The kinetochore-specific dynein adaptor Spindly is identified as a novel substrate of farnesyltransferase in human cells. Farnesylation is required for Spindly accumulation at kinetochores, and nonfarnesylated Spindly delays chromosome congression, providing new mechanistic insight into the biological effect of farnesyltransferase inhibitors.
Abstract
The clinical interest in farnesyltransferase inhibitors (FTIs) makes it important to understand how these compounds affect cellular processes involving farnesylated proteins. Mitotic abnormalities observed after treatment with FTIs have so far been attributed to defects in the farnesylation of the outer kinetochore proteins CENP-E and CENP-F, which are involved in chromosome congression and spindle assembly checkpoint signaling. Here we identify the cytoplasmic dynein adaptor Spindly as an additional component of the outer kinetochore that is modified by farnesyltransferase (FTase). We show that farnesylation of Spindly is essential for its localization, and thus for the proper localization of dynein and its cofactor dynactin, to prometaphase kinetochores and that Spindly kinetochore recruitment is more severely affected by FTase inhibition than kinetochore recruitment of CENP-E and CENP-F. Molecular replacement experiments show that both Spindly and CENP-E farnesylation are required for efficient chromosome congression. The identification of Spindly as a new mitotic substrate of FTase provides insight into the causes of the mitotic phenotypes observed with FTase inhibitors.
INTRODUCTION
A wide variety of proteins, including RAS, pre–lamin A, fungal pheromones, and rhodopsin kinase, depend on posttranslational prenylation for proper localization and activity. Prenylated proteins are modified by either farnesyltransferase (FTase) or geranylgeranyltransferase (GGTase), which covalently link a 15- or 20-carbon isoprenoid moiety to a C-terminal cysteine via a thiolether bond, with farnesyl diphosphate (FPP) and geranylgeranyl diphosphate (GGPP) acting as the lipid donors (Pechlivanis and Kuhlmann, 2006; Nguyen et al., 2010; Zverina et al., 2012). FTase and GGTase type 1 have been proposed to recognize protein substrates containing a C-terminal CAAX box, where C refers to the modified cysteine residue, A is any aliphatic amino acid, and X refers to a subset of amino acids that determine whether the cysteine receives a farnesyl or geranylgeranyl group. Besides facilitating association with membranes, the farnesyl/geranylgeranyl moiety has also been shown to be involved in the regulation and modulation of protein–protein interactions (Hoffman et al., 2000; Rak et al., 2003; Rotblat et al., 2004; Pylypenko et al., 2006; Ignatev et al., 2008).
Oncogenic mutations in the RAS genes constitutively activate GTPase activity, leading to uncontrolled proliferation. Farnesylation is required for the translocation of RAS to the plasma membrane and subsequent activation of cellular signaling. The promise of inhibiting the membrane translocation of oncogenic RAS spurred the development of potent farnesyltransferase inhibitors (FTIs; Crul et al., 2001). However, studies with these inhibitors revealed that RAS undergoes alternative geranylgeranylation that is capable of promoting membrane translocation in the presence of FTIs (Berndt et al., 2011). This has limited the clinical use of FTIs as anticancer agents, although recent evidence suggests that FTIs may be valuable in the treatment of malignant hematological disease (Berndt et al., 2011; Holstein and Hohl, 2012). Of importance, the use of FTIs is not limited to cancer therapeutics: inhibition of FTase has gained interest as an approach for fighting parasites (Eastman et al., 2006), and FTIs have shown promise for the treatment of Hutchinson–Gilford progeria syndrome, a disease caused by the aberrant accumulation of a farnesylated form of lamin A (Yang et al., 2011; Gordon et al., 2014).
The medical relevance of FTIs makes it important to identify the cellular substrates of FTase. Most human tumor cells lines that are sensitive to FTI treatment exhibit a defect in chromosome congression that leads to an accumulation in the mitotic phase of the cell cycle (Moasser et al., 1998; Ashar et al., 2000; Crespo et al., 2001; Schafer-Hales et al., 2007). This effect has been attributed to nonfarnesylated CENP-E and CENP-F, which are the only mitotic substrates of FTase known to date. Both CENP-E, a microtubule plus end–directed kinesin, and CENP-F, a coiled-coil protein implicated in the recruitment of cytoplasmic dynein, localize to mitotic kinetochores, where they are involved in mediating attachments to spindle microtubules that drive chromosome segregation (Weaver et al., 2003; Bomont et al., 2005; Feng et al., 2006; Liang et al., 2007; Vergnolle et al., 2007; Kim et al., 2008). Farnesylation of CENP-E and CENP-F is required for their APC/C-mediated degradation at mitotic exit by the proteasome (Hussein and Taylor, 2002; Gurden et al., 2010). In addition, kinetochore localization of CENP-E and CENP-F has been reported to be impaired after FTase inhibition in some studies (Hussein and Taylor, 2002; Schafer-Hales et al., 2007) but not others (Ashar et al., 2000; Crul et al., 2001; Verstraeten et al., 2011). Thus the contribution from the lack of CENP-E and CENP-F farnesylation to the mitotic phenotype of FTIs remains unclear.
Spindly is a kinetochore-specific adaptor for cytoplasmic dynein (Griffis et al., 2007; Chan et al., 2009; Barisic et al., 2010; Gassmann et al., 2010) that, like CENP-E and CENP-F, localizes to the fibrous corona, the outermost domain of the kinetochore, and is removed from microtubule-attached kinetochores by dynein-mediated transport to spindle poles. Here we identify Spindly as a novel substrate of FTase in human cells. We demonstrate that Spindly physically interacts with FTase and show that farnesylation is required for Spindly accumulation at kinetochores. Functional replacement experiments show that preventing farnesylation of Spindly and CENP-E delays chromosome congression, producing a phenotype similar to that observed in cells treated with FTIs. The identification of Spindly as a novel farnesylated kinetochore protein provides mechanistic insight into the biological effect of FTIs.
RESULTS
The kinetochore protein Spindly is a substrate of farnesyltransferase
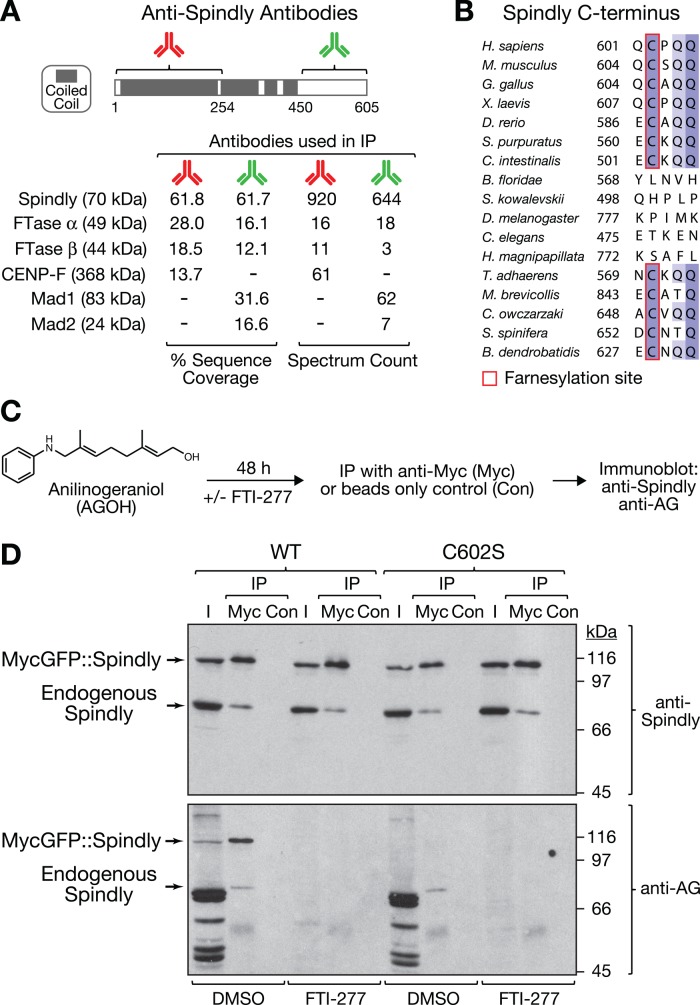
To identify binding partners of human Spindly, we immunoprecipitated Spindly from mitotic HeLa cell extracts using two affinity-purified polyclonal antibodies raised against nonoverlapping epitopes in the N- and C-terminal halves of the protein (Figure 1A). Both antibodies immunoprecipitated Spindly with similar efficiency (Supplemental Figure S1), and mass spectrometric analysis identified the α and β subunits of FTase in a search for proteins that coimmunoprecipitated with both antibodies (Figure 1A). By contrast, the known kinetochore components Mad1-Mad2 and CENP-F were pulled down with only one of the two antibodies (Figure 1A and Supplemental Figure S1). We conclude that a mitotic pool of human Spindly associates with FTase.
FIGURE 1:
Human Spindly interacts with and is modified by farnesyltransferase. (A) Results of mass spectrometric analysis of immunoprecipitates from mitotic HeLa cells with two affinity-purified antibodies raised against nonoverlapping N- and C-terminal epitopes of Spindly (amino acids 2–254 and 450–605). Sequence coverage and spectrum count for selected proteins are shown. (B) Sequence alignment of Spindly C-termini in eukaryotes showing widespread conservation of a putative farnesylation motif that deviates from the canonical CAAX box (Hougland et al., 2009). The amino acid number of the fifth residue from the C-terminus is indicated. The farnesyl cysteine is present in vertebrates (represented by Homo sapiens, Mus musculus, Gallus gallus, Xenopus laevis, Danio rerio). In the two invertebrate chordate groups, the motif is present in tunicates (Ciona intestinalis) but is absent in amphioxus (B. floridae). Of the two closest nonchordate invertebrate relatives of amphioxus, echinoderms (Strongylocentrotus purpuratus) have the motif but hemichordates do not (S. kowalevskii). We could not find the motif in insects (D. melanogaster), nematodes (C. elegans), or cnidaria (H. magnipapillata). However, the motif appears in the primitive multicellular animal Trichoplax adhaerens, the only extant member of Placozoa, as well as in choanoflagellates (Monosiga brevicollis, Salpingoeca spinifera), the closest unicellular relatives of multicellular animals. The motif is also present in the chytrid fungus Batrachochytrium dendrobatidis. (C) Schematic of experimental approach for the detection of farnesylated Spindly based on in vivo labeling with the unnatural farnesyl diphosphate analogue AGOH and AG-specific antibodies. (D) Immunoblots detecting endogenous and transgenic Spindly and corresponding AG signals in cell lysate input fractions (I) and fractions obtained after immunoprecipitation with anti-Myc tag antibody (Myc) or beads-only controls (Con).
Sequence inspection revealed that Spindly contains an atypical variant of the C-terminal CAAX box (Hougland et al., 2009) that is conserved from vertebrates to fungi (Figure 1B). Of interest, the motif appears to have been lost in a subset of phyla, including the invertebrate chordate group amphioxus (Branchiostoma floridae), hemichordates (Saccoglossus kowalevskii), insects (Drosophila melanogaster), nematodes (Caenorhabditis elegans), and cnidaria (Hydra magnipapillata).
The physical association of Spindly with FTase and the conservation of the C-terminal farnesylation motif suggested that Spindly may be modified by FTase. To test this directly, we incubated cells with anilinogeraniol (AGOH; Figure 1C), which is converted to the unnatural, competitive FPP analogue anilinogeranyl diphosphate (AGPP) and incorporated into proteins by FTase (Troutman et al., 2005). The AG moiety in proteins can subsequently be detected with specific polyclonal anti-AG antibodies. Consistent with farnesylated proteins being present at kinetochores, we detected AG signal at kinetochores by immunofluorescence (Supplemental Figure S2A). To assess farnesylation of both endogenous and transgenic Spindly by immunoblot, we used HeLa cell lines in which tetracycline-inducible MycGFP::Spindly constructs were integrated into a specific genomic locus by Flp-mediated DNA recombination (O'Gorman et al., 1991). We had previously implemented this system for the study of Spindly mutants (Gassmann et al., 2010). As a negative control, we mutated the putative farnesylated cysteine residue to serine (C602S). We incubated cells with tetracycline and AGOH for 48 h in the presence or absence of the FTase inhibitor FTI-277 before preparing cell lysates and immunoprecipitating transgene-encoded Spindly with anti-Myc antibodies (Figure 1C). Immunoblotting with anti-Spindly antibodies revealed that endogenous Spindly efficiently coimmunoprecipitated with MycGFP::Spindly (Figure 1D), suggesting that Spindly is present as an oligomer in cells. FTI-277 treatment had no effect on Spindly expression levels, nor did it change the amount of endogenous Spindly that coimmunoprecipitated with Myc-tagged Spindly. Probing the same immunoblot with anti-AG antibodies revealed efficient labeling of multiple farnesylated proteins in total cell lysates (input lanes). MycGFP::SpindlyWT, but not MycGFP::SpindlyC602S, was clearly detectable with anti-AG antibodies in both input and immunoprecipitated fractions. In addition, endogenous Spindly that coimmunoprecipitated with transgene-encoded Spindly could be detected with anti-AG antibodies in both MycGFP::SpindlyWT– and MycGFP::SpindlyC602S–expressing cells. Treatment with FTI-277 abolished all anti-AG signal in cell lysates and immunoprecipitated fractions, demonstrating that AG incorporation into proteins is dependent on FTase activity. We conclude that Spindly is modified by FTase.
We also examined the farnesylation of endogenous CENP-E in human DLD-1 cells and Xenopus egg extracts using the AG labeling approach (Supplemental Figure S2, B and C). Endogenous CENP-E incorporated AG after treatment with dimethyl sulfoxide (DMSO) but not in the presence of FTI-277. In addition, MycGFP-tagged human CENP-E incorporated AG, whereas the farnesylation mutant C2261S did not (Supplemental Figure S2D). Together, these data confirm previous reports that CENP-E is farnesylated (Ashar et al., 2000; Gurden et al., 2010).
Spindly farnesylation is required for its kinetochore localization
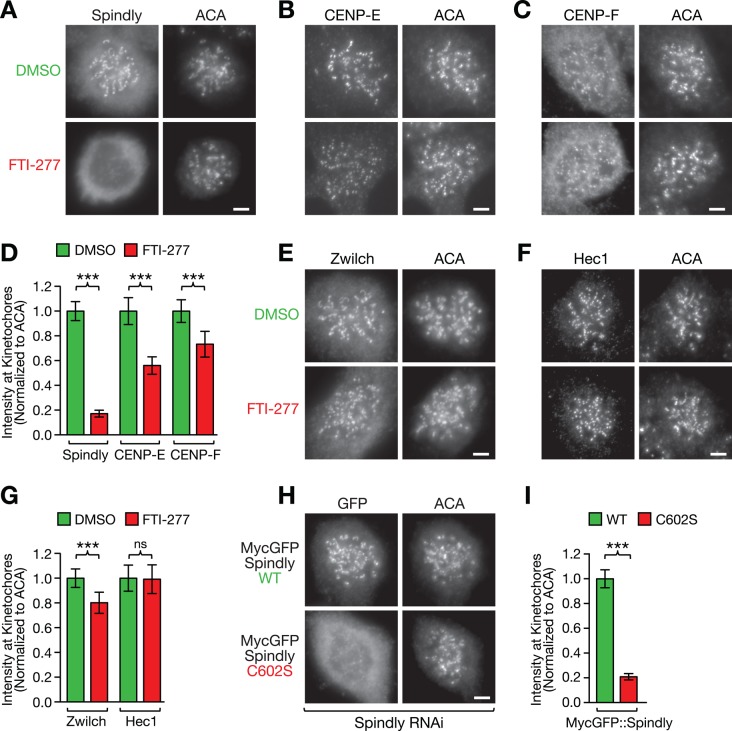
It has been reported that CENP-E and CENP-F require farnesylation for efficient targeting to kinetochores (Hussein and Taylor, 2002; Schafer-Hales et al., 2007). To determine whether Spindly localization to kinetochores requires FTase activity, we treated HeLa and DLD-1 cells with FTI-277 and monitored Spindly localization by immunofluorescence. Microtubules were depolymerized with nocodazole, which causes Spindly and other components of the fibrous corona to accumulate at kinetochores (Hoffman et al., 2001). In DMSO-treated cells arrested in mitosis with nocodazole, Spindly adopted the expected crescent morphology at kinetochores (Figure 2A and Supplemental Figure S3A). By contrast, treatment with FTI-277 reduced Spindly levels at kinetochores to 17 ± 3% (average ± SEM with 95% confidence interval) in HeLa cells and to 17 ± 5% in DLD-1 cells (Figure 2, A and D, and Supplemental Figure S3, A and E). Under the same conditions, CENP-E kinetochore levels were reduced to 56 ± 7% in HeLa cells and 31 ± 7% in DLD-1 cells, whereas CENP-F kinetochore levels were less affected, with a reduction to 73 ± 10% in HeLa cells and 56 ± 10% in DLD-1 cells. (Figure 2, B–D, and Supplemental Figure S3, B, C, and E). Kinetochore levels of Zwilch, a subunit of the ROD/Zw10/Zwilch complex (Karess, 2005; Çivril et al., 2010) that is required for kinetochore recruitment of Spindly, were also reduced by FTI-277 treatment (80 ± 9% in HeLa cells; Figure 2, E and G). By contrast, kinetochore levels of Hec1, a component of the KMN network that forms the core microtubule attachment site at the outer kinetochore (Cheeseman et al., 2006), were not affected by FTI-277 treatment in either cell line (99 ± 12% and 98 ± 12% in HeLa and DLD-1 cells, respectively; Figure 2, F and G, and Supplemental Figure S3, D and E). We conclude that FTase activity is a major requirement for kinetochore targeting of Spindly. CENP-E and CENP-F also require farnesylation for proper kinetochore localization, but Spindly targeting to kinetochores is more severely affected by inhibition of farnesylation.
FIGURE 2:
Farnesylation of Spindly is required for its localization to microtubule-unattached kinetochores. (A–C) HeLa cells immunostained for the kinetochore proteins Spindly (A), CENP-E (B), and CENP-F (C) after treatment for 48 h with 10 μM farnesyltransferase inhibitor FTI-277 or DMSO. Cells were incubated in 1 μM nocodazole for 4 h to maximize the accumulation of the proteins at kinetochores and costained with anti-centromere antibodies (ACAs). Scale bars, 5 μm. (D) Quantification of protein levels at kinetochores in the conditions shown in A–C using immunofluorescence intensity measurements. Each condition represents a total of 100 kinetochore measurements from 20 different cells. Error bars represent the SEM with a 95% confidence interval. The t test was used to determine statistical significance (***p < 0.0001). (E, F) HeLa cells immunostained for the kinetochore proteins Zwilch (E) and Hec1 (F) after treatment for 48 h with 10 μM farnesyltransferase inhibitor FTI-277 or DMSO. Scale bars, 5 μm. (G) Kinetochore level quantification of the conditions in E and F displayed as described for D (ns, not statistically significant). (H) Kinetochore localization of RNAi-resistant, MycGFP-tagged wild-type (WT) and mutant (C602S) Spindly in nocodazole-treated HeLa cells after depletion of endogenous Spindly, visualized by immunofluorescence with an anti-GFP antibody (see Figure 5A for corresponding RNAi immunoblot). Scale bar, 5 μm. (I) Kinetochore level quantification of the condition in H displayed as described for D.
To test directly whether farnesylation of Spindly and CENP-E is required for their recruitment to kinetochores, we examined the localization of farnesylation mutants. In the absence of nocodazole, RNA interference (RNAi)–resistant MycGFP::SpindlyWT localized robustly to prometaphase kinetochores after RNAi-mediated depletion of endogenous Spindly, as expected (Gassmann et al., 2010; Supplemental Figure S4A). By contrast, MycGFP::SpindlyC602S was essentially undetectable at kinetochores of prometaphase cells in the presence of microtubules (Supplemental Figure S4A), confirming the finding of a previous study that discussed the possibility that Spindly is farnesylated (Barisic et al., 2010). However, analysis in cells treated with nocodazole showed that the C602S mutant does retain some ability to target to kinetochores in the absence of microtubule attachment (Figure 2, H and I). Quantification of immunofluorescence signals revealed that the reduction in kinetochore levels of MycGFP::SpindlyC602S (20 ± 3% of MycGFP::SpindlyWT) was similar to the reduction observed for endogenous Spindly after FTI-277 treatment (17 ± 3% of DMSO control). The MycGFP::CENP-EC2261S mutant expressed in DLD-1 cells also showed reduced kinetochore levels (45 ± 13% of MycGFP::CENP-EWT), which were not significantly different from kinetochore levels of MycGFP::CENP-EWT after FTI-277 treatment (33 ± 8% of DMSO control; Supplemental Figure S4, B and C). We conclude that modification by FTase is required for proper kinetochore targeting of Spindly and CENP-E.
Inhibition of farnesylation impairs kinetochore recruitment of dynein and dynactin
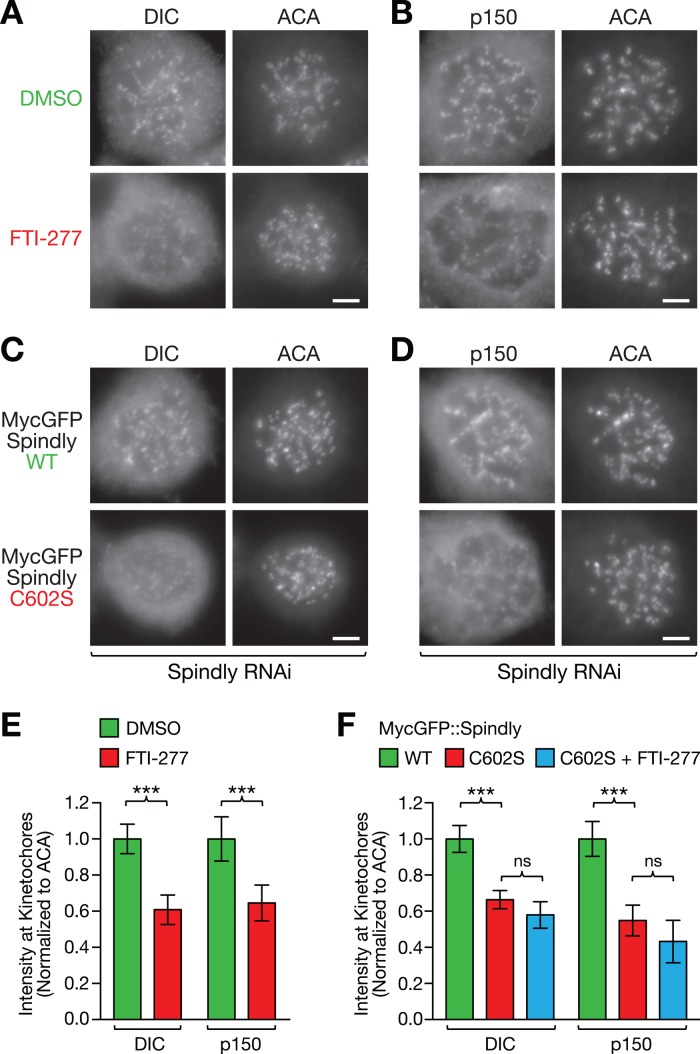
Spindly is required to target dynein and its cofactor dynactin to the kinetochore, predicting that these proteins should be among the kinetochore components whose levels are affected by FTI treatment. Indeed, we observed a significant decrease in kinetochore levels of both dynein (DIC, 61 ± 8% of DMSO control) and dynactin (p150, 65 ± 10% of DMSO control) in nocodazole-treated HeLa cells after incubation with FTI-277 (Figure 3, A, B, and E). We also observed a reduction of kinetochore dynein and dynactin levels in cells expressing MycGFP::SpindlyC602S to 66 ± 5% for dynein and 55 ± 8% for dynactin relative to cells expressing MycGFP::SpindlyWT (Figure 3, C, D, and F). CENP-F is known to contribute to kinetochore dynein recruitment through the dynein cofactor NudE/L (Liang et al., 2007; Stehman et al., 2007; Vergnolle and Taylor, 2007). To assess whether the inhibition of CENP-F farnesylation could be contributing to the decrease of dynein/dynactin levels at kinetochores observed in FTI-treated cells, we added FTI-277 to cells expressing MycGFP::SpindlyC602S. The FTI-277 treatment did not result in a significant additional decrease in dynein/dynactin levels (Figure 3F). We conclude that inhibition of FTase reduces kinetochore levels of dynein/dynactin and that this reduction is primarily a consequence of preventing Spindly farnesylation.
FIGURE 3:
Farnesyltransferase inhibition or prevention of Spindly farnesylation impairs kinetochore recruitment of dynein and its cofactor dynactin. (A–D) HeLa cells immunostained for dynein IC (DIC) and the dynactin subunit p150Glued in cells treated with FTI-277 or DMSO (A, B) and in cells expressing wild-type (WT) and mutant (C602S) MycGFP::Spindly after RNAi-mediated depletion of endogenous Spindly (C, D). Cells were incubated in 1 μM nocodazole for 4 h to maximize the accumulation of the proteins at kinetochores and costained with ACAs. Scale bars, 5 μm. (E, F) Quantification of dynein and dynactin levels at kinetochores in the conditions shown in A–D, as well as in the Spindly mutant C602S after treatment with FTI-277, using immunofluorescence intensity measurements. Each condition represents a total of 100 kinetochore measurements from 20 different cells. Error bars represent the SEM with a 95% confidence interval. The t test was used to determine statistical significance (***p < 0.0001; ns, not significant).
CENP-E farnesylation is important for chromosome segregation
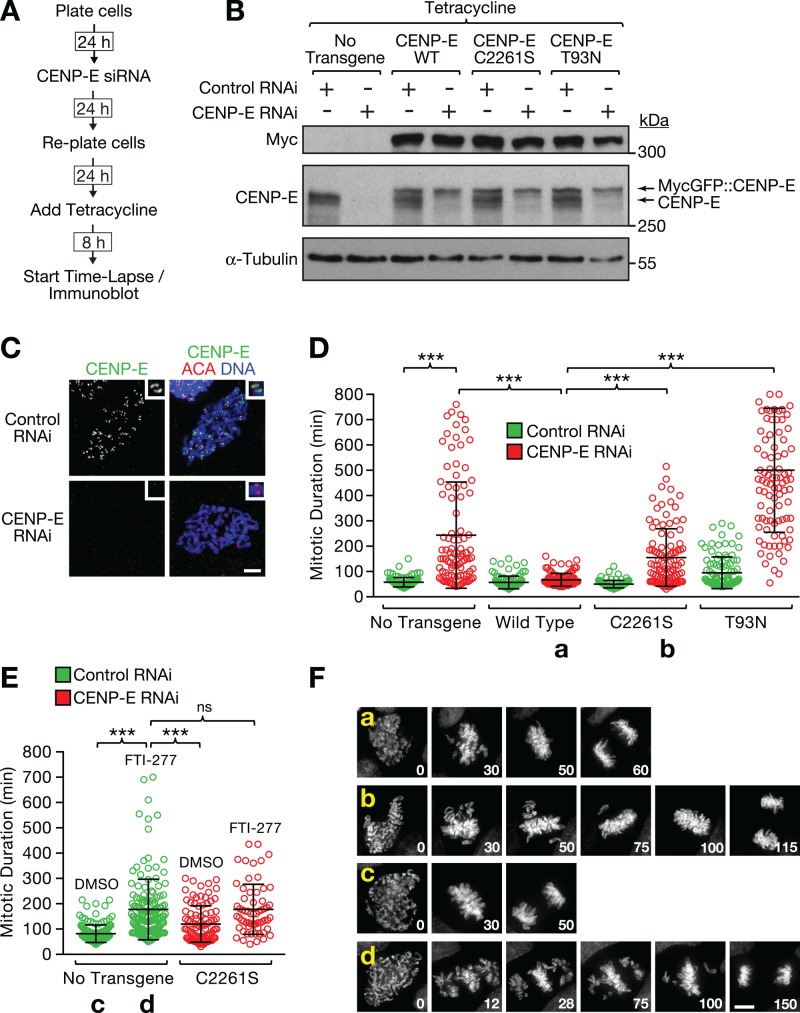
Previous work showed that farnesylation is required for the degradation of CENP-E upon mitotic exit (Gurden et al., 2010). To determine whether CENP-E farnesylation contributes to chromosome segregation, we took advantage of an RNAi-based replacement strategy that we had established for CENP-E in DLD-1 cells (Kim et al., 2010). We knocked down endogenous CENP-E with small interfering RNAs (siRNAs) directed against the 3′ untranslated region of CENP-E mRNA and induced expression of RNAi-resistant CENP-E transgenes with tetracycline (Figure 4A). Immunoblotting confirmed efficient replacement of endogenous CENP-E with the transgene-encoded protein (Figure 4B), and endogenous CENP-E was undetectable at kinetochores in cells treated with CENP-E siRNAs by immunofluorescence (Figure 4C). We then monitored chromosome segregation by time-lapse microscopy in cells coexpressing histone H2B::mRFP. Depletion of CENP-E increased the average time cells spent in mitosis, defined as the interval between nuclear envelope breakdown and the onset of anaphase, from 58 min to 244 min on average (Figure 4D). This mitotic delay was largely rescued by MycGFP::CENP-EWT (67 min on average) but not when we replaced endogenous CENP-E with the motor-dead rigor mutant T93N (500 min on average), consistent with previously published data (Kim et al., 2010). Replacement of endogenous CENP-E with the farnesylation mutant C2261S led to a mitotic delay (120 min on average), which was less pronounced than that observed in cells treated with FTI-277 (177 min on average; Figure 4E). Combining the CENP-E C2261S mutant with FTI-277 treatment resulted in a mitotic delay that was equal to FTI-277 treatment on its own (178 min on average). Inspection of the time-lapse movies confirmed that the mitotic delay in cells treated with FTI-277 was caused by a defect in chromosome congression, similar to what was observed in CENP-E–depleted cells and in cells in which endogenous CENP-E was replaced with the CENP-E C2261S mutant (Figure 4F). These results demonstrate that farnesylation of CENP-E is required for chromosome congression and timely mitotic progression but that inhibition of CENP-E farnesylation alone cannot fully account for the mitotic phenotype observed in cells treated with farnesyltransferase inhibitors.
FIGURE 4:
CENP-E farnesylation is important for chromosome segregation. (A) Experimental approach for the characterization of DLD-1 Flp-In T-Rex cells with integrated RNAi-resistant CENP-E constructs whose expression is inducible with tetracycline. (B) Immunoblot showing successful replacement of endogenous CENP-E with RNAi-resistant MycGFP::CENP-E in DLD-1 cells. C2261S corresponds to the CENP-E farnesylation mutant and T93N to motor-dead “rigor” CENP-E. α-Tubulin was used as a loading control. (C) Immunofluorescence of DLD-1 cells showing loss of kinetochore-localized CENP-E 48 h after transfection with CENP-E siRNA. Cells were costained with ACAs. Scale bar, 5 μm. (D) Quantification of mitotic duration (nuclear envelope breakdown to anaphase onset) from time-lapse fluorescence microscopy experiments conducted with DLD-1 cells expressing histone H2b::mRFP with or without the CENP-E transgenes described in B. Mitotic duration is shown as a scatter plot with the average and SD for the indicated experimental conditions. All conditions were filmed in parallel during the same imaging session using a multiwell chamber. One hundred cells were scored for each condition. The t test was used to determine statistical significance (***p < 0.0001). (E) Quantification of mitotic duration for the indicated conditions from a time-lapse experiment, as described in D (ns, not statistically significant). (F) Selected images from a time-lapse series of DLD-1 cells coexpressing histone H2b::mRFP and CENP-E transgenes showing chromosome congression defects in cells expressing nonfarnesylated CENP-E. Time is indicated in minutes relative to nuclear envelope breakdown (time point 0). Lowercase letters refer to the conditions labeled with the same letters in D and E. Scale bar, 5 μm.
A C-terminal CENP-E fragment (amino acids 1569–2264), called the CENP-E tail, contains the kinetochore-binding domain and causes a pronounced mitotic arrest when overexpressed (Chan et al., 1998; Gurden et al., 2010; Supplemental Figure S5, A and B). The mitotic delay in cells expressing the CENP-E tail in the presence of endogenous CENP-E (1231 min on average) was significantly more severe than in CENP-E–depleted cells (244 min on average; compare Figure 4D with Supplemental Figure S5B), suggesting that the CENP-E tail is a gain-of-function mutant, possibly acting through chronic activation of spindle assembly checkpoint signaling (Mao et al., 2005). Strikingly, we observed that the dominant-negative effect of the CENP-E tail depends entirely on its modification by FTase: expression of the CENP-E tail farnesylation mutant C2261S had no adverse effect on mitotic duration (65 min on average), and treatment with FTI-277 reduced the mitotic duration of cells expressing the CENP-E tail from 1231 min to 210 min on average, which is close to the mitotic duration observed in FTI-277-treated cells (177 min on average; Figure 4E and Supplemental Figure S5, A and B). Furthermore, the C2261S mutation rescued the penetrant lethality of CENP-E tail expression in a clonogenic survival assay (Supplemental Figure S5C). This effect would not have been predicted from the comparatively mild functional consequences of inhibiting farnesylation of wild-type CENP-E and serves to demonstrate that inhibition of farnesylation can dramatically alter toxic properties of mutant proteins.
Preventing farnesylation of Spindly and inhibition of farnesyltransferase activity result in a similar mitotic phenotype
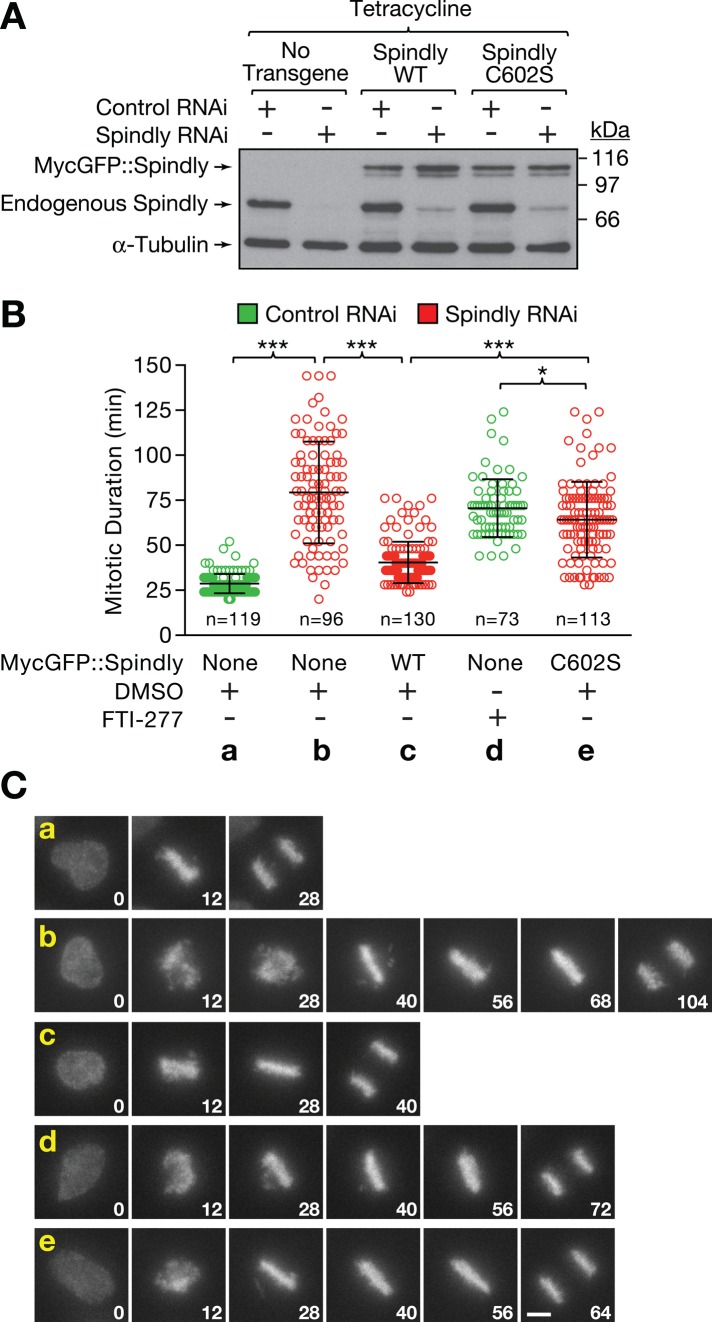
The loss of Spindly from kinetochores after FTI-277 treatment indicated that nonfarnesylated Spindly contributes to the mitotic phenotype of FTase inhibitors. We tested this hypothesis by combining specific depletion of endogenous Spindly with expression of RNAi-resistant MycGFP::Spindly transgenes in HeLa cells coexpressing H2B::mRFP (Figure 5A). As expected from previous work, the mitotic delay induced by depletion of Spindly (79 min on average) was largely rescued by ectopic expression of MycGFP::SpindlyWT (40 vs. 28 min on average for control cells; Gassmann et al., 2010; Figure 5B). Expression of the MycGFP::SpindlyC602S mutant caused a mitotic delay (64 min on average), which was slightly less pronounced than that observed in FTI-277-treated cells (71 min on average). Visualization of chromosome segregation confirmed that the mitotic delay in cells expressing the MycGFP::SpindlyC602S mutant was caused by misaligned chromosomes, a defect characteristic of Spindly depletions (Chan et al., 2009; Barisic et al., 2010; Gassmann et al., 2010; Figure 5C). We conclude that farnesylation of Spindly is required for efficient recruitment of dynein/dynactin to kinetochores and timely chromosome congression.
FIGURE 5:
Preventing farnesylation of Spindly and inhibiting farnesyltransferase result in a similar mitotic delay. (A) Immunoblot showing expression levels of endogenous and RNAi-resistant, transgene-encoded MycGFP::Spindly. Cells were treated with control or Spindly siRNA for 22 h, followed by induction of transgene expression with 0.2 μg/ml tetracycline for 8 h. α-Tubulin was used as a loading control. (B) Quantitative analysis of mitotic duration (nuclear envelope breakdown to anaphase onset) for time-lapse fluorescence microscopy experiments performed with HeLa cells expressing histone H2b::mRFP with or without Spindly transgenes. RNAi and induction of transgene expression was performed as for the immunoblot in A. Data are shown as scatter plots with average and SD. Two independent experiments were performed for each condition, and the total number (n) of cells scored is indicated. The t test was used to determine statistical significance (***p < 0.0001; *p < 0.05). (C) Selected images from the time-lapse experiments described in B, showing chromosome congression defects in cells expressing the Spindly farnesylation mutant C602S. Time is indicated in minutes relative to nuclear envelope breakdown (time point 0). Scale bar, 5 μm.
DISCUSSION
FTase inhibitors (FTIs) are increasingly regarded as promising agents for therapeutic intervention in a variety of diseases, which makes the identification of a complete catalogue of FTase substrates an important goal. Previously it was shown that FTase inhibition leads to mitotic defects that have been largely attributed to a lack of farnesylation of the mitotic proteins CENP-E and CENP-F. Our study now identifies Spindly as a third farnesylated mitotic protein and shows that preventing farnesylation of Spindly is a major contributor to the mitotic phenotype of FTIs.
CENP-E, CENP-F, and Spindly all localize to the fibrous corona at the outer kinetochore, where Spindly and CENP-F are responsible for the recruitment of cytoplasmic dynein and its cofactor dynactin. The fibrous corona thus harbors multiple microtubule-binding activities that are required for efficient chromosome congression. Our analysis shows that FTI treatment selectively perturbs the formation of this kinetochore domain, whereas the core microtubule attachment site (represented in our experiments by Hec1) assembles normally. Although changes in kinetochore morphology are difficult to quantify, we found that the telltale crescent and ring shapes of microtubule-unattached kinetochores (Hoffman et al., 2001) were largely missing when staining for fibrous corona components after FTI-277 treatment. Thus the decreased kinetochore levels of corona components in the presence of FTIs may partially reflect the inability of the fibrous corona domain to undergo expansion. Among the three farnesylated kinetochore components, the localization of Spindly was the most dramatically affected by FTI treatment. We find that the kinetochore recruitment of Spindly requires its farnesylation, raising the possibility that the farnesyl moiety at the Spindly C-terminus is necessary for the interaction with a kinetochore receptor. Spindly kinetochore recruitment depends on the Rod/Zw10/Zwilch complex, and the C-terminus of human Spindly is positioned close to Zwilch, Zw10, and the N-terminus of Rod, as determined by nanometer-scale immunofluorescence mapping experiments (Varma et al., 2013). It will be interesting to investigate whether Spindly farnesylation is required for binding to the Rod/Zw10/Zwilch complex. However, since the critical cysteine required for Spindly farnesylation is not present in all phyla, there are likely other conserved regions of Spindly that participate in its kinetochore targeting. Nevertheless, our work indicates that future biochemical reconstitution efforts of kinetochore complexes involving vertebrate Spindly will need to take into account the key role of its farnesyl group.
There are contradictory reports regarding the contribution of farnesylation to the kinetochore recruitment of CENP-E and CENP-F, as previous studies did not measure kinetochore levels of these proteins (Ashar et al., 2000; Crespo et al., 2001; Hussein and Taylor, 2002; Schafer-Hales et al., 2007; Verstraeten et al., 2011). Our experiments in HeLa and DLD-1 cells demonstrate that both CENP-E and CENP-F require farnesylation for proper localization to microtubule-unattached kinetochores, although their localization is less affected by FTase inhibition than Spindly. We found that inhibition of FTase also affects the localization of the ROD/Zw10/Zwilch complex, which targets to the KMN network upstream of Spindly. The expansion of the fibrous corona in the absence of microtubules is not understood at the molecular level, but it is evident that the expansion step must involve interactions among corona components that are distinct from the interactions that mediate the initial recruitment by the KMN network, which itself does not expand. One interpretation of our results is that the kinetochore accumulation of the ROD/Zw10/Zwilch complex in the absence of microtubules is in part facilitated by interactions with CENP-E, CENP-F, or Spindly, whose kinetochore recruitment is directly affected by FTI treatment.
Our molecular replacement experiments establish that both nonfarnesylated CENP-E and nonfarnesylated Spindly cause chromosome congression defects and contribute to the delay in mitotic progression that is observed after FTI treatment. Assessing the functional significance of CENP-F farnesylation for chromosome segregation will necessitate the development of a molecular replacement approach similar to the one we used here. Of interest, FTI treatment in DLD-1 cells had a more pronounced effect on kinetochore levels of CENP-E and CENP-F than FTI treatment in HeLa cells, yet the mitotic delay caused by FTase inhibition was similar in both cell lines (2.2- and 2.5-fold in DLD-1 and HeLa cells, respectively). Taking this together with the observation that nonfarnesylated Spindly was robustly displaced from kinetochores in both cell lines, we propose that loss of Spindly farnesylation is a major factor contributing to the mitotic phenotype observed in cells treated with FTIs.
MATERIALS AND METHODS
Cells lines and antibodies
Stable isogenic cell lines expressing Spindly and CENP-E transgenes were generated by FRT/Flp-mediated recombination as described previously (Tighe et al., 2004). Full-length Spindly and CENP-E cDNA corresponding to amino acids 1569–2264 of CENP-E (CENP-E tail) were cloned into a pcDNA5/FRT/TO-based vector (Invitrogen, Carlsbad, CA) modified to contain an N-terminal Myc-LAP epitope tag. The LAP tag consists of GFP-TEV-S-peptide (Cheeseman et al., 2004). Affinity-purified rabbit polyclonal antibodies against Spindly fragments comprising amino acids 2–254 (OD173) and 450–605 (OD174) were generated as described previously (Desai et al., 2003).
Cell culture and RNA interference
HeLa and DLD-1 cells were maintained at 37°C in a 5% CO2 atmosphere in DMEM (Thermo Fischer Scientific, Waltham, MA) supplemented with 10% tetracycline-free fetal bovine serum (Clontech, Mountain View, CA), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine. For immunofluorescence, cells were seeded on 12-mm poly-l-lysine–coated coverslips in 12-well plates 24 h before transfection with siRNAs. For live-cell imaging experiments, cells were seeded in a 35-mm glass-bottom dish coated with poly-d-lysine (MatTek, Ashland, MA). Cells were transfected with siRNAs targeting Spindly and the CENP-E 3′ UTR as described previously (Gassmann et al., 2010; Kim et al., 2010). For immunofluorescence of HeLa Flp-In T-Rex cells, transgene expression was induced with 0.2 μg/ml tetracycline 24 h posttransfection, and cells were fixed 20–24 h later. For live-cell imaging of HeLa Flp-In T-Rex cells and for the immunoblot shown in Figure 5A, transgene expression was induced 22 h after transfection, and time-lapse imaging experiments were started 8 h later.
AGOH labeling and drug treatments
For labeling of DLD-1 or HeLa Flp-In T-Rex cells with the farnesol analogue AGOH, a confluent 10-cm dish of unsynchronized cells was split 1:5 into one 15-cm dish. After 24 h, cells were incubated with 30 μM AGOH, 0.2 μg/ml tetracycline, and 10 μM FTI-277 (Sigma-Aldrich, St. Louis, MO) for 40 h before cells were harvested and processed for immunoprecipitation experiments and immunoblotting. To depolymerize microtubules before immunofluorescence, cells were treated with 1 μM nocodazole for 4 h.
Immunoprecipitations and mass spectrometry
For mass spectrometry analysis of Spindly immunoprecipitates, endogenous Spindly was immunoprecipitated with 50 μg of affinity-purified anti-Spindly antibody OD173 or OD174 (covalently coupled to 100 μl of agarose protein A beads [BioRad, Hercules, CA]) from a mitotic HeLa cell extract (30 mg of total protein). The extract was prepared by sonicating nocodazole-arrested cells (harvested by mitotic shake-off) in lysis buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.6, 200 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid [EGTA], 5% glycerol, 0.05% NP-40, 1 mM dithiothreitol [DTT], 5 mM β-glycerophosphate, 400 nM microcystin, and cOmplete protein inhibitor cocktail [Roche, Basel, Switzerland]), followed by sequential rounds of centrifugation in an ultracentrifuge at 20,000 and 50,000 × g. Beads were washed with 6 × 1 ml of lysis buffer and 3 × 1 ml of lysis buffer without detergent. Proteins were eluted from beads with 0.1 M glycine, pH 2.6, and the samples neutralized with 2 M Tris-HCl, pH 8.5. Proteins were precipitated by addition of 1/5 volume trichloroacetic acid (TCA) overnight on ice, and the TCA pellet was washed twice with acetone before air-drying. Mass spectrometry was conducted as described previously (Cheeseman et al., 2004).
For immunoprecipitations from HeLa cells after labeling with the farnesol analogue AGOH, unsynchronized cells (one 15-cm dish per condition) were harvested, washed with 1× phosphate-buffered saline (PBS), and sonicated in 3 ml of lysis buffer (50 mM HEPES, pH 7.4, 200 mM KCl, 1 mM EGTA, 1 mM MgCl2, 0.05% NP-40, 1 mM DTT, 5 mM β-glycerophosphate, and protease inhibitors). The crude lysate was clarified by spinning at 20,000 × g for 10 min, and 1.3 ml of the cleared lysate was incubated with 5 μg of anti-Myc tag antibody (clone 4A6) covalently coupled to 10 μl of agarose protein A beads (or with protein A beads alone as a control) for 2 h at 4°C. After washing of the beads with 3 × 1 ml of lysis buffer, proteins were eluted with SDS–PAGE sample buffer without DTT (50 mM Tris-HCl, pH 6.8, 15% [wt/vol] sucrose, 2 mM EDTA, 3% SDS) at 70°C for 15 min.
Live-cell imaging
For live-cell imaging, growth medium was replaced with CO2-independent medium (Thermo Fischer Scientific) supplemented with 0.2 μg/ml tetracycline immediately before filming and covered with a layer of mineral oil. Time-lapse sequences were recorded on a DeltaVision microscope (Applied Precision, Mississauga, Canada) equipped with an environmental chamber heated to 36–37°C as measured in the dish. Image stacks of 5 × 3 μm were acquired every 4 min for 10 h with a CoolSnap charge-coupled device camera (Photometrics, Tuscon, AZ) and a 40×/numerical aperture (NA) 1.35 U-planApo objective (Olympus, Tokyo, Japan) at 2 × 2 binning.
Indirect immunofluorescence and immunoblotting
For immunofluorescence experiments, cells were fixed immediately after aspiration of the medium with 4% formaldehyde in Phem buffer (60 mM 1,4-piperazinediethanesulfonic acid, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9) for 5 min at room temperature and then permeabilized for 2 min with 0.1% Triton X-100 in Phem buffer and rinsed three times in Phem buffer. For stainings with anti-dynein antibodies, cells were fixed in methanol at −20°C for 45 min, followed by rehydration for 2 × 5 min in PBS/0.5% Triton X-100 and PBS/0.1% Triton X-100. Cells were blocked for 30 min in AbDil solution (4% immunoglobulin G–free bovine serum albumin [Jackson ImmunoResearch, West Grove, PA], 0.1% Triton X-100) and incubated with primary antibody for 1 h at room temperature (overnight at 4°C for anti-dynein), diluted in AbDil (human anti–centromere antibodies [ACAs], 1:1000; rabbit anti-AG, 1:1000; mouse anti-CENP-E 1H12, 1:500; sheep anti-CENP-F, 1:800; mouse anti-dynein IC 70.1, 1:500; mouse anti-dynactin p150, 1:150; goat anti-GFP, 1:15000; mouse anti-Hec1 9G3, 1:1000; rabbit anti-Spindly OD174, 1:5000; rabbit anti-Zwilch, 1:900). After washing for 3 × 5 min in PBS/0.1% Triton X-100, cells were incubated with secondary antibodies conjugated with fluorescent dyes (Alexa 488, rhodamine red X, Cy5 [Jackson ImmunoResearch]). Cells were washed again for 3 × 5 min and mounted in Prolong Gold with 4′,6-diamidino-2-phenylindole stain (Invitrogen). Images were recorded on an Axio Observer microscope (Carl Zeiss, Oberkochen, Germany) at 1 × 1 binning with a 100×/NA 1.46 Plan-Apochromat objective and an Orca Flash 4.0 camera (Hamamatsu Photonics, Hamamatsu, Japan). Maximum intensity projections of z-stacks were imported into Photoshop CS5.1 (Adobe Systems, San Jose, CA) for further processing. For quantification of kinetochore signals, unprocessed 16-bit images were analyzed with Fiji software as described previously (Hoffman et al., 2001). At each kinetochore, the signal intensity of the outer kinetochore component was normalized to the signal intensity of ACAs.
Immunoblotting was performed using standard methods with horseradish peroxidase–coupled secondary antibodies (Jackson ImmunoResearch) and ECL (GE Healthcare, Little Chalfont, United Kingdom) following the manufacturer's instructions. Primary antibodies were rabbit anti-AG (1:1000), rabbit anti-Spindly OD157 (1:3000), mouse anti–α-tubulin (DM1α, 1:1000), rabbit anti-CENP-E (1:1000), mouse anti-Mad1 (1:10), and rabbit anti-Mad2 (1:100).
Statistical analysis
Statistical significance was evaluated with the t test using Prism 6 software (GraphPad, La Jolla, CA).
Supplementary Material
Acknowledgments
We thank Stephen S. Taylor for providing the CENP-F antibody and the parental DLD-1 and HeLa Flp-In T-Rex cell lines, Andrea Musacchio for providing antibodies against Mad1 and Zwilch, Jennifer Meerloo of the University of California, San Diego, Neuroscience Microscopy Shared Facility (NINDS P30 NS047101) for help with live-cell imaging, and Ana Carvalho for critical reading of the manuscript. This work was supported by a Starting Grant from the European Research Council (338410), an EMBO Installation Grant, and funding from the Fundação para a Ciência e a Tecnologia (IF/01015/2013/CP1157/CT0006) to R.G., a W.W. Smith Charitable Trust Research Grant, a March of Dimes Basil O'Conner Scholar Award, a Pew-Stewart Scholar Award, and a Kimmel Scholar Award to A.J.H., grants from the National Institutes of Health to A.D. (GM074215) and D.W.C. (GM29513), and funding from the Ludwig Institute for Cancer Research to A.D. and D.W.C.
Abbreviations used:
- AGOH
anilinogeraniol
- FPP
farnesyl diphosphate
- FTase
farnesyltransferase
- FTI
farnesyltransferase inhibitor
- GGPP
geranylgeranyl diphosphate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1560) on March 25, 2015.
*Present address: Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205.
REFERENCES
- Ashar HR, James L, Gray K, Carr D, Black S, Armstrong L, Bishop WR, Kirschmeier P. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J Biol Chem. 2000;275:30451–30457. doi: 10.1074/jbc.M003469200. [DOI] [PubMed] [Google Scholar]
- Barisic M, Sohm B, Mikolcevic P, Wandke C, Rauch V, Ringer T, Hess M, Bonn G, Geley S. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol Biol Cell. 2010;21:1968–1981. doi: 10.1091/mbc.E09-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. EMBO J. 2005;24:3927–3939. doi: 10.1038/sj.emboj.7600848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, Fava LL, Uldschmid A, Schmitz MHA, Gerlich DW, Nigg EA, Santamaria A. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J Cell Biol. 2009;185:859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çivril F, Wehenkel A, Giorgi FM, Santaguida S, Di Fonzo A, Grigorean G, Ciccarelli FD, Musacchio A. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 2010;18:616–626. doi: 10.1016/j.str.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Crespo NC, Ohkanda J, Yen TJ, Hamilton AD, Sebti SM. The farnesyltransferase inhibitor, FTI-2153, blocks bipolar spindle formation and chromosome alignment and causes prometaphase accumulation during mitosis of human lung cancer cells. J Biol Chem. 2001;276:16161–16167. doi: 10.1074/jbc.M006213200. [DOI] [PubMed] [Google Scholar]
- Crul M, de Klerk GJ, Beijnen JH, Schellens JH. Ras biochemistry and farnesyl transferase inhibitors: a literature survey. Anticancer Drugs. 2001;12:163–184. doi: 10.1097/00001813-200103000-00001. [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Müller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman RT, Buckner FS, Yokoyama K, Gelb MH, Van Voorhis WC. Thematic review series: lipid posttranslational modifications. Fighting parasitic disease by blocking protein farnesylation. J Lipid Res. 2006;47:233–240. doi: 10.1194/jlr.R500016-JLR200. [DOI] [PubMed] [Google Scholar]
- Feng J, Huang H, Yen TJ. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 2006;115:320–329. doi: 10.1007/s00412-006-0049-5. [DOI] [PubMed] [Google Scholar]
- Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LB, Massaro J, D'Agostino RB, Campbell SE, Brazier J, Brown WT, Kleinman ME, Kieran MW, Progeria Clinical Trials Collaborative Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation. 2014;130:27–34. doi: 10.1161/CIRCULATIONAHA.113.008285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurden MDJ, Holland AJ, van Zon W, Tighe A, Vergnolle MA, Andres DA, Spielmann PH, Malumbres M, Wolthuis RMF, Cleveland DW, Taylor SS. Cdc20 is required for the post-anaphase, KEN-dependent degradation of centromere protein F. J Cell Sci. 2010;123:321–330. doi: 10.1242/jcs.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Holstein SA, Hohl RJ. Is there a future for prenyltransferase inhibitors in cancer therapy. Curr Opin Pharmacol. 2012;12:704–709. doi: 10.1016/j.coph.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Hougland JL, Lamphear CL, Scott SA, Gibbs RA, Fierke CA. Context-dependent substrate recognition by protein farnesyltransferase. Biochemistry. 2009;48:1691–1701. doi: 10.1021/bi801710g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein D, Taylor SS. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J Cell Sci. 2002;115:3403–3414. doi: 10.1242/jcs.115.17.3403. [DOI] [PubMed] [Google Scholar]
- Ignatev A, Kravchenko S, Rak A, Goody RS, Pylypenko O. A structural model of the GDP dissociation inhibitor rab membrane extraction mechanism. J Biol Chem. 2008;283:18377–18384. doi: 10.1074/jbc.M709718200. [DOI] [PubMed] [Google Scholar]
- Karess R. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 2005;15:386–392. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Yu W, Li Y, Yu L, Zhang Q, Wang F, Yang Z, Du J, Huang Q, Yao X, Zhu X. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol Biol Cell. 2007;18:2656–2666. doi: 10.1091/mbc.E06-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moasser MM, Sepp-Lorenzino L, Kohl NE, Oliff A, Balog A, Su D-S, Danishefsky SJ, Rosen N. Farnesyl transferase inhibitors cause enhanced mitotic sensitivity to taxol and epothilones. Proc Natl Acad Sci USA. 1998;95:1369–1374. doi: 10.1073/pnas.95.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen UTT, Goody RS, Alexandrov K. Understanding and exploiting protein prenyltransferases. Chembiochem. 2010;11:1194–1201. doi: 10.1002/cbic.200900727. [DOI] [PubMed] [Google Scholar]
- O'Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- Pechlivanis M, Kuhlmann J. Hydrophobic modifications of Ras proteins by isoprenoid groups and fatty acids—more than just membrane anchoring. Biochim Biophys Acta. 2006;1764:1914–1931. doi: 10.1016/j.bbapap.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H, et al. Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. EMBO J. 2006;25:13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- Rotblat B, Niv H, André S, Kaltner H, Gabius H-J, Kloog Y. Galectin-1(L11A) predicted from a computed galectin-1 farnesyl-binding pocket selectively inhibits Ras-GTP. Cancer Res. 2004;64:3112–3118. doi: 10.1158/0008-5472.can-04-0026. [DOI] [PubMed] [Google Scholar]
- Schafer-Hales K, Iaconelli J, Snyder JP, Prussia A, Nettles JH, El-Naggar A, Khuri FR, Giannakakou P, Marcus AI. Farnesyl transferase inhibitors impair chromosomal maintenance in cell lines and human tumors by compromising CENP-E and CENP-F function. Mol Cancer Ther. 2007;6:1317–1328. doi: 10.1158/1535-7163.MCT-06-0703. [DOI] [PubMed] [Google Scholar]
- Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007;178:583-594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117:6339–6353. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- Troutman JM, Roberts MJ, Andres DA, Spielmann HP. Tools to analyze protein farnesylation in cells. Bioconjug Chem. 2005;16:1209–1217. doi: 10.1021/bc050068+. [DOI] [PubMed] [Google Scholar]
- Varma D, Wan X, Cheerambathur D, Gassmann R, Suzuki A, Lawrimore J, Desai A, Salmon ED. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J Cell Biol. 2013;202:735–746. doi: 10.1083/jcb.201304197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle MAS, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Verstraeten VLRM, Peckham LA, Olive M, Capell BC, Collins FS, Nabel EG, Young SG, Fong LG, Lammerding J. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc Natl Acad Sci USA. 2011;108:4997–5002. doi: 10.1073/pnas.1019532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Bonday ZQ, Putkey FR, Kops GJPL, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Chang SY, Ren S, Wang Y, Andres DA, Spielmann HP, Fong LG, Young SG. Absence of progeria-like disease phenotypes in knock-in mice expressing a non-farnesylated version of progerin. Hum Mol Genet. 2011;20:436–444. doi: 10.1093/hmg/ddq490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zverina EA, Lamphear CL, Wright EN, Fierke CA. Recent advances in protein prenyltransferases: substrate identification, regulation, and disease interventions. Curr Opin Chem Biol. 2012;16:544–552. doi: 10.1016/j.cbpa.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.