Abstract
This year marks the 35th anniversary of the isolation of 23 SEC genes. These genes all encode key regulators of the secretory pathway, and much of our knowledge of the secretory pathway is based on this initial discovery. The identification of the SEC genes is a result of combining genetics, biochemistry, and electron microscopy in a very clever way. Scientists have been busy ever since seeking to understand the function and regulation of these genes and to identify further key players in the process. Although most of the machinery acting along the secretory pathway is known and its function generally understood, knowledge of regulation of the pathway under various conditions is still scarce and will keep researchers busy for years to come.
Scientists have been fascinated by cells since their discovery in the 17th century—how they function and how they communicate with their neighbors and environment. However, research efforts were hindered for a long time by the lack of suitable tools with which to interrogate cellular function. Then in the 1930s and 1940s transmission electron microscopy (TEM) was invented and developed, which paved the way to investigating the structure of cellular organelles through the imaging of thin sections. TEM permitted for the first time a view of cells at low-nanometer resolution, well beyond the resolution of light microscopy (∼0.2 μm). At the time, this technological advance generated probably as much hype as there has been in recent years for superresolution light microscopy techniques.
In spite of the ability to see ribosomes, the endoplasmic reticulum (ER), the Golgi apparatus, lysosomes, and vesicles in thin sections, only limited information could be gained about the dynamics of the secretory pathway. George Palade pioneered a technique in which he combined pulse-chase labeling with electron microscopy, revealing how proteins were transported along the secretory pathway (Jamieson and Palade, 1968, 1971). However, no connection was available linking observable static TEM images to the identity of the players that would enable the cell to form organelles and promote the communication between them.
In parallel, another powerful approach to understanding cellular function was the development of cell fractionation and biochemical assays. These methods allowed the identification of cellular processes and their reconstitution in vitro. Major insights were gained into mitochondrial function, DNA replication, RNA and protein synthesis, and lipid biosynthesis, just to name a few. Nonetheless, although proteins responsible for certain enzymatic activities could be purified, the identity of most cellular players remained elusive in the pregenomic era.
The third pillar for the interrogation of cellular functions came through genetics. In the late 1960s to early 1970s, genetic screens for conditional, temperature-sensitive mutants revealed mutants that would arrest at particular points in the cell cycle (Hartwell, 1967; Hartwell et al., 1970; Nurse, 1975; Nurse et al., 1976).
This was around the time that Randy Schekman set up his lab at the University of California in Berkeley. Schekman had been trained as a biochemist working on DNA replication but became interested in membranes, organelles, and secretion, inspired by work of George Palade, Albert Claude, and Christian De Duve. It was also still a time when a young assistant professor did not necessarily have to continue on and further develop the research theme from his previous experience but could start something completely new. However, already back then, the National Institutes of Health refused to fund Schekman's first grant application because he had no experience working with yeast and had no preliminary data. When Palade visited Berkeley and met Schekman soon after they had initiated the project, Palade was surprised to learn that yeast cells make glycoproteins.
Fascinated by the success of genetic screens for conditional mutants in yeast, Schekman, together with his first graduate student, Peter Novick, set out to screen for temperature-sensitive mutants that would be defective in secretion. As so often happens, the first, initial and clever idea for the screen did not work out. However, Schekman and Novick were not so easily discouraged and came up with another idea. They took temperature-sensitive mutants from the initial screen and measured the accumulation of secretory enzymes in cells using simple colorimetric assays of invertase and acid phosphatase activity (Novick and Schekman, 1979). When they looked more carefully, by TEM, at some mutants with defects in enzyme secretion, they realized the mutant cells were full of vesicles and accumulated internal membranes. The very first mutant, sec1-1, was found to block the fusion of transport vesicles with the plasma membrane (Novick and Schekman, 1979). From this observation that secretion mutants accumulated proteins and membranes inside the cell (Novick and Schekman, 1979) they suspected that mutant yeast cells would possess different physicochemical properties than wild-type cells. Indeed, Novick could show that mutant cells displayed an increase in buoyant density. The difference provided a powerful screening procedure through which secretory mutants could be enriched by gradient centrifugation while complementation groups were determined by classical genetics. In 1980, Novick, Field, and Schekman published the landmark paper in which they reported the discovery of 23 complementation groups involved in posttranslational events in the secretory pathway in Saccharomyces cerevisiae (Novick et al., 1980). Within 1 year, Novick, Ferro, and Schekman managed to assign the 23 genes to specific steps along the secretory pathway (Novick et al., 1981; Figure 1). Not only had they discovered 23 genes involved in secretion, but they had also elegantly combined three very prominent investigation tools: electron microscopy, biochemistry, and genetics. This powerful mix of approaches enabled Schekman and many coworkers over the years to craft an impressive picture describing the path of a nascent secretory protein from when it leaves the ribosome and enters the ER until its discharge at the plasma membrane. The realization in the 1980s and 1990s that the basic transport machineries are conserved from yeast to human made yeast THE model of choice to identify genes involved not only in secretion but also in endocytosis and lysosomal/vacuolar sorting pathways (Riezman, 1985; Bankaitis et al., 1986; Chvatchko et al., 1986; Rothman and Stevens, 1986; Rothman et al., 1989; Robinson et al., 1988).
FIGURE 1:
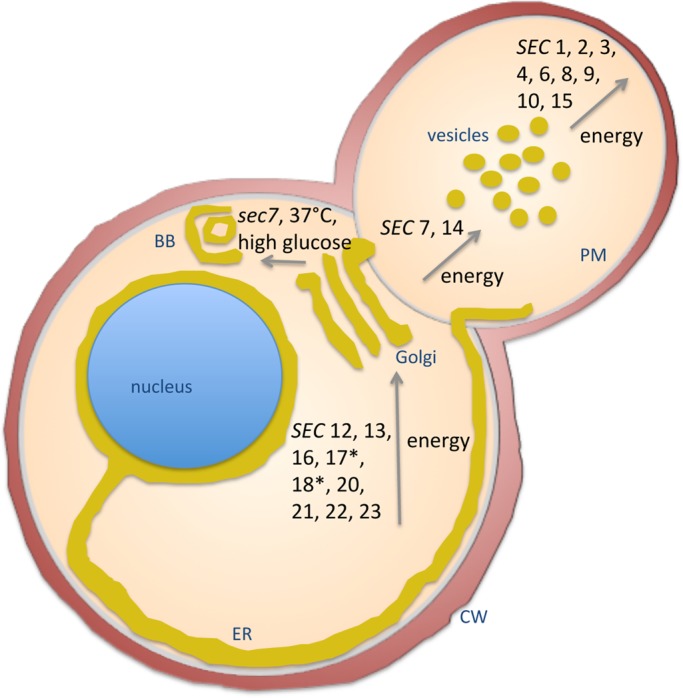
Yeast secretory pathway. BB, Berkeley body; CW, cell wall; ER, endoplasmic reticulum; PM, plasma membrane; SEC, wild-type gene product; sec, mutant gene product. Asterisks denote gene products that were found later to function along the entire secretory pathway, in fact, at all membrane fusion steps. Redrawn from Novick et al. (1981).
After the identification of the mutants and cloning of the genes, Schekman went back to his roots in biochemistry, in keeping with the immortal words of Richard Feynman, “what I cannot create, I do not understand.” The mantra in the lab for years to come was reconstitution— the in vitro establishment of transport processes such as polypeptide translocation into the ER and formation of transport vesicles. Schekman's lab over the years came up with a number of in vitro systems that faithfully recapitulated transport processes and greatly influenced our current understanding of the secretory pathway.
What happened to the brave graduate student Peter Novick? He was hooked on the secretory pathway: after his postdoc on yeast actin, he went on to discover, among other things, a Rab GTPase and its activator, the exocyst complex, and the soluble N-ethylmaleimide–sensitive factor attachment protein receptors, promoting fusion of secretory vesicles with the plasma membrane (Salminen and Novick, 1987; Goud et al., 1988; Bowser and Novick, 1991; Brennwald et al., 1994; TerBush and Novick, 1995; Walch-Solimena et al., 1997). Most of theses components were among the 23 gene products identified in the original screen, and the mutations blocked fusion of secretory vesicles with the plasma membrane (Figure 1).
The functions of all 23 gene products that Novick and Schekman initially identified are now generally understood, and crystal structures are available for many of them, providing insights into the basic machineries that operate along the secretory pathway. In spite of all this knowledge, we are only starting to understand the regulation and fine-tuning of the secretory pathway to ensure proper protein and lipid distribution and to maintain protein and lipid homeostasis upon aging, stress, and environmental changes.
It all started with this sec1-1 mutant filled with secretory vesicles some 35 years ago, and it will take us perhaps the same amount of time to fully understand the regulation of intracellular transport pathways.
Acknowledgments
I thank R. Schekman and I. G. Macara for comments on the manuscript. This work was supported by the Swiss National Science Foundation (31003A_141207).
Abbreviations used:
- ER
endoplasmic reticulum
- TEM
transmission electron microscopy.
Footnotes
REFERENCES
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser R, Novick P. Sec15 protein, an essential component of the exocytotic apparatus, is associated with the plasma membrane and with a soluble 19.5S particle. J Cell Biol. 1991;112:1117–1131. doi: 10.1083/jcb.112.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Chvatchko Y, Howald I, Riezman H. Two yeast mutants defective in endocytosis are defective in pheromone response. Cell. 1986;46:355–364. doi: 10.1016/0092-8674(86)90656-2. [DOI] [PubMed] [Google Scholar]
- Goud B, Salminen A, Walworth NC, Novick PJ. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Reid B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci USA. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson JD, Palade GE. Intracellular transport of secretory proteins in the pancreatic exocrine cell. IV. Metabolic requirements. J Cell Biol. 1968;39:589–603. doi: 10.1083/jcb.39.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson JD, Palade GE. Synthesis, intracellular transport, and discharge of secretory proteins in stimulated pancreatic exocrine cells. J Cell Biol. 1971;50:135–158. doi: 10.1083/jcb.50.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Howald I, Stevens TH. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989;8:2057–2065. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]


