Two UNC-87 isoforms with seven calponin-like repeats are expressed widely in muscle and nonmuscle cells in Caenorhabditis elegans. They bind to actin and myosin and inhibit actomyosin motility in vitro. unc-87 mutation enhances contraction of nonstriated muscle in vivo, suggesting that UNC-87 isoforms are negative regulators of actomyosin contractility.
Abstract
Calponin-related proteins are widely distributed among eukaryotes and involved in signaling and cytoskeletal regulation. Calponin-like (CLIK) repeat is an actin-binding motif found in the C-termini of vertebrate calponins. Although CLIK repeats stabilize actin filaments, other functions of these actin-binding motifs are unknown. The Caenorhabditis elegans unc-87 gene encodes actin-binding proteins with seven CLIK repeats. UNC-87 stabilizes actin filaments and is essential for maintenance of sarcomeric actin filaments in striated muscle. Here we show that two UNC-87 isoforms, UNC-87A and UNC-87B, are expressed in muscle and nonmuscle cells in a tissue-specific manner by two independent promoters and exhibit quantitatively different effects on both actin and myosin. Both UNC-87A and UNC-87B have seven CLIK repeats, but UNC-87A has an extra N-terminal extension of ∼190 amino acids. Both UNC-87 isoforms bind to actin filaments and myosin to induce ATP-resistant actomyosin bundles and inhibit actomyosin motility. UNC-87A with an N-terminal extension binds to actin and myosin more strongly than UNC-87B. UNC-87B is associated with actin filaments in nonstriated muscle in the somatic gonad, and an unc-87 mutation causes its excessive contraction, which is dependent on myosin. These results strongly suggest that proteins with CLIK repeats function as a negative regulator of actomyosin contractility.
INTRODUCTION
Muscle contractile apparatuses are highly organized, with properly arranged actin and myosin filaments, together with regulatory components for contractility (Clark et al., 2002). To produce efficient contractile forces, bipolar myosin filaments and actin filaments with proper orientations need to be assembled into sarcomeric patterns. Additional structural and regulatory proteins support integrity and dynamics of the contractile structures and modulate the actin–myosin interaction (Squire, 1997; Ono, 2010). Such mechanisms are important not only in muscle but also in nonmuscle cells when transient or persistent contractile apparatuses are required to support cell migration, cell division, and morphogenesis (Pollard and Cooper, 2009). However, how actomyosin-based contractile structures are assembled and regulated in vivo is not fully understood.
Calponin is a cytoskeletal protein that is expressed in both muscle and nonmuscle cells and has biochemical activities to stabilize actin filaments and regulate actomyosin contractility (Rozenblum and Gimona, 2008). Of note, calponin has been characterized as a major component of smooth muscle thin filaments in vertebrates (Takahashi et al., 1988). In vitro, calponin binds to actin filaments and myosin and inhibits actomyosin ATPase in a Ca2+-calmodulin–regulated manner (Abe et al., 1990; Winder and Walsh, 1990). Consistently, in skinned smooth muscle cells, calponin is required for maintaining a relaxed state by inhibiting contractility caused by unphosphorylated myosin (Malmqvist et al., 1997). However, in vivo, calponin-knockout mice show only mild alterations in smooth muscle contractility (Matthew et al., 2000; Takahashi et al., 2000; Yoshimoto et al., 2000). This may be due to the presence of functionally overlapping proteins such as caldesmon (Hodgkinson, 2000) and/or structurally related proteins such as SM22/transgelin (Pearlstone et al., 1987).
Vertebrate calponins have one calponin-homology (CH) domain in the N-terminus and three calponin-like (CLIK) repeats (or motifs) in the C-terminus (Rozenblum and Gimona, 2008). Although CH domains are found in a number of signaling or cytoskeletal proteins and function as an actin-binding domain in many cases, the CH domain of calponin does not bind to actin (Gimona and Mital, 1998; Galkin et al., 2006). Instead, the CLIK repeats in the C-terminus of calponins are essential for binding to actin filaments (Gimona and Mital, 1998). The number of CLIK repeats in naturally occurring proteins correlates with the strength of actin-binding. SM22/transgelin, which has only one CLIK repeat, binds to actin at lower affinity than calponin, which has three CLIK repeats (Shapland et al., 1993). UNC-87 in the nematode Caenorhabditis elegans is a unique protein with seven CLIK repeats and no CH domain (Goetinck and Waterston, 1994a), which binds to actin filaments with high affinity (Kranewitter et al., 2001). Of importance, proteins with multiple CLIK repeats stabilize actin filaments and prevent depolymerization in vitro (Kake et al., 1995; Yamashiro et al., 2007). Studies in mammalian cells have shown a correlation between the number of CLIK repeats and the actin-stabilizing function (Gimona et al., 2003) and as a consequence to inhibit cell migration and cytokinesis (Lener et al., 2004).
The biochemical analysis of C. elegans UNC-87 protein unambiguously demonstrated the function of the CLIK repeat as an actin-binding motif (Kranewitter et al., 2001). The C. elegans unc-87 gene is essential for proper sarcomeric actin organization in larval and adult striated muscle but not for initial sarcomere assembly in embryos (Goetinck and Waterston, 1994a, b). UNC-87 competes with actin-depolymerizing factor (ADF)/cofilin for binding to actin filaments and protects actin filaments from severing by ADF/cofilin (Yamashiro et al., 2007). Although the actin-stabilizing role of UNC-87 is similar to that of tropomyosin (Ono and Ono, 2002; Yu and Ono, 2006), UNC-87 and tropomyosin apparently have opposite roles in sarcomere organization in striated muscle (Yamashiro et al., 2007), suggesting that UNC-87 has an additional, uncharacterized function other than actin-filament stabilization. Furthermore, the initial analysis of the unc-87 gene identified two gene products with different N-terminal sequences, but functional differences between the two isoforms are unknown. In this study, we report that the two UNC-87 isoforms are expressed in different tissues in C. elegans and bind to both actin and myosin. This results in inhibition of actomyosin motility and formation of ATP-resistant actomyosin bundles. Genetic analysis shows that UNC-87B is a negative regulator of myosin-dependent contractility of smooth muscle–like cells in the somatic gonad. These results reveal a novel function of UNC-87 isoforms as regulators of actomyosin contractility.
RESULTS
Two alternative promoters drive expression of two UNC-87 isoforms in a tissue-specific manner
The original report of the molecular characterization of the unc-87 gene demonstrated that two isoforms are expressed from the unc-87 gene by an alternative choice of the first two exons (Goetinck and Waterston, 1994a; Supplemental Figure S1, A–C). According to WormBase (www.wormbase.org), the large and small isoforms have been designated UNC-87A (565 amino acids) and UNC-87B (374 amino acids), respectively. To maintain consistency, the downstream first exon used in UNC-87A is designated exon 1A, and the upstream exon used in UNC-87B is designated exon 1B (Supplemental Figure S1A). To determine the mechanism of isoform-specific expression, we examined whether exon 1A or 1B is selectively expressed by independent promoters. We hypothesized that the flanking upstream sequence of each first exon contains promoter activity. To test this, we fused 2-kb upstream sequences from exons 1A and 1B with green fluorescent protein (GFP) as a reporter (Supplemental Figure S1, D and E) and examined their promoter activity in vivo.
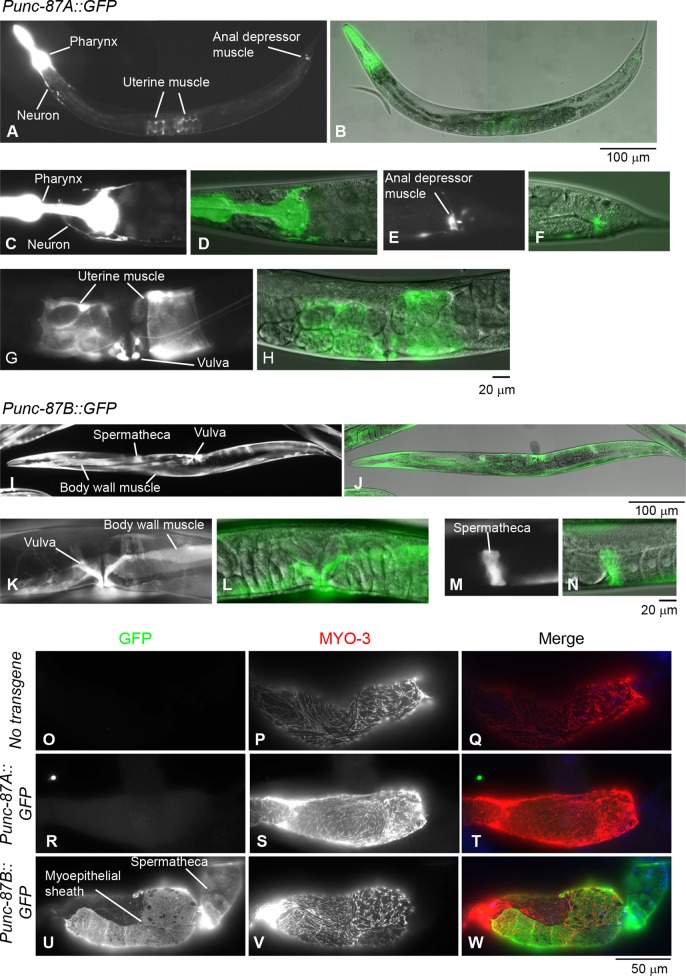
The reporter analysis showed that expression of exons 1A and 1B was driven by separate promoters in a tissue-specific manner. The 2-kb upstream sequence from exon 1A, which is entirely downstream of exon 1B (Supplemental Figure S1D; Punc-87A::GFP), promoted expression of GFP in the pharynx (Figure 1, A–D), anal depressor muscle (Figure 1, A, E, and F), uterine muscle (Figure 1, A, G, and H), vulva (Figure 1, G and H), and unidentified neurons in the head and the ventral region (Figure 1, A, C, and D). By contrast, the 2-kb upstream sequence from exon 1B (Supplemental Figure S1E; Punc-87B::GFP) promoted expression of GFP in the body wall muscle (Figure 1, I–L), spermatheca (Figure 1, I, J, M, and N), and vulva (Figure 1, I–L). Because the hermaphroditic gonads were obscured by strong GFP signals in the body wall muscle, expression of GFP was further analyzed in dissected gonads by immunofluorescence staining for GFP and MYO-3, a myosin heavy chain expressed in the myoepithelial sheath (Ardizzi and Epstein, 1987). The results showed predominant expression of GFP from the unc-87B promoter both in the myoepithelial sheath (MYO-3 positive) and the spermatheca (MYO-3 negative; Figure 1, U–W) but not from the unc-87A promoter (Figure 1, R–T) or in the gonad with no transgene (Figure 1, O–Q). These results suggest that separate promoters for unc-87A and unc-87B are tissue specific in a mutually exclusive manner except for the vulva and control expression of the two UNC-87 isoforms.
FIGURE 1:
Promoter analysis of unc-87A and unc-87B. (A–N) Promoter activity of unc-87A (A–H) and unc-87B (I–N) was examined using GFP as a reporter. Each fluorescence micrograph of GFP (black and white) is paired with a differential interference contrast image overlaid with green fluorescence of GFP. Images of whole worms with their heads on the left (A, B, I, and J) and magnified views of representative tissues (C–H, K–N) are shown. Note that the micrograph in C is somewhat overexposed for GFP in the pharynx to demonstrate relatively weak GFP in the neurons. (O–W) Expression of GFP in hermaphrodite gonads with no transgene (O–Q), Punc-87A::GFP (R–T), or Punc-87B::GFP (U–W) was examined by immunostaining of dissected gonads for GFP (O, R, and U) and MYO-3 as a marker of myoepithelial sheath (P, S, and V). Merged images are shown in Q, T, and W (green, GFP; red, MYO-3; blue, DNA [not shown as individual images]). Scale bars, 100 μm (whole worms; A, B, I, and J), 20 μm (magnified tissues; C–H,K–N), 50 μm (dissected gonads; O–W).
Previous studies reported that unc-87 mutations cause disorganized sarcomeric actin filaments in the body wall muscle (Goetinck and Waterston, 1994a, b), but the nature of mutation sites has not been characterized at a molecular level. We determined mutation sites in two unc-87 alleles: unc-87(e1216) shows mild defects in body wall muscle, and unc-87(e1459) shows very severe defects (Goetinck and Waterston, 1994a). We sequenced the genomic sequences and found that unc-87(e1216) had a point mutation (G to A) at the splice donor site in exon 2 (Supplemental Figure S1F) and unc-87(e1459) had two point mutations in exon 2, a missense mutation (A207V in UNC-87A or A16V in UNC-87B) and a nonsense mutation (W225stop in UNC-87A or W34stop in UNC-87B; Supplemental Figure S1F). Therefore these mutations are located in common exons for UNC-87A and UNC-87B and should affect both isoforms. unc-87(e1216) expresses reduced levels of UNC-87 proteins (Goetinck and Waterston, 1994a), suggesting that the mutation reduces efficiency of splicing but does not completely inhibit protein expression, thereby acting as a weak loss-of-function allele. By contrast, unc-87(e1459) does not express detectable UNC-87 proteins (Goetinck and Waterston, 1994a), indicating that this is a null or a strong loss-of-function mutant.
UNC-87A, a longer isoform, exhibits stronger actin-bundling activity than UNC-87B
Previous biochemical studies on UNC-87 were done using UNC-87B, a shorter isoform (Kranewitter et al., 2001; Yamashiro et al., 2007), and characterization of UNC-87A has not been reported. To determine whether the two UNC-87 isoforms have any different biochemical properties, we produced and purified recombinant UNC-87A protein and compared it biochemically with UNC-87B.
UNC-87B has been shown to bind to F-actin and bundle the filaments (Kranewitter et al., 2001). Similarly, UNC-87A bound to F-actin, as determined by cosedimentation assays using ultracentrifugation (Figure 2A), and bundled actin filaments, as determined by sedimentation assays using low-speed centrifugation (Figure 2B). In the cosedimentation assays using ultracentrifugation, UNC-87A bound to F-actin so strongly that nearly all UNC-87A cosedimented with F-actin at low UNC-87A concentrations (Figure 2A; 1.1 μM UNC-87A to 10 μM actin), which makes it difficult to estimate its affinity for F-actin. In the sedimentation assays using low-speed centrifugation, in which only bundled actin filaments precipitate, UNC-87A induced bundle formation of actin filaments (10 μM actin) at >1.1 μM (Figure 2C, black circles). By comparing these data with the published data on UNC-87B (Yamashiro et al., 2007), we see that UNC-87A showed much stronger actin-bundling activity than UNC-87B, in particular, at a low concentration range (<2.5 μM; Figure 2C).
FIGURE 2:

UNC-87 isoforms bundle actin filaments. (A) F-actin (10 μM) was incubated with or without 1.1 μM UNC-87A for 30 min and ultracentrifuged to sediment all F-actin. Supernatants (s) and pellets (p) were analyzed by SDS–PAGE. UNC-87A alone in the absence of F-actin was also examined in the same manner, and the are results shown on the right. Molecular weight markers (M) are shown on the left. Positions of UNC-87A (U) and actin (A) are indicated on the right. (B) F-actin (10 μM) was incubated without or with 1.1 or 2.2 μM UNC-87A for 30 min and centrifuged at low speed to sediment only bundled F-actin. Supernatants (s) and pellets (p) were analyzed by SDS–PAGE. (C) Actin-filament bundling at various concentrations of UNC-87A (black circles) or UNC-87B (white circles) was examined by low-speed centrifugation as described in B, and percentages of actin in the pellets were plotted as a function of UNC-87 concentration. Data for UNC-87B are derived from Yamashiro et al. (2007). Data are average ± SD from three independent experiments.
UNC-87 isoforms bind to both actin and myosin and inhibit actomyosin ATPase and motility
To explore a novel function of UNC-87 proteins, we examined their interaction with myosin. We purified myosin from C. elegans (Ce-myosin) and found that Ce-myosin by itself remained highly soluble even under low-salt conditions (27 mM KCl), as determined by low-speed centrifugation (18,000 × g, 30 min; Figure 3, A and B), under which rabbit skeletal muscle myosin (R-myosin) readily precipitated (unpublished data). However, UNC-87A induced precipitation of Ce-myosin and coprecipitated with Ce-myosin (Figure 3A), although UNC-87A remained in the supernatants in the absence of Ce-myosin (Figure 3A). These results strongly suggest that UNC-87A bound to Ce-myosin and induced formation of bundles or aggregates of Ce-myosin. UNC-87B also induced precipitation of Ce-myosin (Figure 3B). Comparison of the myosin precipitation at various concentrations of UNC-87A and UNC-87B showed that UNC-87A exhibited stronger activity than UNC-87B (Figure 3C).
FIGURE 3:

UNC-87 isoforms bind to myosin. (A, B) Ce-myosin (0.19 μM) was incubated without or with various concentrations of UNC-87A (A) or UNC-87B (B) for 30 min and centrifuged at low speed. UNC-87A or UNC-87B alone in the absence of F-actin was also examined in the same manner. Supernatants (s) and pellets (p) were analyzed by SDS–PAGE. Molecular weight markers (M) are shown on the left. Positions of UNC-87A (UA), UNC-87B (UB), and myosin heavy chain (MHC) are indicated on the right. (C) The Ce-myosin sedimentation assays were performed at various concentrations of UNC-87A (black circles) or UNC-87B (white circles). Percentages of myosin in the pellet were quantified by SDS–PAGE and densitometry and plotted as a function of concentration of UNC-87 isoforms. Data are average ± SD from three independent experiments.
The myosin-binding activity of UNC-87 isoforms prompted us to test whether UNC-87 proteins can cross-link actin filaments with myosin. We tested this by observing morphology of fluorescently labeled actin filaments by microscopy in the presence or absence of UNC-87 proteins and/or Ce-myosin (Figure 4). In the absence of Ce-myosin or UNC-87, only single actin filaments were detected (Figure 4A), and ATP did not alter the morphology of actin filaments (Figure 4B). Ce-myosin induced formation of short, needle-like bundles of actin filaments in the absence of ATP (Figure 4C), but these bundles were nearly completely disassembled in the presence of ATP (Figure 4D). These are consistent with conserved properties of conventional myosin: actin-filament bundling by two-headed myosin molecules and ATP-dependent dissociation of myosin heads from actin filaments. By contrast, both UNC-87A (Figure 4E) and UNC-87B (Figure 4I) induced formation of much larger and longer actin bundles than Ce-myosin–induced bundles (Figure 4C), and UNC-87-induced actin bundles were resistant to ATP (Figure 4, F and J). Of interest, when both Ce-myosin and UNC-87 proteins were simultaneously present, more-robust formation of actin bundles was observed (Figure 4, G and K). Actin bundles were much larger in the presence of both Ce-myosin and UNC-87 than in the presence of Ce-myosin or UNC-87 alone. In addition, a number of unbundled single actin filaments remained in the presence of UNC-87A or UNC-87B alone (Figure 4, E and I), but unbundled filaments were almost undetectable in the presence of both Ce-myosin and UNC-87A or UNC-87B (Figure 4, G and K), suggesting that most of the actin filaments were bundled by the synergistic effects of Ce-myosin and UNC-87A or UNC-87B. We also found that UNC-87 isoforms induced different forms of actin bundles in the presence of Ce-myosin (Figure 4, G and K). UNC-87A and Ce-myosin induced very large aggregates of actin bundles (Figure 4G), whereas UNC-87B and Ce-myosin induced large but dispersed actin bundles (Figure 4K). These actin bundles formed by Ce-myosin and UNC-87 isoforms were resistant to ATP (Figure 4, H and L), suggesting that UNC-87 proteins play a role in maintaining actomyosin structures when the ATP-sensitive actin–myosin association and dissociation occur.
FIGURE 4:
UNC-87 isoforms induce large actomyosin bundles. DyLight 549–labeled actin filaments (2 μM) were incubated without (A–D) or with 1.0 μM UNC-87A (E–H) or 1.0 μM UNC-87B (I–L) in the absence (left two columns) or presence of 0.19 μM Ce-myosin (right two columns) and in the absence (A, C, E, G, I, and K) or presence (B, D, F, H, J, and L) of 5 mM ATP. The actin filaments were directly observed by fluorescence microscopy. Bar, 20 μm.
Next we examined how UNC-87 proteins affect actomyosin ATPase activity. As reported previously (Harris et al., 1977; Tanii et al., 1985), Ce-myosin had much lower ATPase activity than R-myosin, and actin modestly enhanced the ATPase activity (Figure 5, A and C). Actin activation of Ce-myosin ATPase was 13–28% (Figure 5, B and D). Both UNC-87A (Figure 5, A and B) and UNC-87B (Figure 5, C and D) inhibited activation of myosin ATPase by actin. Because actin activation of Ce-myosin ATPase was modest, we also examined the effect of UNC-87 isoforms on actin-activated R-myosin ATPase, in which actin activation of R-myosin ATPase was >20-fold (Figure 5, E and F). Under these conditions, both UNC-87A (Figure 5E) and UNC-87B (Figure 5F) strongly inhibited actin-activated R-myosin ATPase. In cosedimentation assays with R-myosin, both UNC-87 isoforms only weakly interacted with R-myosin (unpublished data), suggesting that the strong inhibition of actin-activated myosin ATPase is due to strong interactions between actin and UNC-87A or UNC-87B.
FIGURE 5:
UNC-87 isoforms inhibit actomyosin ATPase. (A–D) ATPase activity of Ce-myosin (0.44 μM) was determined in the absence or presence of 1.0 μM actin and 0.96 μM UNC-87A (A, B) or 3.3 μM UNC-87B (C, D). ATPase activity (A, C) and percentage activation as compared with Ce-myosin alone (B, D). (E, F) ATPase activity of R-myosin (0.35 μM) was determined in the absence or presence of 1.0 μM actin and 0.96 μM UNC-87A (E) or 3.3 μM UNC-87B (F). Data are average ± SD from three independent experiments.
The inhibitory functions of UNC-87 isoforms on actomyosin ATPase strongly suggest that they also inhibit actomyosin contractility. We examined effects of UNC-87A and UNC-87B on actomyosin motility by in vitro motility (gliding) assays (Figure 6). DyLight 549–labeled actin filaments were attached to myosin on a glass surface, incubated with a buffer with or without UNC-87A or UNC-87B for 2 min, and then exposed to a buffer containing ATP with no UNC-87 proteins to initiate motility. In Figure 6, A–C, actin filaments at time 0 and after 4 s were pseudocolored by red and green, respectively, and overlaid in single images. In control with no UNC-87 proteins, most actin filaments were displaced, and overlaps between red and green filaments were minimal (Figure 6A). By contrast, both UNC-87A (Figure 6B) and UNC-87B (Figure 6C) at 2.5 μM significantly increased populations of yellow (red plus green) filaments, indicating that actomyosin motility was inhibited by the UNC-87 isoforms. Quantitative analysis of the velocity of the filament movement showed that 2.5 μM UNC-87A or UNC-87B significantly inhibited actomyosin motility (Figure 6D). At 1.0 μM, both UNC-87A and UNC-87B had weaker effects on actomyosin motility (Figure 6D) and did not cause statistically significant changes in the mean values as estimated by one-way analysis of variance. However, it should be noted that velocity of the bottom 25th percentile was reduced to less than half by 1.0 μM UNC-87A (0.133 μm/s) or UNC-87B (0.147 μm/s) as compared with control (0.302 μm/s), suggesting that populations of slow-moving filaments were increased by 1.0 μM UNC-87A or UNC-87B. Pairwise comparisons between UNC-87A and UNC-87B at the same concentrations showed no statistically significant differences between the two isoforms. Overall this biochemical analysis strongly suggests that UNC-87 isoforms are negative regulators of actomyosin contraction.
FIGURE 6:
UNC-87 isoforms inhibit actomyosin motility in vitro. In vitro motility (gliding) of DyLight 549–labeled actin filaments was examined on coverglasses coated with intact rabbit skeletal muscle myosin. Myosin-bound actin filaments were incubated with buffer only (A, control) or buffer with 2.5 μM UNC-87A (B) or 2.5 μM UNC-87B (C) for 2 min. Motility was initiated by a buffer containing 1 mM ATP without UNC-87 proteins, and pseudocolored micrographs of actin filaments at time 0 (red) and after 4 s (green) were overlaid. Bar, 5 μm. (D) Box plots of velocity of actin filaments in the motility assays. Fifty randomly selected filaments from three separate recordings were quantified for each condition. Boxes represent a range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. The data were analyzed by one-way analysis of variance; *p < 0.05 as compared with control. Pairwise comparisons between UNC-87A and UNC-87B at the same concentrations showed no significant differences.
unc-87 mutations cause excessive contraction in the somatic gonad
To determine whether the inhibitory roles of UNC-87 proteins for actomyosin ATPase and motility are functionally significant in vivo, we examined muscle contractility in unc-87 mutant worms. Previous studies showed that unc-87 mutant worms move much more slowly than wild-type worms, which is primarily due to defects in the body wall muscle (Goetinck and Waterston, 1994a). However, body wall muscle cells are firmly attached to relatively rigid cuticles (Ono, 2014), and extent of muscle contraction could not be easily quantitated. In addition, unc-87 mutation causes disorganization of sarcomeres in body wall muscle and very likely disturbs muscle contractility. Therefore we analyzed contractility of the myoepithelial sheath in the somatic gonad, which is nonstriated muscle responsible for ovulation and surrounded only by soft basement membranes (Ono et al., 2007). Our promoter-reporter analysis indicated that UNC-87B was expressed in the myoepithelial sheath (Figure 1U). Immunostaining using anti–UNC-87 showed that the UNC-87B protein localized along the actin filaments (Figure 7, A and B) and partially overlapped with the regions where MYO-3 myosin was localized (Figure 7, C and D), indicating that UNC-87B is in locations where it can interact with both actin and myosin. This localization pattern of UNC-87B was distinct from that of another actin-binding protein, kettin (Ono et al., 2006), which is concentrated at the proximal regions of the actin filaments (Figure 7, E–H), and that of vinculin, which is concentrated at the dense bodies and links actin filaments to the membrane (Figure 7, I–L). These localization patterns of UNC-87B suggest that it can regulate actin–myosin interaction.
FIGURE 7:
UNC-87 is associated with actin filaments in the myoepithelial sheath of the somatic gonad. Myoepithelial sheath in dissected gonads from wild-type worms were triple immunostained for UNC-87 (A, E, and I), actin (B, F, and J), and MYO-3 (C), kettin (G), or vinculin (K). Merged images are shown in D, H, and L (green, UNC-87; blue, actin; red, MYO-3, kettin, or vinculin). Bar, 10 μm.
FIGURE 8:
unc-87 mutation induces excessive actomyosin contraction in the myoepithelial sheath of the somatic gonad. Dissected gonads from wild type (A–D), unc-54(s95) (E–H), unc-87(e1459) (I–L), or unc-87(e1459) unc-54(s95) (M–P) were immunostained for MYO-3 to visualize the myoepithelial sheath at a low magnification (left column) or for both actin and MYO-3 to characterize actomyosin organization at a high magnification (right three columns). Merged images of actin (red) and MYO-3 (green) are shown in D, H, L, and P. The proximal region of the gonad is oriented to the right. Quantitative data on the length of the myoepithelial sheath are shown in Table 1. Bars, 10 μm.
The myoepithelial sheath is present in the proximal gonad, as marked by expression of the MYO-3 myosin heavy chain (Figure 8; Ardizzi and Epstein, 1987). In wild type, the myoepithelial sheath had nonstriated networks of actin filaments and covered most of the proximal region of the gonad (Ono et al., 2007; Figure 8A). In unc-87(e1459) (a null or severe loss-of-function mutant), the MYO-3–positive regions were much smaller than those of wild type (compare Figure 8, A and I). Measurement of the length of MYO-3–positive region showed that unc-87(e1459) had >50% shorter myoepithelial sheath than wild type (Figure 8, A and I, and Table 1). Both wild type and unc-87(e1459) had six myoepithelial sheath cells, excluding the possibility that the number of cells made the difference. Actin and myosin (MYO-3) were organized into networks in both wild type (Figure 8, B–D) and unc-87(e1459) (Figure 8, J–L). However, myosin filaments appeared more densely aligned in unc-87(e1459) (Figure 8K) than in wild type (Figure 8C), which is consistent with the contraction of the actomyosin network in unc-87(e1459). To determine whether this shortening is due to actomyosin contractility, we examined the effects of a mutation in unc-54, a gene encoding another major myosin heavy chain expressed in the myoepithelial sheath (Ardizzi and Epstein, 1987). unc-54(s95) is a missense mutation near the ATP-binding site in the myosin head and reduces muscle contractility without altering organization of the contractile apparatus (Moerman et al., 1982; Dibb et al., 1985). unc-54(s95) single mutant had slightly shorter myoepithelial sheath (Figure 8E and Table 1) and unc-87(e1459) unc-54(s95) double mutant had significantly longer myoepithelial sheath than unc-87(e1459) single mutant (compare Figure 8, I and M), indicating that the myosin mutation suppressed the Unc-87 phenotype. The unc-54(s95) mutation did not affect the integrity of the actomyosin networks (Figure 8, F–H), and the myosin filaments in unc-87(e1459) unc-54(s95) double mutant (Figure 8O) exhibited a more relaxed appearance than those in unc-87(e1459) (Figure 8K). Thus, these genetic evidence strongly suggests that unc-87B is a negative regulator of actomyosin contractility in vivo.
TABLE 1:
Effects of unc-87 and unc-54 mutations on the length of the myoepithelial sheath.
| Genotype | Length (μm) | n |
|---|---|---|
| Wild type | 150 ± 29 | 31 |
| unc-54(s95) | 131 ± 18 | 48 |
| unc-87(e1459) | 74.8 ± 21 | 35 |
| unc-87(e1459) unc-54(s95) | 125 ± 20 | 50 |
DISCUSSION
In this study, we found that two UNC-87 isoforms are expressed in a tissue-specific manner by two separate promoters driving two alternative first exons and that the two UNC-87 isoforms bind to both actin and myosin to inhibit actomyosin motility and form ATP-insensitive actin–myosin bundles. The inhibitory role of UNC-87 isoforms for actomyosin contractility was further demonstrated in vivo: unc-87 mutants showed excessive contraction of the myoepithelial sheath, a smooth muscle–like tissue in the somatic gonad. In addition to the previously reported function of UNC-87B to stabilize actin filaments (Kranewitter et al., 2001; Yamashiro et al., 2007), the present study revealed another function of UNC-87 to regulate actomyosin contractility. In vertebrate smooth muscle, similar dual functions have been demonstrated in vitro for calponin (Winder et al., 1998), caldesmon (Hemric and Chalovich, 1988; Velaz et al., 1989), and fesselin/synaptopodin-2 (Schroeter and Chalovich, 2005), but their in vivo functions are not clearly understood. UNC-87A and UNC-87B are the only known actin-bundling proteins in C. elegans that also inhibit actomyosin contractility. Our biochemical and genetic studies in C. elegans suggest that UNC-87 isoforms are functional homologues of these vertebrate proteins and regulate actin filament stability and actomyosin contractility in vivo.
The two UNC-87 isoforms with different N-terminal sequences are expressed in different tissues. Our biochemical analysis shows that UNC-87A, with a longer N-terminus, has more robust actin-bundling and stronger myosin-binding activities than UNC-87B, with a shorter N-terminus, suggesting that these quantitative differences in their activities are important for their tissue-specific functions. UNC-87B is predominantly expressed in the body wall muscle and the somatic gonad, where it regulates actin organization (Goetinck and Waterston, 1994a), actin filament stability (Yamashiro et al., 2007), and contractility (this study). Therefore the relatively weak activity of UNC-87B might be optimal for actin regulation in these striated and nonstriated muscles. By contrast, the in vivo function of UNC-87A is unknown because a specific mutation for unc-87A is not available. The mutations in unc-87(e1216) and unc-87(e1459) are located in exon 2, which is common to both UNC-87 isoforms (Supplemental Figure S1F). Therefore UNC-87A should also be ablated in these mutants. However, we were not able to detect a phenotype in tissues in which expression of UNC-87A was detected (unpublished data). This might be due to a weak phenotype or a phenotype other than actin organization and contractility, which we might have overlooked. Another possibility is that a functionally overlapping actin-binding protein compensates for the function of UNC-87A.
UNC-87A and UNC-87B commonly have seven CLIK repeats but are different in their N-terminal sequences. UNC-87A (565 amino acids) has an extra ∼190 amino acids with no homology to known functional protein sequences, which is absent in UNC-87B (374 amino acids; Goetinck and Waterston, 1994a). Although this N-terminal sequence of UNC-87A is most likely responsible for the quantitative differences in the activities of the UNC-87 isoforms, a role of this unique region is unknown. If this region binds to F-actin or myosin, it may cooperate with the C-terminal CLIK repeats to enhance binding of UNC-87A to F-actin or myosin. Alternatively, the N-terminal sequence may allosterically enhance an actin- or myosin-binding function of the C-terminus. Previous functional studies on recombinant proteins containing different numbers of CLIK repeats showed that an increase in the number of CLIK repeats is correlated with an increase in actin-stabilizing functions in vitro (Kranewitter et al., 2001) and in cultured cells (Gimona et al., 2003; Lener et al., 2004). CLIK repeats appear to be intrinsically unstructured in solution (Rozenblum and Gimona, 2008), and the unique sequence of the UNC-87A may have a function similar to CLIK repeats even in the absence of sequence similarity.
Myosin binding and inhibition of actomyosin ATPase are novel functions of UNC-87 isoforms. Our biochemical experiments show that both UNC-87 isoforms can mediate formation of ATP-resistant actomyosin bundles. Therefore UNC-87 proteins can also function not only as a regulator of actomyosin contractility, but also as a stabilizer of actomyosin structural integrity. These activities of UNC-87 isoforms are indeed consistent with the previously reported unc-87 mutant phenotypes. In body wall muscle, sarcomeric actin filaments are disorganized in unc-87 mutants, but this phenotype can be suppressed by a mutation in the unc-54 myosin heavy chain (Goetinck and Waterston, 1994b), suggesting that a loss of the inhibitory function of UNC-87B for actomyosin contractility can be compensated by reduction in the contractile function of myosin itself. Alternatively, UNC-87B may normally stabilize sarcomeres against mechanical stress by actin–myosin cross-linking, and reduced contractility due to the myosin mutation can suppress mechanical damage on sarcomeres in unc-87 mutants.
The myoepithelial sheath of the C. elegans somatic gonad is nonstriated muscle and essential for oocyte maturation and ovulation (Kim et al., 2013). However, the mechanism of regulation of myoepithelial sheath contractility is poorly understood. Unlike vertebrate smooth muscle, the troponin–tropomyosin system is required for contractility of myoepithelial sheath (Myers et al., 1996; Ono and Ono, 2004; Obinata et al., 2010). When the two troponin I isoforms, TNI-1 and UNC-27, are depleted, the myoepithelial sheath becomes excessively contracted (Obinata et al., 2010), which is similar to the phenotype of unc-87 mutants. Thus, in addition to troponin I, which is well established as an inhibitor of actomyosin interaction (Squire and Morris, 1998), UNC-87B is also important for contractile regulation in the myoepithelial sheath. However, the functional difference between troponin and UNC-87B is unknown. The troponin complex is regulated by calcium (Ebashi, 1984), but UNC-87B is not likely to be regulated by calcium or calcium/calmodulin because of a lack of recognizable sequence for binding to these ligands. Therefore troponin and UNC-87B may function as calcium-dependent and -independent regulators, respectively.
UNC-87B was associated with actin filaments in the myoepithelial sheath but did not appear to have an essential role in assembly or maintenance of the nonstriated actin networks. Previously we showed that ADF/cofilin (UNC-60A; Ono et al., 2008) and two actin-interacting protein 1 isoforms (UNC-78 and AIPL-1; Ono and Ono, 2014) are essential for assembly of the nonstriated actin networks in the myoepithelial sheath. These proteins enhance actin filament dynamics by promoting actin filament disassembly (Ono and Benian, 1998; Ono et al., 2004, 2011; Yamashiro et al., 2005; Mohri et al., 2006). Because UNC-87B strongly inhibits actin filament severing by ADF/cofilin in vitro, it should be interesting to determine whether UNC-87B and/or a functionally similar protein antagonizes ADF/cofilin and actin-interacting protein 1 during assembly and maintenance of nonstriated actin networks.
In conclusion, our study demonstrates that C. elegans UNC-87 isoforms are actin-associated inhibitors of actomyosin contractility in vitro and in vivo. In addition to the previously characterized role of UNC-87B as a stabilizer of actin filaments, these functions are important for contractility of striated and nonstriated muscles. These functions have been proposed for vertebrate proteins, including calponin, caldesmon, and fesselin/synaptopodin-2, mostly from biochemical studies, but have not been clearly demonstrated in vivo. This could be partly due to functional redundancy among functionally similar proteins. The C. elegans genome does not have caldesmon or fesselin/synaptopodin-2, and its relatively simple system allowed us to determine the biological significance of the CLIK-repeat protein. Furthermore, we detected tissue-specific expression of UNC-87 isoforms in both muscle and nonmuscle cells, suggesting that UNC-87A or UNC-87B is involved in actin-dependent events in a variety of cell types. Similarly, calponins and calponin-like proteins in other organisms should also function in a similar manner to UNC-87 isoforms using CLIK repeats as actin-binding motifs.
MATERIALS AND METHODS
Nematode strains
The following strains were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN) and used in this study: wild-type strain N2, CB1216 unc-87(e1216), CB1459 unc-87(e1459), RW1524 unc-87(e1459) unc-54(s95), and RW5008 unc-54(s95). The mutant strains have been published previously (Moerman et al., 1982; Goetinck and Waterston, 1994a, b). All mutants were used as homozygotes. Nematodes were grown under standard conditions at 20°C on Nematode Growth Medium agar plates for phenotypic analysis or at room temperature in liquid Nematode Growth Medium for myosin preparation as described (Stiernagle, 2006).
Promoter-reporter analysis
Promoter::GFP constructs for analysis of promoters for UNC-87 isoforms were made using fusion PCR as previously described (Hobert, 2002). For unc-87A, 2038 base pairs of genomic DNA containing 2008 base pairs upstream plus 30 base pairs downstream from the initiation codon of unc-87A was amplified by PCR and fused in-frame with the GFP-coding cassette (GFP plus let-858 3′-untranslated region) from pPD118.20 (kindly provided by A. Fire, Stanford University, Stanford, CA). For unc-87B, 2421 base pairs of genomic DNA containing 2406 base pairs upstream plus 15 base pairs downstream from the initiation codon of unc-87B was amplified by PCR and fused in-frame with the same GFP-coding cassette. The fusion-PCR constructs were injected together with pRF4, a dominant rol-6 transformation marker, into the syncytial region of the gonad of wild-type worms. GFP-positive F1 worms were isolated, and F2 worms that stably inherited the transgenes were selected to establish transgenic lines. Worms were anesthetized in M9 buffer containing 0.1% tricaine and 0.01% tetramisole for 30 min, mounted on 2% agarose pads, and observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope (Nikon Instruments, Tokyo, Japan).
Proteins
Rabbit muscle actin was prepared from rabbit muscle acetone powder (Pel-Freeze Biologicals, Rogers, AR) as described (Pardee and Spudich, 1982). Rabbit muscle myosin was purified from rabbit back muscle as described (Perry, 1955). C. elegans myosin was purified from liquid-grown N2 strain as described (Tanii et al., 1985). Recombinant UNC-87B was expressed in Escherichia coli and purified as described (Kranewitter et al., 2001). To produce recombinant UNC-87A, a full-length protein coding sequence for UNC-87A was amplified by PCR from the cDNA clone yk1701a7 (kindly provided by Yuji Kohara, National Institute of Genetics, Mishima, Japan) and cloned into pET-3a (EMD Millipore, Billerica, MA). Recombinant UNC-87A with no additional tag was expressed in E. coli BL21 (DE3) and purified in the same procedure for purification of UNC-87B (Kranewitter et al., 2001). Protein concentrations were determined by the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA).
Actin sedimentation assays
F-actin (10 μM) was incubated with various concentrations of UNC-87A or UNC-87B in F-buffer (0.1 M KCl, 2 mM MgCl2, 1 mM dithiothreitol [DTT], 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 7.5) for 30 min at room temperature. The reactions were centrifuged either at low speed (18,000 rpm for 10 min using a Beckman Microfuge) to examine actin bundling or at high speed (80,000 rpm for 20 min using a Beckman TL-100 ultracentrifuge with a TLA-100 rotor) to examine F-actin binding. The supernatants and pellets were adjusted to the same volumes and analyzed by SDS–PAGE. Gels were stained with Coomassie Brilliant Blue R-250 (National Diagnostics, Atlanta, GA) and scanned by an Epson V700 scanner at 300 dots/inch, and the band intensity was quantified by ImageJ (National Institutes of Health, Bethesda, MD).
Myosin sedimentation assays
Ce-myosin (0.19 μM) was incubated with various concentrations of UNC-87A or UNC-87B in a buffer containing 27 mM KCl, 1 mM MgCl2, 1 mM DTT, and 20 mM imidazole-HCl, pH 7.5, for 30 min at room temperature. The reactions were centrifuged at 18,000 × g for 30 min using a Beckman Microfuge. The supernatants and pellets were fractionated, adjusted to the same volumes, and analyzed by SDS–PAGE. Quantitative analysis was done in the same manner as described for actin sedimentation assays.
Direct observation of actin bundles by fluorescence microscopy
Rabbit muscle G-actin (8 μM) was copolymerized with DyLight 549–labeled G-actin (2 μM) (Liu et al., 2010) in F-buffer for 2 h at room temperature. The labeled actin filaments (final 2 μM actin) were incubated with myosin (0.19 μM), UNC-87A (1.0 μM), and/or UNC-87B (1.0 μM) in the presence or absence of 5 mM ATP in F-buffer for 10 min at room temperature, mounted on nitrocellulose-coated coverslips, and observed by epifluorescence using a Nikon TE2000 inverted microscope.
Myosin ATPase assays
Myosin ATPase activity was determined by a colorimetric method as described previously (Obinata and Sato, 2012) in a modified buffer containing 27 mM KCl, 1 mM MgCl2, and 20 mM imidazole-HCl, pH 7.5. Ce-myosin and R-myosin were used at 0.44 and 0.35 μM, respectively. Actin (1.0 μM), UNC-87A (0.96 μM), and/or UNC-87B (3.3 μM) were added in some reactions.
In vitro motility assays
In vitro motility of actomyosin was examined at room temperature as described by Sellers (2001) with modifications. Intact rabbit skeletal muscle myosin was diluted to 0.3 mg/ml in a buffer containing 0.3 M NaCl, 2 mM MgCl2, and 20 mM Tris-HCl, pH 8.5, and applied to a flow cell that was made with a nitrocellulose-coated glass coverslip (No. 1, 35 × 50 mm), an 18-mm square No. 1 coverslip, and Scotch double-sided tapes as spacers. The cell was sequentially blocked by 1 mg/ml bovine serum albumin (BSA) in wash buffer (27 mM KCl, 5 mM MgCl2, 0.1 mM ethylene glycol tetraacetic acid, 20 mM imidazole-HCl, pH 7.5) and 5 μM unlabeled F-actin and 1 mM ATP in wash buffer (for blocking nonmotile myosin). DyLight 549–labeled actin filaments (26% labeled) were diluted to 0.4 μM actin in wash buffer and incubated in the flow cell for 2 min. Wash buffer with or without UNC-87A or UNC-87B was infused in the cell and incubated for 2 min. Motility was initiated by adding wash buffer containing 1 mM ATP, 0.7% methylcellulose, and 34 mM DTT. Time-lapse images of fluorescent actin filaments were recorded for 10 s using a Nikon Eclipse TE2000 inverted microscope with a Plan Apo 60× (oil; numerical aperture [NA] 1.40) objective and IPLab imaging software (BD Biosciences, San Jose, CA). Displacement of filaments was quantified using ImageJ, and statistical analysis and graphing were performed using SigmaPlot 12.0 (Systat Software, San Jose, CA).
Sequencing
Genomic DNA fragments for the unc-87 gene were amplified by PCR from purified total genomic DNA from each unc-87 mutant strain, and all exons were sequenced using custom primers. Sequencing was performed on at least two independent PCR products to ensure that there were no PCR-induced errors.
Immunofluorescence microscopy
Gonads were dissected from adult hermaphrodites on polylysine-coated glass slides as described previously (Ono et al., 2007). For quantification of the length of the myoepithelial sheath, young adults were selected for experiments. They were fixed by methanol at −20ºC for 5 min and washed by phosphate-buffered saline (PBS) and primary antibodies in PBS containing 1% BSA. After being washed with PBS, they were treated with fluorophore-labeled secondary antibodies, followed by washing with PBS.
Primary antibodies used were rabbit anti-GFP polyclonal (Rockland, Limerick, PA), guinea pig anti–UNC-87 polyclonal (Yamashiro et al., 2007), rabbit anti-actin polyclonal (Cytoskeleton, Denver, CO), mouse anti–MYO-3 monoclonal (5-6; Miller et al., 1983), mouse anti-kettin monoclonal (MH44; Francis and Waterston, 1985), and mouse anti-vinculin monoclonal (MH24; Francis and Waterston, 1985). Secondary antibodies used were Alexa 488–labeled goat anti–guinea pig immunoglobulin G (IgG), Alexa 488–labeled goat anti-mouse IgG, Alexa 488–labeled goat anti-rabbit IgG, and Alexa 647–labeled goat anti-rabbit IgG from Life Technologies (Carlsbad, CA) and Cy3-labeled goat anti-mouse IgG from Jackson ImmunoResearch (West Grove, PA).
Samples were mounted with ProLong Gold (Life Technologies) and observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40× (dry; NA 0.60) or Plan Apo 60× (oil; NA 1.40) objective. Images were captured by a SPOT RT monochrome charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) and processed by IPLab imaging software (BD Biosciences) and Photoshop CS3 (Adobe, San Jose, CA). Quantification of the myoepithelial sheath lengths was performed using ImageJ.
Supplementary Material
Acknowledgments
We thank Jung Hee Woo for technical assistance in sequencing the unc-87 mutation sites. Monoclonal antibody 5–6 (developed by Henry Epstein, University of Texas Medical Branch, Galveston, TX) and MH24 (developed by Robert Waterston, University of Washington, Seattle, WA) were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA. Monoclonal antibody MH44 was obtained from Robert Waterston. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from the National Institutes of Health (R01 AR48615) to S.O. and from the Ministry of Education, Science, and Culture, Japan (Grant 23570097) to T. O.
Abbreviations used:
- ADF
actin-depolymerizing factor
- Ce-myosin
Caenorhabditis elegans myosin
- CH
calponin homology
- CLIK
calponin-like
- GFP
green fluorescent protein
- R-myosin
rabbit skeletal muscle myosin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1483) February 25, 2015.
REFERENCES
- Abe M, Takahashi K, Hiwada K. Effect of calponin on actin-activated myosin ATPase activity. J Biochem. 1990;108:835–838. doi: 10.1093/oxfordjournals.jbchem.a123289. [DOI] [PubMed] [Google Scholar]
- Ardizzi JP, Epstein HF. Immunochemical localization of myosin heavy chain isoforms and paramyosin in developmentally and structurally diverse muscle cell types of the nematode Caenorhabditis elegans. J Cell Biol. 1987;105:2763–2770. doi: 10.1083/jcb.105.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Dibb NJ, Brown DM, Karn J, Moerman DG, Bolten SL, Waterston RH. Sequence analysis of mutations that affect the synthesis, assembly and enzymatic activity of the unc-54 myosin heavy chain of Caenorhabditis elegans. J Mol Biol. 1985;183:543–551. doi: 10.1016/0022-2836(85)90170-6. [DOI] [PubMed] [Google Scholar]
- Ebashi S. Ca2+ and the contractile proteins. J Mol Cell Cardiol. 1984;16:129–136. doi: 10.1016/s0022-2828(84)80701-4. [DOI] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Fattoum A, Walsh MP, Egelman EH. The CH-domain of calponin does not determine the modes of calponin binding to F-actin. J Mol Biol. 2006;359:478–485. doi: 10.1016/j.jmb.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Gimona M, Kaverina I, Resch GP, Vignal E, Burgstaller G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol Biol Cell. 2003;14:2482–2491. doi: 10.1091/mbc.E02-11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M, Mital R. The single CH domain of calponin is neither sufficient nor necessary for F-actin binding. J Cell Sci. 1998;111:1813–1821. doi: 10.1242/jcs.111.13.1813. [DOI] [PubMed] [Google Scholar]
- Goetinck S, Waterston RH. The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J Cell Biol. 1994a;127:79–93. doi: 10.1083/jcb.127.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetinck S, Waterston RH. The Caenorhabditis elegans UNC-87 protein is essential for maintenance, but not assembly, of bodywall muscle. J Cell Biol. 1994b;127:71–78. doi: 10.1083/jcb.127.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris HE, Tso MY, Epstein HF. Actin and myosin-linked calcium regulation in the nematode Caenorhabditis elegans. Biochemical and structural properties of native filaments and purified proteins. Biochemistry. 1977;16:859–865. doi: 10.1021/bi00624a008. [DOI] [PubMed] [Google Scholar]
- Hemric ME, Chalovich JM. Effect of caldesmon on the ATPase activity and the binding of smooth and skeletal myosin subfragments to actin. J Biol Chem. 1988;263:1878–1885. [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hodgkinson JL. Actin and the smooth muscle regulatory proteins: a structural perspective. J Muscle Res Cell Motil. 2000;21:115–130. doi: 10.1023/a:1005697301043. [DOI] [PubMed] [Google Scholar]
- Kake T, Kimura S, Takahashi K, Maruyama K. Calponin induces actin polymerization at low ionic strength and inhibits depolymerization of actin filaments. Biochem J. 1995;312:587–592. doi: 10.1042/bj3120587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Spike C, Greenstein D. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv Exp Med Biol. 2013;757:277–320. doi: 10.1007/978-1-4614-4015-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranewitter WJ, Ylanne J, Gimona M. UNC-87 is an actin-bundling protein. J Biol Chem. 2001;276:6306–6312. doi: 10.1074/jbc.M009561200. [DOI] [PubMed] [Google Scholar]
- Lener T, Burgstaller G, Gimona M. The role of calponin in the gene profile of metastatic cells: inhibition of metastatic cell motility by multiple calponin repeats. FEBS Lett. 2004;556:221–226. doi: 10.1016/s0014-5793(03)01401-7. [DOI] [PubMed] [Google Scholar]
- Liu Z, Klaavuniemi T, Ono S. Distinct roles of four gelsolin-like domains of Caenorhabditis elegans gelsolin-like protein-1 in actin filament severing, barbed end capping, and phosphoinositide binding. Biochemistry. 2010;49:4349–4360. doi: 10.1021/bi100215b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmqvist U, Trybus KM, Yagi S, Carmichael J, Fay FS. Slow cycling of unphosphorylated myosin is inhibited by calponin, thus keeping smooth muscle relaxed. Proc Natl Acad Sci USA. 1997;94:7655–7660. doi: 10.1073/pnas.94.14.7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew JD, Khromov AS, McDuffie MJ, Somlyo AV, Somlyo AP, Taniguchi S, Takahashi K. Contractile properties and proteins of smooth muscles of a calponin knockout mouse. J Physiol. 2000;529:811–824. doi: 10.1111/j.1469-7793.2000.00811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Plurad S, Waterston RH, Baillie DL. Mutations in the unc-54 myosin heavy chain gene of Caenorhabditis elegans that alter contractility but not muscle structure. Cell. 1982;29:773–781. doi: 10.1016/0092-8674(82)90439-1. [DOI] [PubMed] [Google Scholar]
- Mohri K, Ono K, Yu R, Yamashiro S, Ono S. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol Biol Cell. 2006;17:2190–2199. doi: 10.1091/mbc.E05-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CD, Goh PY, Allen TS, Bucher EA, Bogaert T. Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans. J Cell Biol. 1996;132:1061–1077. doi: 10.1083/jcb.132.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinata T, Ono K, Ono S. Troponin I controls ovulatory contraction of non-striated actomyosin networks in the C. elegans somatic gonad. J Cell Sci. 2010;123:1557–1566. doi: 10.1242/jcs.065060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinata T, Sato N. Comparative studies on troponin, a Ca2+-dependent regulator of muscle contraction, in striated and smooth muscles of protochordates. Methods. 2012;56:3–10. doi: 10.1016/j.ymeth.2011.09.026. [DOI] [PubMed] [Google Scholar]
- Ono S. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton (Hoboken) 2010;67:677–692. doi: 10.1002/cm.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Regulation of structure and function of sarcomeric actin filaments in striated muscle of the nematode Caenorhabditis elegans. Anat Rec. 2014;297:1548–1559. doi: 10.1002/ar.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Benian GM. Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J Biol Chem. 1998;273:3778–3783. doi: 10.1074/jbc.273.6.3778. [DOI] [PubMed] [Google Scholar]
- Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Ono S, Nomura K, Hitosugi S, Tu DK, Lee JA, Baillie DL, Ono K. The two actin-interacting protein 1 genes have overlapping and essential function for embryonic development in Caenorhabditis elegans. Mol Biol Cell. 2011;22:2258–2269. doi: 10.1091/mbc.E10-12-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ono S. Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol Biol Cell. 2004;15:2782–2793. doi: 10.1091/mbc.E04-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ono S. Two actin-interacting protein 1 isoforms function redundantly in the somatic gonad and are essential for reproduction in Caenorhabditis elegans. Cytoskeleton (Hoboken) 2014;71:36–45. doi: 10.1002/cm.21152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yamashiro S, Ono S. Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J Cell Sci. 2008;121:2662–2670. doi: 10.1242/jcs.034215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yu R, Mohri K, Ono S. Caenorhabditis elegans kettin, a large immunoglobulin-like repeat protein, binds to filamentous actin and provides mechanical stability to the contractile apparatuses in body wall muscle. Mol Biol Cell. 2006;17:2722–2734. doi: 10.1091/mbc.E06-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yu R, Ono S. Structural components of the nonstriated contractile apparatuses in the Caenorhabditis elegans gonadal myoepithelial sheath and their essential roles for ovulation. Dev Dyn. 2007;236:1093–1105. doi: 10.1002/dvdy.21091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Pearlstone JR, Weber M, Lees-Miller JP, Carpenter MR, Smillie LB. Amino acid sequence of chicken gizzard smooth muscle SM22 alpha. J Biol Chem. 1987;262:5985–5991. [PubMed] [Google Scholar]
- Perry SV. Myosin adenosinephosphatase. Methods Enzymol. 1955;2:582–588. [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblum GT, Gimona M. Calponins: adaptable modular regulators of the actin cytoskeleton. Int J Biochem Cell Biol. 2008;40:1990–1995. doi: 10.1016/j.biocel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Schroeter MM, Chalovich JM. Fesselin binds to actin and myosin and inhibits actin-activated ATPase activity. J Muscle Res Cell Motil. 2005;26:183–189. doi: 10.1007/s10974-005-9009-6. [DOI] [PubMed] [Google Scholar]
- Sellers JR. In vitro motility assay to study translocation of actin by myosin. Curr Protoc Cell Biol. 2001 doi: 10.1002/0471143030.cb1302s00. Chapter 13, Unit 13.12. [DOI] [PubMed] [Google Scholar]
- Shapland C, Hsuan JJ, Totty NF, Lawson D. Purification and properties of transgelin: a transformation and shape change sensitive actin-gelling protein. J Cell Biol. 1993;121:1065–1073. doi: 10.1083/jcb.121.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire JM. Architecture and function in the muscle sarcomere. Curr Opin Struct Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- Squire JM, Morris EP. A new look at thin filament regulation in vertebrate skeletal muscle. FASEB J. 1998;12:761–771. doi: 10.1096/fasebj.12.10.761. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Hiwada K, Kokubu T. Vascular smooth muscle calponin. A novel troponin T-like protein. Hypertension. 1988;11:620–626. doi: 10.1161/01.hyp.11.6.620. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yoshimoto R, Fuchibe K, Fujishige A, Mitsui-Saito M, Hori M, Ozaki H, Yamamura H, Awata N, Taniguchi S, et al. Regulation of shortening velocity by calponin in intact contracting smooth muscles. Biochem Biophys Res Commun. 2000;279:150–157. doi: 10.1006/bbrc.2000.3909. [DOI] [PubMed] [Google Scholar]
- Tanii I, Osafune M, Arata T, Inoue A. ATPase characteristics of myosin from nematode Caenorhabditis elegans purified by an improved method. Formation of myosin-phosphate-ADP complex and ATP-induced fluorescence enhancement. J Biochem. 1985;98:1201–1209. doi: 10.1093/oxfordjournals.jbchem.a135386. [DOI] [PubMed] [Google Scholar]
- Velaz L, Hemric ME, Benson CE, Chalovich JM. The binding of caldesmon to actin and its effect on the ATPase activity of soluble myosin subfragments in the presence and absence of tropomyosin. J Biol Chem. 1989;264:9602–9610. [PubMed] [Google Scholar]
- Winder SJ, Allen BG, Clement-Chomienne O, Walsh MP. Regulation of smooth muscle actin-myosin interaction and force by calponin. Acta Phys Scand. 1998;164:415–426. doi: 10.1111/j.1365-201x.1998.tb10697.x. [DOI] [PubMed] [Google Scholar]
- Winder SJ, Walsh MP. Smooth muscle calponin. Inhibition of actomyosin MgATPase and regulation by phosphorylation. J Biol Chem. 1990;265:10148–10155. [PubMed] [Google Scholar]
- Yamashiro S, Gimona M, Ono S. UNC-87, a calponin-related protein in C. elegans, antagonizes ADF/cofilin-mediated actin filament dynamics. J Cell Sci. 2007;120:3022–3033. doi: 10.1242/jcs.013516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Mohri K, Ono S. The two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry. 2005;44:14238–14247. doi: 10.1021/bi050933d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto R, Hori M, Takahashi K, Taniguchi SI, Katsuki M, Ozaki H, Karaki H. Ca2+-Sensitization of contraction in the h1 calponin-deficient smooth muscle. Jpn J Pharm. 2000;84:474–475. doi: 10.1254/jjp.84.474. [DOI] [PubMed] [Google Scholar]
- Yu R, Ono S. Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton. 2006;63:659–672. doi: 10.1002/cm.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.