DNA damage induces Chk2/Mnk-dependent mitotic and developmental blocks in early Drosophila embryos. The Mnk-FHA domain and its phosphopeptide-binding ability play essential dual functions in mediating the embryonic DNA damage response: activating Mnk upon DNA damage and recruiting Mnk to multiple cellular structures independently of DNA damage.
Abstract
DNA damage responses, including mitotic centrosome inactivation, cell-cycle delay in mitosis, and nuclear dropping from embryo cortex, maintain genome integrity in syncytial Drosophila embryos. A conserved signaling kinase, Chk2, known as Mnk/Loki, is essential for the responses. Here we demonstrate that functional EGFP-Mnk expressed from a transgene localizes to the nucleus, centrosomes, interkinetochore/centromere region, midbody, and pseudocleavage furrows without DNA damage and in addition forms numerous foci/aggregates on mitotic chromosomes upon DNA damage. We expressed EGFP-tagged Mnk deletion or point mutation variants and investigated domain functions of Mnk in vivo. A triple mutation in the phosphopeptide-binding site of the forkhead-associated (FHA) domain disrupted normal Mnk localization except to the nucleus. The mutation also disrupted Mnk foci formation on chromosomes upon DNA damage. FHA mutations and deletion of the SQ/TQ-cluster domain (SCD) abolished Mnk transphosphorylations and autophosphorylations, indicative of kinase activation after DNA damage. A potent NLS was found at the C-terminus, which is required for normal Mnk function. We propose that the FHA domain in Mnk plays essential dual functions in mediating embryonic DNA damage responses by means of its phosphopeptide-binding ability: activating Mnk in the nucleus upon DNA damage and recruiting Mnk to multiple subcellular structures independently of DNA damage.
INTRODUCTION
In the canonical cell cycle, DNA lesions cause activation of cell cycle checkpoints that delay or arrest the cell cycle before mitotic entry. However, during early Drosophila embryonic cleavage divisions, which feature rapid S/M cycles (Foe and Alberts, 1983), DNA lesions do not arrest the cell cycle before mitotic entry and are easily carried over into mitosis and trigger a unique set of embryonic DNA damage responses. During the syncytial blastoderm stage, when somatic precursor nuclei align and divide at the embryo cortex, DNA damage causes centrosome inactivation and cell cycle delay during mitosis and nuclear dropping from the embryo cortex during interphase. The centrosome inactivation is a mitosis-specific loss of microtubule nucleation at centrosomes and is associated with release of γ-tubulin-ring complex (γTuRC), leading to anastral spindle formation. The cell cycle delay during cleavage divisions upon DNA damage occurs during mitosis, in contrast to cell cycle delay/arrest, which mainly occurs during G1, S, or G2 phase during the canonical cell cycle. Nuclei that have undergone DNA damage drop from the cortex into the interior of the embryo; therefore they are not incorporated into the embryo proper (Sullivan et al., 1993; Sibon et al., 1997, 1999, 2000; Takada et al., 2003, 2007; Varmark et al., 2010).
A tumor suppressor, Chk2, is one of the key effector kinases in DNA damage signaling pathways and the Drosophila orthologue of Chk2 is essential for embryonic DNA damage responses (Takada et al., 2003, 2007). In mammals, DNA lesions, such as double-strand breaks (DSBs), trigger recruitment of DNA damage sensors and mediators to the sites of DNA lesions, including the Mre11/Rad50/Nbs1 complex, ataxia-telangiectasia mutated (ATM), and ATM and Rad3-related (ATR). ATM and ATR are phosphatidylinositide-3 kinase–like protein kinases that act at the site of DNA lesions to phosphorylate and activate downstream effector kinases such as Chk1 and Chk2. The activated effector kinases phosphorylate downstream effector proteins that are involved in various cellular processes (Bartek et al., 2001; Su, 2006; Antoni et al., 2007). The main target of Chk1 is Cdc25, which removes inhibitory phosphate from cyclin-dependent kinases (Sanchez et al., 1997). Chk2 phosphorylates diverse substrates, including Cdc25, E2F1, p53, BRCA1, PP2A, HuR, and Tau, and affects a wide range of cellular processes, including cell cycle checkpoints, apoptosis, DNA repair, RNA processing, transcription, and cytoskeleton (Bartek et al., 2001; Dozier et al., 2004; Zhang et al., 2004; Pommier et al., 2006; Abdelmohsen et al., 2007; Antoni et al., 2007; Iijima-Ando et al., 2010).
Drosophila Chk2 is encoded by loki (Masrouha et al., 2003), alternatively known as mnk (Oishi et al., 1998). Mnk is dispensable for normal development; however, mnk-null mutants fail to trigger proper responses to DNA damage (Xu et al., 2001; Abdu et al., 2002; Peters et al., 2002; Masrouha et al., 2003; Takada et al., 2003; Brodsky et al., 2004; Klattenhoff et al., 2007; Bakhrat et al., 2010). Embryos mutant for grp, which encodes Chk1, progress into mitosis before completing S phase due to lack of a functional replication checkpoint during the later syncytial blastoderm stage (Sibon et al., 1997) and show defects such as centrosome inactivation, cell cycle delay in mitosis, and nuclear dropping (Sibon et al., 2000). grp embryos accumulate DNA damage that triggers Mnk activation. mnk-null mutation rescues those defects in grp mutants without restoring normal S-phase length or removing DNA damage (Takada et al., 2007). In grp embryos, inhibitory phosphorylation of Tyr15 on mitotic kinase Cdc2, which is required for terminating the rapid cleavage cell cycle at the midblastula transition (MBT), is inhibited (Sibon et al., 1999; Takada et al., 2007). grp embryos undergo additional rounds of cleavage divisions and do not initiate high-level zygotic transcription and cellularization at the MBT (Sibon et al., 1997). These developmental blocks are rescued by mnk-null mutation (Takada et al., 2007). Several other maternal-effect mutant embryos show mitotic and developmental defects in early embryos similar to grp embryos, which are either partially or completely rescued by mnk-null mutation, indicating that these mutant embryos spontaneously produce DNA damage and trigger Mnk-dependent DNA damage responses (Sibon et al., 1999; Stumpff et al., 2004; Rickmyre et al., 2007; Takada et al., 2007; Benoit et al., 2009; Merkle et al., 2009; Sakurai et al., 2011; Pushpavalli et al., 2013). Iampietro et al. (2014) recently reported that Chk2/Mnk phosphorylates the Drosophila stem loop–binding protein (SLBP) and that the phosphorylation leads to degradation of SLBP and nuclear retention of specific mRNAs, including histone. They propose that these result in DNA damage–induced nuclear fallout/dropping. To acquire further understanding of embryonic DNA damage responses, additional Mnk substrates need to be identified.
The domain structure of Chk2 is conserved throughout evolution (Bartek et al., 2001). It contains an N-terminal SQ/TQ-cluster domain (SCD), a forkhead-associated (FHA) domain, and a C-terminal serine/threonine kinase domain. A functional nuclear localization signal (NLS) was found in the C-terminal region of human Chk2 (CHEK2; Zannini et al., 2003). The SCD is a preferred phosphorylation site by ATM and ATR and was found in many DNA damage signaling proteins, such as Chk1, Chk2, p53, and NBS1 (Traven and Heierhorst, 2005). The FHA domain binds to specific phosphothreonine-containing peptides and has been identified in a variety of prokaryotic and eukaryotic proteins, including kinases, phosphatases, transcription factors, and motor proteins (Mahajan et al., 2008). ATM phosphorylates human Chk2 at Thr-68 within the SCD upon DNA damage (Matsuoka et al., 2000; Ahn et al., 2002). This phosphorylation promotes dimerization or oligomerization of human Chk2 (Schwarz et al., 2003). Based on structural studies, a model was proposed that dimerization of Chk2 is mediated by a binding of the FHA domain to the phosphorylated SCD, which induces phosphorylation of two threonine residues within the kinase T-loop, leading to catalytic activation of the kinase (Oliver et al., 2006; Cai et al., 2009). Dimerization of Chk2 also induces autophosphorylation of several residues throughout the molecule; some of them regulate the kinase activity (King et al., 2006, 2007; Lovly et al., 2008). In this model, the main function of the Chk2-FHA domain is described as binding to the phosphorylated SCD to produce Chk2 dimer/oligomer for kinase activation. However, the FHA domain binding is most likely not limited to the phospho-SCD, and the FHA domain may have additional roles to be unraveled. On the basis of our in vivo study of Drosophila Chk2/Mnk function, we discuss how the FHA domain of Chk2/Mnk may bind to more diverse targets, allowing dynamic recruitment of Chk2/Mnk throughout the cell.
RESULTS
EGFP–Mnk localizes to the nucleus, centrosomes, interkinetochore/centromere region, midbody, and pseudocleavage furrows without DNA damage
We previously showed by immunostaining of fixed embryos that Mnk weakly localized to centrosomes and the spindle and that DNA damage increased the level of Mnk at these structures (Takada et al., 2003). Drosophila syncytial cleavage divisions feature very rapid cell cycles (9–20 min), and DNA damage responses occur within a few minutes after damage. This rapid kinetics makes it difficult to use fixed embryos to fully elucidate subcellular localization of Mnk at all stages of cell cycle and localization changes after DNA damage. To perform more direct observations, we produced transgenic fly lines that express enhanced green fluorescent protein (EGFP)–tagged Mnk for live analyses.
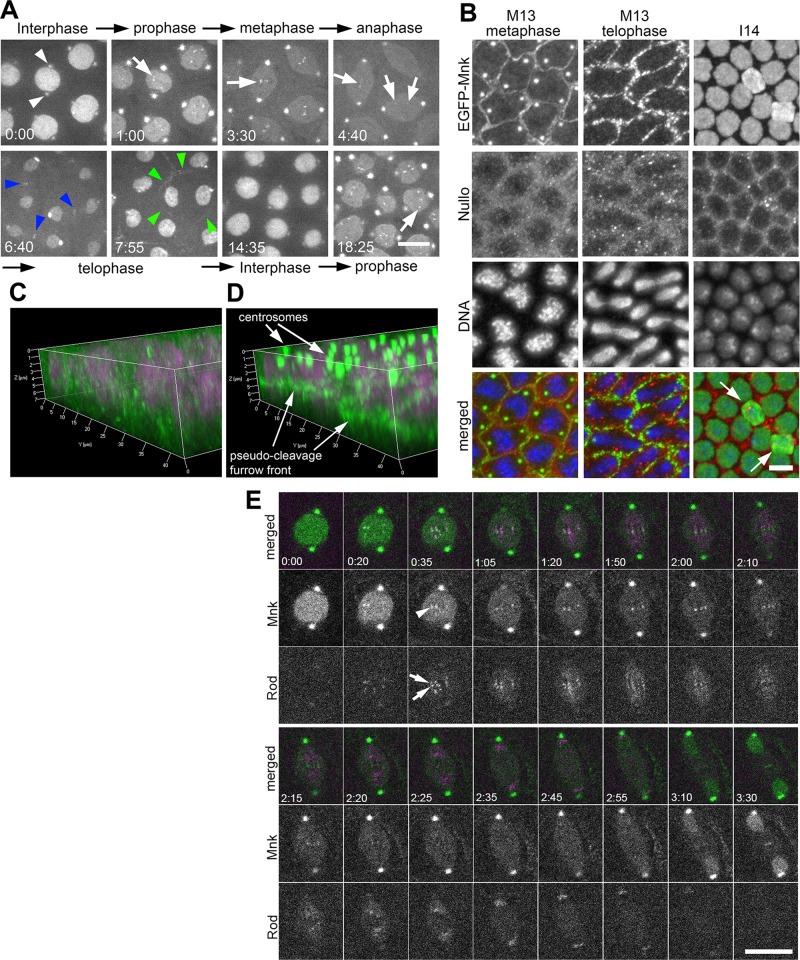
We first examined EGFP-Mnk localization without DNA damage. Figure 1A shows that EGFP-Mnk localizes to the nucleus and centrosomes (white arrowheads) during interphase. The EGFP-Mnk signal on centrosomes intensifies at nuclear envelope breakdown (NEB) and remains throughout mitosis. During interphase, however, the signal gradually decreases and becomes significantly weaker. In prophase-to-prometaphase nuclei, EGFP-Mnk forms several small dots that vigorously move around within the nucleus (white arrows). In metaphase, these dots align at the metaphase plate. As spindles elongate and sister chromosomes segregate in anaphase, each dot stretches and splits into two parts that move toward opposite poles and then disappear (Supplemental Movie S1). The behavior of the dots suggested that EGFP-Mnk was associated with the kinetochore or centromere. Note that EGFP-Mnk dots never appeared as a closely associated pair of dots, which represent sister kinetochores, as shown with kinetochore components such as CID, Polo, Rod, and Mad2 (Basto et al., 2004; Buffin et al., 2005; Schuh et al., 2007; McCleland et al., 2009).
FIGURE 1:
EGFP-Mnk localizes to the nucleus, centrosomes, the interkinetochore/centromere region, the midbody, and pseudocleavage furrows without DNA damage during the syncytial blastoderm stage in Drosophila embryos. (A) Selected frames from a time-lapse spinning-disk confocal movie on a single z-section (Supplemental Movie S1). From cycle 11 interphase to cycle 12 prophase. White arrowheads indicate EGFP signals on centrosomes adjacent to the nucleus during interphase. White arrows indicate EGFP signals on centromeres. Blue arrowheads indicate EGFP signals on the midbody. Green arrowheads indicate EGFP signal on a pseudocleavage furrow surrounding a dividing nucleus. Elapsed time is shown in minutes:seconds. Scale bar, 10 μm. (B) Localization of EGFP-Mnk on pseudocleavage furrows, centrosomes, and interphase nuclei in fixed mnk mutant embryos expressing EGFP-Mnk. Embryos were probed with anti-GFP antibody, anti-Nullo antibody, and DAPI. Each panel presents a maximum projection of z-stacks (M13 metaphase Δz = 7.13 μm; M13 telophase Δz = 5.99 μm; I14 Δz = 8.84 μm). In merges, green indicates anti-EGFP staining, red indicates anti-Nullo staining, and blue indicates DAPI staining. White arrows indicate two nuclei that are dropping from the cortex likely due to spontaneous DNA damage. These nuclei have more EGFP signals than surrounding nuclei (I14). Scale bar, 10 μm. (C) Three-dimensional (3D) reconstruction of Nullo (green) and DNA (magenta) staining. (D) A 3D reconstruction of EGFP-Mnk (green) and DNA (magenta) staining. C and D are reconstructed from the same confocal scanning data shown in B, M13 metaphase. (E) EGFP-Mnk localizes to the interkinetochore/centromere region from prometaphase to anaphase. Frames were selected from a time-lapse confocal movie of an embryo coexpressing EGFP-Mnk and mRFP-Rod (Supplemental Movie S21). White arrows indicate mRFP-Rod that localizes on sister kinetochores, and white arrowhead indicates EGFP-Mnk that localizes between the two sister kinetochores (time, 0:35). Elapsed time is shown in minutes:seconds. Scale bar, 10 μm.
To investigate whether these dots were associated with kinetochores or centromeres, we expressed EGFP-Mnk and monomeric red fluorescent protein (mRFP)–Rough Deal (Rod) together and simultaneously followed their behavior (Figure 1E and Supplemental Movie S21). Rod is a component of a Rod/ZW10 complex required for the metazoan spindle checkpoint and recruitment of dynein/dynactin to the kinetochore. Rod localizes to the outer kinetochore from prometaphase to metaphase and robustly streams along microtubules from the time of bipolar microtubule attachment until anaphase onset (Basto et al., 2004; Buffin et al., 2005). Our time-lapse confocal recordings show that EGFP-Mnk (indicated by arrowhead at time 0:35 in Figure 1E) localizes to the region between sister kinetochores that are marked with mRFP-Rod as two closely associated dots (indicated by arrows at time 0:35 in Figure 1E) from prophase to early anaphase. When mRFP-Rod streams after bipolar attachment, EGFP-Mnk continues to localize to the center between streaming mRFP-Rod signals (time 1:20–2:10 in Figure 1E). During anaphase, when chromosomes and mRFP-Rod signal start to migrate to spindle poles, the EGFP-Mnk signal slightly stretches, then separates into two, and each part follows mRFP-Rod signal toward the poles (Supplemental Movies S1 and S21). We conclude that EGFP-Mnk localizes not to kinetochores, but to interkinetochore/centromere regions from prophase to anaphase.
EGFP-Mnk also localizes weakly on spindles from prometaphase to anaphase and transiently concentrates around chromosomes and at the midbody during telophase (blue arrowheads). In addition, EGFP-Mnk forms circles around each spindle during mitosis (green arrowheads), which connect to each other and form a mesh-like structure between spindles. The mesh-like structure initially forms on the cortex at the beginning of mitosis, moves into the embryo between spindles, reaching to a depth deeper than spindle thickness at metaphase, and then moves back to the cortex and disintegrates at telophase (Supplemental Movie S1). This observation suggests that EGFP-Mnk localizes to pseudocleavage furrows. Pseudocleavage furrows are actin-cytoskeleton–associated membrane structures that form in Drosophila embryos before cellularization. They initially form at the cortex around each spindle in prophase, invaginate into the embryo, reaching to 10-μm depth at prometaphase, and then retract back to the surface and dissipate by the end of mitosis, preventing contact between neighboring spindles during mitosis (Stevenson and Theurkauf, 2000; Stevenson et al., 2001, 2002; Mazumdar and Mazumdar, 2002; Silverman-Gavrila et al., 2008).
To examine closely EGFP-Mnk on pseudocleavage furrows, we fixed embryos by boiling (Postner and Wieschaus, 1994) and immunostained them with anti-GFP and anti-Nullo antibodies (Figure 1B). Boiling fixation preserved EGFP-Mnk on centrosomes, at pseudocleavage furrows, and in the nucleus but not on the interkinetochore region and the midbody. Nullo was reported to localize to pseudocleavage furrows and cellularization furrows (Postner and Wieschaus, 1994). In cycle 13 metaphase, EGFP-Mnk localized to circles around spindles similar to Nullo (M13 metaphase), but it appeared as narrow, distinct lines compared with Nullo antibody staining. In cycle 13 telophase, the EGFP-Mnk lines were irregular and discontinuous, and Nullo protein started to disperse (M13 telophase). During cycle 14 interphase, EGFP-Mnk exclusively localized to the nucleus, whereas Nullo localized to cellularization furrows (I14). Three-dimensional reconstructions of GFP, Nullo, and 4′,6-diamidino-2-phenylindole (DAPI) staining of M13 metaphase (Figure 1B) revealed that Nullo protein on pseudocleavage furrows distributed broadly from the cortex to ∼6 μm into the embryo (Figure 1C), whereas EGFP-Mnk was restricted to a 2-μm-thick band at 4- to 6-μm depth from the cortex, the area close to the leading edge of pseudocleavage furrows (Figure 1D).
EGFP-Mnk restores DNA damage responses in mnk-null mutant embryos during the syncytial blastoderm stage
We next asked whether EGFP-Mnk is functional to mediate DNA damage responses during the syncytial blastoderm stage in the absence of endogenous Mnk. We previously showed that injection of bleomycin, a radiomimetic drug that directly binds to DNA and causes DSBs (Povirk, 1996), triggers nuclear dropping, centrosome inactivation, and mitotic delay in wild-type but not in mnk mutant embryos (Takada et al., 2003).
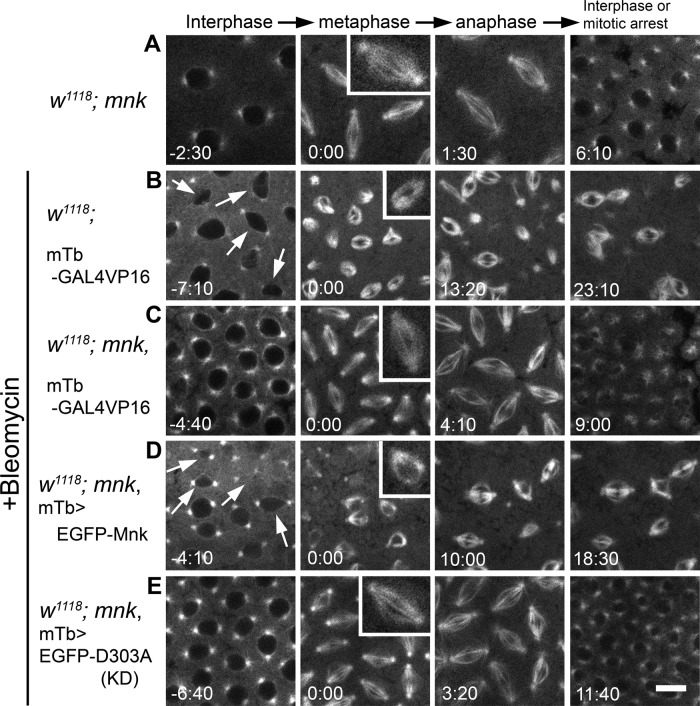
As previously described (Takada et al., 2003), mnk-null embryos injected with only rhodamine-tubulin showed normal spindle formation and cell cycle progression (Figure 2A and Supplemental Movie S2). Bleomycin injection into embryos that were wild type for mnk consistently caused severe nuclear dropping, centrosome inactivation, and mitotic delay/arrest (Figure 2B and Supplemental Movie S3). During the interphase immediately after injection, nuclei started to drop from the cortex (indicated by arrows). At the beginning of mitosis, centrosomes lost the ability to nucleate microtubules, and anastral spindles formed (inset). Centrosomes regained the ability to nucleate microtubules at anaphase and asters reformed but often detached from a spindle, as described previously (Sibon et al., 2000). The length of this mitosis lasted at least 28 min (measured from NEB to the end of recording in anaphase; Supplemental Table S1). It was much longer than normal mitosis, which takes only 5–6 min (from NEB to nuclear envelope formation [NEF]) during the syncytial stage in wild-type or mnk mutant embryos (Foe and Alberts, 1983; Sibon et al., 1997; Takada et al., 2007). On the other hand, mnk-null mutant embryos injected with bleomycin showed normal spindle formation and timely exit from mitosis (Figure 2C and Supplemental Movie S4). Spindles elongated and pulled sister chromatids toward opposite poles in anaphase; however, segregation failed, and dumbbell-shaped nuclei formed at the end of mitosis. DNA damage appears to inhibit sister chromatid separation. All nuclei exited from mitosis on time, and no nuclear dropping occurred in interphase.
FIGURE 2:
EGFP-Mnk restores DNA damage responses in mnk mutant embryos during the syncytial blastoderm stage. All frames are from the rhodamine channel. (A) An mnk mutant embryo injected with rhodamine-tubulin. Normal spindles formed (inset), and a timely exit from mitosis occurred. Selected frames from Supplemental Movie S2. (B) An embryo wild type for mnk carrying a P[mTb-GAL4VP16] transgene was injected with bleomycin and rhodamine-tubulin. Anastral spindles formed (inset), and mitotic delay (or arrest) occurred. Arrows indicate dropping nuclei from the cortex. Selected frames from Supplemental Movie S3. (C) An mnk mutant embryo carrying a P[mTb-GAL4VP16] transgene was injected with bleomycin and rhodamine-tubulin. Normal spindles formed (inset), and a timely exit from mitosis occurred. Selected frames from Supplemental Movie S4. (D) An mnk mutant embryo expressing EGFP-Mnk was injected with bleomycin and rhodamine-tubulin. Anastral spindles formed (inset), and mitotic delay/arrest occurred. Selected frames from Supplemental Movie S5. Arrows indicate dropping nuclei from the cortex. (E) An mnk-null embryo expressing EGFP-D303A was injected with bleomycin and rhodamine-tubulin. Normal spindles formed (inset), and a timely exit from mitosis occurred. Selected frames from Supplemental Movie S6. Elapsed time is shown in minutes:seconds. Scale bar, 10 μm.
When EGFP-Mnk was expressed in mnk mutant embryos, we observed nuclear dropping and severe centrosome inactivation after bleomycin injection (Figure 2D and Supplemental Movie S5). The mitosis lasted >22 min (from NEB to the end of recording in anaphase). Six mnk embryos expressing EGFP-Mnk were injected. All of the injected embryos showed centrosome inactivation and nuclear dropping. Five of them showed significant mitotic delay. The average time from NEB to anaphase (the end of recordings) was 14.44 min (n = 6; detailed data are shown in Supplemental Table S1). We conclude that EGFP-Mnk expression restores wild-type DNA damage responses in mnk-null mutant embryos. The D303A mutation was previously reported to make Mnk kinase inactive (Peters et al., 2002). The mnk mutant embryos expressing EGFP-D303A (EGFP-Mnk with D303A mutation) showed no centrosome inactivation, mitotic delay, or nuclear dropping after bleomycin injection. In spite of normal spindle formation, chromosome segregations failed and dumbbell-shaped nuclei formed, as we saw in mnk mutant embryos. Nuclei exited from mitosis on time, and no nuclear dropping was observed in interphase (Figure 2E and Supplemental Movie S6). Five mnk embryos expressing EGFP-D303A were injected. None of them showed centrosome inactivation, nuclear dropping, or significant mitotic delay. The average mitosis length from NEB to NEF was 6.13 ± 1.16 min (n = 5; Supplemental Table S1). We conclude that kinase activity is necessary for Mnk to mediate embryonic DNA damage responses.
EGFP-Mnk restores developmental block at the midblastula transition in mnk,grp double-mutant embryos
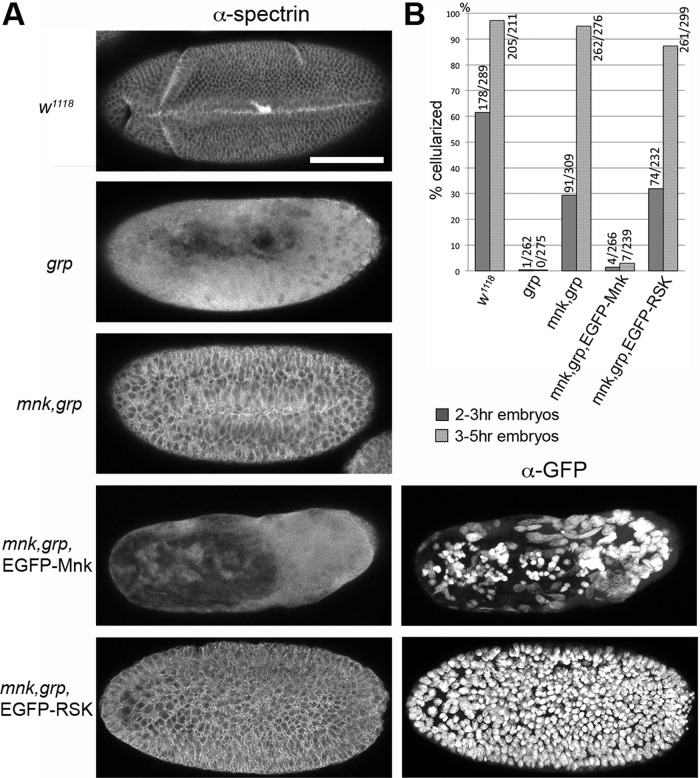
In Drosophila, zygotic control of development begins as the blastoderm cellularizes. Functional null mutation in grp, which encodes an essential component of the DNA replication checkpoint, blocks development at the MBT (Sibon et al., 1997). We previously showed that this developmental block was the embryonic response to DNA damage accumulated in grp mutant embryos by the end of syncytial stage and that Mnk was essential for the response. To investigate whether EGFP-Mnk was functional to mediate the developmental block at the MBT, we examined cellularization, which is the first morphological event that requires zygotic gene expression (Schejter and Wieschaus, 1993; Postner and Wieschaus, 1994). Spectrin is a component of actin-membrane structures and localizes to the cellular membrane surrounding cortical nuclei on the embryo cortex when an embryo cellularizes (Byers et al., 1987). As shown in Figure 3A, the majority of 3- to 5-h-old wild-type embryos cellularized and gastrulated (w1118), whereas the grp mutant did not (grp; Sibon et al., 1997). The mnk-null mutation suppressed the cellularization and gastrulation defects associated with grp mutation, as described previously (mnk,grp). Cells in mnk,grp embryos are larger than in wild-type embryos due to chromosome segregation failures during late syncytial blastoderm divisions (Takada et al., 2007). EGFP-Mnk expression in mnk,grp embryos inhibited cellularization (mnk,grp, EGFP-Mnk; α-spectrin). On the other hand, expression of EGFP-RSK, which carries a triple mutation in the FHA domain and appears to be kinetically inactive (described later in Figures 7 and 8), did not inhibit cellularization in mnk,grp embryos (mnk,grp, EGFP-RSK; α-spectrin). Anti-GFP staining revealed that massive nuclear dropping occurred in the embryo expressing EGFP-Mnk (mnk,grp, EGFP-Mnk; α-GFP) but not in the embryo expressing EGFP-RSK (mnk,grp, EGFP-RSK; α-GFP). We counted cellularized and noncellularized embryos in 2- to 3- and 3- to 5-h-old embryo collections of each genotype (Figure 3B). In 2- to 3-h-old collections, 29.4% of mnk,grp embryos cellularized, whereas only 1.5% of mnk,grp embryos expressing EGFP-Mnk cellularized similar to grp mutant (0.38%), and 31.9% of mnk,grp embryos expressing EGFP-RSK cellularized. In 3- to 5-h-old collections, 94.9% of mnk,grp embryos cellularized; however, only 2.92% of mnk,grp embryos expressing EGFP-Mnk cellularized, similar to grp mutant (0%), whereas 87.3% of mnk,grp embryos expressing EGFP-RSK cellularized. These results indicate that EGFP-Mnk, but not EGFP-RSK, mimics endogenous Mnk function and causes DNA damage–induced developmental block at the MBT in the presence of grp mutation.
FIGURE 3:
EGFP-Mnk, but not EGFP-RSK, restores developmental block at the midblastula transition in mnk,grp double-mutant embryos. Cellular membranes were stained with anti-spectrin antibody to examine cellularization. (A) Representative phenotype for each genotype in collections of 3- to 5-h-old embryos. The majority of wild-type (w1118) embryos, mnk,grp double-mutant embryos, and mnk,grp embryos expressing EGFP-RSK cellularized and started to gastrulate. The majority of grp mutant embryos and mnk,grp double-mutant embryos expressing EGFP-Mnk did not show membrane structures stained with α-spectrin. EGFP-Mnk and EGFP-RSK localize to the interphase nucleus (Figures 1 and 7). Severe nuclear aggregation and dropping occurred in mnk,grp embryos expressing EGFP-Mnk, whereas all nuclei stayed on the cortex in mnk,grp embryos expressing EGFP-RSK (α-GFP). Scale bar, 100 μm. (B) Percentages of cellularized embryos in 2- to 3- and 3- to 5-h-old collections. Number of cellularized/total counted embryos is indicated at the top of each data bar.
FIGURE 7:
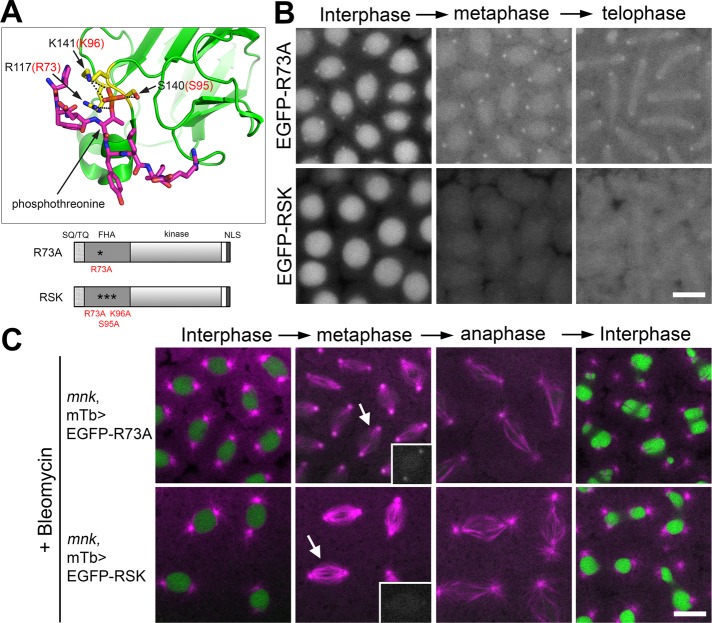
The phosphopeptide-binding ability of the FHA domain is required for Mnk localization to key mitotic structures. (A) The 3D structure of part of the human Chk2 FHA domain (green) and a synthetic phosphopeptide HFDpTYLI (magenta) based on an x-ray structural study by Li et al. (2002; Protein Data Bank ID: 1GXC). Prepared using PyMOL (www.pymol.org). The phosphate of pThr is shown in orange. Three conserved residues (R117, S140, and K141) that form hydrogen bonds (black dotted lines) with the phosphate are shown in yellow. Corresponding amino acid numbers of the three residues in Drosophila Mnk are shown in red (R73, S95, K96). Schematic diagrams of Mnk variants that carry R73A or RSK mutation are shown below the 3D structure. (B) R73A mutation greatly reduced localization of the molecule to centrosomes, interkinetochores/centromeres, and the midbody and disrupted localization to pseudocleavage furrows. RSK mutation completely disrupted localization of the molecule to all the mitotic structures. Frames were selected from Supplemental Movies S17 (EGFP-R73A) and S18 (EGFP-RSK). Scale bar, 10 μm. (C) EGFP-R73A and EGFP-RSK are not functional to restore centrosome inactivation, mitotic delay, and nuclear dropping responses after bleomycin injection in mnk mutant embryos. EGFP signals are shown in green and rhodamine signals in magenta. Frames were selected from Supplemental Movies S19 (mnk, mTb>EGFP-R73A) and S20 (mnk, mTb>EGFP-RSK). Insets, EGFP signals on spindles indicated by arrows. Scale bar,10 μm.
FIGURE 8:
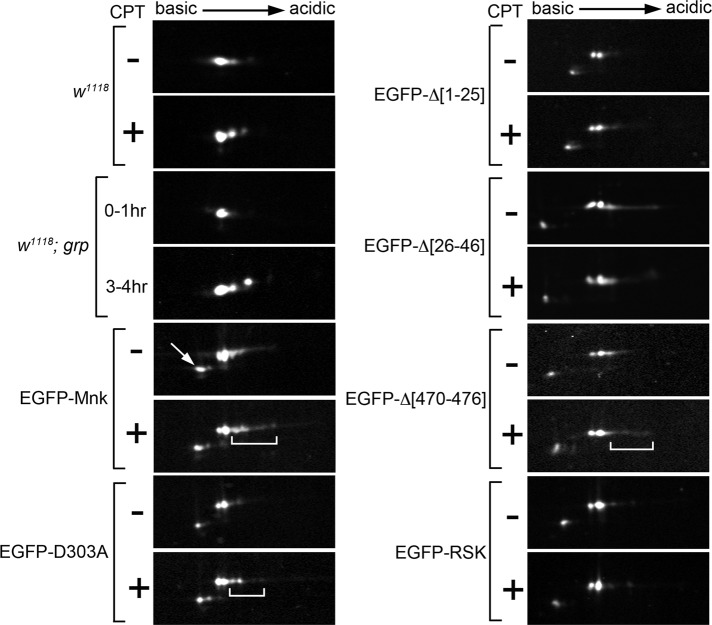
The FHA domain and its phosphopeptide-binding ability are required for Mnk phosphorylation. Endogenous Mnk and EGFP-Mnk variants in embryonic lysates were analyzed by 2D electrophoresis and Western blot. Endogenous Mnk in w1118 and w1118; grp embryos was detected with anti-Mnk antibody. EGFP-Mnk variants expressed in mnk mutant embryos were detected with anti-GFP antibody. Lysates were made without (–) and with (+) CPT treatment, except for grp embryos. (w1118) CPT treatment increased acidic protein spots. Note that acidic spots show upward mobility shift. (w1118; grp) Single Mnk protein spot appeared in 0- to 1-h-old embryo lysate. Additional acidic spots appeared in 3- to 4-h-old embryos. Note that the acidic spots show upward mobility shift. (EGFP-Mnk) EGFP-Mnk was expressed in mnk mutant embryos. CPT treatment increased acidic protein spots (bracket). (EGFP-D303A) EGFP-D303A was expressed in mnk mutant embryos. CPT treatment moderately increased acidic protein spots (bracket). (EGFP-Δ[1-25]) EGFP-Δ[1-25] was expressed in mnk mutant embryos. CPT treatment did not change appearance of protein spots. (EGFP-Δ[26-46]) EGFP-Δ[26-46] was expressed in mnk mutant embryos. CPT treatment did not increase acidic spots. (EGFP-Δ[470-476]) EGFP-Δ[470-476] was expressed in mnk mutant embryos. CPT treatment slightly increased acidic spots (bracket). (EGFP-RSK) EGFP-RSK was expressed in mnk mutant embryos. CPT treatment did not increase acidic protein spots. Anti-EGFP Western blot detected minor protein spots with slightly higher pI and lower molecular weight (indicated by arrow in EGFP-Mnk). These are likely truncated products of each EGFP-Mnk variant.
Quantitative Western blot analyses (Supplemental Figure S3) showed that the ratio of the EGFP-Mnk amount expressed in mnk embryos to the endogenous Mnk amount in wild-type (w1118) embryos was estimated to be 1.54 ± 0.59 (n = 6, independent blots), the ratio of EGFP-D303A amount to endogenous Mnk amount was 1.38 ± 0.54 (n = 4, independent blots), and the ratio of EGFP-RSK amount to endogenous Mnk amount was 3.72 ± 0.80 (n = 5, independent blots), indicating that EGFP-Mnk and EGFP-D303A proteins were expressed at slightly increased level relative to the endogenous Mnk and that EGFP-RSK is moderately overexpressed relative to the endogenous Mnk level. Taken together with the fact that EGFP-Mnk restores DNA damage responses both during the syncytial blastoderm stage and at the MBT in the absence of endogenous Mnk, we conclude that EGFP-Mnk mimics endogenous Mnk behavior and function, and the inabilities of EGFP-D303A and EGFP-RSK to restore the embryonic DNA damage responses were due to functional inactivation of the molecule.
The NLS at the C-terminus is required for active import of Mnk into the nucleus
To understand how Mnk is recruited to multiple cellular structures in such a specific manner, we looked for domain(s) of Mnk that are required for Mnk localization. We created a series of EGFP-tagged Mnk variants, each carrying a point mutation or a deletion, and expressed them individually in mnk-null embryos. We first examined localizations of these tagged molecules without DNA damage (Figure 4). EGFP-D303A, a kinase-inactive mutant (Peters et al., 2002), localizes similarly to EGFP-Mnk, indicating that the kinase activity is not necessary for normal Mnk localization (Figure 4, A and B, and Supplemental Movie S7). Deletion of the SQ/TQ-cluster domain (SCD) also did not affect Mnk localization (Figure 4C and Supplemental Movie S8).
FIGURE 4:
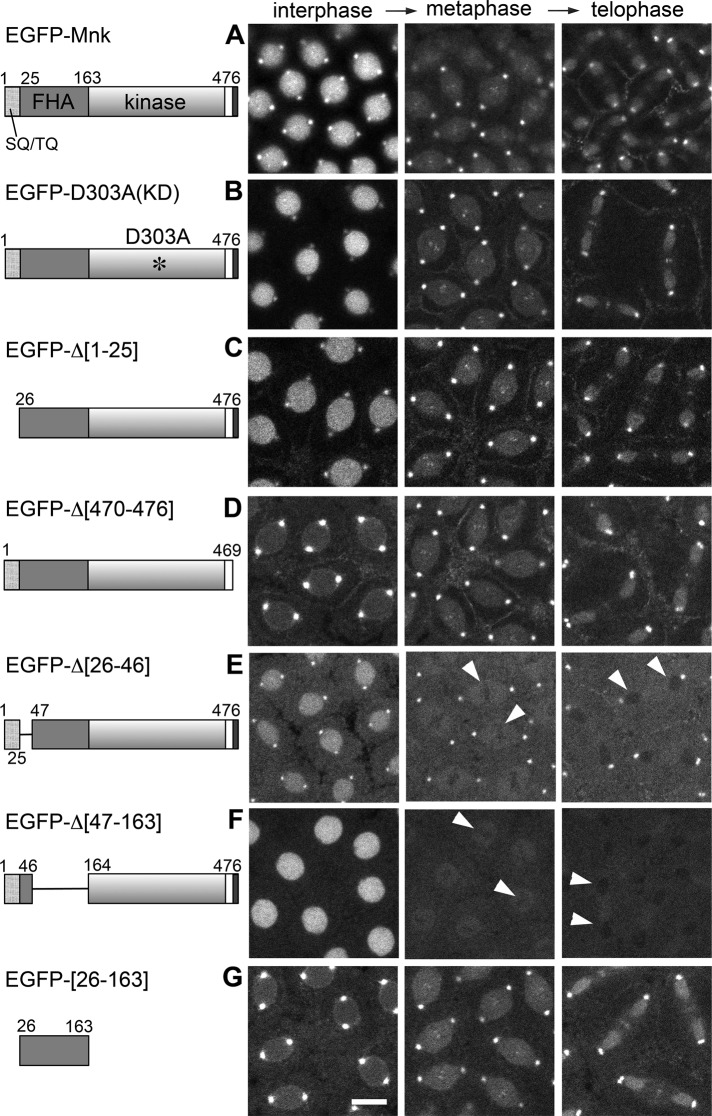
Localization of EGFP-Mnk variants without DNA damage in syncytial blastoderm– stage Drosophila embryos. EGFP was conjugated at the N-terminus of each Mnk variant. Left, schematic diagrams. EGFP was omitted. Selected frames from time-lapse laser-scanning confocal microscope movies that captured EGFP signal during cycle 11 or 12. (A) EGFP-Mnk localizes to the nucleus, centrosomes, interkinetochore/centromeres, the midbody, and pseudocleavage furrows without DNA damage as shown in Figure 1A and Supplemental Movie S1. (B) EGFP-D303A localizes similarly to EGFP-Mnk (Supplemental Movie S7). (C) EGFP-Δ[1-25] localizes similarly to EGFP-Mnk (Supplemental Movie S8). (D) EGFP-Δ[470-476] is not actively transported into the interphase nucleus but localizes to centrosomes, interkinetochore/centromeres, the midbody, and pseudocleavage furrows (Supplemental Movie S9). (E) EGFP-Δ[26-46] localizes to the nucleus, centrosomes, and pseudocleavage furrows but not to interkinetochore/centromeres and the midbody. It is excluded from the area close to mitotic chromosomes (white arrowheads; Supplemental Movie S10). (F) EGFP-Δ[47-163] localizes to the nucleus but not to centrosomes, interkinetochore/centromeres, the midbody, and pseudocleavage furrows. It is excluded from the area close to mitotic chromosomes (white arrowheads; Supplemental Movie S11). (G) EGFP-[26-163] localizes to centrosomes, interkinetochore/centromeres, the midbody, and pseudocleavage furrows but is not actively transported into the nucleus (Supplemental Movie S12). Transgenic proteins were expressed by nos-GAL4VP16 (A) or mTb-GAL4VP16 (B–G) driver in w1118 (A) or mnk mutant (B–G) embryos. Scale bar, 10 μm.
EGFP-Mnk is efficiently imported into the nucleus, and the centrosomal signal gradually decreases during interphase (Figure 1A and Supplemental Movie S1), implying the presence of a potent NLS in Mnk. Kosugi et al. (2009) proposed six classes of NLS consensus that potentially bound to importin α, based on a random peptide library screening assay. We noted that the C-terminal 26 amino acids of Mnk (KRLMKLDGMEIEEENFLEPPTKRSRR; GenBank: BAA28755.1; AAF53867.2) match a bipartite NLS consensus (KRX10-20K(KR)X(K/R)). The linker (underlined) between the first KR and the C-terminal basic stretch fulfills optimal characteristics of the consensus; the central region of the linker is acidic, and the terminal region contains proline residues. We deleted the C-terminal seven amino acids of Mnk (PTKRSRR), which overlaps the C-terminal basic stretch of the bipartite consensus and matches a monopartite NLS consensus (P/R)XXKR*(K/R) (asterisk indicates any amino acid except for Asp or Glu). This deletion disrupted active nuclear transport of the molecule (EGFP-Δ[470-476]). The intensity of nuclear EGFP signal remained similar to or slightly higher than the cytoplasmic EGFP in interphase. The EGFP signal was slightly concentrated around the nuclear envelope, suggesting that it was unable to enter the nucleus. The deletion did not affect localization of the molecule to centrosomes and other mitotic structures (Figure 4D and Supplemental Movie S9). We conclude that a potent NLS resides at the C-terminus of Mnk.
The FHA domain is essential and sufficient for Mnk localization to mitotic structures
The FHA domain of human Chk2 folds into 11 β-strands that form two β-sheets that lie on top of each other. Loops between β3-β4, β4-β5, β6-β7, and β10-β11 constitute a phosphopeptide-binding surface (Li et al., 2002; Mahajan et al., 2008; Cai et al., 2009).
Deletion of amino acids 26–46 from the Mnk-FHA domain, which includes a β1-strand based on an alignment of Mnk against human Chk2 (Supplemental Figure S1), did not affect localization of the molecule to the nucleus, centrosomes, or pseudocleavage furrows (Figure 4E and Supplemental Movie S10). However, it caused exclusion of the molecule from the vicinity of mitotic chromosomes (arrowheads in Figure 4E). The exclusion disrupted localization of EGFP-Δ[26-46] to the interkinetochore/centromere region and around chromosomes during telophase. This variant also did not localize to the midbody during telophase.
Deletion of amino acids 47–163 from the Mnk-FHA domain disrupted localization of the molecule to centrosomes and other mitotic structures, whereas it did not affect nuclear localization (Figure 4F and Supplemental Movie S11). This deletion includes loops that constitute the phosphopeptide-binding site (Li et al., 2002; Supplemental Figure S1). EGFP-Δ[47-163] was excluded from the area surrounding mitotic chromosomes, similar to EGFP-Δ[26-46] (arrowheads in Figure 4F), suggesting that at least a part of amino acids 47–163 may be required for Mnk to draw near to mitotic chromosomes. Of importance, as we describe later, mutations in the phosphopeptide-binding site of the FHA domain do not cause such exclusion.
Finally, we demonstrated that the entire FHA domain (amino acids 26–163) tagged with EGFP localized to centrosomes, interkinetochore/centromeres, the midbody, and pseudocleavage furrows, similar to EGFP-Mnk. It was not actively imported into the nucleus, likely due to a lack of the C-terminal part of Mnk, which contains the NLS (Figure 4G, EGFP-[26-163]; Supplemental Movie S12). The FHA domain that lacks amino acids 26–46 (EGFP-[47-163]) localized similarly to EGFP-[26-163], except that it was excluded from the vicinity of mitotic chromosomes and did not localize to centromeres and the midbody (unpublished data). Taken together, our results indicate that the FHA domain is essential and sufficient for localization to centrosomes, interkinetochore/centromeres, the midbody, and pseudocleavage furrows and that the N-terminus of the FHA domain is essential for Mnk to be in the vicinity of mitotic chromosomes.
EGFP-Mnk forms foci on mitotic chromosomes with DNA damage
We next examined localization of EGFP-Mnk with DNA damage. mnk mutant embryos expressing EGFP-Mnk were injected with bleomycin and rhodamine-tubulin. As shown in Figure 2D, EGFP-Mnk mediates DNA damage responses, including centrosome inactivation, mitotic delay, and nuclear dropping, similarly to endogenous Mnk. A representative spindle shown in Figure 5A (frames were selected from Supplemental Movie S5) is anastral and still in an anaphase-like state 16 min after NEB (time 16:30). EGFP-Mnk localized to the centrosomes and the nucleus in interphase and was recruited more to the centrosomes at NEB (time 0:00). It also localized to pseudocleavage furrows during mitosis, although the structure of pseudocleavage furrows was significantly disrupted (Supplemental Movie S5). Chromosome segregations failed, and the midbody did not form. We found that EGFP-Mnk also formed numerous foci/aggregates on mitotic chromosomes (Figure 5A time 2:30–16:30).
FIGURE 5:
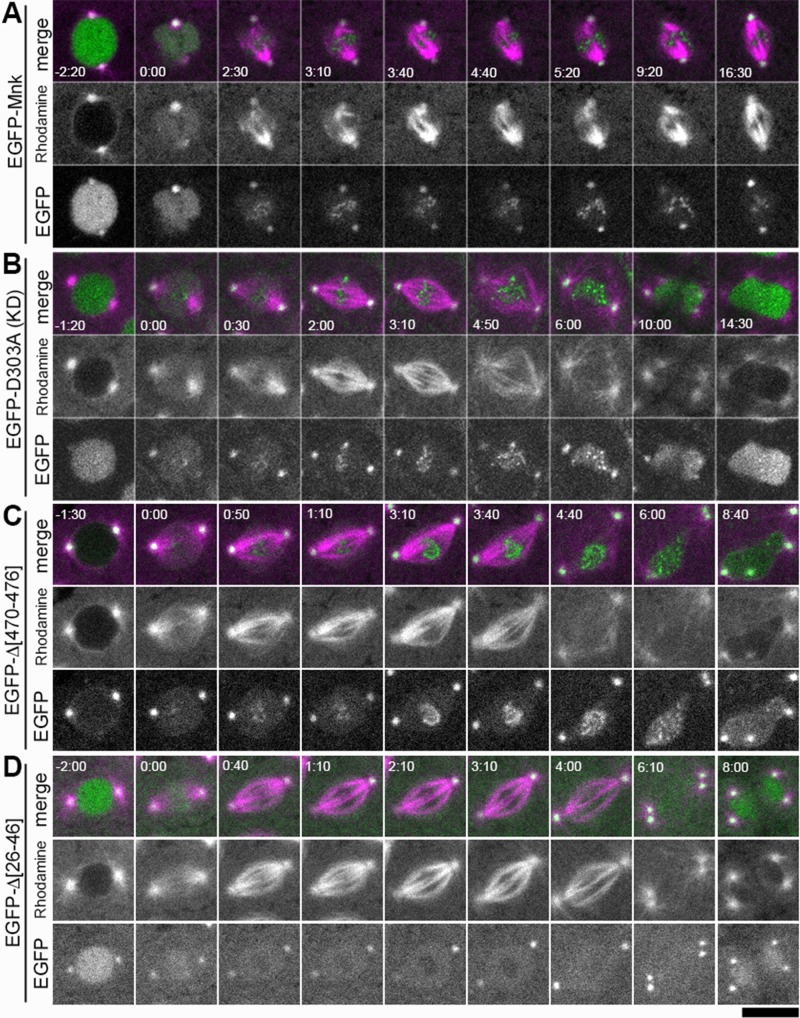
Localization of EGFP-Mnk and Mnk variants with DNA damage. Bleomycin and rhodamine-tubulin were injected into transgenic embryos expressing each EGFP-Mnk variant in mnk mutant background. EGFP is shown in green and rhodamine is shown in magenta in merged images. (A) EGFP-Mnk restored centrosome inactivation and mitotic delay in mnk mutant. Anastral spindles formed (time 2:30–9:20). EGFP-Mnk localized to centrosomes and formed foci/aggregates on mitotic chromosomes (time 2:30–16:30). Frames were selected from Supplemental Movie S5. (B) EGFP-D303A did not restore centrosome inactivation and mitotic delay in mnk mutant. Normal spindles formed (time 2:00–3:10). Mitotic delay did not occur; however, chromosome segregation failed, and connected dumbbell-shaped nuclei formed. The midbody did not form (time 10:00–14:30). EGFP-D303A localized to centrosomes and formed foci/aggregates on mitotic chromosomes (time 0:30–6:00) similarly to EGFP-Mnk. Frames were selected from Supplemental Movie S6. (C) EGFP-Δ[470-476] did not restore centrosome inactivation and mitotic delay in this mnk mutant embryo. EGFP-Δ[470-476] was not actively imported into the nucleus. It localized to centrosomes and formed foci/aggregates on mitotic chromosomes (time 0:50–4:40) similarly to EGFP-Mnk or EGFP-D303A. Frames were selected from Supplemental Movie S16. (D) EGFP-Δ[26-46] did not restore centrosome inactivation and mitotic delay in mnk mutant. EGFP-Δ[26-46] localized to the nucleus and centrosomes. It was excluded from the vicinity of mitotic chromosomes (time 0:40–4:00) as shown without DNA damage (Figure 4E). It did not make foci/aggregates on mitotic chromosomes. Frames were selected from Supplemental Movie S14. Elapsed time is shown in minutes:seconds. NEB is set as time 0:00. Scale bar, 10 μm.
Some of the chromosomal foci/aggregates with DNA damage may associate with the interkinetochore/centromere region (Figure 5A) as we observed without DNA damage (Figure 1E and Supplemental Movie S21). We injected bleomycin into embryos that express both EGFP-Mnk and mRFP-Rod to see whether EGFP-Mnk foci are in the interkinetochore/centromere region after DNA damage. However, bleomycin injection somehow severely disrupted structure of the embryos, and the mRFP-Rod signal became too weak to follow (unpublished data). Without DNA damage, a limited number of EGFP-Mnk dots align at spindle equator at metaphase (Figure 1, A and E, and Supplemental Movies S1 and S21). Only three to five dots are visible per spindle without DNA damage. Our time-lapse confocal recordings may not capture all the centromeres at each time point because the recordings are on single z-section, not a projection of multiple z-sections. However, with DNA damage, significantly more foci appear—for example, at least 10–15 foci at time 5:20 in Figure 5A—and they spread on mitotic chromosomes from anaphase to the end of mitosis (compare Figure 1A, time 3:30–4:05, and Figure 5A, time 3:10–16:30; Supplemental Movies S1 and S5). This suggests that most of the DNA damage–induced foci do not associate with centromeres.
The SCD and the FHA domain are essential for Mnk function to mediate DNA damage responses
To investigate which structural domain of Mnk is important for localization and Mnk function after DNA damage, we performed bleomycin injection into mnk mutant embryos that express EGFP-Mnk deletion variants, each of them lacking a specific structural domain. We looked at localization of each EGFP-Mnk variant and simultaneously examined whether it induces centrosome inactivation, mitotic delay, and nuclear dropping (Figures 5, B–D, and 6).
FIGURE 6:
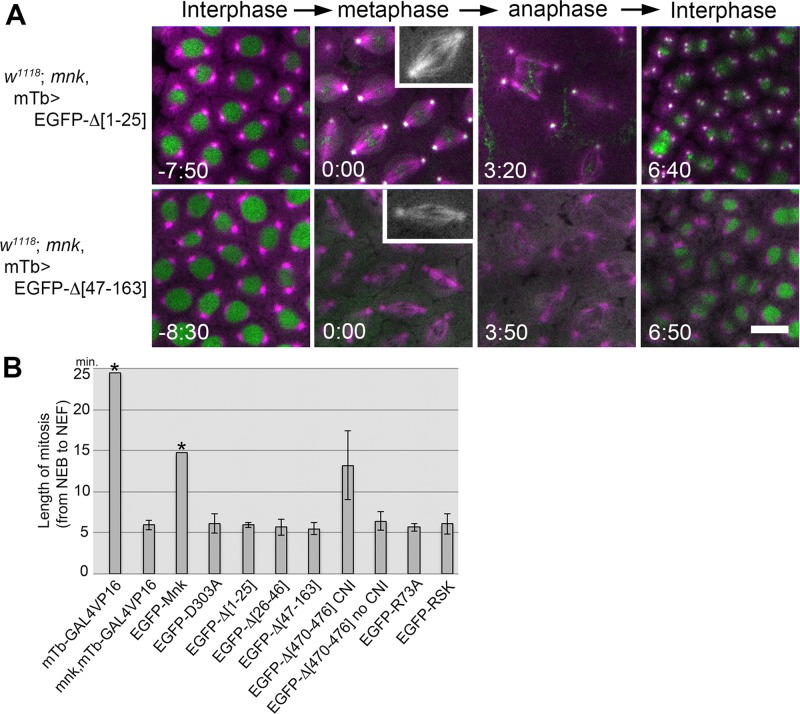
The SCD, the FHA domain, and the NLS are required for Mnk function to mediate centrosome inactivation, mitotic delay, and nuclear dropping. (A) Expression of EGFP-Δ[1-25] (deletion of the SCD; Supplemental Movie S13) and EGFP-Δ[47-163] (deletion of the major part of the FHA domain; Supplemental Movie S15) in mnk mutant embryos did not restore centrosome inactivation (CNI), mitotic delay, and nuclear dropping responses after bleomycin injection. Bleomycin and rhodamine-tubulin were injected. EGFP-Δ[1-25] localized similarly to EGFP-Mnk and formed foci/aggregates on mitotic chromosomes. EGFP-Δ[47-163] localized to the nucleus but did not localized to centrosomes and pseudocleavage furrows and was excluded from the vicinity of mitotic chromosomes (Supplemental Movie S15). Rhodamine is shown in magenta, and EGFP is shown in green. Elapsed time is shown in minutes:seconds. Bar, 10 μm. (B) Average length of mitosis (from NEB to NEF) in mnk mutant embryos expressing EGFP-Mnk or EGFP-Mnk variant after bleomycin and rhodamine-tubulin injection. mTb-GAL4VP16 driver was used to express EGFP fusion proteins. mTb-GAL4VP16 : w1118; P[matalpha4-GAL4-VP16] embryos that were used as the wild-type control (n = 2). mnk, mTb-GAL4VP16 : w1118; mnk P[matalpha4-GAL4-VP16] embryos that were used as the mnk mutant control (n = 4). EGFP-Mnk : mnk mutant embryos expressing EGFP-Mnk (n = 6). EGFP-D303A : mnk mutant embryos expressing EGFP-D303A (n = 5). EGFP-Δ[1-25] : mnk mutant embryos expressing EGFP-Δ[1-25] (n = 8). EGFP-Δ[26-46] : mnk mutant embryos expressing EGFP-Δ[26-46] (n = 5). EGFP-Δ[47-163] : mnk mutant embryos expressing EGFP-Δ[47-163] (n = 6). EGFP-Δ[470-476] CNI : mnk mutant embryos expressing EGFP-Δ[470-476], which showed CNI and mitotic delay (n = 2). EGFP-Δ[470-476] no CNI : mnk mutant embryos expressing EGFP-Δ[470-476], which showed no CNI and mitotic delay (n = 8). EGFP-R73A : mnk mutant embryos expressing EGFP-R73A (n = 5). EGFP-RSK : mnk mutant embryos expressing EGFP-RSK (n = 6). Average mitosis length from NEB to the end of recording is shown for the data bar with asterisk, as these embryos showed significant mitotic delay, and recordings were stopped during anaphase before NEF. Error bars indicate SDs. Injection results are summarized in Supplemental Table S1.
EGFP-D303A (kinase-inactive Mnk) did not mediate centrosome inactivation, mitotic delay, and nuclear dropping, as shown in Figure 2E. Figure 5B shows a representative nucleus that shows normal spindle formation (time 2:00–3:10) and timely exit from mitosis (time 6:00). EGFP-D303A localizes similarly to EGFP-Mnk with DNA damage. It forms foci/aggregates that spread on the chromosomes (time 2:00–4:50; also see Supplemental Movie S6). When the nucleus exits from mitosis, the foci disperse (time 10:00–14:30).
The NLS deletion mutant (EGFP-Δ[470-476]) did not cause centrosome inactivation or significant mitotic delay in the majority of bleomycin-injected embryos (8 of 10 injected embryos). A representative mitosis from one of the embryos (Figure 5C; frames from Supplemental Movie S16) shows normal spindle formation (time 0:50–3:10) and timely exit from mitosis (time 4:40–6:00). EGFP-Δ[470-476] also localizes similarly to EGFP-Mnk except that it is not actively imported into the nucleus in interphase (time −1:30). It also forms foci/aggregates on mitotic chromosomes, and the foci appear to cover the entire chromosomal region (time 2:30–4:40). Two of 10 mnk mutant embryos expressing EGFP-Δ[470-476] showed centrosome inactivation and mitotic delay after bleomycin injection. They exited mitosis >11 min after NEB (Supplemental Table S1).
The FHA-domain N-terminal deletion mutant (EGFP-Δ[26-46]) did not mediate centrosome inactivation or mitotic delay. EGFP-Δ[26-46] localizes to centrosomes and pseudocleavage furrows, as well as to the nucleus; however, it does not form any foci/aggregates on chromosomes (Supplemental Movie S14). The molecule is excluded from the vicinity of chromosomes (Figure 5D, time 0:40–4:00; Supplemental Movie S14), similar to what we observed without DNA damage (Figure 4E and Supplemental Movie S10). Another FHA-domain-deletion mutant lacking the major part of FHA domain (EGFP-Δ[47-163]), which includes the phosphopeptide-binding site, also did not mediate DNA damage responses (Figure 6A). EGFP-Δ[47-163] did not localize to centrosomes and mitotic structures, similar to what we observed without DNA damage (Figure 4F and Supplemental Movie S9) and did not form any foci/aggregates on mitotic chromosomes with DNA damage (Figure 6A and Supplemental Movie S11). We conclude that the FHA domain is required not only for proper Mnk localization to centrosomes, centromeres, the midbody, and pseudocleavage furrows, but also for formation of foci/aggregates on mitotic chromosomes with DNA damage.
Finally, the SCD deletion mutant (EGFP-Δ[1-25]) was also incompetent to induce DNA damage responses; however, EGFP-Δ[1-25] localized similarly to EGFP-Mnk (Figure 6A and Supplemental Movie S13).
We performed bleomycin and rhodamine-tubulin injection into multiple embryos for each genotype and calculated average mitotic length after injection (Figure 6B and Supplemental Table S1). Wild-type control embryos (mTb-GAL4VP16), as well as mnk embryos expressing EGFP-Mnk (EGFP-Mnk), showed significant lengthening of mitosis compared with mnk mutant (mnk, mTb-GAL4VP16). The recordings were stopped during anaphase-like state before NEF in these delayed embryos. Therefore the data bars (marked by asterisks) indicate average time from NEB to the end of recordings. For all other genotypes except for embryos that express EGFP-Δ[470-476], embryos did not show significant mitotic delay, and the data bars indicate average length of mitosis from NEB to NEF. Among the embryos expressing EGFP-Δ[470-476], 8 of 10 did not show DNA damage responses (EGFP-Δ[470-476] no CNI); however, two embryos showed centrosome inactivation and significant mitotic delay (EGFP-Δ[470-476] CNI). The average mitotic lengths of the two groups are shown separately. We conclude that the SCD and the FHA domain of Mnk are essential for DNA damage responses during the syncytial blastoderm stage and the NLS is required for Mnk to fully elicit its function.
The phosphopeptide-binding ability of the FHA domain is essential for Mnk localization and function to mediate DNA damage responses
To examine whether the phosphopeptide-binding ability of the FHA domain is required for Mnk localization and function, we introduced point mutations in the phosphopeptide-binding site. On the basis of a structural study of human Chk2 with synthetic phosphopeptide complex (Li et al., 2002) and an alignment of Mnk-FHA domain against human Chk2-FHA domain (Figure 7A and Supplemental Figure S1; Cai et al., 2009), we first replaced a conserved arginine (R73) on the β3-β4 loop with alanine (Figure 7A). This arginine in human Chk2 (R117) forms a hydrogen bond with the pThr phosphate in the binding phosphopeptide, and an alanine substitution severely reduces phosphopeptide binding (Li et al., 2002). This mutation did not affect nuclear localization but significantly reduced localization of the molecule to centrosomes, centromere, the midbody, and pseudocleavage furrows (Figure 7B, EGFP-R73A; Supplemental Movie S17). We next introduced two additional alanine substitutions on conserved serine 95 and lysine 96 residues (R95 and K96). Corresponding human Chk2 residues (S140 and K141) form four additional hydrogen bonds with the pThr phosphate (Li et al., 2002; Figure 5A). This triple mutation completely abolished Mnk localization to the mitotic structures but did not affect the nuclear localization (Figure 7B, EGFP-RSK; Supplemental Movie S18). Both EGFP-R73A and EGFP-RSK are slightly concentrated on spindles and not excluded from the vicinity of mitotic chromosomes (Figure B, metaphase and telophase, and C, insets; Supplemental Movies S17–S20), unlike the FHA deletion mutants EGFP-Δ[26-46] and EGFP-Δ[47-163] (Figure 4, E and F). Nevertheless, they did not form DNA damage–induced foci on mitotic chromosomes (Figure 7C, insets; Supplemental Movies S19 and S20), similarly to EGFP-Δ[26-46] and EGFP-Δ[47-163] (Figures 5D and 6A).
Both EGFP-R73A and EGFP-RSK did not restore DNA damage responses in mnk mutant embryos (Figure 7C and Supplemental Movies S19 and S20). Average mitotic length (from NEB to NEF) in embryos expressing EGFP-R73A after bleomycin injection was 5.67 ± 0.47 min (n = 5), and in embryos expressing EGFP-RSK, it was 6.06 ± 1.21 min (n = 6; Figure 6B and Supplemental Table S1). Taking the results together, we conclude that the phosphopeptide-binding ability of the FHA domain is essential for recruiting Mnk to centrosomes, interkinetochore/centromere, the midbody, and pseudocleavage furrows, independently of DNA damage. It is also essential for Mnk to form foci/aggregates on mitotic chromosomes upon DNA damage and mediate the embryonic DNA damage responses. The N-terminus of the FHA domain, but not the phosphopeptide-binding ability of the FHA domain, is essential for Mnk to be in close vicinity of mitotic chromosomes.
Is Mnk localization to key mitotic structures essential for the embryonic DNA damage responses? To answer this question, we conjugated human histone H2B to the EGFP-Mnk N-terminus to tether the molecule to chromatin. In cultured human cells, it was shown that Chk2 is highly mobile in the nucleus irrespective of DNA damage, and forced immobilization of Chk2 by conjugating histone H2B to GFP-Chk2 impaired its stimulating effects on p53-dependent transcription (Lukas et al., 2003). We sought to disrupt normal Mnk localization to mitotic structures without disrupting FHA-domain function and nuclear localization. Unexpectedly, expression of the H2B-EGFP-Mnk blocked female oogenesis at 25ºC, the temperature we normally use for transgene expression, and we could not collect enough embryos to analyze. At lower induction temperature, we obtained a small number of embryos with low transgenic protein expression. We confirmed that the fusion protein was tethered to chromatin throughout the cell cycle and did not mediate centrosome inactivation, mitotic delay, and nuclear dropping after bleomycin injection (Supplemental Figure S2 and Supplemental Movie S22). However, we were able to inject only two embryos with low-level transgene expression, which was insufficient to determine whether the H2B-EGFP-Mnk is functional.
The FHA mutations block phosphorylations of Mnk upon DNA damage
Human Chk2 is known to autophosphorylate multiple sites, including two threonines in the kinase activation loop. It becomes active after initial transphosphorylation on Thr-68 within the SCD by ATM upon DNA damage (Ahn et al., 2000, 2002; Lee and Chung, 2001; Oliver et al., 2006). Multiple phosphorylations, indicative of kinase activation, can be detected by mobility shifts on SDS–PAGE (Ahn et al., 2000, 2002). Drosophila Mnk was similarly reported to undergo mobility shifts on SDS–PAGE due to phosphorylations upon DNA damage (Brodsky et al., 2004). To determine whether EGFP-Mnk variants used in this study could become catalytically active upon DNA damage, we examined whether each EGFP-Mnk variant was phosphorylated in embryos after camptothecin (CPT) treatment. CPT inhibits topoisomerase I (Pommier et al., 1998) and thereby causes DSBs and efficiently induces embryonic DNA damage responses (Sibon et al., 2000; Takada et al., 2003).
EGFP-Mnk has a higher molecular weight, and it is difficult to see mobility shifts on simple SDS–PAGE. Therefore we performed two-dimensional (2D) gel electrophoresis to detect isoelectric point (pI) shifts of EGFP-Mnk and EGFP-Mnk variants (Figure 8). When the protein is multiply phosphorylated, the phosphorylated isoforms appear as multiple protein spots horizontally aligned over small intervals on 2D electrophoresis (Farrell et al., 1978). To test whether the 2D system detects endogenous Mnk phosphorylations, we performed anti-Mnk Western blot after 2D electrophoresis of wild-type embryonic extracts without and with CPT treatment. We found that acidic isoforms of Mnk increased, indicating that phosphorylation of Mnk occurred with CPT treatment (w1118). The acidic isoforms exhibit small upward shifts on the second dimension, consistent with a previous report that phosphorylated Mnk run slightly more slowly on SDS–PAGE (Brodsky et al., 2004). We also examined endogenous Mnk phosphorylations in grp mutant embryos. grp mutation causes spontaneous accumulation of DNA damage by the end of the syncytial blastoderm stage, which phosphorylates and activates Mnk (Takada et al., 2007). We found that Mnk was not phosphorylated in the early syncytial stage (w1118; grp 0–1 h); however, it was phosphorylated around the time of MBT (w1118; grp 3–4 h) when grp embryos manifest mitotic and developmental defects (Sibon et al., 1997). The acidic isoforms that appear in 3- to 4-h-old embryos exhibit small upward mobility shifts on the second dimension (w1118; grp 3–4 h).
We next examined EGFP-Mnk variants expressed in mnk mutant embryos by anti-GFP Western blot after 2D electrophoresis. EGFP-Mnk showed some acidic isoforms even without CPT treatment; however, CPT treatment further increased acidic spots (denoted with a bracket; EGFP-Mnk), indicating that DNA damage–dependent phosphorylation occurred. We predicted that deletion of the SCD should abolish the priming transphosphorylation by ATM and the following autophosphorylation. Consistent with our prediction, CPT treatment did not cause any change in phosphorylation status of EGFP-Δ[1-25]. EGFP-D303A, a kinase-inactive Mnk (Peters et al., 2002), showed increase of acidic spots with CPT treatment (denoted with a bracket) to a lesser extent than with EGFP-Mnk. This suggests that transphosphorylations by ATM upon DNA damage might occur; however, autophosphorylations, which require Mnk's own kinase activity, might not. EGFP-Δ[470-476] showed a slight increase of acidic spots with CPT treatment (denoted by a bracket). Although this variant is not actively imported into the nucleus, some molecules still reside there (Figure 4D and Supplemental Movie S9); therefore some phosphorylation and activation of EGFP-Δ[470-476] might occur in the nucleus upon DNA damage. Our functional rescue experiments consistently showed that EGFP-Δ[470-476] mediated DNA damage responses, such as centrosome inactivation and mitotic delay, at lower efficiency than with EGFP-Mnk—in only 2 of 10 injected embryos (Figure 6B and Supplemental Table S1). We found that deletion of the N-terminal FHA domain (EGFP-Δ[26-46]) abolished phosphorylations after CPT treatment. As we showed earlier, EGFP-Δ[26-46] is excluded from the vicinity of mitotic chromosomes irrespective of DNA damage (Figures 4E and 5D); therefore it does not form foci/aggregates on mitotic chromosomes upon DNA damage (Figure 5D). We hypothesize that the damage-induced EGFP-Mnk foci form around DNA damage sites, where a number of proteins are phosphorylated (Polo and Jackson, 2011), as formation of foci is dependent on the phosphopeptide-binding ability of the FHA domain (Figure 7). EGFP-Δ[26-46] does not draw near to DNA damage sites, where activated ATM waits to phosphorylate downstream molecules (Falck et al., 2005; You et al., 2005). This may be why the priming transphosphorylation and subsequent autophosphorylations did not occur on EGFP-Δ[26-46]. Finally, we found that EGFP-RSK did not show damage-induced phosphorylations, similar to EGFP-Δ[1-25] and EGFP-Δ[26-46]. The RSK mutation is a triple mutation that disrupts the phosphopeptide-binding ability of the FHA domain. As we showed, EGFP-RSK does not form foci on mitotic chromosomes upon DNA damage (Figure 7C). This probably means that EGFP-RSK is not recruited to DNA damage sites to be phosphorylated by ATM. Consistent with our hypothesis, EGFP-RSK did not show transphosphorylations and autophosphorylations with CPT treatment (Figure 8).
DISCUSSION
DNA damage signaling pathways consist of cascades of protein interactions and phosphorylations (Bartek et al., 2001; Su, 2006). To alter multiple cellular functions promptly in rapid cell cycles in response to DNA damage, it is important to have a mechanism that quickly sends DNA damage signals out from the nucleus to spread throughout the cytoplasm. In this study, we showed the dynamic behavior of Drosophila Chk2/Mnk in live embryos (Figures 1 and 5). We showed that Mnk is recruited to multiple mitotic structures without DNA damage and additionally forms chromosomal foci with DNA damage by means of an active mechanism that requires the phosphopeptide-binding ability of the FHA domain (Figure 7 and Supplemental Movies S17–S20). Our results indicate that Mnk itself is physically transported from the nucleus to the mitotic structures, transducing a DNA damage signal. This would indeed explain how the DNA damage signal rapidly reaches specific cellular machineries.
The FHA domain is found in a variety of signaling molecules, and its major function is to bind specific phosphothreonine-containing sequences (Durocher and Jackson, 2002; Mahajan et al., 2008). We hypothesize that many proteins that localize to centrosomes, interkinetochore/centromere, the midbody, and pseudocleavage furrows are phosphorylated via a DNA damage-independent mechanism. Among such proteins, specific protein phosphorylations may be recognized and bound by the Mnk-FHA domain and therefore trap Mnk to the structure. With DNA damage, chromosomes accumulate a number of phosphoproteins around DNA damage sites (Polo and Jackson, 2011). Mnk may bind to specific phosphoproteins through the FHA domain and form numerous chromosomal foci (Figures 5 and 6). We could not detect clear Mnk foci in the interphase nucleus (Figure 5). This may be because foci are not visible unless chromosomes are condensed or Mnk is too mobile to make foci in the interphase nucleus, irrespective of DNA damage, as described for mammalian Chk2 (Lukas et al., 2003).
Of note, the Mnk recruitment to each structure appears to be controlled by the cell cycle. For example, the amount of Mnk protein on centrosomes is most abundant immediately after NEB, persists during mitosis, and gradually decreases during interphase (Figure 1A and Supplemental Movie S1). The phosphorylation status of Mnk-FHA domain–binding proteins on the centrosome probably changes in a cell cycle–dependent manner. An example of mitosis-specific FHA interaction with its target molecule was shown for human Ki-67/MKI67 and nucleolar protein NIFK (Byeon et al., 2005). NIFK is phosphorylated at Thr-234 by GSK3 only when it is first phosphorylated at Thr-238 by the CDK1 mitotic kinase. The FHA domain of Ki-67/MKI67 binds to the NIFK peptide, which is phosphorylated at multiple sites, including Thr-234 and Thr-238. This kind of mechanism would control the FHA domain–dependent protein interaction in a cell cycle–dependent manner.
We found another intriguing function of the Mnk-FHA domain that resides within the N-terminal part and allows Mnk to be found in the vicinity of mitotic chromosomes regardless of DNA damage. When the N-terminal 20 amino acids were deleted from the FHA domain, the molecule was excluded from the vicinity of mitotic chromosomes (Figure 4E and Supplemental Movie S10) and therefore did not localize to centromeres or form foci on mitotic chromosomes upon DNA damage (Figures 4E and 5D and Supplemental Movies S10 and S14). Disrupting the phosphopeptide-binding ability of the FHA domain by point mutations did not cause this exclusion (Figure 7B and Supplemental Movies S17 and S18). It is important to understand the mechanism of how this short peptide brings Mnk close to chromosomes and whether other FHA domains in different proteins have similar function.
Robust recruitment of Mnk to the centrosomes at the beginning of mitosis (Figure 1A and Supplemental Movie S1) exactly correlates with the timing of when centrosome inactivation occurs after DNA damage. This suggests direct involvement of Mnk for the release of the major microtubule nucleator γTuRC and other components from the centrosomes (Sibon et al., 2000). Mnk localization to the interkinetochore/centromere region (Figure 1E and Supplemental Movie S21) and the observations that DNA damage causes mitotic delay (Figures 2, 5A, and 6B and Supplemental Table S1) suggest that Mnk may directly affect spindle assembly checkpoint function, similar to Aurora B kinase, but only when DNA damage exists, unlike Aurora B. Aurora B localizes to centromeres during mitosis (Adams et al., 2001) and is involved in the spindle assembly checkpoint (Ditchfield et al., 2003; Santaguida et al., 2011). Mnk localization to pseudocleavage furrows is restricted to the region close to leading edge (Figure 1D) and appears to be highly specific, as it is completely abolished by a point mutation in the FHA domain (R73A; Figure 7B and Supplemental Movie S17). Pseudocleavage furrows prevent contact between neighboring spindles during mitosis (Stevenson et al., 2001). It may also create a physical barrier around each spindle that prevents free diffusion of cellular signals such as a DNA damage signal initiated from one nucleus (Takada et al., 2003). When DNA damage occurs, the structure of pseudocleavage furrows becomes significantly disrupted, and neighboring spindles often fuse (Figure 2, B and D; Sibon et al., 2000). Mnk on pseudocleavage furrows may be involved in the disruption of the furrows upon DNA damage.
To examine whether normal Mnk localization is essential for mitotic DNA damage responses and interphase nuclear dropping, we sought to disrupt Mnk localization without disrupting FHA domain function and nuclear localization, which are necessary for Mnk kinase activation. We produced transgenic flies that express chromatin-tethered EGFP-Mnk (H2B-EGFP-Mnk; Supplemental Figure S2). However, expression of H2B-EGFP-Mnk blocked oogenesis at the temperature we normally use for transgene expression, and we could not obtain enough embryos to analyze. To clarify the localization–function relationship of Mnk and understand molecular mechanisms of Mnk-mediated DNA damage responses, we need to identify binding partners of Mnk through the FHA domain on each structure. Previous reports that the FHA domain is required for human Chk2 interaction with downstream substrates such as p53, BRCA1, and Cdc25A (Falck et al., 2001a, b; Li et al., 2002) suggest that the phosphoproteins that bind to Mnk through the FHA domain could also be Mnk substrates.
We propose that when DNA damage occurs, Mnk is first activated in the nucleus during interphase, spreads to the cytoplasm at NEB, and localizes to the various mitotic structures because of the phosphopeptide-binding ability of the FHA domain. Then, on the mitotic structures, activated Mnk phosphorylates effector proteins on-site, which should effectively alter centrosome function, cell cycle progression, and pseudocleavage furrow function in mitosis.
We showed that the phosphopeptide-binding ability of the FHA domain is required for Mnk's transphosphorylations and autophosphorylations, indicative of catalytic activation of the kinase (Figure 8). This is consistent with mammalian Chk2-FHA domain function, which is required for Chk2 dimerization and thus for Chk2 autophosphorylation/activation (Oliver et al., 2006; Cai et al., 2009). Together our results show that the FHA domain of Chk2/Mnk plays two essential roles in mediating embryonic DNA damage responses: activating the kinase upon DNA damage and recruiting the kinase to multiple cellular machineries. We are interested to know whether the FHA domains in a number of other signaling molecules play a role in protein localization in the cell.
MATERIALS AND METHODS
Drosophila stocks
The w1118; P[GAL4::VP16-nos.UTR]CG6325MVD1 and w*; P[matalpha4-GAL-VP16]V2H transgenic fly lines were provided by the Bloomington Drosophila Stock Center at Indiana University (Bloomington, IN). w1118; mnkP6 was used as mnk-null mutant (Brodsky et al., 2004). w1118; grpfsA4/CyO (Sibon et al., 1997) was used as grp mutant. w1118 was used as wild-type control. The mRFP-Rod transgenic line on the X chromosome was provided by R. Karess (CNRS, Université Paris Diderot, France).
Transgenic fly production
The Gateway cloning system (Thermo Fisher Scientific, Waltham, MA) was used to create transgenic fly lines that express EGFP-tagged Mnk and EGFP-tagged Mnk variants. pGUSgw-Mnk, which contained the Mnk coding sequence (long-form Dmnk; Oishi et al., 1998) flanked by lambda integrase attB1 and attB2 sites was kindly provided by M. Brodsky (University of Massachusetts Medical School; Brodsky et al., 2004). BP recombination between pGUSgw-Mnk and donor vector pDONR201 produced entry clone pENT-Mnk. LR recombination between pENT-Mnk and destination vector pUASp-NTEGFPgw (Cook et al., 2004) produced pUASp-EGFP-Mnk fly transformation plasmid. Mnk single-mutation variants pENT-D303A (aspartic acid 303 was replaced with alanine) and pENT-R73A (arginine 73 was replaced with alanine) and a triple-mutation variant pENT-RSK (arginine 73, serine 95, and lysine 96 were replaced with alanine) were created by QuikChange site-directed mutagenesis (Agilent Technologies, Santa Clara, CA). Mnk deletion variants Δ[1-25], Δ[470-476], [26-163], and [47-163] were amplified by PCR from pENT-Mnk. Forward primers with an attB1 site and reverse primers with a stop codon and an attB2 site were used. Resulting PCR fragments were recombined with pDONR221 to produce pENT-Δ[1-25], pENT-Δ[470-476], pENT-[26-163], and pENT-[47-163]. To produce pENT-Δ[47-163], two PCR fragments that contain amino acids 1–46 and 164–476 were ligated. To produce pENT-Δ[26-46], the amino acid 47–476 fragment of Mnk was amplified using a forward primer with an attB1 site and the N-terminal 75 nucleotides of Mnk encoding amino acids 1–25 and a reverse primer with an attB2 site and then recombined with pDONR221. LR recombination between each entry plasmid described and a destination vector pUASp-NTEGFPgw produced germline transformation plasmids pUASp-EGFP-D303A, pUASp-EGFP-R73A, pUASp-EGFP-RSK, pUASp-EGFP-Δ[1-25], pUASp-EGFP-Δ[470−476], pUASp-EGFP-Δ[26-46], pUASp-EGFP-Δ[47-163], pUASp-EGFP-[47−163], and pUASp-EGFP-[26-163], respectively. To produce pUASp-H2B-EGFP-Mnk, human histone H2B coding nucleotides without stop codon were PCR amplified and inserted into the KpnI site located at the 5′ end of EGFP in pUASp-EGFP-Mnk. P element–mediated germline transformation was performed as previously described (O'Connor and Chia, 1993). DNA injections into w1118 embryos were performed by BestGene (Chino Hills, CA) and Rainbow Transgenic Flies (Camarillo, CA).
Confocal imaging of live embryos
EGFP-Mnk was expressed from a transgene that is controlled by the GAL4-responsive promoter UASp (Rørth, 1998; Duffy, 2002). We used fly lines that maternally express GAL4VP16 protein, an enhanced version of GAL4 (Sadowski et al., 1988), under the control of either nanos or mat-αTub67C promoter (nos-GAL4VP16 and mTb-GAL4VP16; Van Doren et al., 1998; Wakefield et al., 2000). Live imaging of embryos that express EGFP-Mnk or one of the EGFP-Mnk variants was carried out as previously described, with some modifications (Takada et al., 2003; Takada and Cha, 2011). Briefly, embryos were collected for 2 h on a grape juice/agar plate at 25ºC, immediately hand dechorionated, mounted on a glue-coated coverglass, and covered with halocarbon oil 700 (Sigma-Aldrich, St. Louis, MO). The coverglass was placed on the stage of a Zeiss Observer Z1 inverted fluorescence microscope equipped with Zeiss LSM710 laser-scanning confocal system (Carl Zeiss, Jena, Germany). EGFP signals were captured with α Plan-Apochromat 100×/1.46 oil differential interference contrast (DIC) objective lens (Carl Zeiss) at 488-nm excitation at room temperature. Time-lapse movies were recorded with a 10-s interval through two to four rounds of syncytial blastoderm divisions. Image acquisitions and analyses were performed with ZEN software (Carl Zeiss) and ImageJ (National Institutes of Health, Bethesda, MD; Schneider et al., 2012). Throughout the recording, z-axis focus was manually adjusted to the position of centrosomes in most recordings. Some time-lapse movies were obtained on a Zeiss Axiovert 200M inverted microscope equipped with UltraView LCI spinning-disk confocal imaging system (PerkinElmer, Waltham, MA) and Orca-ER CCD camera (Hamamatsu Photonics, Hamamatsu, Japan) with Plan-Apochromat 100×/1.40 oil DIC objective lens (Carl Zeiss) at 488-nm excitation with 5-s intervals. Image acquisitions and analyses were performed with MetaMorph (Molecular Devices, Sunnyvale, CA) and ImageJ.
For simultaneous observation of EGFP-Mnk and mRFP-Rod, yw P[mRFP-Rod] (Buffin et al., 2005) flies were crossed with w1118; mnkP6 P[UASp-EGFP-Mnk] P[matalpha4-GAL-VP16]V2H, and cages were set up with flies that carried yw P[mRFP-Rod]/w1118/Y; mnkP6 P[UASp-EGFP-Mnk] P[matalpha4-GAL-VP16]V2H/+. We collected 0- to 2-h-old embryos and imaged them live by laser scanning confocal microscopy as described. Time-lapse movies were recorded with 5-s intervals.
DNA damage injection
Embryo injection to cause DNA damage was performed as previously described (Takada et al., 2003; Takada and Cha, 2011). Briefly, 0- to 2-h-old embryos were hand dechorionated, slightly dehydrated on Dri-rite (W. A. Hammond, Xenia, OH) for 2–4 min, mounted on a glue-coated coverglass, and covered with Halocarbon oil 700 (Sigma-Aldrich). The embryos on the coverglass were placed on Zeiss Observer Z1 inverted fluorescence microscope. For functional rescue experiments, 25 μg/ml bleomycin (Sigma-Aldrich), which consistently causes nuclear dropping, severe centrosome inactivation, and mitotic delay in wild-type embryos, was coinjected with 5 mg/ml rhodamine-tubulin (Cytoskeleton, Denver, CO) in BRB80 (80 mM Na–1,4-piperazinediethanesulfonic acid, pH 6.9, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, and 1 mM GTP). Injection was manually performed with a glass needle connected to a 10-ml syringe with a narrow tube and mounted on a micromanipulator. Rhodamine signal representing microtubule structures and EGFP signal representing localization of EGFP-tagged protein were captured simultaneously starting immediately after the injection with a Zeiss LSM710 laser-scanning confocal system and α Plan-Apochromat 100×/1.46 Oil DIC objective lens with 10-s intervals. Image acquisitions and analyses were performed with ZEN and ImageJ.
Immunofluorescence
We fixed 0- to 3-h embryos by a boiling method described previously (Postner and Wieschaus, 1994) and stained as previously described (Theurkauf, 1994). For primary antibodies, anti-GFP antibody (ab290, rabbit polyclonal; Abcam, Cambridge, MA) at 1:1000 and anti-Nullo antibody (5C3-12, mouse monoclonal, Developmental Studies Hybridoma Bank, Iowa City, IA; Postner and Wieschaus, 1994) at 1:5 dilution were used. Alexa 488–anti-rabbit immunoglobulin G (IgG) and Alexa 555–anti-mouse IgG (Thermo Fisher Scientific) were used at 1:500 as secondary antibodies. DNA was stained with 0.5 μg/ml DAPI (Sigma-Aldrich). Immunostained embryos were cleared, mounted on slide glass, and sealed with a coverglass. The cortical areas of the embryos were scanned at 488-, 561-, and 405-nm excitations by Zeiss LSM710 laser-scanning confocal microscope system with 100×/1.46 oil α Plan-Apochromat objective lens. Image acquisitions and analyses were performed with ZEN and ImageJ.
Cellularization assay
To examine whether EGFP-Mnk or EGFP-RSK expression restores DNA damage–induced developmental block at the MBT in mnkP6 grpfsA4 embryos, fly lines that carry mnkP6 and grpfsA4 mutations and P[UASp-EGFP-Mnk] or P[UASp-EGFP-RSK] transgene on the second chromosome were produced by recombination. We tried to produce a fly line that carries P[UASp-EGFP-D303A] (only one transgenic line is available) with mnkP6 and grpfsA4 mutations on the second chromosome as kinase inactive control, but such a triple recombinant could not be obtained, likely due to the position of the transgene insertion. Therefore we used P[UASp-EGFP-RSK] as an alternative inactive Mnk control (Figures 7 and 8). The resulting w1118; mnkP6 grpfsA4, P[UASp-EGFP-Mnk]/CyO or w1118; mnkP6 grpfsA4 P[UASp-EGFP-RSK]/CyO was crossed with w1118; mnkP6 grpfsA4 P[matalpha4-GAL-VP16]V2H. Small cages were set up for embryo collections with flies that carried the mnkP6 grpfsA4 P[UASp-EGFP-Mnk]/mnkP6 grpfsA4 P[matalpha4-GAL-VP16]V2H and mnkP6 grpfsA4 P[UASp-EGFP-RSK]/mnkP6 grpfsA4 P[matalpha4-GAL-VP16]V2H genotypes. At the same time, cages were set up for embryo collections for w1118 (wild-type control), w1118; grpfsA4 mutant, and w1118; mnkP6 grpfsA4 mutant. We collected 2- to 3- and 3- to 5-h-old embryos for each genotype and fixed them with formaldehyde and methanol as described previously (Theurkauf, 1994). The fixed embryos were immunostained with anti-GFP antibody at 1:1000 dilution (ab290; Abcam) and anti-spectrin at 1:10 dilution (3A9; Developmental Studies Hybridoma Bank; Byers et al., 1987), followed by Alexa 488–anti-rabbit IgG and Alexa 555–anti-mouse IgG secondary antibodies. Numbers of cellularized and noncellularized embryos were counted for each genotype with tetramethylrhodamine isothiocyanate channel on a Zeiss Observer.Z1 immunofluorescence microscope. Embryos were also imaged for GFP and Spectrin by a Zeiss LSM710 laser-scanning confocal microscope system using a 20×/0.8 Plan-Apochromat objective lens. Image acquisitions and analyses were performed with ZEN and ImageJ.
Quantitative Western blot
We collected 0- to 3-h-old mnk mutant embryos that express EGFP-Mnk, EGFP-D303A, and EGFP-RSK at 25ºC, homogenized them, and boiled them in SDS–PAGE sample buffer at a concentration of 1 embryo/μl. As controls, w1118 and mnkP6 embryos were collected at 25ºC. The embryo samples were loaded onto 7.5% SDS–PAGE gel (15 embryos/lane) and transferred onto a polyvinylidene fluoride (PVDF) membrane. Precision Plus Kaleidoscope Standards (Bio-Rad, Hercules, CA) were used as molecular weight markers. The membrane was probed with anti–α-tubulin antibody (DM1A; used at 1:15,000; Sigma-Aldrich) to detect endogenous α-tubulin, and anti-Mnk antibodies (Mnk173 used at 1:1000, or Chk2C used at 1:2500; Takada et al., 2007) to detect endogenous Mnk in w1118 control and EGFP-Mnk variants in transgenic embryos. The membrane was then washed and probed with IRDye 800–anti-rabbit IgG and IRDye 700–anti-mouse IgG secondary antibodies (used at 1:10,000; Rockland, Limerick, PA), washed again, and scanned on the Odyssey infrared fluorescence imaging system (Li-Cor, Lincoln, NE). To quantify expression level, intensities of EGFP-Mnk variant bands in transgenic embryos and endogenous Mnk band in control w1118 embryos were measured using Odyssey or Image Studio software (Li-Cor). They were then divided by the intensity of α-tubulin in the same sample to standardize loading variations. To determine the ratio of EGFP-Mnk variants to endogenous Mnk, a standardized value of each EGFP-Mnk variant was divided by the standardized value of endogenous Mnk. Multiple Western blots were performed and quantified (n = 6 for EGFP-Mnk, n = 4 for EGFP-D303A, and n = 5 for EGFP-RSK). Average value and SD were calculated for each construct.
Antibody production and purification
DNA fragments that encode amino acids 1–173 of Mnk were synthesized with an optimized sequence for bacterial expression (Lok1-173; Bio Basic, Markham, Canada). The synthesized DNA was inserted into pE-SUMO amp vector (LifeSensors, Malvern, PA; pE-SUMO-Lok1-173) and transformed into BL21(DE3)-competent cells (New England BioLabs, Ipswich, MA). The recombinant His-SUMO-Lok1-173 protein was expressed and purified on a nickel-nitriloacetic acid (Ni-NTA) Superflow Cartridge (Qiagen) using a Bio Logic chromatography system (Bio-Rad). Histidine (His)-SUMO-Lok1-173 protein was cleaved with recombinant SUMO protease Ulp1 between His-SUMO and Lok1-173. The cleaved sample was loaded on to an open Ni-NTA column, Lok1-173 fragment was collected in the flowthrough fraction, and the buffer was exchanged to phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4). The protein fraction was lyophilized and used as antigen. The antigen was injected into two rabbits. Antisera were collected, and IgG fraction was purified using Protein-A beads (Thermo Fisher Scientific). Anti–Lok1-173 antibody (Mnk173) was affinity purified from the purified IgG fraction using Lok1-173 recombinant protein coupled to AminoLink Plus resin (Thermo Fisher Scientific).
Two-dimensional electrophoresis and Western blot
w1118 and transgenic 2- to 3-h-old embryos were collected and dechorionated. To prepare minus DNA damage samples, dechorionated embryos were vigorously mixed in octane/methanol to remove vitelline membrane, washed, counted under a dissecting microscope, homogenized in 2D sample buffer (7 M urea, 2 M thiourea, 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 0.002% bromophenol blue, and DeStreak reagent [GE Healthcare Life Sciences, Pittsburgh, PA]) at a concentration of 1 embryo/μl buffer, and sonicated. To prepare plus DNA damage samples, dechorionated embryos were incubated for 20 min in a two-phase mixture of octane and Robb's medium (Robb, 1969) containing 20 μM camptothecin (Takada et al., 2003) on a horizontal shaker before completely removal of vitelline membranes and processed in the same way as the minus DNA damage samples. For grp mutant, 0- to 1- and 3- to 4-h-old embryos were collected and processed without camptothecin treatment. The samples were individually loaded on rehydrated 7-cm IPG DryStrips (pH 6–11), and isoelectric focusing was carried out on an Ettan IPGphore3 isoelectric focusing unit (GE Healthcare Life Sciences). The strips were equilibrated in equilibration buffer (6 M urea, 75 mM Tris-HCl, pH 8.8, 29.3% [vol/vol] glycerol, 2% SDS, 0.002% bromophenol blue) supplemented with 10 mg/ml dithiothreitol and then in equilibration buffer supplemented with 25 mg/ml iodoacetamide. Each strip was loaded on a 7% SDS–PAGE minigel. Precision Plus Kaleidoscope Standards were loaded at the side of the gel as molecular weight markers. Proteins resolved on 2D gels were transferred onto a PVDF membrane. EGFP-Mnk variants were expressed in mnk-null embryos and detected with anti-GFP antibody (A11122; used at 1:1000; Thermo Fisher Scientific), and endogenous Mnk in control w1118 or grp mutant embryos was detected with anti-Mnk antibody (Mnk173, used at 1:1000), followed by IRDye 800–goat anti-rabbit secondary antibody (used at 1:10,000). The membranes were scanned at 700 and 800 nm on the Odyssey infrared fluorescence imaging scanner using Image Studio software. Molecular weight marker bands were detected at 700 nm.
Supplementary Material
Acknowledgments
We thank Michael O'Connor, William Theurkauf, Helen Piwnica-Worms, and Scott Selleck for support of this research, Aidan Peterson for manuscript editing, Hideki Aihara for protein purification help, Tom Hays for the use of the spinning-disk confocal microscope and discussion, and Ross Cagan and Hiroshi Nakato for the use of laboratory space and equipment. We thank Roger Karess for mRFP-Rod transgenic flies and Michael Brodsky for the pGUSgw-Mnk plasmid. GAL4 fly stocks were obtained from the Bloomington Drosophila Stock Center (National Insitutes of Health Grant P40OD018537). The monoclonal antibody 5C3-12 developed by M. A. Postner and E. F. Wieschaus and 3A9 developed by T. J. Byers and coworkers were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biology, Iowa City, IA. This study was supported by research and equipment grants from the Minnesota Medical Foundation to S.T.
Abbreviations used:
- CPT
camptothecin
- DSB
double-strand break
- EGFP
enhanced green fluorescent protein
- FHA
forkhead-associated
- MBT
midblastula transition
- mRFP
monomeric red fluorescent protein
- NEB
nuclear envelope breakdown
- NEF
nuclear envelope formation
- NLS
nuclear localization signal
- SCD
SQ/TQ-cluster domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1238) on March 25, 2015.
REFERENCES
- Abdelmohsen K, Pullmann R, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdu U, Brodsky M, Schüpbach T. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr Biol. 2002;12:1645–1651. doi: 10.1016/s0960-9822(02)01165-x. [DOI] [PubMed] [Google Scholar]
- Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Ahn JY, Li X, Davis HL, Canman CE. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem. 2002;277:19389–19395. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin. Nat Rev Cancer. 2007;7:925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- Bakhrat A, Pritchett T, Peretz G, McCall K, Abdu U. Drosophila Chk2 and p53 proteins induce stage-specific cell death independently during oogenesis. Apoptosis. 2010;15:1425–1434. doi: 10.1007/s10495-010-0539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Falck J, Lukas J. CHK2 kinase—a busy messenger. Nat Rev Mol Cell Biol. 2001;2:877–886. doi: 10.1038/35103059. [DOI] [PubMed] [Google Scholar]
- Basto R, Scaerou F, Mische S, Wojcik E, Lefebvre C, Gomes R, Hays T, Karess R. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr Biol. 2004;14:56–61. doi: 10.1016/j.cub.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol. 2005;15:856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- Byeon IJ, Li H, Song H, Gronenborn AM, Tsai MD. Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67. Nat Struct Mol Biol. 2005;12:987–993. doi: 10.1038/nsmb1008. [DOI] [PubMed] [Google Scholar]
- Byers TJ, Dubreuil R, Branton D, Kiehart DP, Goldstein LS. Drosophila spectrin. II. Conserved features of the alpha-subunit are revealed by analysis of cDNA clones and fusion proteins. J Cell Biol. 1987;105:2103–2110. doi: 10.1083/jcb.105.5.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Chehab NH, Pavletich NP. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol Cell. 2009;35:818–829. doi: 10.1016/j.molcel.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozier C, Bonyadi M, Baricault L, Tonasso L, Darbon JM. Regulation of Chk2 phosphorylation by interaction with protein phosphatase 2A via its B' regulatory subunit. Biol Cell. 2004;96:509–517. doi: 10.1016/j.biolcel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Falck J, Lukas C, Protopopova M, Lukas J, Selivanova G, Bartek J. Functional impact of concomitant versus alternative defects in the Chk2-p53 tumour suppressor pathway. Oncogene. 2001a;20:5503–5510. doi: 10.1038/sj.onc.1204811. [DOI] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuåsen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001b;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- Farrell PJ, Hunt T, Jackson RJ. Analysis of phosphorylation of protein synthesis initiation factor eIF-2 by two-dimensional gel electrophoresis. Eur J Biochem. 1978;89:517–521. doi: 10.1111/j.1432-1033.1978.tb12556.x. [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Iampietro C, Bergalet J, Wang X, Cody NA, Chin A, Lefebvre FA, Douziech M, Krause HM, Lécuyer E. Developmentally regulated elimination of damaged nuclei involves a Chk2-dependent mechanism of mRNA nuclear retention. Dev Cell. 2014;29:468–481. doi: 10.1016/j.devcel.2014.03.025. [DOI] [PubMed] [Google Scholar]
- Iijima-Ando K, Zhao L, Gatt A, Shenton C, Iijima K. A DNA damage-activated checkpoint kinase phosphorylates tau and enhances tau-induced neurodegeneration. Hum Mol Genet. 2010;19:1930–1938. doi: 10.1093/hmg/ddq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JB, Gross J, Lovly CM, Piwnica-Worms H, Townsend RR. Identification of protein phosphorylation sites within Ser/Thr-rich cluster domains using site-directed mutagenesis and hybrid linear quadrupole ion trap Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:3443–3451. doi: 10.1002/rcm.3223. [DOI] [PubMed] [Google Scholar]
- King JB, Gross J, Lovly CM, Rohrs H, Piwnica-Worms H, Townsend RR. Accurate mass-driven analysis for the characterization of protein phosphorylation. Study of the human Chk2 protein kinase. Anal Chem. 2006;78:2171–2181. doi: 10.1021/ac051520l. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- Lee CH, Chung JH. The hCds1 (Chk2)-FHA domain is essential for a chain of phosphorylation events on hCds1 that is induced by ionizing radiation. J Biol Chem. 2001;276:30537–30541. doi: 10.1074/jbc.M104414200. [DOI] [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9:1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- Lovly CM, Yan L, Ryan CE, Takada S, Piwnica-Worms H. Regulation of Chk2 ubiquitination and signaling through autophosphorylation of serine 379. Mol Cell Biol. 2008;28:5874–5885. doi: 10.1128/MCB.00821-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- Mahajan A, Yuan C, Lee H, Chen ES, Wu PY, Tsai MD. Structure and function of the phosphothreonine-specific FHA domain. Sci Signal. 2008;1:re12. doi: 10.1126/scisignal.151re12. [DOI] [PubMed] [Google Scholar]
- Masrouha N, Yang L, Hijal S, Larochelle S, Suter B. The Drosophila chk2 gene loki is essential for embryonic DNA double-strand-break checkpoints induced in S phase or G2. Genetics. 2003;163:973–982. doi: 10.1093/genetics/163.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar A, Mazumdar M. How one becomes many: blastoderm cellularization in Drosophila melanogaster. Bioessays. 2002;24:1012–1022. doi: 10.1002/bies.10184. [DOI] [PubMed] [Google Scholar]
- McCleland ML, Shermoen AW, O'Farrell PH. DNA replication times the cell cycle and contributes to the mid-blastula transition in Drosophila embryos. J Cell Biol. 2009;187:7–14. doi: 10.1083/jcb.200906191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle JA, Rickmyre JL, Garg A, Loggins EB, Jodoin JN, Lee E, Wu LP, Lee LA. no poles encodes a predicted E3 ubiquitin ligase required for early embryonic development of Drosophila. Development. 2009;136:449–459. doi: 10.1242/dev.027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M, Chia W. P element-mediated germ-line transformation of Drosophila. Methods Mol Biol. 1993;18:75–85. doi: 10.1385/0-89603-245-0:75. [DOI] [PubMed] [Google Scholar]
- Oishi I, Sugiyama S, Otani H, Yamamura H, Nishida Y, Minami Y. A novel Drosophila nuclear protein serine/threonine kinase expressed in the germline during its establishment. Mech Dev. 1998;71:49–63. doi: 10.1016/s0925-4773(97)00200-1. [DOI] [PubMed] [Google Scholar]
- Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH. Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J. 2006;25:3179–3190. doi: 10.1038/sj.emboj.7601209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M, DeLuca C, Hirao A, Stambolic V, Potter J, Zhou L, Liepa J, Snow B, Arya S, Wong J, et al. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc Natl Acad Sci USA. 2002;99:11305–11310. doi: 10.1073/pnas.172382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Weinstein JN, Aladjem MI, Kohn KW. Chk2 molecular interaction map and rationale for Chk2 inhibitors. Clin Cancer Res. 2006;12:2657–2661. doi: 10.1158/1078-0432.CCR-06-0743. [DOI] [PubMed] [Google Scholar]
- Postner MA, Wieschaus EF. The nullo protein is a component of the actin-myosin network that mediates cellularization in Drosophila melanogaster embryos. J Cell Sci. 1994;107:1863–1873. doi: 10.1242/jcs.107.7.1863. [DOI] [PubMed] [Google Scholar]
- Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- Pushpavalli SN, Sarkar A, Ramaiah MJ, Chowdhury DR, Bhadra U, Pal-Bhadra M. Drosophila MOF controls Checkpoint protein2 and regulates genomic stability during early embryogenesis. BMC Mol Biol. 2013;14:1. doi: 10.1186/1471-2199-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickmyre JL, Dasgupta S, Ooi DL, Keel J, Lee E, Kirschner MW, Waddell S, Lee LA. The Drosophila homolog of MCPH1, a human microcephaly gene, is required for genomic stability in the early embryo. J Cell Sci. 2007;120:3565–3577. doi: 10.1242/jcs.016626. [DOI] [PubMed] [Google Scholar]
- Robb JA. Maintenance of imaginal discs of Drosophila melanogaster in chemically defined media. J Cell Biol. 1969;41:876–885. doi: 10.1083/jcb.41.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Okado M, Ito F, Kawasaki K. Anaphase DNA bridges induced by lack of RecQ5 in Drosophila syncytial embryos. FEBS Lett. 2011;585:1923–1928. doi: 10.1016/j.febslet.2011.04.074. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Vernieri C, Villa F, Ciliberto A, Musacchio A. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 2011;30:1508–1519. doi: 10.1038/emboj.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schejter ED, Wieschaus E. bottleneck acts as a regulator of the microfilament network governing cellularization of the Drosophila embryo. Cell. 1993;75:373–385. doi: 10.1016/0092-8674(93)80078-s. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Schwarz JK, Lovly CM, Piwnica-Worms H. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol Cancer Res. 2003;1:598–609. [PubMed] [Google Scholar]
- Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Laurençon A, Hawley R, Theurkauf WE. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr Biol. 1999;9:302–312. doi: 10.1016/s0960-9822(99)80138-9. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- Silverman-Gavrila RV, Hales KG, Wilde A. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol Biol Cell. 2008;19:3735–3744. doi: 10.1091/mbc.E08-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson V, Hudson A, Cooley L, Theurkauf WE. Arp2/3-dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr Biol. 2002;12:705–711. doi: 10.1016/s0960-9822(02)00807-2. [DOI] [PubMed] [Google Scholar]
- Stevenson VA, Kramer J, Kuhn J, Theurkauf WE. Centrosomes and the Scrambled protein coordinate microtubule-independent actin reorganization. Nat Cell Biol. 2001;3:68–75. doi: 10.1038/35050579. [DOI] [PubMed] [Google Scholar]
- Stevenson VA, Theurkauf WE. Actin cytoskeleton: putting a CAP on actin polymerization. Curr Biol. 2000;10:R695–697. doi: 10.1016/s0960-9822(00)00712-0. [DOI] [PubMed] [Google Scholar]
- Stumpff J, Duncan T, Homola E, Campbell SD, Su TT. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr Biol. 2004;14:2143–2148. doi: 10.1016/j.cub.2004.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Fogarty P, Theurkauf W. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development. 1993;118:1245–1254. doi: 10.1242/dev.118.4.1245. [DOI] [PubMed] [Google Scholar]
- Takada S, Cha BJ. In vivo live-analysis of cell cycle checkpoints in Drosophila early embryos. Methods Mol Biol. 2011;782:75–92. doi: 10.1007/978-1-61779-273-1_7. [DOI] [PubMed] [Google Scholar]
- Takada S, Kelkar A, Theurkauf WE. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 2003;113:87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- Takada S, Kwak S, Koppetsch BS, Theurkauf WE. grp (chk1) replication-checkpoint mutations and DNA damage trigger a Chk2-dependent block at the Drosophila midblastula transition. Development. 2007;134:1737–1744. doi: 10.1242/dev.02831. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 1994;44:489–505. doi: 10.1016/s0091-679x(08)60928-0. [DOI] [PubMed] [Google Scholar]
- Traven A, Heierhorst J. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays. 2005;27:397–407. doi: 10.1002/bies.20204. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Varmark H, Kwak S, Theurkauf WE. A role for Chk2 in DNA damage induced mitotic delays in human colorectal cancer cells. Cell Cycle. 2010;9:312–320. doi: 10.4161/cc.9.2.10418. [DOI] [PubMed] [Google Scholar]
- Wakefield JG, Huang JY, Raff JW. Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr Biol. 2000;10:1367–1370. doi: 10.1016/s0960-9822(00)00776-4. [DOI] [PubMed] [Google Scholar]
- Xu J, Xin S, Du W. Drosophila Chk2 is required for DNA damage-mediated cell cycle arrest and apoptosis. FEBS Lett. 2001;508:394–398. doi: 10.1016/s0014-5793(01)03103-9. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannini L, Lecis D, Lisanti S, Benetti R, Buscemi G, Schneider C, Delia D. Karyopherin-alpha2 protein interacts with Chk2 and contributes to its nuclear import. J Biol Chem. 2003;278:42346–42351. doi: 10.1074/jbc.M303304200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN, Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.