Abstract
Background
Regulation of gene expression in trypanosomatids is mainly posttranscriptional. Tight regulation of mRNA stability and access to polysomes allows Trypanosoma cruzi to adapt to different environmental conditions during its life cycle. Posttranscriptional regulation requires association between mRNAs and specific proteins to form mRNP complexes. Proteins that lack a canonical RNA-binding domain, such as eukaryotic elongation factor-1α (EF-1α), may also associate with mRNPs. EF-1α is conserved in many organisms, and it plays roles in many cellular processes other than translation, including RNA transport, the cell cycle, and apoptosis.
Results
In a previous study, EF-1α was found associated with mRNP-forming mRNAs in polysome-free fractions both in epimastigotes growing under normal conditions and in nutritionally stressed parasites. This finding suggested the possibility that EF-1α has a non-canonical function. Thus, we investigated the dynamics of EF-1α in association with T. cruzi epimastigote mRNAs under normal and stressed nutritional conditions. EF-1α is expressed throughout the parasite life cycle, but it shows a slight decrease in protein levels in the metacyclic trypomastigote form. The protein is cytoplasmically localized with a granular pattern in all forms analyzed. Following puromycin treatment, EF-1α migrated with the heaviest gradient fractions in a sucrose polysome profile, indicating that its association with large protein complexes was independent of the translation machinery. We next characterized the EF-1α-associated mRNAs in unstressed and stressed epimastigotes. We observed that specific subsets of mRNAs were associated with EF-1α-mRNPs in unstressed or stressed epimastigotes. Some mRNAs were identified in both physiological conditions, whereas others were condition-specific. Gene ontology analysis identified enrichment of gene sets involved in single-organism metabolic processes, amino acid metabolic processes, ATP and metal ion binding, glycolysis, glutamine metabolic processes, and cobalt and iron ion binding.
Conclusion
These results indicate that in T. cruzi, as in other eukaryotes, EF-1α may play a non-canonical cellular role. We observed the enrichment of functionally related transcripts bound to EF-1α in normal growth conditions as well as in nutritionally stressed cell indicating a potential role of EF-1α mRNP in stress response.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0436-2) contains supplementary material, which is available to authorized users.
Keywords: Trypanosoma cruzi, Gene expression, mRNPs, EF-1 alpha, Non-canonical function
Background
The life cycle of Trypanosoma cruzi involves two intermediary hosts and at least three well-defined developmental stages that are based on morphology and biological characteristics [1, 2]. The metacyclogenesis process, in which replicating epimastigotes transform into infective and non-replicating metacyclic trypomastigotes, occurs in the midgut of the invertebrate triatomine host and can be mimicked in vitro under chemically defined conditions [3, 4]. Interestingly, the differentiation of epimastigotes is triggered by nutritional stress, and different sets of genes are expressed in normally growing epimastigotes and in nutritionally stressed epimastigotes [5].
The mechanisms of gene expression regulation in T. cruzi present some peculiarities. In the absence of typical RNA polymerase II promoter regions, transcription begins within the divergent strand-switch region and occurs in both directions. As a result, dozens to hundreds of genes with unrelated functions are transcribed in the same polycistronic unit, which upon processing results in different levels of protein production [6, 7]. These features demonstrate that the regulation of gene expression in these organisms is mainly post-transcriptional and is based on mechanisms involving the localization, translation and degradation of mRNAs [6, 8, 9]. In recent years, several mechanisms have been described as possible strategies for gene expression regulation.
The translation is a very important step of gene expression regulation. The ribosome profiling was developed in order to have a better view of the mRNAs that are actually engaged to translation and how is the relevance of the translational control [10]. For Trypanosoma brucei and Plasmodium falciparum, it has been shown that translation has a very important role in gene expression regulation in these parasites [11–13]. Another mechanism is the organization of mRNAs into post-transcriptional operons or regulons, where mRNAs with related functions are associated with similar sets of proteins in mRNPs, thereby enabling their coordinated expression [5, 14–17]. Another possible mechanism is the association of RNAs into RNA granules [18–20] that ultimately determine the fate of mRNAs within the cell [5, 14–17].
Elongation factor 1α (EF-1α) binds GTP, and the GTP-bound form interacts with aminoacyl-tRNA and carries it to the A site of the ribosome [21]. Control of EF-1α protein levels is important for normal cell function, as EF-1α acts as a regulator of the cell cycle. Accordingly, EF-1α overexpression is associated with many tumor types [22]. EF-1α has also been described as an actin-binding protein in several species [23, 24]. In addition, other non-canonical functions have been assigned to EF-1α because of its involvement in various cellular processes, including metabolism [25], cytoskeletal organization [26], oncogenic transformation [27, 28], apoptosis and anti-apoptosis [29, 30], and gene expression [31]. It has also been demonstrated that EF-1α plays a role in connecting the translation machinery with the cytoskeleton, thus permitting mRNA localization in chicken embryo fibroblasts. EF-1α associates with F-actin; the resulting complex binds to beta-actin mRNA and anchors it in the cellular protrusions of the fibroblast [31]. EF-1α is also found in RNA granules, which are transported by microtubules in neurons [32].
In T. cruzi, EF-1α [23] was previously shown to bind mRNAs not only in polysomes but also in polysome-free fractions, both in exponentially growing epimastigotes and in parasites under nutritional stress [33]. The association of EF-1α with mRNA complexes that are not associated with the protein translation apparatus permitted us to better characterize EF-1α mRNPs by identifying their associated transcripts. This approach yielded further insights into the role of EF-1α mRNPs in the regulation of T. cruzi gene expression.
Results and discussion
T. cruzi EF-1α genomic organization and protein characteristics
T. cruzi EF-1α is organized as a multicopy gene family that includes 11 tandem copies on chromosome 5 [34]. The EF-1α protein has three domains. Domain I is responsible for GTP binding, domain II is responsible for recognizing the 3' end of the tRNA, and domain III connects the aminoacyl-tRNA to the ribosome (Fig. 1a) [35]. The T. cruzi EF-1α protein has an estimated molecular weight of 43 kDa, and it is highly conserved among trypanosomes and other eukaryotes, with 95 % identity when compared to trypanosomes and 76 % identity when compared to Homo sapiens.
Fig. 1.
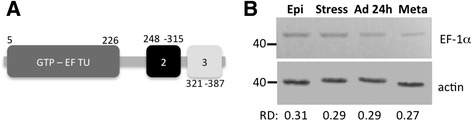
Scheme of the EF-1α protein from T. cruzi. (a) The domains are indicated; the GTP-binding domain is dark grey, the second domain is black and the third domain is light grey. The numbers indicate the positions of the domains within the protein. (b) Western-blot analysis of T. cruzi extracts with antisera against EF-1α and actin. Epi: exponentially growing normal epimastigotes. Stress: epimastigotes under nutritional stress. Ad 24 h: epimastigotes adhered to the substrate. Meta: metacyclic trypomastigotes. Ten μg of protein was loaded into each lane. Top panel: anti-EF-1α antibody signal (1:300). Bottom panel: anti-β-actin used for normalization (1:500). The relative densities of the bands (RD) are indicated. The molecular mass standards are indicated (kDa)
EF-1α protein expression is controlled throughout the parasite life cycle (Fig. 1b). Accordingly, proteomic data have shown that EF-1α protein levels decrease in metacyclic trypomastigotes compared to epimastigotes [36]. All analyzed forms of EF-1α showed a granular distribution throughout the cytoplasm of the parasite, with a decrease of signal in metacyclic trypomastigotes that corroborated the proteomic and western blot results (Fig. 2). The immunofluorescence controls are shown in the Additional file 4: Figure S1.
Fig. 2.
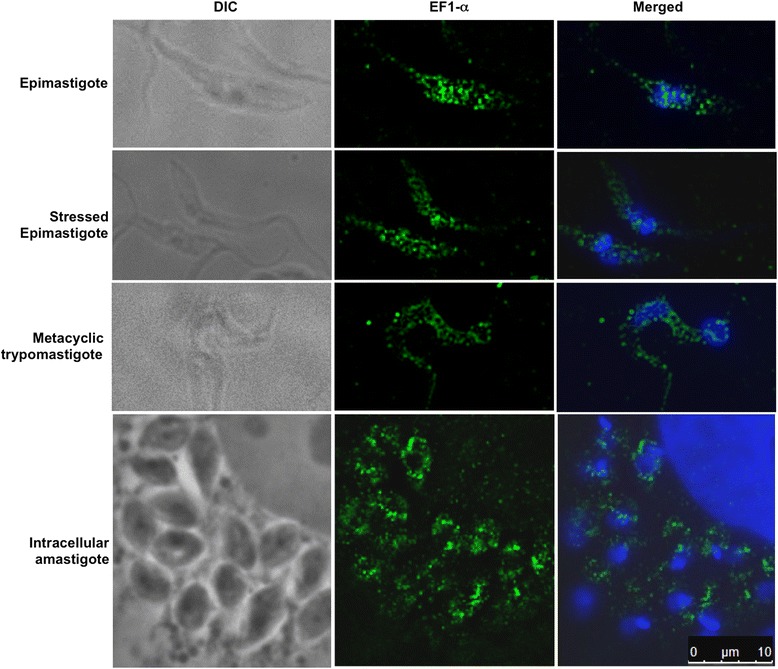
Immunolocalization assay. Cells were incubated with anti-EF-1α antibody (1:300). Immune complexes were detected by reaction with Alexa 488-labelled goat anti-rabbit antibody (1:400). Kinetoplasts and nuclei were stained with DAPI, and the obtained image was merged with the EF-1α image. Bar = 10 μm
EF-1α is present in large mRNP complexes that are not associated with translation machinery
To investigate the distribution of the EF-1α protein in complexes, we analyzed its sedimentation profile in sucrose density gradients (15–55 %). Protein extracts of fractions from unstressed and stressed epimastigotes treated with cycloheximide or puromycin were analyzed by western blot using antisera against the EF-1α protein and the ribosomal protein S7 as a control (Fig. 3).
Fig. 3.
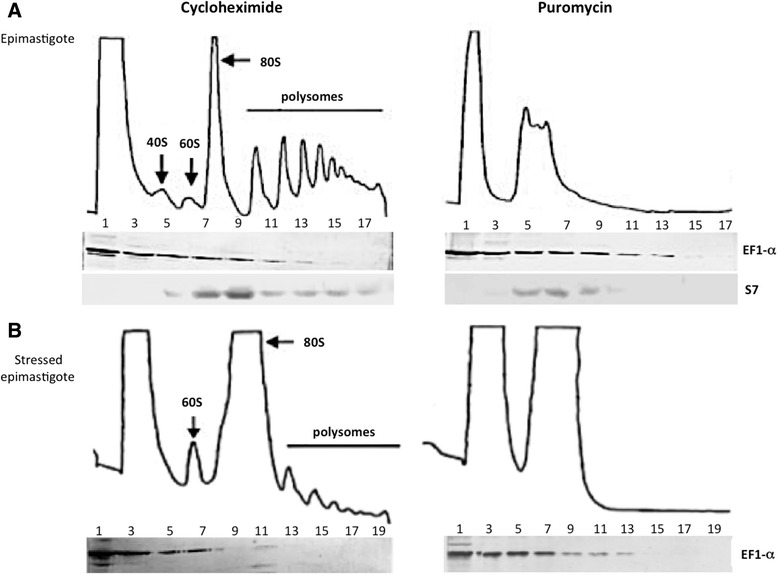
Polysome fractionation. (a) Unstressed epimastigote polysome profile. (b) Stressed epimastigote polysome profile. Fractions were analyzed by western blotting with antiserum against EF-1α (1:300 dilution) and S7 ribosomal protein (1:500). The numbers in the fractions are related to: 1 and 3 – free fraction, 5 – 40 S, 7 – 60 S, 9-17 monosomes to heavy polysomes
A typical polysome sedimentation profile was obtained from epimastigotes treated with cycloheximide, a drug that inhibits the function of the translation factor eEF2. eEF2 acts in the translocation of peptidyl-tRNA from the A site to the P site of the ribosome. Thus, cycloheximide treatment blocks translation elongation and maintains the association between ribosomes and mRNA. The EF-1α protein was detected in light fractions and in fractions corresponding to as many as 3 ribosomes (Fig. 3a). Alternatively, for polysome dissociation analysis, parasites were treated with puromycin, a drug that causes the premature release of the polypeptide chain from the ribosome. Interestingly, even after polysome dissociation by puromycin, the distribution of EF-1α in the gradient was unchanged. EF-1α migrated with the heaviest fractions under these conditions, indicating a translation machinery-independent association with large complexes (Fig. 3a).
Nutritional stress is a trigger for T. cruzi differentiation [3, 4]. In nutritionally stressed epimastigotes treated with cycloheximide, the polysome profile was characteristic of stressed cells. The polysomes were smaller and less abundant than in normal culture conditions. EF-1α protein was also present over a range of lighter to heavier fractions, but much less EF-1α was detected in the heavier fractions in stressed conditions than in unstressed epimastigotes, a finding that reflects the lower translational activity in stressed conditions (Fig. 3). The EF-1α migration pattern in stressed parasites after puromycin treatment was similar to that of unstressed puromycin-treated epimastigotes (Fig. 3b).
The results of the polysome sedimentation analysis suggested that, in addition to its association with ribosomes during translation, EF-1α is associated with heavier complexes that co-sediment with polysomes but are not necessarily associated with the translation machinery. This type of profile is typical of RNA granules that consist of mRNP complexes that are not involved in translation. One example is the mRNP complex involving the T. cruzi protein TcDHH1, which forms large complexes associated with mRNAs that co-sediment with polysomes but that are connected to the storage or degradation of associated transcripts [37]. EF-1α has a possible non-canonical function that very likely involves the formation of non-translatable mRNP complexes.
mRNAs associated with EF-1α mRNPs
Next, we aimed to investigate whether EF-1α was involved in mRNP complexes and to characterize the associated mRNAs in unstressed and nutritionally stressed epimastigotes. The ribonomic assay was performed with biological triplicates and was followed by RNA-seq. The EF-1α immunoprecipitation was confirmed by western blot showing the protein in the elution fraction (Additional file 5: Figure S2). Rabbit pre-immune serum under the same conditions was used as a control. The sequencing data were deposited in the NCBI Sequence Read Archive (SRA) database under accession number SRP051179. We used an RPKM (Reads Per Kilo base per Million) value of at least 50, an FDR (False Discovery Rate) ≤ 0.01 and a minimum of a four-fold change with respect to the control (pre-immune serum) as a threshold for selecting mRNAs that were associated with EF-1α-containing mRNPs. Using these criteria we identified 85 transcripts that were enriched for EF-1α mRNP by at least fourfold over the control (anti-EF-1α vs. pre-immune control in epimastigotes extracts) (Additional file 1: Table S1). In stressed parasites, 133 mRNAs were identified as enriched in mRNPs in comparison to the control (anti-EF-1α vs. pre-immune control in stressed epimastigotes extracts) (Additional file 2: Table S2). The RNA-seq data was validated by qRT-PCR (Additional file 6: Figure S3).
We observed specific subsets of mRNAs that were associated with EF-1α mRNPs in unstressed or stressed epimastigotes. Some transcripts were identified in both physiological conditions, while others were specific to a given condition. A possible functional correlation among the enriched transcripts was investigated using the Gene Ontology (GO) categories of the DAVID functional annotation and Blast2GO tools [38, 39] (Fig. 4 and Table 1). Upon grouping the mRNAs on the basis of the GO terms, we noticed the enrichment of terms specific to biological processes in epimastigotes and stressed epimastigotes. In unstressed epimastigotes, there was a slight enrichment in terms related to ATP binding and metal ion binding (Fig. 4 and Table 1). However, for nutritionally stressed epimastigotes, it was possible to identify a greater number of GO categories, such as single-organism metabolic processes, amino acid metabolic processes, glycolysis, glutamine metabolic processes, cobalt ion binding and iron ion binding (Fig. 4 and Table 1). The glycolysis pathway in trypanosomatids is very similar to other eukaryotes, with the exception of two main differences: some enzymes in the pathway are compartmentalized within the glycosome, and inhibitory control of hexokinase and phosphofructokinase is absent [40–42]. The compartmentalization of the enzymes seems to be important for parasite survival [43, 44]. In T. brucei, expression of phosphoglycerate kinase and triose phosphate isomerase in the cytoplasm rather than in the glycosome leads to parasite death [43, 44]. Interestingly, the mRNAs that encode the glycolytic enzymes enclosed in glycosomes were found to associate with EF-1α. Enolase, fructose-biphosphate aldolase, glucose-6-phosphate isomerase, aldolase-1 epimerase, hexokinase and alcohol dehydrogenase transcripts were also found associated with EF-1α-containing mRNPs (Additional file 1: Table S1 and Additional file 2: Table S2). It is tempting to speculate that EF-1α associates with a subset of transcripts to transport them to the vicinity of glycosomes. Such transport would allow localized translation and more efficient transport into the organelle, hence minimizing the number of active enzymes in the cytoplasm. We also analyzed the expression patterns of some transcripts that were associated with EF-1α by using the transcriptome data of the T. cruzi life cycle available at tritrypdb.org [45] (Fig. 5). An observable pattern of expression was shared by all the transcripts; namely, they are upregulated in epimastigotes (as in the case of glucose-6-phosphate isomerase, isocitrate dehydrogenase and lanosterol 14-alpha-demethylase), and they all encode enzymes associated with carbohydrate and lipid metabolism (Fig. 5).
Fig. 4.
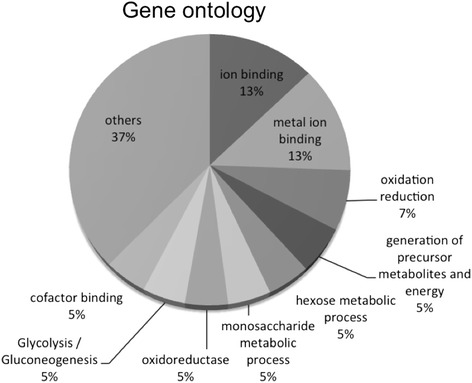
Gene Ontology pie chart. The percentage indicates the enrichment for the GO terms in the transcripts associated to EF-1α in epimastigotes and stressed epimastigotes
Table 1.
Gene ontology terms for EF-1α associated transcripts
| Gene ontology | Count | Enrichment score | p-value | Benjamini |
|---|---|---|---|---|
| Epimastigote | ||||
| Nucleotide binding | 8 | 0.83 | 1.00E-01 | 9.80E-01 |
| Kinase | 5 | 0.83 | 1.50E-01 | 9.70E-01 |
| ATP binding | 6 | 0.56 | 1.90E-01 | 9.80E-01 |
| Transition metal ion binding | 3 | 0.1 | 7.20E-01 | 9.90E-01 |
| Stressed epimastigote | ||||
| Generation of precursor metabolites and energy | 7 | 2.84 | 4.30E-02 | 3.00E-04 |
| Monosaccharide metabolic process | 6 | 2.84 | 4.50E-02 | 2.50E-03 |
| Glutamine family amino acid metabolic process | 4 | 1.59 | 5.20E-02 | 1.10E-03 |
| Proteolysis | 7 | 1.07 | 9.70E-01 | 3.10E-01 |
Fig. 5.
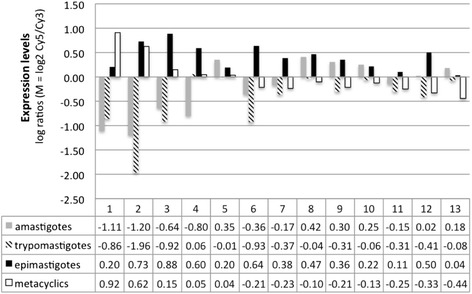
mRNA expression levels of the T. cruzi life cycle stages (trypomastigotes, amastigotes, epimastigotes, and metacyclic trypomastigotes) determined by microarray analysis [45]. Six hybridizations were performed and consisted of three dye-swap experiments from three independent samples (biological replicates). In each case, the experimental sample was from a single life-cycle stage and the control sample was an equal mixture of all four life-cycle stages [45]. Expression values for 2-channel microarray experiments are shown as log ratios (M = log2 Cy5/Cy3) [45]. The numbers refer to different mRNAs: 1- TcCLB.510879.130 (hypothetical protein); 2- TcCLB.503975.50 (hypothetical protein); 3- TcCLB.509599.130 (serine/threonine protein kinase); 4- TcCLB.504163.60 (hypothetical protein); 5- TcCLB.509455.114 (hypothetical protein); 6- TcCLB.504005.54 (hypothetical protein); 7- TcCLB.508719.70 (flagellum targeting protein Kharon 1); 8- TcCLB.511365.80 (mitochondrial carrier protein); 9- TcCLB.506825.200 (hypothetical protein); 10- TcCLB.506529.324 (hypothetical protein); 11- TcCLB.506529.508 (glucose-6-phosphate isomerase); 12- TcCLB.510535.100 (cysteine C peptidase CPC); 13- TcCLB.511801.60 (mitochondrial DEAD-box helicase)
The RNA regulon theory, which states that mRNAs encoding proteins with related functions are associated with specific mRNP complexes [6, 16, 17] could explain the results obtained for EF-1α in T. cruzi, however further experiments need to be performed in order to validate this hypothesis for EF-1α. One example of a regulon in T. cruzi is the TcZC3H39-mRNP, which binds a specific subset of mRNAs that encode the cytochrome oxidase C complex. Interestingly, this association occurs in response to stress [5]. A comparison of transcripts associated with EF-1α and with TcZC3H39 show no target overlap, indicating that the transcripts associated with each protein form distinct mRNPs in the cell.
The stress triggers a global rearrangement of the metabolism in the cell, including the translation machinery. Some RNAs shift from the polysomes to RNA granules as p-bodies or stress granules while others remain associated to polysomes and are actively translated, for example heat shock proteins and chaperones, as a translational reprogramming necessary in order to rapidly respond to the stress condition [19, 20]. The shifts observed on the targets associated to EF1-α protein suggest that this protein might play a non-canonical role in T.cruzi through participation in distinct mRNP complexes, such as its potential role in localized translation in epimastigotes or translation repression in parasites under nutritional stress conditions (Additional file 1: Table S1, Additional file 2: Table S2 and Additional file 6: Figure S3).
The mRNA and protein levels in a cell are not necessarily correlated. Recent data using polysome-profiling techniques allow a more precise quantification of the mRNAs actively engaged in translation [46–48]. The transcripts stability is not sufficient to explain the level of the encoded proteins since this is a reflex of translation efficiency. However, the role of RNA binding proteins associated to mRNP complexes cannot be underestimated since they ultimately define the fate of a given mRNA as being translated, stored or degraded.
Conclusions
Specific subsets of mRNAs were enriched in EF-1α-containing mRNPs, indicating a non-canonical role for this protein in T. cruzi. We are aware that EF-1α is a rather abundant and positively charged cytoplasmic protein that might bind nonspecifically to mRNAs. However, the experimental conditions used in the previous work (isolation of polysomal and non-polysomal mRNPs with 300 mM NaCl) minimize spurious and non-specific binding between proteins and mRNAs [33]. Sucrose gradient sedimentation analysis showed that EF-1α migrated to the heavier fractions of the gradient irrespective of whether polysomes were dissociated with puromycin, indicating its association with large mRNP complexes. The mRNAs bound to EF-1α mRNPs were enriched for metabolic processes; notably, six of these transcripts encoded glycosomal proteins that are involved in the glycolysis pathway. This result indicates that in T. cruzi, as well as in other eukaryotes, EF-1α might have additional functions other than in translation. The results presented here allow us to hypothesize that EF-1α acts by associating with transcripts that must be localized to specific cellular sites for translation and during the stress response might be acting in mRNPs that associates to translation repression. Our findings shed new light on the understanding of post-transcriptional regulation in T. cruzi.
Methods
EF-1α cloning and antibody production
The complete coding sequence of the EF-1α gene (TcCLB.510119.9 and TcCLB508949.4) was amplified by PCR using the forward primer ATGGGGAAGGACAAGGTG and the reverse primer GGACTTGATCGACTTGGG. The amplicon was cloned into the pDNOR vector from the Gateway platform. The gene was then sub-cloned into the pDEST15 expression vector to allow recombinant His-tagged protein purification. The recombinant protein was expressed in an Escherichia coli DH5α pLysS strain and purified using an electron affinity column. The purified recombinant protein was then inoculated into rabbits to obtain polyclonal antibodies [3].
Parasite cultures
T. cruzi Dm 28c epimastigotes were cultured in liver infusion tryptose (LIT) medium at 28 °C. Epimastigotes in the late exponential growth phase were obtained from five-day cultures (density of 5 × 107 parasites ml−1). For stress conditions, epimastigotes from five-day cultures were harvested by centrifugation at 7,000 × g for 5 min at 10 °C and then incubated for 2 h at 28 °C in TAU medium (190 mM NaCl, 17 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 8 mM phosphate buffer, pH 6.0) at a density of 5 × 108 parasites ml−1. The assay for the parasite life cycle was conducted as previously described [3, 4].
Western blot analysis
For western blot analysis, T. cruzi extracts were separated by SDS-PAGE on a 15 % polyacrylamide gel; the separated bands were then transferred to a nitrocellulose membrane. Nonspecific binding sites were blocked by incubating the membrane with 5 % non-fat milk powder and 0.1 % Tween 20 in PBS for 30 min. The membrane was then incubated for 1 h with specific antibodies against EF-1α diluted at 1:500, thoroughly washed in PBS, and incubated with goat alkaline phosphatase-conjugated anti-rabbit IgG (Sigma) diluted at 1:10,000. The color reaction was developed using 5-bromo-4-chloro-3-indozyl phosphate and nitroblue tetrazolium (Promega). Quantification of bands was performed using ImageJ software (Bethesda, MD, USA).
Immunofluorescence and imaging
Immunofluorescence assays were conducted as previously described [46]. Image capture and deconvolution were performed using the Leica AF6000 Modular System with LAS AF 3.x.
Sucrose density gradient separation
T. cruzi polysomes were isolated on sucrose gradients. Cells (1 × 109) were incubated with 100 μg ml−1 cycloheximide for 10 min or with 2 mM puromycin for one hour. They were kept on ice for 5 min before being pelleted by centrifugation (7,000 × g for 5 min at 4 °C) and washed with cold TKM buffer (10 mM Tris, pH 7.4, 300 mM KCl and 10 mM MgCl2). The cell pellet was resuspended in 900 μl TKM supplemented with 10 μg ml−1 heparin, 10 μM E-64, and 1:100 EDTA-free protease cocktail (Roche). The suspension was transferred to a new tube containing 100 μl of lysis buffer (TKM supplemented with 10 % (v/v) NP-40 and 2 M sucrose), and homogenized by repeated passages through a pipette. Lysis was monitored using phase-contrast microscopy. The lysate was centrifuged at 18,000 × g at 4 °C for 5 min. The cleared supernatant (500 μl; equivalent to 5 × 108 cells) was layered onto linear 15-to-55 % sucrose density gradients prepared in TKM buffer supplemented with inhibitors (10 μM E-64, 1 mM PMSF, and 1 mg ml−1 heparin) and centrifuged at 4 °C for 2 h at 365,000 × g in a Beckman SW41 rotor. After centrifugation, 500-μl fractions were collected using the ISCO gradient fractionation system.
EF-1α immunoprecipitation RNA-seq
To identify mRNAs associated with EF-1α protein or complexes, rabbit anti-EF-1α antibody (15 μl) was incubated with 50 μl of goat anti-rabbit magnetic beads (New England Biolabs) and 40 U/ml RNAse OUT (Invitrogen) for 16 h at 4 °C with shaking. Cytoplasmic extract corresponding to 1 × 109 cells was then incubated with the beads. The beads were then washed three times with IMP1 buffer. The immunoprecipitated RNAs were eluted and purified with the RNeasy® (Qiagen) kit by using the “Animal Cells I” protocol in the manufacturer’s manual but with an additional column-based DNase treatment step. The purified RNAs were then subjected to deep sequencing using the SOLiD 3 plus platform. The experiments were performed in triplicate and the rabbit pre-immune serum was used as a control in the same conditions as the immune serum.
In silico data analysis
The sequencing data obtained were analyzed using the CLC Genomics Workbench© v 5.5.1. The reads were trimmed on the basis of quality using a threshold phred score of 15. The T. cruzi reference genome used for mapping was obtained from the NCBI database (AAHK01), and the alignment was performed with the following parameters: additional upstream and downstream sequences of 100 bases; minimum number of reads, 10; maximum number of mismatches, 2; nonspecific match limit, −2; use of colorspace encoding. The minimum similarity of the reads mapped to the reference genome was 70 %. We selected possible targets of EF-1α mRNPs by using the beta binomial statistical test [47], with a p-value-corrected FDR of ≤ 1 % and a minimum four-fold change with respect to the control (pre-immune serum) as the threshold for significance.
Functional annotation
To conduct the gene ontology term enrichment analysis used for the classification of transcripts and to conduct Kegg pathway map analysis we used the DAVID annotation tool (david.abcc.ncifcrf.gov/home.jsp) [48] and Blast2GO [38]. For enrichment analysis and functional annotation, the statistical p-value was based on the EASE score, a modified one-tailed Fisher’s exact test p-value. An EASE value of 0 represents perfect enrichment, while values of up to 0.05 were considered to indicate strong enrichment in the annotation categories considered.
Acknowledgments
This work was supported by the Conselho Nacional do Desenvolvimenmto Científico e Tecnológico (CNPq), Fundação Oswaldo Cruz (Fiocruz, PAPES), Fundação Araucária and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). SG is a research fellow from CNPq.
We thank Vanessa Huf Sane for excellent technical assistance and Drs. Bruno Dallagiovanna and Alejandro Correa for helpful discussions.
Additional files
RNAs associated to EF-1a in epimastigotes.
RNAs associated to EF-1a in stressed epimastigotes.
Supplemental methods.
Controls of the immunofluorescence assay.
EF-1a immunoprecipitation western-blot.
Validation of the RNA-seq by qRT-PCR.
Primers used in this study.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SG: experimental design, interpretation of data and manuscript preparation; LRA and CO: experimental design and procedures, interpretation of data and manuscript preparation. All authors approved the final manuscript.
References
- 1.de Souza W, de Carvalho TM, Barrias ES. Review on Trypanosoma cruzi: Host Cell Interaction. Int J Cell Biol. 2010. doi:10.1155/2010/295394. [DOI] [PMC free article] [PubMed]
- 2.de Souza W. Trypanosoma cruzi-Host Cell Interaction. Front Immunol. 2014;5:339. doi: 10.3389/fimmu.2014.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonaldo MC, Souto-Padron T, de Souza W, Goldenberg S. Cell-substrate adhesion during Trypanosoma cruzi differentiation. J Cell Biol. 1998;106:1349–58. [DOI] [PMC free article] [PubMed]
- 4.Contreras VT, Salles JM, Thomas N, Morel CM, Goldenberg S. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol Biochem Parasitol. 1985;16(3):315–27. [DOI] [PubMed]
- 5.Alves LR, Oliveira C, Mörking PA, Kessler RL, Martins ST, Romagnoli BA, Marchini FK, Goldenberg S. The mRNAs associated to a zinc finger protein from Trypanosoma cruzi shift during stress conditions. RNA Biol. 2014;11(7):921–33. doi: 10.4161/rna.29622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldenberg S, Avila AR. Aspects of Trypanosoma cruzi stage differentiation. Adv Parasitol. 2011;75:285–305. doi: 10.1016/B978-0-12-385863-4.00013-7. [DOI] [PubMed] [Google Scholar]
- 7.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156(2):93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Alves LR, Guerra-Slompo EP, de Oliveira AV, Malgarin JS, Goldenberg S, Dallagiovanna B. mRNA localization mechanisms in Trypanosoma cruzi. PLoS One. 2013;8(12):e81375. doi: 10.1371/journal.pone.0081375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21(8):1881–8. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caro F, Ahyong V, Betegon M, DeRisi JL. Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. Elife 2014, 3. [DOI] [PMC free article] [PubMed]
- 12.Vasquez JJ, Hon CC, Vanselow JT, Schlosser A, Siegel TN. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res. 2014;42(6):3623–37. doi: 10.1093/nar/gkt1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen BC, Ramasamy G, Vasconcelos EJR, Ingolia NT, Myler PJ, Parsons M. Extensive stage-regulation of translation revealed by ribosome profiling of Trypanosoma brucei. BMC Genomics. 2014;15:911. doi: 10.1186/1471-2164-15-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keene JD. Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol. 2007;72:157–65. doi: 10.1101/sqb.2007.72.013. [DOI] [PubMed] [Google Scholar]
- 15.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8(7):533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 16.Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics. 2009;10:495. doi: 10.1186/1471-2164-10-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouellette M, Papadopoulou B. Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol. 2009;8(11):100. doi: 10.1186/jbiol203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–8. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 19.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4(9):a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8(1):9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen L, Andersen GR, Knudsen CR, Kinzy TG, Nyborg J. Crystallization of the yeast elongation factor complex eEF1A-eEF1B alpha. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 1):159–61. doi: 10.1107/S0907444900015559. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki H, Patel V, Wang H, Ensley JF, Gutkind JS, Yeudall WA. Growth factor-sensitive molecular targets identified in primary and metastatic head and neck squamous cell carcinoma using microarray analysis. Oral Oncol. 2006;42(3):240–56. doi: 10.1016/j.oraloncology.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Murray JW, Edmonds BT, Liu G, Condeelis J. Bundling of actin filaments by elongation factor 1 alpha inhibits polymerization at filament ends. J Cell Biol. 1996;135(5):1309–21. doi: 10.1083/jcb.135.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12(9):772–8. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 25.Lau J, Castelli LA, Lin EC, Macaulay SL. Identification of elongation factor 1alpha as a potential associated binding partner for Akt2. Mol Cell Biochem. 2006;286(1-2):17–22. doi: 10.1007/s11010-005-9006-5. [DOI] [PubMed] [Google Scholar]
- 26.Liu G, Tang J, Edmonds BT, Murray J, Levin S, Condeelis J. F-actin sequesters elongation factor 1alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996;135(4):953–63. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thornton S, Anand N, Purcell D, Lee J. Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J Mol Med (Berl) 2003;81(9):536–48. doi: 10.1007/s00109-003-0461-8. [DOI] [PubMed] [Google Scholar]
- 28.Tatsuka M, Mitsui H, Wada M, Nagata A, Nojima H, Okayama H. Elongation factor-1 alpha gene determines susceptibility to transformation. Nature. 1992;359(6393):333–6. doi: 10.1038/359333a0. [DOI] [PubMed] [Google Scholar]
- 29.Kato MV, Sato H, Nagayoshi M, Ikawa Y. Upregulation of the elongation factor-1alpha gene by p53 in association with death of an erythroleukemic cell line. Blood. 1997;90(4):1373–8. [PubMed] [Google Scholar]
- 30.Blanch A, Robinson F, Watson IR, Cheng LS, Irwin MS. Eukaryotic translation elongation factor 1-alpha 1 inhibits p53 and p73 dependent apoptosis and chemotherapy sensitivity. PLoS One. 2013;8(6):e66436. doi: 10.1371/journal.pone.0066436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Grant WM, Persky D, Latham VM, Singer RH, Condeelis J. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol Biol Cell. 2002;13(2):579–92. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18(1):251–65. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alves LR, Avila AR, Correa A, Holetz FB, Mansur FC, Manque PA, de Menezes JP, Buck GA, Krieger MA, Goldenberg S. Proteomic analysis reveals the dynamic association of proteins with translated mRNAs in Trypanosoma cruzi. Gene. 2010;452(2):72–8. doi: 10.1016/j.gene.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 34.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309(5733):409–15. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 35.Negrutskii BS, El'skaya AV. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acid Res Mol Biol. 1998;60:47–78. doi: 10.1016/S0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- 36.de Godoy LM, Marchini FK, Pavoni DP, Rampazzo RC, Probst CM, Goldenberg S, Krieger MA. Quantitative proteomics of Trypanosoma cruzi during metacyclogenesis. Proteomics. 2012;12(17):2694–703. doi: 10.1002/pmic.201200078. [DOI] [PubMed] [Google Scholar]
- 37.Holetz FB, Alves LR, Probst CM, Dallagiovanna D, Marchini FK, Manque P, Buck G, Krieger MA, Correa A, Goldenberg S. Protein and mRNA content of TcDHH1-containing mRNPs in Trypanosoma cruzi. Febs Journal. 2010;277(16):3415–3426. doi: 10.1111/j.1742-4658.2010.07747.x. [DOI] [PubMed] [Google Scholar]
- 38.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Tálon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 40.Cazzulo JJ. Energy metabolism in Trypanosoma cruzi. Subcell Biochem. 1992;18:235–57. doi: 10.1007/978-1-4899-1651-8_7. [DOI] [PubMed] [Google Scholar]
- 41.Maugeri DA, Cannata JJ, Cazzulo JJ. Glucose metabolism in Trypanosoma cruzi. Essays Biochem. 2011;51:15–30. doi: 10.1042/bse0510015. [DOI] [PubMed] [Google Scholar]
- 42.Parsons M. Glycosomes: parasites and the divergence of peroxisomal purpose. Mol Microbiol. 2004;53(3):717–24. doi: 10.1111/j.1365-2958.2004.04203.x. [DOI] [PubMed] [Google Scholar]
- 43.Blattner J, Helfert S, Michels P, Clayton C. Compartmentation of phosphoglycerate kinase in Trypanosoma brucei plays a critical role in parasite energy metabolism. Proc Natl Acad Sci U S A. 1998;95(20):11596–600. doi: 10.1073/pnas.95.20.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helfert S, Estévez AM, Bakker B, Michels P, Clayton C. Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei. Biochem J. 2001;357(Pt 1):117–25. doi: 10.1042/0264-6021:3570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minning TA, Weatherly DB, Atwood J, Orlando R, Tarleton RL. The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics. 2009;10:370. doi: 10.1186/1471-2164-10-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holetz FB, Correa A, Ávila AR, Nakamura Krieger MA, Goldenberg S. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem Biophys Res Commun. 2007;356(4):1062–7. doi: 10.1016/j.bbrc.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 47.Baggerly KA, Deng L, Morris JS, Aldaz CM. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics. 2003;19(12):1477–83. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- 48.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]


