foci formed at a faster rate as compared with stationary phase (Figure 1A and Supplemental Figure S1C). Prolonged glucose depletion led to an increase in cell size without affecting cellular intactness. We confirmed Fas1 foci formation upon glucose depletion by immunofluorescence against a C-terminally myc-tagged Fas1 to exclude the possibility of FAS sequestration being artificially caused by tagging with fluorescent probes (Supplemental Figure S1D). To test whether FAS sequestration is specific, we monitored the localization of Rpl25-GFP (protein of the large ribosomal subunit) and Eno1-GFP (a glycolytic enzyme) upon glucose depletion (Supplemental Figure S2, A and B). Staining of Rpl25-GFP and Eno1-GFP remained diffuse, demonstrating specificity of FAS foci formation.
FIGURE 1:
Lipid biosynthetic enzymes sequester into foci upon prolonged glucose depletion. (A) Fas1-mcherry accumulates into multiple foci upon prolonged glucose depletion. Cells were grown to log phase in SD medium and shifted to medium lacking glucose (SC medium). Maximum-intensity projections of z-stack images are shown. (B) S. cerevisiae cells coexpressing either Fas1-mcherry and Acc1-GFP, Fas2-GFP and Pis1-3Xmcherry, Fas2-GFP and Psd1-3Xmcherry, or Fas1-GFP and Erg6-mcherry were grown at 30°C to either log phase or were depleted of glucose for 4 d. Single-plane images are shown. Scale bars: 2 μm.
To gain insight into the molecular nature of the FAS foci and their subcellular localization, we performed correlative fluorescence microscopic and electron tomographic analysis of glucose depletion-induced Fas1-mcherry foci. In all 10 electron tomograms with predicted Fas-mcherry localization, Fas1 foci represent ribosome-free zones in the cytosol, indicating tight packing of Fas1-mcherry molecules within foci (Supplemental Figure S2C).
We next analyzed whether other enzymes involved in fatty acid and phospholipid biosynthesis are also sequestered during starvation and whether they potentially cocluster with FAS foci (Figure 1B). We monitored potential foci formation of cytosolic Acc1, which synthesizes the FAS substrate malonyl-CoA, and the enzymes Pis1 (ER resident), Psd1 (mitochondria resident), and Erg6 (present in lipid droplets), which use fatty acids, the products of FAS activity, for phospholipid and lipid droplet biosynthesis. Genomic C-terminal fusions of the above enzymes to either GFP or mcherry were generated and analyzed by fluorescence microscopy. Functionality and correct localization of all fusions was ensured (Acc1, Pis1), given that some of these genes are essential or were reported by others (Erg6, Psd1) (Jacquier et al., 2011; Voss et al., 2012). While all tested enzymes showed their typical subcellular localization in log-phase cells, they accumulated into punctate or rod-like (Acc1) structures during glucose starvation (Figure 1B). Thus sequestration of lipid biosynthetic enzymes during starvation extends beyond FAS. During the time course of starvation, Pis1-3xmcherry, Psd1-3xmcherry, and Erg6-GFP precede the sequestration of cytosolic Acc1-GFP and Fas1-mcherry (Supplemental Figure S2D) into punctate structures. Thus sequestration of enzymes involved in phospholipid and fatty acid biosynthesis show temporal differences, with the latter sequestering more slowly. Importantly, all tested enzymes, while being sequestered upon starvation, never colocalized (Figure 1B), indicating that clustering of different, functionally related enzymes during starvation is not a general phenomenon.
FAS foci are not substrates of protein quality-control machineries
Because lipid biosynthetic enzymes, although individually sequestered upon starvation, are spatially separated from each other, it is unlikely that sequestration enhances phospholipid biosynthesis. We therefore envisioned two alternative explanations. First, the observed sequestrations might represent misfolded protein aggregates, in line with previous work (O'Connell et al., 2014), which reported that a majority of starvation-induced protein deposits represent aggregates of misfolded conformers. Second, the sequestration serves to spatially confine the enzymes from one another and perhaps from their substrates, which might represent an adaptive mechanism to regulate the flux of metabolites.
We first analyzed the possibility of sequestrations of lipid biosynthetic enzymes representing misfolded protein aggregates. Apart from sequestering during starvation, FAS is also known to form aggregates upon exposure to oxidative stress, such as arsenite, H2O2, or menadione treatment (Cabiscol et al., 2000; Tyedmers et al., 2010; Jacobson et al., 2012). Thus, being prone to sequestration under a variety of environmental stress conditions and due to its importance in lipid metabolism, FAS serves as an ideal model substrate for a systematic analysis.
We next analyzed the relationship between starvation-induced FAS sequestrations and misfolded protein aggregates. In yeast, the AAA+ protein disaggregase Hsp104 is specifically targeted, with high efficiency, to misfolded protein aggregates for solubilization and refolding of aggregated proteins in cooperation with the Hsp70 system (Saibil, 2013). Hsp104-GFP is thus a reliable and widely used marker for misfolded protein aggregates (Specht et al., 2011). Fluorescence microscopic analysis revealed that Hsp104-GFP forms multiple foci in stationary-phase cells, which, however, do not colocalize with Fas1-mcherry foci, indicating that Fas1 sequestrations are not misfolded protein aggregates (Figure 2A). Fas1-mcherry also formed foci in quiescent hsp42Δ cells, which fail to form cytosolic misfolded protein aggregates (Specht et al., 2011), further supporting a nonaggregate-like nature of FAS sequestrations (Figure 2B).
FIGURE 2:
Starvation-induced sequestration of FAS is active and not a misfolded protein aggregate. (A) S. cerevisiae cells expressing Fas1-mcherry and Hsp104-GFP were depleted of glucose (day 4) or grown to stationary phase (day 10). Single z-plane images (top panel) or maximum-intensity projections of z-stack images (bottom panel) are shown. Scale bars: 2 μm. (B) S. cerevisiae wt or hsp42Δ cells expressing Fas1-mcherry were grown to stationary phase for 10 d at 30°C, and FAS foci formation was monitored. Maximum-intensity projections of z-stack images are shown. Scale bars: 2 μm. (C) S. cerevisiae cells expressing Fas1-GFP and Ape1-mcherry were glucose depleted (day 4). Maximum-intensity projections of z-stack images are shown. Scale bars: 2 μm. (D) S. cerevisiae cells expressing Fas2-GFP were grown at 30°C either to log phase or were depleted of glucose for 4 d to induce Fas2-GFP foci. Total cell lysates (T) were separated into soluble (S) and insoluble (P) fractions by centrifugation. Distribution of Fas2-GFP was determined by Western blot analysis using GFP-specific antibodies. (E) FAS activity was determined in total and soluble cell fractions of log-phase and glucose-starved cells. Activity determined in presence of the FAS inhibitor cerulenin indicates background conversion of NADPH by other cellular components. SDs are given (n = 2).
Different types of misfolded proteins tend to coaggregate (Kaganovich et al., 2008; Specht et al., 2011). We therefore addressed the question of how FAS sequestration relates to sequestrations reported for other enzymes (Laporte et al., 2008; Narayanaswamy et al., 2009; Noree et al., 2010). Colocalization analysis using Gln1-mcherry, Ura7-GFP, Pre6-GFP, and Ade4-GFP revealed that Fas1-mcherry/Fas1-GFP are sequestered at a site distinct from the sequestrations of the other markers (Supplemental Figure S3A), unlike misfolded proteins. Moreover, Fas1-mcherry (similar to Gln1-mcherry) is sequestered much later during stationary phase than Ura7-GFP, Pre6-GFP, and Ade4-GFP (Supplemental Figure S3B).
We also considered the possibility that FAS foci are not substrates for the Hsp104 disaggregase but are cleared from cells by autophagy. Coexpression of Fas1-GFP and Ape1-mcherry, a marker for the preautophagosomal structure (PAS; Suzuki et al., 2013), did not reveal colocalization of these proteins upon starvation (Figure 2C). Together these findings suggest that FAS foci do not represent accumulation of misfolded proteins subjected to the protein quality-control machinery.
FAS foci are assemblies of active enzymes
To directly test whether the sequestered FAS enzymes retained or lost enzymatic activity, we monitored the activity of FAS in crude cell lysates. The activity assay was based on the observation that the majority of Fas2-GFP foci could be efficiently pelleted by centrifugation of lysates from starved cells, while diffuse Fas2-GFP remained entirely in the soluble fraction of lysed log-phase cells (Figure 2D). Comparing FAS activity of total cell extracts with that of soluble fractions upon centrifugation thus allows the activity status of Fas2-GFP within foci to be deciphered. In the case of log-phase cells, most of the activity of the Fas2-GFP determined in the total extracts was retained in the soluble fraction, correlating with the presence of diffuse Fas2-GFP fluorescence (Figure 2, D and E). FAS activity in total extracts of starved cells was comparable with that of extracts of log-phase cells. However, FAS activity in extracts of starved cells was largely lost in the supernatant upon centrifugation and dropped to background activity levels, which were also detected when the FAS inhibitor cerulenin was added (Figure 2, D and E). Together these findings demonstrate that FAS sequestrations formed during starvation contain active enzyme and do not represent misfolded protein aggregates.
Altered subcellular localization of FAS does not abolish its sequestration
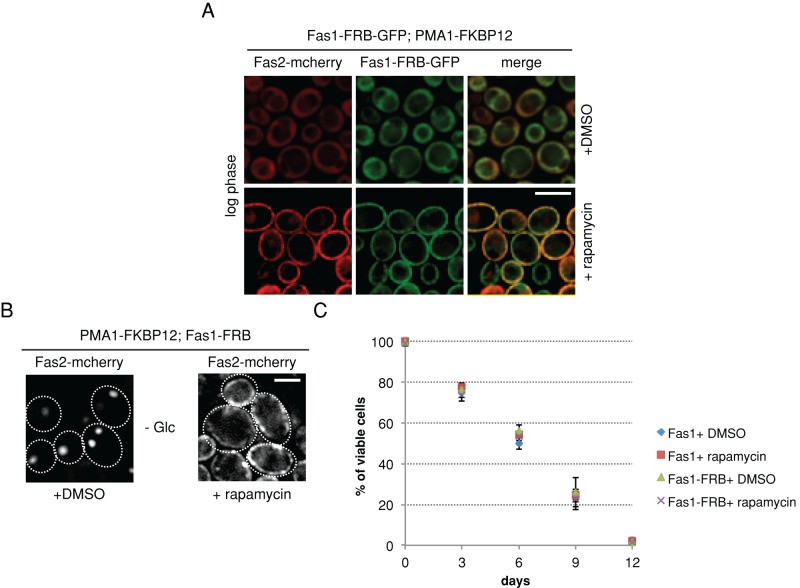
FAS foci being enzymatically active raises the possibility that FAS sequestration aids in a spatially confined synthesis of fatty acids within the cytosol. We envisioned that, in this scenario, relocalizing FAS to a different subcellular localization might affect cellular viability. To test this possibility, we exploited the “anchor-away technique” (Haruki et al., 2008), wherein any freely diffusing protein can be targeted to a particular cellular compartment in an inducible manner upon addition of the drug rapamycin. For this approach, Fas1 was genetically C-terminally fused to the FKBP12-rapamycin–binding domain (FRB)-GFP protein in a rapamycin-resistant tor1-1 mutant strain, which expresses the plasma membrane protein Pma1 genetically tagged to FKBP12 (12 kDa FK506-binding protein) at its C-terminus. In addition, we genetically tagged Fas2 with mcherry at its C-terminus to follow the cellular localization of the entire FAS complex. Both tagged FAS subunits, Fas1-FRB-GFP and Fas2-mcherry, showed cytoplasmic localization during logarithmic growth, but were quantitatively retargeted to the plasma membrane within minutes upon addition of rapamycin (Figure 3A). This collective retargeting of both FAS subunits allowed us to subsequently monitor Fas2-mcherry as a readout for localization of the entire FAS complex.
FIGURE 3:
Altered subcellular localization of FAS does not abolish its sequestration. (A and B) Rapamycin-resistant S. cerevisiae cells expressing PMA1-FKBP12, Fas1-FRB-GFP, and Fas2-mcherry were grown to log phase in SD medium (A) or were glucose depleted (B). Cells were treated with either DMSO or rapamycin. In rapamycin-treated cells, both Fas1-FRB-GFP and Fas2-mcherry are targeted to PMA1-FKBP12 at the plasma membrane, while both subunits retain cytoplasmic localization in DMSO-treated control cells (A). On glucose starvation, control cells (+DMSO) form cytosolic Fas2-mcherry foci, while rapamycin addition keeps Fas2-mcherry at the membrane (B). (C) In rapamycin-resistant S. cerevisiae cells coexpressing PMA1-FKBP12 and Fas2-mcherry, Fas1 was either genomically tagged FKBP12 or not. Both strains were grown to log phase in SD medium and depleted of glucose by transferring them to SC medium. DMSO (control) or rapamycin was added upon glucose depletion. Cell viability was measured on different days of starvation by Sytox green staining–based FACS sorting of dead cells (C).
Cells were grown to logarithmic phase and subsequently starved of glucose with or without addition of rapamycin. In untreated control cells, cytosolic FAS foci had already formed after 1 d of glucose depletion, while in rapamycin-treated cells, Fas2-mcherry remained anchored away at the plasma membrane (Figure 3B). Faster sequestration of FAS in starved cells and enlarged vacuoles observed in the log-phase cells are likely due to the mutation in TOR1, as described previously (Michaillat et al., 2012). Notably, under starvation conditions, the membrane staining by Fas2-mcherry became less smooth, and membrane-bound FAS foci were apparent. Thus, altering the localization of FAS did not abolish its sequestration into foci. As a control, we coexpressed plasma membrane–bound Hxt3-GFP (hexose transporter) and Fas2-mcherry in yeast cells expressing Fas1-FRB and Pma1-FKBP12. On starvation and rapamycin addition, Hxt3-GFP fluorescence also became less smooth, and foci became apparent (Supplemental Figure S4). Hxt3-GFP and Fas2-mcherry foci did not colocalize, indicating that their respective protein sequestrations are specific. We currently cannot exclude that Pma1-FKBP12 influences FAS foci formation at the plasma membrane. The experimental setup, however, still allowed us to test for physiological consequences upon prevention of FAS foci formation in the cytosol.
We tested the viability of cells at different time points using fluorescence-activated cell sorting (FACS) based on a sensitive dead cell–staining dye (Sytox green) (Figure 3C). We did not observe any significant effect on cell viability upon alteration of the site of FAS sequestration.
We therefore conclude that the primary purpose of sequestering FAS, and perhaps by extension other enzymes of lipid biosynthesis, is likely to confine them spatially, presumably restricting their access to one another and to substrates.
Starvation-induced sequestrations of lipid biosynthetic enzymes are reversible
To further characterize sequestration of lipid biosynthetic enzymes, we determined the protein dynamics in FAS foci by fluorescence loss in photobleaching (FLIP) experiments. Here a small area of stationary-phase cells distant to the Fas1-mcherry foci was continuously bleached, and the fluorescence intensity of the foci was monitored in parallel. Fluorescence intensity of the Fas1-mcherry foci dropped rapidly over the period of bleaching, indicating a rapid exchange of Fas1-mcherry molecules between the foci and the cytosol, also distinguishing FAS foci from immobile cytosolic aggregates of misfolded proteins (Specht et al., 2011; Figure 4A).
FIGURE 4:
Starvation-induced sequestrations of lipid biosynthetic enzymes are reversible. (A) FLIP measurements of Fas1-mcherry foci formed in stationary-phase cells. A cytosolic area was bleached, and loss of fluorescence intensities of Fas1-mcherry foci was determined. In bleaching controls, Fas1-mcherry fluorescence intensities of adjacent cells were followed. SEs are given (n = 20). (B) Fas1-mcherry sequestration is reversible—glucose alone is sufficient to trigger the resolubilization of Fas1-mcherry foci. S. cerevisiae cells harboring Fas1-mcherry foci after growth to stationary phase were resuspended in either PBS buffer, fresh SD medium, fresh SC medium, or PBS buffer containing 2% (wt/vol) glucose. CHX was added to block new protein synthesis. Maximum-intensity projections of z-stack images are shown. (C) Fas1-mcherry foci resolubilization is Hsp104 independent. Glucose-starved (day 4) or stationary-phase (day 10) hsp104Δ cells harboring Fas1-mcherry foci were treated with PBS containing glucose (2% wt/vol) and CHX. Scale bars: 2 μm. (D and E) ER, mitochondria, and other cytosolic enzyme sequestrations are reversible. PBS containing glucose (2% wt/vol) and CHX was added to glucose-starved cells (day 4) harboring Psd1-3Xmcherry (D), Pis1-3XGFP (E), or Acc1-GFP (F). Changes in localization were monitored at the indicated time points. Maximum-intensity projections of z-stack images are shown for D and E. Scale bars: 2 μm.
We next analyzed the fate of starvation-induced FAS sequestration upon replenishment of nutrients. FAS foci formed in stationary yeast cells rapidly dissolved upon addition of fresh complete medium (SD medium), and Fas1-mcherry redistributed uniformly in the cytosol (Figure 4B). Foci disintegration was complete after 120 min in most cells. Simultaneous addition of cycloheximide (CHX) did not prevent FAS foci disintegration, demonstrating that solubilization does not rely on protein synthesis and that the regained diffuse Fas1-mcherry fluorescence originates from the starvation-induced sequestrations (Figure 4B). Fas1-mcherry levels remained unaffected upon glucose and CHX addition, indicating that changes in Fas1-mcherry fluorescence intensities are caused by changes in localization (foci vs. diffuse; Supplemental Figure S5, A and B).
To determine the minimal nutrient requirement for the solubilization of Fas1-mcherry foci, we performed a series of experiments wherein either one or a combination of media components were added to the starved cells. Glucose alone was sufficient to trigger the solubilization of Fas1-mcherry foci, while other media components individually or in combination did not (Figure 4B). A nonmetabolizable glucose analogue, 2,4-deoxyglucose, did not trigger solubilization, indicating that metabolizing glucose is the prerequisite for disintegration of Fas1-mcherry foci (unpublished data). Solubilization of Fas1-mcherry foci upon glucose addition was not perturbed in hsp104Δ cells, confirming further that FAS foci are not misfolded protein aggregates (Figure 4C).
Given that FAS foci formation and dissolution are driven by nutrient availability, we monitored FAS sequestration in mutants of major nutrient-sensing pathways—TOR and Snf1 signaling pathways (tor1∆ and reg1∆), and in rho-minus cells that are defective in ATP production by respiration. In all the mutant strains tested, we observed a substantially faster sequestration of FAS as compared with wild-type (wt) cells (Supplemental Figure S6A). FAS foci were still reversed by the addition of glucose, demonstrating that only the kinetics of formation but not the dissolution of FAS foci is changed in the mutant cells (Supplemental Figure S6B). Sequestrations of other tested lipid biosynthetic enzymes (Pis1, Psd1, Acc1) dissolved at similar kinetics to regain their typical localizations upon addition of glucose (Figure 4, D–F). Levels of all tested enzymes remained constant upon glucose and CHX addition, indicating that fluorescence intensities are caused by changes in localization (foci vs. diffuse; Supplemental Figure S5, C–E).
These data support the involvement of mechanisms driven by nutrient limitation. Taken together, sequestrations of FAS and other lipid biosynthetic enzymes represent reversible enzyme assemblies.
Reversible reorganization of the ER and mitochondria during starvation
Not only cytosolic but also ER- and mitochondria-localized lipid biosynthetic enzymes accumulated into punctate structures upon prolonged glucose starvation (Figure 1B). We envisioned two possibilities to explain this observation. First, the lipid biosynthetic enzymes dissociate from their normal localization under unperturbed conditions to accumulate into punctate structures without the overall organization of the ER and mitochondria being affected. Second, ER and mitochondria themselves undergo spatial reorganization that causes redistribution of their resident enzymes.
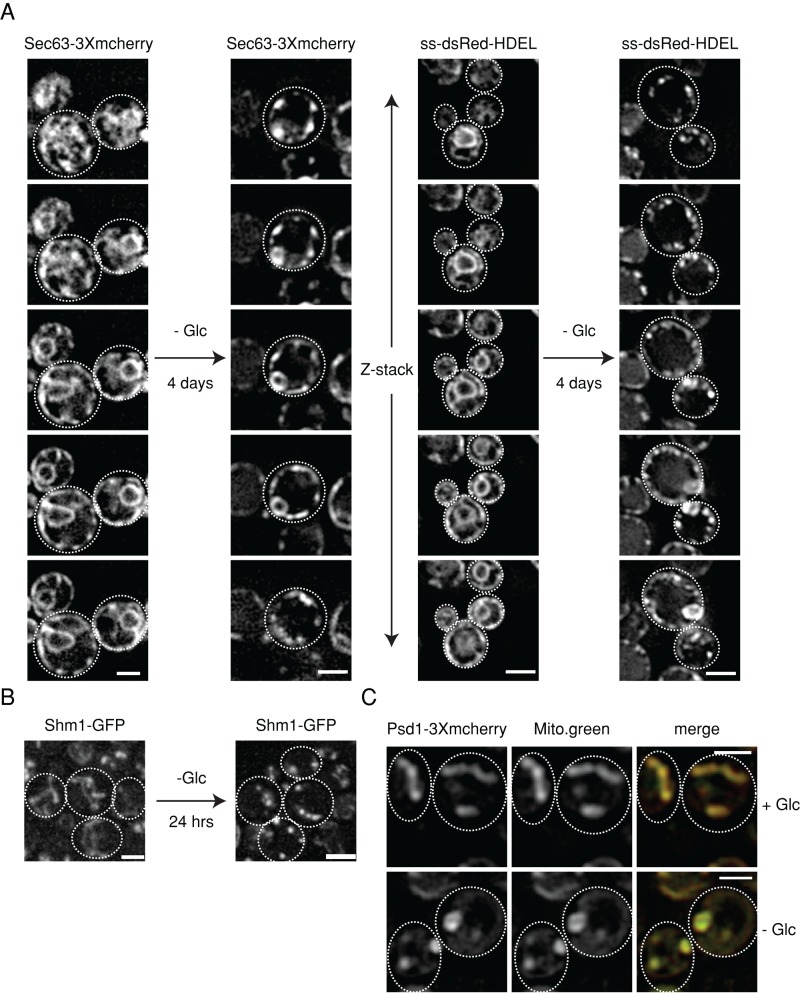
To differentiate between these possibilities, we stained ER and mitochondria with specific dyes (DiOC6 and MitoTracker green, respectively). DiOC6-stained ER appeared continuous upon glucose depletion in contrast to punctuate localization of Pis1-3Xmcherry (Supplemental Figure S7), suggesting that Pis1-3Xmcherry is sequestered to specific subdomains of the ER. To test whether other ER-resident proteins also change localization, we monitored Sec63-3xmcherry, ss-dsRed-HDEL (staining nuclear and cortical ER), and Rtn1-mcherry (staining only cortical ER) (Figures 5A and 6A). All tested ER proteins accumulated into punctate structures. Coexpression of Pis1-3Xmcherry and Sec63-GFP revealed little colocalization of starvation-induced foci, indicating that the observed protein sequestrations are specific (Supplemental Figure S7).
FIGURE 5:
Reversible reorganization of ER and mitochondria underlies the sequestration of their respective phospholipid biosynthetic enzymes upon starvation. (A) S. cerevisiae cells expressing the ER markers Sec63-3Xmcherry or ss-dsRed-HDEL were grown at 30°C to log phase or depleted of glucose (4 d). Changes in protein localizations were determined. The z-stacks of the imaged ER markers are shown. (B) S. cerevisiae cells expressing the mitochondrial marker Shm1-GFP were grown at 30°C to log phase or depleted of glucose (day 1). Changes in protein localizations were determined. Maximum-intensity projections of the z-stack images are shown. (C) Glucose starvation leads to reorganization of mitochondria. S. cerevisiae cells expressing Psd1-3Xmcherry were grown to log phase and starved of glucose for 1 d. Mitochondria were stained by MitoTracker green and colocalized with Psd1-3Xmcherry foci upon starvation. Scale bar: 2 μm.
FIGURE 6:
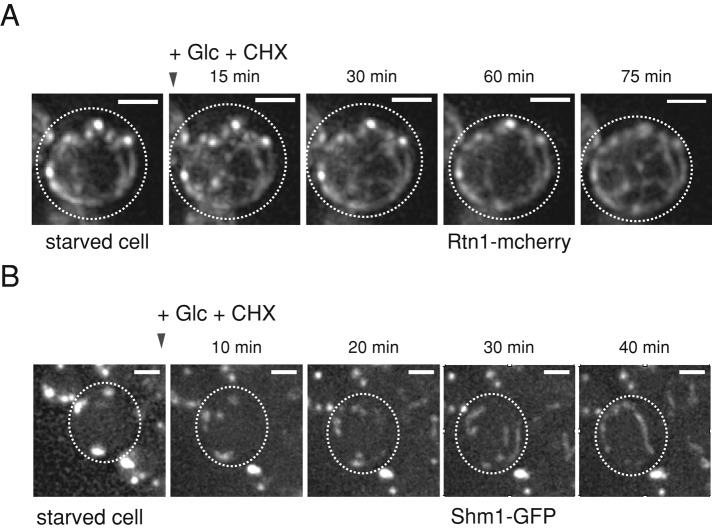
Sequestration of ER-resident proteins and mitochondrial morphological changes are reversible. (A) S. cerevisiae cells expressing the peripheral ER marker Rtn1-mcherry were depleted of glucose to induce punctate organization of the ER. Cells were resuspended in PBS buffer containing 2% (wt/vol) glucose and CHX, and changes in Rtn1-mcherry localizations were recorded at the indicated time points. (B) S. cerevisiae cells expressing Shm1-GFP were glucose starved to induce punctate organization of mitochondria. Cells were resuspended in PBS buffer containing 2% (wt/vol) glucose and CHX, and changes in protein localizations were recorded at the indicated time points. Scale bars: 2 μm.
Changes in mitochondrial organization upon glucose depletion were determined by expressing the marker protein Shm1-GFP and MitoTracker green staining (Figure 5, B and C). Shm1-GFP showed accumulation into punctate structures similar to the enzyme Psd1 (Figure 1B). Global reorganization of mitochondria upon glucose depletion was confirmed by MitoTracker green staining, demonstrating that changes in mitochondrial morphology underlie the observed changes in Psd1-3Xmcherry localization (Figure 5C).
We next tested whether the reorganization of mitochondria and ER proteins tested is reversible upon addition of glucose. The punctate structures of ER (Rtn1) and mitochondria (Shm1) markers regained their normal spatial organization within 120 min upon glucose addition (Figure 6, A and B), similar to the Pis1 (Figure 4D) and Psd1 (Figure 4E) enzymes. Levels of Rtn1-mcherry remained unaffected upon glucose and CHX addition, indicating that changes in Rtn1-mcherry fluorescence intensities are caused by changes in localization (foci vs. diffuse; Supplemental Figure S5F).
Interorganelle contact sites are reversibly lost upon starvation
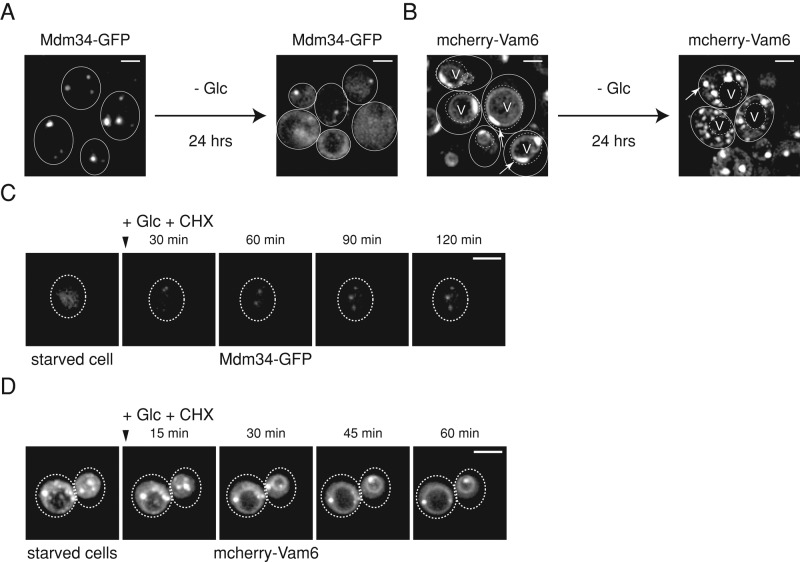
Organelles communicate with one another through contact sites (Elbaz and Schuldiner, 2011; Prinz, 2014). Mitochondria are dynamic organelles that are physically linked to the ER and vacuoles through the ERMES and vCLAMP contact sites, respectively (Kornmann et al., 2009; Elbaz-Alon et al., 2014; Honscher et al., 2014). Given that we observed global reorganization of ER and mitochondria, we tested whether the ERMES and vCLAMP contact sites were affected. We used genomically tagged Mdm34-GFP and mcherry-Vam6 as established markers for ERMES and vCLAMP, respectively (Kornmann et al., 2009; Elbaz-Alon et al., 2014; Honscher et al., 2014). The fusions harbored their typical localizations during logarithmic growth with dense foci of Mdm34-GFP with almost no cytosolic staining (Figure 7A, left panel) and vacuolar membrane and patch-like staining of mcherry-Vam6 (Figure 7B, left panel). However, upon 24 h of glucose depletion, both contact sites were severely affected, with mcherry-Vam6 losing its vacuolar membrane localization to accumulate into numerous cytosolic foci (Figure 7B, right panel), while Mdm34-GFP foci were drastically reduced in number and intensity owing to their acquired cytosolic localization (Figure 7A, right panel). Accordingly, Mdm34-GFP levels partially shifted from the insoluble to the soluble cell fraction upon starvation (Supplemental Figure S8A). Foci formation of mcherry-Vam6 was specific, as vacuoles, stained by either FM4-64 or Vph1-GFP, did not exhibit morphological changes upon glucose depletion (Supplemental Figure S8, B and C) Thus, concomitant with changes of ER and mitochondrial proteins, the contact site markers relocalize to the cytosol.
FIGURE 7:
ERMES and vCLAMP interorganelle contact sites are reversibly lost upon starvation coinciding with ER and mitochondrial reorganization. (A and B) Starvation leads to loss of ERMES and vCLAMP interorganelle contact sites. S. cerevisiae cells expressing Mdm34-GFP (A) or mcherry-Vam6 (B) were grown to log phase and depleted of glucose. Changes in localization of the markers were monitored before and after glucose depletion (day 1). (C and D) ERMES and vCLAMP sites are reestablished upon glucose addition. PBS containing 2% (wt/vol) glucose and CHX was added to starved (day 1) S. cerevisiae cells expressing either Mdm34-GFP (C) or mcherry-Vam6 (D). Changes in localization were monitored at indicated time points. Maximum-intensity projections of z-stack images are shown. Scale bars: 2 μm.
We next tested whether the contact sites can be reformed upon replenishment of glucose. Indeed, both Mdm34-GFP and mcherry-Vam6 regained their normal localizations (Figure 7, C and D), indicating re-formation of ERMES and vCLAMP. mcherry-Vam6 levels remained constant upon glucose and CHX addition, indicating that changes in mcherry-Vam6 fluorescence intensity are caused by changes in localization (foci vs. diffuse; Supplemental Figure S5G). Thus starvation-induced alterations in dynamics of ER proteins and mitochondria coincide with reversible loss of ERMES and vCLAMP.
Starvation causes qualitative and quantitative alterations in the lipid profile
De novo synthesis of phospholipids is compartmentalized in yeast, with phosphatidylcholine (PC), phosphatidylserine (PS), and phosphatidylinositol (PI) being synthesized in the ER (Carman and Han, 2011). Enzymes involved in phosphatidylethanolamine (PE) biosynthesis, however, are localized to the mitochondrial inner membrane (Psd1) and the vacuolar membrane (Psd2) (Trotter and Voelker, 1995). They catalyze conversion of PS to PE, thus necessitating shuttling of PS between the ER, mitochondria, and vacuole. ERMES and vCLAMP together are known to aid in PS flux across these organelles and thereby play a vital role in phospholipid homeostasis (Kornmann et al., 2009; Elbaz-Alon et al., 2014; Honscher et al., 2014; summarized in Figure 8A). Given that we observed loss of ERMES and vCLAMP upon 24 h of glucose depletion, we next tested whether their loss is reflected in alterations in lipid profile.
FIGURE 8:
Starvation-induced qualitative and quantitative alterations in lipid profiles. (A) Phospholipid biosynthesis pathways involve enzymes in the ER, mitochondria, and vacuole, thus necessitating transport of substrates between the compartments. Etn: ethanolamine; Cho: choline; CDP-DAG: cytidine diphosphate diacylglycerol; DAG: diacylglycerol. (B) S. cerevisiae cells were grown to log phase or starved of glucose. Total cell lysates were prepared at the indicated time points of glucose starvation, and lipids were analyzed by mass spectrometry. Relative levels of different phospholipid species were determined. Values from 10 independent biological replicates are plotted. §, *, and # refer to p < 0.05, t test compared with 0 h.
We therefore performed a detailed correlative lipidomic analysis of yeast cells subjected to glucose starvation, which to our knowledge is the first analysis of this kind to be performed. Total lipids were extracted from cells starved of glucose for different time periods (0–96 h) and analyzed quantitatively by mass spectrometry (Figure 8B; Supplemental Table S1). The data suggest that, upon glucose starvation, species-specific changes in phospholipids occurred. Total levels of PE sharply dropped within 1 d of glucose depletion, and PC levels decreased eventually during the starvation period. In contrast, levels of PI increased upon glucose starvation, whereas levels of PS remained unchanged (Figure 8B). These alterations are qualitatively consistent with those observed in mutants lacking both ERMES and vCLAMP (Elbaz-Alon et al., 2014). The more pronounced decrease in PE levels observed in the previous study (Elbaz-Alon et al., 2014) could be attributed to deletion of Psd2 activity on top of loss of contact sites. Phosphatidic acid (PA) is a common precursor for all phospholipid biosynthesis. The decreased flux of PS due to loss of contact sites might result in accumulation of PA levels leading to increased PI biosynthesis, an observation also made in a previous report (Elbaz-Alon et al., 2014). Thus loss of ERMES and vCLAMP coincides with global reorganization of ER and mitochondria, resulting in qualitative alterations in lipid profile.
DISCUSSION
The sequestration of a multiplicity of cellular factors is a widespread consequence of periods of scarcity in yeast cells. It is a matter of great debate whether starvation-induced protein sequestrations represent storage assemblies, strategies to aid in enhanced substrate channeling, or misfolded protein aggregates (An et al., 2008; Narayanaswamy et al., 2009; Noree et al., 2010; O'Connell et al., 2014; Petrovska et al., 2014). Most studies focused on individual enzymes of specific metabolic pathways without providing insights into the significance of their sequestrations to their respective metabolic pathways.
Here, in order to obtain functional insights into sequestration of enzymes involved in lipid homeostasis, we followed spatial localization of major fatty acid and phospholipid biosynthetic enzymes upon prolonged glucose starvation. The Fas2 subunit of FAS was reported to accumulate into foci in stationary-phase cells (Narayanaswamy et al., 2009). We find that this observation extends to the other FAS subunit, Fas1, as well, and both subunits cosequester, indicating that intact FAS complexes are sequestered. Interestingly, this phenomenon is not limited to FAS but extends to other enzymes of fatty acid, ergosterol, and phospholipid biosynthesis localized to the cytosol (Acc1), lipid droplets (Erg6), ER (Pis1), and mitochondria (Psd1). Importantly, these sequestrations do not cocluster, negating the possibility of cosequestrations to aid in efficient synthesis by enhanced substrate channeling as has been suggested for other enzymes previously shown to cocluster (An et al., 2008; Noree et al., 2010).
To characterize the nature of deposits of lipid biosynthetic enzymes, we chose FAS as a model owing to its importance in lipid metabolism and given its tendency to sequester under a variety of conditions (Cabiscol et al., 2000; Tyedmers et al., 2010; Jacobson et al., 2012). We demonstrate that FAS foci are not misfolded protein aggregates based on the observations that 1) they do not colocalize with the Hsp104 disaggregase, 2) they are dynamic and rapidly dissolve upon glucose addition in an Hsp104-independent process, and 3) they retain enzymatic activity. Thus nutrient limitation does not seem to globally affect protein folding, resulting in aggregation of a wide variety of metabolic enzymes, as recently suggested (O'Connell et al., 2014). Accordingly, the starvation-induced deposits of all tested proteins were unique and did not intermix, suggesting that sequestrations of distinct proteins follow specific pathways but are not caused by misfolding, which should lead to the coaggregation observed in many studies (Kaganovich et al., 2008; Tyedmers et al., 2010; Specht et al., 2011).
FAS foci formation might serve to restrict accessibility of FAS to its substrates during periods of scarcity or to increase efficiency of localized fatty acid synthesis. However, altering the site of FAS sequestration (Figure 3) does not affect cellular viability upon starvation, arguing against a role of FAS sequestration for site-specific localized enzymatic activity. Rather, the specific, nonmixing sequestration of lipid biosynthetic enzymes tested supports a role of these foci in spatially confining enzymes and thereby limiting access to their potential substrates, possibly regulating lipid homeostasis. Similarly, starvation-induced Gln1 rods and proteasomal granules have been suggested to represent storage compartments (Petrovska et al., 2014), implicating a general cellular strategy to sequester native proteins during times of scarcity. Given that lipid biosynthetic enzymes are known to interact with one another, aiding in metabolic flux under nutrient-rich conditions (Durek and Walther, 2008), we speculate that FAS sequestration and, by extension, sequestration of other lipid biosynthetic enzymes, while retaining enzymatic activity, might lower their accessibility to one another and their substrates by restricting their local availability and thereby altering lipid flux. Protein sequestration upon starvation might additionally protect the sequestered proteins from bulk degradation by autophagy. Accordingly, Fas1-GFP foci did not colocalize with PAS (Figure 2C). On addition of glucose, availability of FAS and other lipid biosynthetic enzymes are restored rapidly for lipid biosynthesis supporting the resumption of growth.
The trigger for sequestration of FAS and other lipid biosynthetic enzymes remains unknown. We also noticed that starvation-induced sequestration of different protein species occurs with different kinetics and deposition sites do not colocalize. This suggests the existence of various triggers and related pathways to initiate and control protein sequestrations. While changes in cytosolic pH have been identified as a sufficient trigger for formation of Gln1 rods and proteasomal storage granules during the stationary phase (Peters et al., 2013; Petrovska et al., 2014), they are unlikely to trigger sequestrations of FAS and other lipid biosynthetic enzymes, given that FAS is sequestered at a much later time point as compared with either 1) pH drop–induced translocation of the ER membrane–bound transcriptional repressor of phospholipid biosynthesis Opi1 into the nucleus in minutes after glucose depletion or 2) Gln1 rod formation (Young et al., 2010; Petrovska et al., 2014). We consider it likely that metabolite(s) feeding into lipid metabolism provide a starvation signal that, sensed by the enzymes, triggers foci formation. Given the different kinetics of enzyme sequestrations observed, different pathways or thresholds might be involved. The identification of cellular factors or intrinsic sequence and structural features of sequestered enzymes is required for a mechanistic understanding of starvation-induced protein sequestration.
We also show strong reversible reorganization of mitochondria that underlies punctate structure formation. Various ER proteins tested, including ER-resident lipid biosynthetic enzymes, also exhibit specific changes of localization upon glucose depletion. Similarly, the ER exit site marker Sec16 reversibly accumulates into dense punctate structures in Drosophila cells upon depletion of serum and amino acids (Zacharogianni et al., 2011). However, these massive morphological changes of various ER proteins during glucose starvation have not been previously reported. Mitochondrial fragmentation, similar to our observation of punctate organization, in response to glucose depletion has been reported for mammalian cells (Rambold et al., 2011). Also, Opa1-mediated modulation of mitochondrial cristae structure in response to starvation has been recently described in mammalian cells (Patten et al., 2014), further supporting possible conservation of our observations to higher eukaryotes. Therefore organellar reorganization seems to be a common theme in adaptation of cells to starvation. Our findings therefore extend the known starvation-induced localization changes from individual enzymes to entire organelles, showing that the cellular response to starvation occurs at different levels of cellular organization.
Recently vCLAMP contact sites between mitochondria and vacuoles were described in yeast; together with ERMES, they were proposed to play vital roles in phospholipid biosynthesis (Elbaz-Alon et al., 2014; Honscher et al., 2014). The cellular metabolic state was shown to reversibly regulate the formation of these contact sites (Honscher et al., 2014). Yeast cells lacking both contact sites and vacuole-localized Psd2 exhibited a significant drop in PC and PE levels and increased PI levels (Elbaz-Alon et al., 2014). Our observed changes in ER and mitochondrial morphology coincide with reversible loss of ERMES and vCLAMP contact sites reflected by changes in levels of specific lipids that are qualitatively identical to those previously reported (Elbaz-Alon et al., 2014). Moreover, yeast cells lacking the ERMES complex (mdm34Δ) and having an altered ER structure due to mutations in ER-shaping proteins (rtn1Δ yop1Δ) exhibit defects in PS shuttling that correlates with reduced PE steady-state levels (Voss et al., 2012), changes that are qualitatively similar to our own observations during glucose starvation. Interestingly, similar to our observations of the vCLAMP marker Vam6, which accumulates into reversible punctate structures in the cytosol upon glucose starvation, Nvj2, which marks the nucleus–vacuole contact sites, also accumulates into similar punctate structures upon exposure of cells to DTT (Breker et al., 2013). Thus alteration in interorganelle contact sites by spatial confinement of their constituent factor(s) into cytosolic deposits seems to be a conserved mechanism for cellular adaptation to stress.
Taken together, our data show regulation of lipid homeostasis upon glucose starvation at three different levels, reversible 1) spatial confinement of enzymes, 2) mitochondrial and ER protein reorganization, and 3) loss of organelle contact sites. These changes likely regulate flux through the lipid biosynthetic pathways to efficiently modulate lipid homeostasis in response to changing nutrient availability. Direct evidence for a functional relationship remains to be explored, and other mechanisms, such as those involving regulation of activity or stability of cellular factors involved in phospholipid biosynthesis and flux, might also contribute to the observed alterations. We suggest that the sequestration of enzymes allows regulation of lipid homeostasis without affecting the enzymatic properties of individual enzymes such that, upon entry into favorable growth conditions, cells can quickly alter their lipid flux by simply relocalizing their enzymes.
MATERIALS AND METHODS
Yeast strains, media, and growth conditions
mcherry- and GFP-tagged chromosomal constructs or gene knockouts were created using optimized PCR-amplified cassettes (Janke et al., 2004). Supplemental Table S2 lists all strains used in this study; Supplemental Table S3 includes all plasmids used. Synthetic medium (SD medium) was used for cell growth at 30°C. For glucose starvation, cells were washed twice with water and transferred to SD medium lacking glucose. For replenishment of nutrients, stationary-phase or glucose-depleted cells were harvested by centrifugation, resuspended in either SD medium or SC medium or phosphate-buffered saline (PBS) buffer with or without glucose (2% wt/vol). In all these cases, CHX was added to a final concentration of 100 μg/ml. In some experiments, 3 mM guanidiniume hydrochloride was added to inhibit Hsp104 activity.
Anchor-away and viability assays
Cells coexpressing Pma1-FKBP12 and Fas2-mcherry with either Fas1-FRB or untagged Fas1 were glucose depleted as described. Either dimethyl sulfoxide (DMSO) or rapamycin (1 μg/ml) was added to the cells immediately after glucose starvation. Localization of Fas2-mcherry was monitored within minutes to ensure that rapamycin treatment led to anchoring-away of FAS. Subsequently, aliquots of cultures were taken on different days of starvation, and viability was measured by Sytox green–based sorting of dead cells. Briefly, 1 OD600 of cells were harvested and resuspended in PBS buffer. A 5 mM Sytox green solution (Invitrogen, Darmstadt, Germany) was used to stain cells at 10,000 times dilution for 10 min at room temperature. The cells were harvested by centrifugation, washed in PBS, and immediately analyzed by FACS (BD FACS CantoTM II) using a 488-nm laser. A total of 20,000 events were measured at each time point per repetition. Dead cells being stained by the dye can be separated, and the percentage of viable cells can therefore be calculated.
Image acquisition, processing, and data analysis
Cells were treated as indicated, harvested by centrifugation, and resuspended in PBS. Optical sections of 0.2 μm were acquired to image the whole cell volume using a wide-field system (xcellence IX81, Olympus) equipped with a Plan-Apochromat 100×/1.45 NA oil-immersion objective and an EMCCD camera (Hamamatsu ORCA-R2 or Hamamatsu ImagEM Enhanced [C9100-13] or Hamamatsu EM-CCD [C9100-02]). Acquired z-stacks were deconvolved with xcellence software using the Wiener Filter. All further processing of digital images was performed with ImageJ (http://imagej.nih.gov/ij).
Time-lapse microscopy
For sample preparation, a round-bottom dish was coated with concanavalin A (ConA). Cells were adhered on the ConA-coated coverslip for 15 min, unbound cells were removed by washing, and a final volume of 3 ml of PBS buffer containing 2% (wt/vol) glucose and 100 μg/ml of CHX was added. Image acquisition was started simultaneously, and images were acquired using the xcellence IX61 microscope using a Plan-Apochromat 100×/1.45 NA oil-immersion objective with autofocusing hardware during the course of the experiment. Fourteen optical sections of 0.2 μm were acquired to image the whole cell volume. Stacks were deconvolved as described.
Immunofluorescence
For immunostaining, cells were fixed with 4% (vol/vol) p-formaldehyde/100 mM KPi (Sigma Aldrich) for 1 h before cell wall digestion with 500 μg/ml Zymolase T-100 in wash buffer (1.2 M sorbitol/100 mM KPi pH 6.5) supplemented with 20 mM β-mercaptoethanol at 30°C. Spheroblasts were attached to poly-lysine–coated cover slides, permeabilized by being washed three times with 1% Triton X-100/100 mM KPi (pH 6.4), and blocked for 1 h with 1% (wt/vol) bovine serum albumin (BSA) in 100 mM KPi (pH 6.4). Mouse anti-myc antibody (clone 9E10) at a dilution of 1:500 in blocking buffer was applied to spheroplasts, which were incubated for 2 h at room temperature. Spheroplasts were rinsed in blocking buffer three times. Alexa Fluor 488 goat anti-mouse immunoglobulin G (H+L) secondary antibody (Invitrogen) was applied, and spheroplasts were incubated at room temperature for 1 h. Following secondary antibody incubation, spheroplasts were embedded in 55% glycerol and stained.
ER, mitochondria, and vacuole staining
ER of glucose-starved cells (OD600 = 1) was stained with DiOC6 (5 ng/ml) for 10 min and washed three times in PBS buffer before being imaged with a GFP filter. Mitochondria were stained by MitoTracker green (50 nM, Invitrogen) for 20 min at 30°C, washed three times with PBS buffer, and imaged using a GFP filter under the fluorescence microscope. Vacuoles were imaged by staining cells with FM4-64 (20 μM; Invitrogen) for 15 min at 30°C. The dye was washed out, and cells were incubated on either complete medium (for log-phase cultures) or media lacking glucose (for starved cells) and incubated for 60 min at 30°C. Cells were imaged using an mcherry filter under the fluorescence microscope.
FLIP measurements
For FLIP analysis, cells were grown to stationary phase and immobilized on ConA-coated glass-bottom culture dishes. FLIP measurements were performed at 25°C for 20 individual cells. A defined area was bleached 27 times for 380 s each time. Laser intensities for bleaching were set to 100% for the 561-nm laser. Laser settings for imaging were set to 3% for the 561-nm laser. Measurements were performed at 25°C on a confocal microscope (LSM 780; Carl Zeiss) equipped with a 63×/1.40 NA oil objective lens and 561-nm laser lines for mcherry image acquisition and photobleaching. The resulting loss of fluorescence in the region of interest (ROI) as a function of time provides a measure of the relative exchange rate with the bleached cytoplasmic fraction of molecules. Quantification of FLIP experiments was performed using ZEN 2010 software (Carl Zeiss). The fluorescence intensity of the background (Fbg), the ROI at a particular time point (Ft), and the fluorescence intensity before the first bleach (F0) were determined. For calculating the loss of fluorescence at a particular time point, the formula (Ft − Fbg)/(F0 − Fbg) × 100 was used. Curves represent the mean of 20 cells and the corresponding SE.
Correlative fluorescence microscopy and electron tomography
Sample preparation, data acquisition, and the correlation procedure were essentially done as previously described (Kukulski et al., 2011, 2012), with minor modifications. Cultures grown under starvation conditions (4 d) and, in parallel, to log phase, of yeast cells expressing Fas1-mCherry and Sec63-GFP were pelleted by vacuum filtration (McDonald, 2007), loaded into aluminum planchettes (0.1/0.2 mm), covered with the flat side of aluminum planchettes (0.3 mm; Engineering Office M. Wohlwend, Sennwald, Switzerland), and cryo-fixed using a BAL-TEC HPM-010 high-pressure freezer. Samples were freeze substituted with 0.1% uranyl acetate in acetone, embedded in Lowicryl HM20 resin, and sectioned according to a previously published protocol (Kukulski et al., 2011). EM grids with 320-nm resin sections were incubated section face down for 10 min on 15 μl drops of 50-nm TetraSpeck microspheres (Life Technologies, Carlsbad, CA) diluted 100 times in PBS. Grids were washed with water and blotted three times. For fluorescence microscopy, EM grids were placed between two coverslips in a layer of water, as depicted in Kukulski et al. (2012). Imaging was done on an Olympus IX81 microscope with a 100×/1.45 NA objective and an Orca-ER camera (Hamamatsu). An X-Cite 120 PC lamp (EXFO) was used for fluorescence imaging. Excitation filters for GFP, mCherry, and blue channel were 470/22, 556/20, and 377/50 nm, respectively. Emission filters were 520/35, 624/40, and 520/35 nm, respectively. The blue channel was recorded for distinguishing the TetraSpeck from colocalizing GFP and mCherry signals. Fifteen-nanometer protein A–coupled gold beads were adhered to both faces of the grids as fiducial markers for tomographic reconstructions, and sections were stained with Reynolds lead citrate for 15 min. Electron tomographic tilt series were acquired using SerialEM software (Mastronarde, 2005) on a FEI TF30 microscope operated at 300 kV and equipped with a FEI 4k Eagle CCD camera, using a dual-axis tomography holder (Fischione Model 2040). Dual-axis tilt series were acquired from −60 to 60° at 1° increments and 1.18-nm pixel size. For fiducial-based correlation, lower-magnification single-axis tilt series were acquired from −60 to 60° at 3° increments and 2.53-nm pixel size. The IMOD software was used for tomogram reconstruction (Kremer et al., 1996). For location of fluorescence signals in electron tomograms, the correlation procedure described in Kukulski et al. (2012) was applied. In brief, virtual slices in which TetraSpeck microspheres on the section surface were visible were selected from the low-magnification tomogram and averaged using ImageJ. Centroids of TetraSpeck signals in the same channel as the fluorescence signal of interest (RFP for Fas1-mCherry) were determined with subpixel accuracy and assigned to the corresponding positions in the averaged EM image using the MATLAB Control Point Selection Tool. Using TetraSpeck microspheres as fiducial markers permitted direct correlation to the fluorescence channel of interest, avoiding the need to correct for drift between fluorescence images (Kukulski et al., 2011). The transform between fluorescence and electron microscopic images was determined based on the pairs of coordinates and was used to map the subpixel fitted centroid of the fluorescence spot of interest onto the low-magnification electron tomogram. In a second step, the coordinates of the predicted fluorescence spot position were transformed to the high-magnification tomogram in an analogous procedure based on the 15-nm gold beads.
Protein solubility and FAS activity assay
Log-phase or glucose-depleted yeast cells were harvested by centrifugation, resuspended in 50 mM Tris/HCl, 500 mM NaCl (pH 8.5) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF], 5 μg/ml leupeptin, 10 μg/ml pepstatin, 8 μg/ml aprotinin, protease inhibitor mix FY [Serva]) and flash frozen in liquid nitrogen before pulverization by repeated mixer milling (MM 400 [Retsch]; 30 Hz, 2 min). Cell lysates were precleared by centrifugation at 3000 × g for 5 min, and total protein concentration of supernatants was measured by Bradford (Bio-Rad) before the soluble and insoluble fractions were separated by centrifugation at 16,000 × g for 20 min. The soluble fraction was removed and its protein concentration measured, while the pellets were washed once with 50 mM Tris/HCl 150 mM NaCl (pH 8.5) supplemented with protease inhibitors, centrifuged at 16,000 × g for 20 min, and resuspended in 50 mM Tris/HCl, 500 mM NaCl, 8 M urea, 2% (wt/vol) SDS, 2 mM DTT (pH 8.5) supplemented with protease inhibitors. SDS–PAGE and Western blotting with anti–YFP antisera were used to analyze distribution of Fas1-GFP to soluble or insoluble fractions.
For determining FAS activity (Matias et al., 2011), total lysates and supernatants of log-phase and glucose-depleted cells were added to 700 μl of FAS activity buffer (0.1 M potassium phosphate buffer pH 6.5, 2.5 mM EDTA, 10 mM cysteine, 0.3 mg/ml BSA, 0.24 mM acetyl CoA, 0.15 mM NADPH), and activity was measured by addition of malonyl-CoA (0.28 mM) with conversion of NADPH to NADP at 340 nm as a readout using a spectrophotometer (Shimadzu UV-1601; Matias et al., 2011). Cerulenin (20 μM), a specific inhibitor of FAS, was used to monitor specificity of the activity measured.
Cell fractionation
S. cerevisiae log-phase and glucose-starved cells (24 h) were harvested, washed in water, and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 5 μg/ml leupeptin, 10 μg/ml pepstatin, 8 μg/ml aprotinin, protease inhibitor mix FY [Serva]). PMSF (1 mM) was added before cell lysis. Glass beads (0.1 mm) were added, and cells were lysed using the FastPrep-24 Instrument (MP Biomedicals). Lysates were centrifuged at 1300 × g to remove nonlysed cells. Protein concentrations of the lysate (T) were measured by Bradford, and an aliquot of the lysate was centrifuged at 16,000 × g for 15 min. Protein concentrations of the resulting supernatants (S) were measured by Bradford. The pellets were washed with water once and resuspended in 50 mM Tris/HCl (pH 8.5), 500 mM NaCl, 8 M urea, 2% (wt/vol) SDS, 2 mM DTT (P). Equal amounts of total, supernatant, and pellet fractions were loaded on a gel and blotted using the appropriate antibodies.
Lipid analysis
Lipids were extracted and analyzed by mass spectrometry as previously described (da Silveira Dos Santos et al., 2014). Briefly, 25 OD of cells were resuspended in 1.5 ml of extraction solvent (ethanol, water, diethyl ether, pyridine, and 4.2 N ammonium hydroxide [15:15:5:1:0.018, vol/vol]). A mixture of internal standards (7.5 nmol 17:0/14:1 PC, 7.5 nmol 17:0/14:1 PE, 6.0 nmol 17:0/14:1 PI, 4.0 nmol 17:0/14:1 PS, 1.2 nmol C17:0-ceramide, and 2.0 nmol C8-glucosylceramide) and 250 μl of glass beads were added; the sample was vortexed vigorously (Multi-tube vortexer; Lab-tek International, Christchurch, New Zealand) for 5 min and incubated at 60°C for 20 min. Cell debris was pelleted by centrifugation at 1800 × g for 5 min, and the supernatant was collected. The extraction was repeated once, and the supernatants were combined and dried under a stream of nitrogen or under vacuum in a Centrivap (Labconco Corporation, Kansas City, MO). Half of the sample was used for ceramide and complex sphingolipid analysis, in which we performed an extra step to deacylate glycerophospholipids using monomethylamine reagent (methanol, water, n-butanol, methylamine solution [4:3:1:5, vol/vol]; Cheng et al., 2001). For desalting, both lipid extracts were resuspended in 300 μl of water-saturated butanol and sonicated for 5 min. LC-MS grade water (150 μl) was added and samples were vortexed and centrifuged at 3200 × g for 10 min to induce phase separation. The upper phase was collected, and the process was repeated twice. The combined upper phases were dried and kept at −80°C until analysis. For glycerophospholipid and sphingolipid analysis by electro-spray ionization mass spectrometry (ESI-MS/MS), extracts were resuspended in 500 μl of chloroform:methanol (1:1, vol/vol) and diluted in chloroform:methanol:water (2:7:1, vol/vol/vol) and chloroform:methanol (1:2, vol/vol) containing 5 mM ammonium acetate for positive and negative mode, respectively. A Triversa Nanomate (Advion, Ithaca, NY) was used to infuse samples with a gas pressure of 30 psi and a spray voltage of 1.2 kV on a TSQ Vantage (ThermoFisher Scientific, Waltham, MA). The mass spectrometer was operated with a spray voltage of 3.5 kV in positive mode and 3 kV in negative mode. The capillary temperature was set to 190°C. Multiple-reaction monitoring mass spectrometry was used to identify and quantify lipid species (Guan et al., 2010). Data were converted and quantified relative to standard curves of internal standards, which were spiked in before extraction. Values are averages ±SD of three independent samples.
Supplementary Material
Acknowledgments
We thank Matthias Meurer, Michael Knop, Yasuhiro Araki, Yoshinori Ohsumi, and Sebastian Schuck for sharing plasmids and yeast strains. W.K. was a postdoctoral researcher in the laboratory of John A. G. Briggs, where the correlative light and electron microscopy experiments were performed. W.K. was supported by a fellowship from the Swiss National Science Foundation. This study made use of the EMBL Electron Microscopy Core Facility. H.G.S. was supported by the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology. This work was supported by a Deutsche Forschungsgemeinschaft grant (SFB1036) to A.M. and B.B. and by grants from the NCCR Chemical Biology, SystemsX.ch, and the Swiss National Science Foundation to H.R.
Abbreviations used:
- BSA
bovine serum albumin
- CHX
cycloheximide
- ConA
concanavalin A
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- ERMES
ER–mitochondria encounter structure
- FACS
fluorescence-activated cell sorting
- FAS
fatty acid synthetase
- FLIP
fluorescence loss in photobleaching
- FRB
FKBP12-rapamycin–binding domain
- GFP
green fluorescent protein
- PA
phosphatidic acid
- PAS
preautophagosomal structure
- PBS
phosphate-buffered saline
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- ROI
region of interest
- vCLAMP
vacuole and mitochondria patch
- wt
wild-type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1559) on March 11, 2015.
*Current address: Division of Cell Biology, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.
REFERENCES
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol. 2013;200:839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu Rev Biochem. 2011;80:859–883. doi: 10.1146/annurev-biochem-060409-092229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Park TS, Fischl AS, Ye XS. Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol Cell Biol. 2001;21:6198–6209. doi: 10.1128/MCB.21.18.6198-6209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira Dos Santos AX, Riezman I, Aguilera-Romero MA, David F, Piccolis M, Loewith R, Schaad O, Riezman H. Systematic lipidomic analysis of yeast protein kinase and phosphatase mutants reveals novel insights into regulation of lipid homeostasis. Mol Biol Cell. 2014;25:3234–3246. doi: 10.1091/mbc.E14-03-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Walther D. The integrated analysis of metabolic and protein interaction networks reveals novel molecular organizing principles. BMC Sys Biol. 2008;2:100. doi: 10.1186/1752-0509-2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Galdieri L, Mehrotra S, Yu S, Vancura A. Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS J Integr Biol. 2010;14:629–638. doi: 10.1089/omi.2010.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Guan XL, Riezman I, Wenk MR, Riezman H. Yeast lipid analysis and quantification by mass spectrometry. Methods Enzymol. 2010;470:369–391. doi: 10.1016/S0076-6879(10)70015-X. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Honscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Jacobson T, Navarrete C, Sharma SK, Sideri TC, Ibstedt S, Priya S, Grant CM, Christen P, Goloubinoff P, Tamas MJ. Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J Cell Sci. 2012;125:5073–5083. doi: 10.1242/jcs.107029. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods Cell Biol. 2012;111:235–257. doi: 10.1016/B978-0-12-416026-2.00013-3. [DOI] [PubMed] [Google Scholar]
- Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J Cell Biol. 2011;192:949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Salin B, Daignan-Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Matias AC, Marinho HS, Cyrne L, Herrero E, Antunes F. Biphasic modulation of fatty acid synthase by hydrogen peroxide in Saccharomyces cerevisiae. Arch Biochem Biophys. 2011;515:107–111. doi: 10.1016/j.abb.2011.08.009. [DOI] [PubMed] [Google Scholar]
- McDonald K. Cryopreparation methods for electron microscopy of selected model systems. Methods Cell Biol. 2007;79:23–56. doi: 10.1016/S0091-679X(06)79002-1. [DOI] [PubMed] [Google Scholar]
- Michaillat L, Baars TL, Mayer A. Cell-free reconstitution of vacuole membrane fragmentation reveals regulation of vacuole size and number by TORC1. Mol Biol Cell. 2012;23:881–895. doi: 10.1091/mbc.E11-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JD, Tsechansky M, Royall A, Boutz DR, Ellington AD, Marcotte EM. A proteomic survey of widespread protein aggregation in yeast. Mol Biosyst. 2014;10:851–861. doi: 10.1039/c3mb70508k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten DA, Wong J, Khacho M, Soubannier V, Mailloux RJ, Pilon-Larose K, MacLaurin JG, Park DS, McBride HM, Trinkle-Mulcahy L, et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 2014;33:2676–2691. doi: 10.15252/embj.201488349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LZ, Hazan R, Breker M, Schuldiner M, Ben-Aroya S. Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J Cell Biol. 2013;201:663–671. doi: 10.1083/jcb.201211146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovska I, Nuske E, Munder MC, Kulasegaran G, Malinovska L, Kroschwald S, Richter D, Fahmy K, Gibson K, Verbavatz JM, Alberti S. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife. 2014;3:e02409. doi: 10.7554/eLife.02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J Cell Biol. 2014;205:759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Voelker DR. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- Zacharogianni M, Kondylis V, Tang Y, Farhan H, Xanthakis D, Fuchs F, Boutros M, Rabouille C. ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 2011;30:3684–3700. doi: 10.1038/emboj.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.