Abstract
Recently, we hypothesized that silencing suppression activity gained by a viral replicative helicase led to the emergence of the second helicase possessing activity of the viral silencing suppressor and/or movement protein (MP). Our hypothesis accounted for the evolutionary origin of the specialized ‘triple gene block’ (TGB) in plant virus genomes encoding the MPs TGB1, TGB2, and TGB3 required for viral cell-to-cell transport through plasmodesmata. Here, we used public transcriptome databases to identify previously unrecognized viruses. The analysis of novel viral genomes further supported the previously proposed scenario of TGB origin and evolution, which included the following steps. First, the accessory helicase gene could have been acquired by horizontal gene transfer (HGT) presumably occured independently in different virus groups. Second, the TGB2 gene evolved by HGT or autonomization of the C-terminal transmembrane domain found in at least one TGB1 helicase. Third, the TGB3 gene has most likely emerged in the genomic block consisting of the TGB1 and TGB2 genes.
Keywords: plant virus, virus genome, RNA helicase, evolution, movement protein, triple gene block
Introduction
For replicative RNA/DNA synthesis, plant viruses encode enzymes belonging to one of three helicase superfamilies (SFs), SF-I, SF-II, and SF-III (Koonin and Dolja, 1993; Iyer et al., 2004; Hickman and Dyda, 2005; Jankowsky and Fairman, 2007; Byrd and Raney, 2012; Gilhooly et al., 2013). Participation of replicative helicases of all three SFs in virus cell-to-cell movement and silencing suppression is documented (Wang et al., 2012, 2014; Sorel et al., 2014). Particularly, the replicative SF-I RNA helicase domain of distantly related tobamoviruses may play an important role in the suppression of silencing (Kubota et al., 2003; Ding et al., 2004; Csorba et al., 2007; Wang et al., 2012). Recently, assuming the participation of plant virus helicases in non-replicative functions, we proposed a hypothetic scheme, which explains evolving the triple gene block (TGB) in plant viruses (Morozov and Solovyev, 2012). TGB is a module consisting of three genes termed TGB1, TGB2, and TGB3, which encode movement proteins (MPs) necessary for viral cell-to-cell transport. TGB2 and TGB3 encode integral membrane proteins, whereas the TGB1 protein contains a helicase domain, which belongs to a diverged lineage of viral SF-I helicases (Morozov and Solovyev, 2003; Verchot-Lubicz et al., 2010; Solovyev et al., 2012). In agreement with the earlier view that the TGB1 protein can be regarded as an “accessory” helicase evolved after duplication of a replicative RNA helicase (Koonin and Dolja, 1993), we hypothesized that gaining new silencing suppression function by a replicative RNA helicase could precede subsequent helicase domain duplication concomitant with its autonomization, which can occur both in cis (in the context of the same viral genome) or in trans (implying transfer to a foreign virus genome). These events may result in evolving specialized second helicase possessing the activity of viral silencing suppressor (VSR), or, taking into account a tight link between viral movement and silencing suppression, both suppression and movement functions (Burgyán and Havelda, 2011; Morozov and Solovyev, 2012; Pumplin and Voinnet, 2013). Examples of such helicases are provided by potexvirus and carlavirus TGB1 proteins having the VSR function additional to their cell-to-cell movement function (Bayne et al., 1995; Senshu et al., 2011). Further evolution could be accompanied by loss of VSR function by the replicative helicase. We assumed that further TGB1 specialization as a dedicated MP could be accompanied by acquisition of TGB2 and TGB3 genes facilitating the TGB1-mediated cell-to-cell transport (Morozov and Solovyev, 2012).
Based on sequence comparisons and functional studies, two types of TGB were distinguished, a ‘potex-like’ TGB encoded by filamentous viruses of the families Alphaflexiviridae and Betaflexiviridae and a ‘hordei-like TGB’ found in rigid rod-shaped viruses of the family Virgaviridae and the unassigned genus Benyvirus. Apart from these well-characterized TGB-containing viruses, the second “accessory” helicase gene was identified in genomes of a number of diverse unclassified plant viruses of different genome organization and particle morphology, particularly, Nicotiana velutina mosaic virus (NVMV) and Hibiscus green spot virus (HGSV; Morozov and Solovyev, 2003, 2012; Verchot-Lubicz et al., 2010).
In recent years, the identification of previously unknown viral genomes has been greatly accelerated with high-throughput sequencing technologies. New-generation sequencing of plant transcriptomes often gives rise, in addition to species-specific libraries of mRNA sequences assembled from many individual reads (Johnson et al., 2012; Xie et al., 2014), to virus-like RNA assemblies (VLRA) corresponding to genomic RNAs of viruses infecting these host plants. Information obtained from VLRA, full-length or even partial, can be used to discover new viral proteins and domains. As a result, numerous novel genotypes of eukaryotic viruses were identified in last several years (Rosario and Breitbart, 2011; Cook et al., 2013; Junglen and Drosten, 2013; Roossinck, 2015). In this paper, in an attempt to further resolve potential evolutionary relations between distant TGB-containing viruses, we performed new comparative sequence analyses of “accessory” helicases encoded in previously sequenced plant virus genomes, including NVMV and HGSV, and VLRAs found in public databases, particularly, the 1000 Plants (1KP) project (Matasci et al., 2014).
Nicotiana Velutina Mosaic Virus
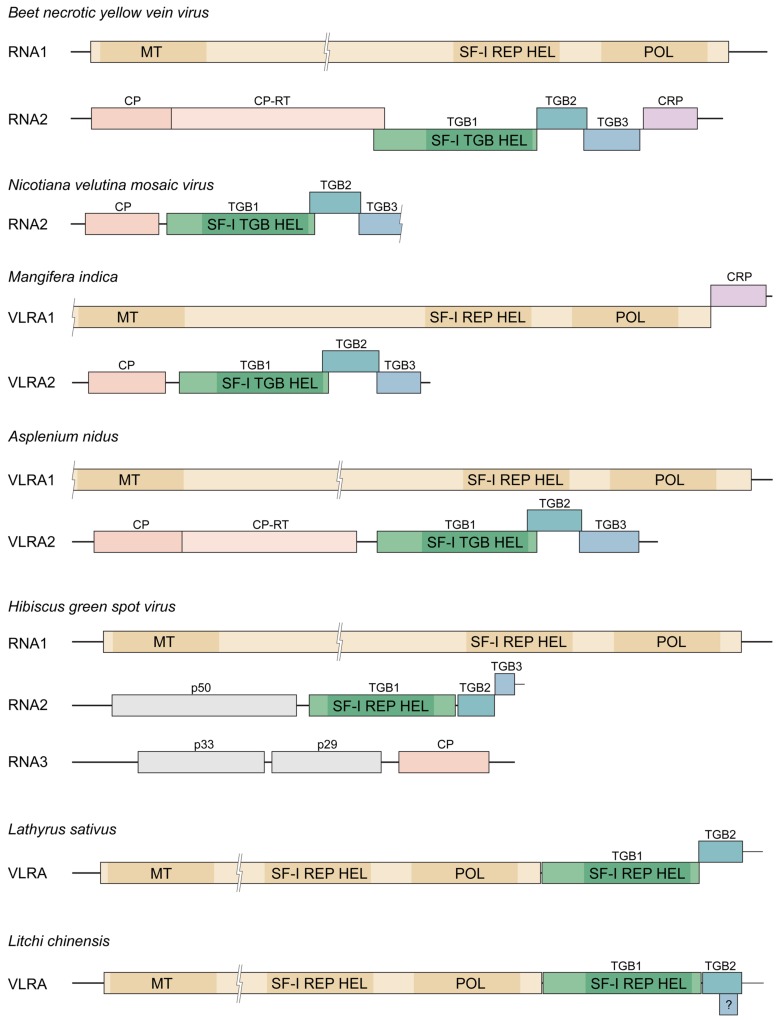
Nicotiana velutina mosaic virus genome consists of RNA1 (8 Kb) and RNA2 (3 Kb), which are encapsidated in rigid, rod-shaped particles (Randles and Rohde, 1990). NVMV RNA1 sequence is unavailable, while incomplete sequence of RNA2 revealed four open reading frames (ORFs; (Figure 1). The 5′-terminal ORF1 encodes a protein showing relatively weak but significant similarity to coat proteins (CPs) of viruses of the genus Benyvirus such as Beet necrotic yellow vein virus (BNYVV), whereas the ORF2 protein was also found to have closest relationship to benyvirus TGB1 “accessory” helicases (identity 28–30%; Randles and Rohde, 1990). NVMV ORF3 protein represents typical TGB2 protein with two terminal hydrophobic regions and highly conserved hydrophilic signature in the central part. Although the NVMV ORF4 was sequenced only partly, the encoded protein was found to have benyvirus-like TGB3 organization with two hydrophobic regions at N- and C-termini (Randles and Rohde, 1990; Morozov and Solovyev, 2003). However, NVMV TGB3 had only marginal (if any) sequence relationship to benyvirus TGB3 proteins in its central hydrophilic region. It should be noted that this region is variable even in known benyvirus TGB3 sequences (Kondo et al., 2013).
FIGURE 1.
Triple gene block (TGB) in genomes of different plant viruses and virus-like RNA assemblies (VLRAs). Genes are shown as boxes, and molecular masses of the encoded proteins with unknown function are indicated. TGB is shown in green. Genes of replicative proteins are shown in yellow. The locations of conserved protein sequence domains of methyltransferase (MT), helicase (HEL) and polymerase (POL) are indicated by shading. Other domains in replicative proteins are not shown. “SF-I REP HEL” denotes the superfamily I helicase domain typical for replicase proteins, “SF-I TGB HEL” – the superfamily I helicase domain typical for TGB1 proteins. CP, coat protein gene; CP-RT, readthrough domain of CPs; CRP, genes of cysteine-rich proteins. “?” indicates the short open reading frame (ORF) in Lc-VLRA potentially representing TGB3 gene.
TBLASTN search of NCBI transcriptome shotgun assembly (TSA) database with NVMV CP and ORF2 protein as queries revealed their significant similarity to proteins encoded by a single VLRA (size 2523 nucleotides) of mango (Mangifera indica; Table 1). This mango VLRA (referred to as Mi-VLRA2) contains four ORFs (Figure 1); ORF1 and ORF2 code for proteins similar to NVMV CP and TGB1, respectively, whereas ORF3-encoded protein can be aligned with the NVMV TGB2 protein (identity 47%; Table 1). The mango VLRA2 ORF4 protein has the typical TGB3 organization with the N- and C-terminal hydrophobic segments (Solovyev et al., 2012), but only marginal sequence similarity to the NVMV TGB3 protein that is not surprising in view of low sequence conservation between benyvirus and NVMV TGB3 proteins. Thus Mi-VLRA2 is similar to NVMV RNA2 both in genetic organization and the encoded proteins.
Table 1.
Comparisons of amino acid sequence identities of the proteins encoded by Triple gene block (TGB) containing virus genomes and virus-like RNA assemblies (VLRAs).
| Query open reading frames (ORF) | Subject ORF | E-value | Maximal amino acid identity (%) | Nucleotide accession numbers |
|---|---|---|---|---|
| Mangifera indica Mi-VLRA1 | NCBI GBCV01010547 | |||
| Replication protein∗∗ | Rice stripe necrosis benyvirus Replication protein | 3e-171 | 61 | NCBI EU099844 |
| Burdock mottle benyvirus Replication protein | 3e-167 | 58 | NCBI AB818898 | |
| Beet necrotic yellow vein benyvirus Replication protein | 3e-166 | 57 | NCBI EU330453 | |
| Beet soil-borne mosaic benyvirus Replication protein | 8e-165 | 56 | NCBI JF513082 | |
| Mangifera indica Mi VLRA2 | NCBI GBCV01004715 | |||
| Coat protein (CP) | Nicotiana velutina mosaic virus (NVMV) CP | 7e-26 | 40 | NCBI D00906 |
| Rice stripe necrosis benyvirus CP | 2e-07 | 30 | NCBI EU099845 | |
| Beet necrotic yellow vein benyvirus CP | 5e-04 | 26 | NCBI AY771345 | |
| Beet virus Qpomovirus CP | 0.003 | 25 | NCBI EU785970 | |
| TGB1 | NVMV TGB1 | 3e-42 | 33 | NCBI D00906 |
| Burdock mottle benyvirus TGB1 | 4e-28 | 33 | NCBI AB818899 | |
| Rice stripe necrosis benyvirus TGB1 | 1e-27 | 31 | NCBI EU099845 | |
| Beet necrotic yellow vein benyvirus TGB1 | 3e-23 | 28 | NCBI X75575 | |
| Beet soil-borne mosaic benyvirus TGB1 | 3e-23 | 27 | NCBI JF513083 | |
| TGB2∗ | NVMV TGB2 | 3e-13 | 52 | NCBI D00906 |
| Potato mop-top pomoviru TGB2 | 1e-06 | 34 | NCBI JX885628 | |
| Broad bean necrosis pomovirus TGB2 | 1e-04 | 32 | NCBI D86638 | |
| Asplenium nidus An-VLRA1 | 1KP database PSKY-2058769 | |||
| Replication protein∗∗ | Rice stripe necrosis benyvirus Replication protein | 2e-153 | 34 | NCBI EU099844 |
| Burdock mottle benyvirus Replication protein | 5e-143 | 33 | NCBI AB818898 | |
| Beet necrotic yellow vein benyvirus Replication protein | 5e-139 | 31 | NCBI EU330453 | |
| Beet soil-borne mosaic benyvirus Replication protein | 3e-136 | 31 | NCBI JF513082 | |
| A. nidus An-VLRA2 | 1KP database PSKY-2058768 | |||
| CP | Peanut clump pecluvirus CP | 3.3 | 27 | NCBI NP620028 |
| Ribgrass mosaic virus CP | 2.1 | 31 | NCBI CAJ13878 | |
| Tobacco mosaic virus CP | 3.2 | 26 | NCBI AAA46587 | |
| TGB1 | Burdock mottle benyvirus TGB1 | 1e-29 | 35 | NCBI AB818899 |
| Rice stripe necrosis benyvirus TGB1 | 8e-29 | 31 | NCBI EU099845 | |
| Beet soil-borne mosaic benyvirus TGB1 | 4e-24 | 31 | NCBI JF513083 | |
| Beet necrotic yellow vein benyvirus TGB1 | 3e-23 | 31 | NCBI X75575 | |
| NVMV TGB1 | 2e-22 | 29 | NCBI D00906 | |
| TGB2∗ | Beet necrotic yellow vein benyvirus TGB2 | 5e-10 | 41 | NCBI AY682694 |
| Rice stripe necrosis benyvirus TGB2 | 5e-10 | 44 | NCBI EU099845 | |
| Burdock mottle benyvirus TGB2 | 1e-07 | 36 | NCBI AB818899 | |
| Litchi chinensis Lc-VLRA | 1KP database WAXR-2010981 | |||
| TGB1 | Hibiscus green spot virus (HGSV) TGB1 | 4e-36 | 37 | NCBI HQ852053 |
| Burdock mottle benyvirus Replication HEL | 1e-13 | 32 | NCBI AB818898 | |
| Rice stripe necrosis benyvirus Replication HEL | 3e-11 | 28 | NCBI EU099844 | |
| Beet soil-borne mosaic benyvirus Replication HEL | 1e-10 | 29 | NCBI JF513082 | |
| Beet necrotic yellow vein benyvirus Replication HEL | 2e-09 | 28 | NCBI EU330453 | |
| Ferret hepatitis E virus Replication HEL | 2e-08 | 25 | NCBI AB890001 | |
| Replication protein∗∗∗ | Beet soil-borne mosaic benyvirus Replication protein | 5e-110 | 32 | NCBI JF513082 |
| Burdock mottle benyvirus Replication protein | 5e-70 | 31 | NCBI AB818898 | |
| Lathyrus sativus Ls-VLRA | NCBI GBSS01016353 | |||
| TGB1 | HGSV TGB1 | 2e-37 | 34 | NCBI HQ852053 |
| Rice stripe necrosis benyvirus Replication HEL | 2e-10 | 29 | NCBI EU099844 | |
| Beet necrotic yellow vein benyvirus Replication HEL | 8e-10 | 27 | NCBI EU330453 | |
| Beet soil-borne mosaic benyvirus Replication HEL | 4e-09 | 27 | NCBI JF513082 | |
| Replication protein∗∗∗ | Burdock mottle benyvirus Replication protein | 9e-87 | 33 | NCBI AB818898 |
| Beet necrotic yellow vein benyvirus Replication protein | 6e-72 | 34 | NCBI EU330453 |
∗more than 60% query coverage.
∗∗POL domain only.
∗∗∗MET, HEL, and POL domains.
Interestingly, NVMV TGB1 was additionally found to have a distant similarity to a protein encoded by another mango VLRA1 (size 4934 nucleotides). Mi-VLRA1 contains two ORFs (Figure 1). The 5′-terminal large ORF includes methyl/guanylyltransferase (MET), cysteine protease (PRO), SF-I helicase (HEL), and RNA polymerase (POL) domains similar to those in the replication protein of benyviruses (Table 1), whereas the 3′-terminal ORF show distant relationship to benyvirus cysteine-rich proteins (data not shown). We speculate that Mi-VLRA1 and Mi-VLRA2 may represent a new genome type of TGB-possessing viruses having larger and smaller genome components organized similarly to pecluvirus RNA1 and hordeivirus RNAβ, respectively, (Morozov and Solovyev, 2003). In the absence of NVMV RNA1 sequence, possible relation of this putative mango virus to NVMV remains to be investigated. We found no similarity of nucleotide sequences at the 5′ and 3′ termini between longer and shorter mango VLRAs that may be a result of incomplete sequencing of terminal regions.
TBLASTN search of 1KP database (www.onekp.com), which contains around 1000 plant transcriptomes, with NVMV ORF2 protein as a query showed significant similarity to a single VLRA (size 4138 nucleotides) of fern Asplenium nidus (Table 1). This fern VLRA (referred to as An-VLRA2) contains five ORFs similar to benyvirus RNA2 genes in both sequences of encoded proteins and linear arrangement excepting the lack of 3′-terminal CRP gene in An-VLRA2 (Figure 1; Kondo et al., 2013). The encoded proteins are related also to those of beny-, pomo-, and pecluviruses (Table 1). Strikingly, the putative TGB3 protein encoded by fern An-VLRA2, despite the characteristic benyvirus-type TGB3 organization with two terminal hydrophobic segments (Solovyev et al., 2012), showed a weak similarity to the central hydrophilic region of TGB2 proteins encoded by potex- and carlaviruses rather than to any known TGB3 protein (data not shown). This observation reinforces our previous conclusion on possible diverse evolutionary origins of TGB3 sequences in distant viruses (Morozov and Solovyev, 2003).
Nicotiana velutina mosaic virus TGB1 protein was also found to have a distant similarity to a protein encoded by another A. nidus An-VLRA1 (size 6960 nucleotides). The fern An-VLRA1 contains a single ORF (Figure 1), which is obviously similar to the replication protein of benyviruses in its HEL, PRO, and POL domains (Table 1). We speculate that the two fern VLRAs represent a new viral genome. As in the case of mango VLRAs, we found no similarity of nucleotide sequences at the 5′ and 3′ termini between longer and shorter fern VLRAs.
Hibiscus Green Spot Virus
Hibiscus green spot virus (proposed genus Higrevirus) genome consists of three RNAs which are encapsidated in short, bacilliform particles of 30–50 nm. These RNAs are designated as RNA1 (8.35 Kb), RNA2 (3.17 Kb), and RNA3 (3.11 Kb; Melzer et al., 2012). HGSV RNA1 has a single ORF encoding the replicase protein, which contains MET, PRO, SF-I helicase (HEL), and POL domains (Figure 1) and shows most significant similarity to replicases of plant cileviruses, furoviruses, and pomoviruses as well as insect negeviruses (Melzer et al., 2012; Vasilakis et al., 2013). RNA2 possesses four ORFs including three TGB genes, whereas RNA3 contains at least three ORFs (Figure 1; Melzer et al., 2012). The HGSV TGB1 helicase is very distantly related to other TGB1 proteins and shows more similarity to the SF-I replicative helicases of the genus Benyvirus (Figure 1). HGSV TGB2 is distantly related to other TGB2 proteins; short HGSV TGB3 (47 aa in length) contains two long hydrophobic segments with extremely small central hydrophilic region and shows no similarity to any other TGB3 protein (Solovyev et al., 2012).
TBLASTN searches of NCBI and 1KP databases with HGSV TGB1 protein as a query revealed significant similarity (higher than to any viral sequences) to two long VLRAs of plants Lathyrus sativus (7970 nucleotides) and Litchi chinensis (7388 nucleotides; Table 1; Figure 1). HGSV TGB2 protein was also best aligned with the proteins encoded by these Lc-VLRA and Ls-VLRA (data not shown). Despite sequence similarity to HGSV (TGB proteins) and benyviruses (HEL–POL domains), it should be noted that VLRAs of L. sativus and L. chinensis represent another type of genome organization, which is more similar to TGB-containing viruses of the families Alphaflexiviridae and Betaflexiviridae (Morozov and Solovyev, 2003). However, the most remarkable features of the Ls-VLRA and Lc-VLRA are (1) the sequence relation of the TGB2 signatures in the central hydrophilic region (conserved also in HGSV TGB2) to the TGB3 of As-VLRA2 (data not shown) and (2) the absence of TGB3-like ORFs downstream of TGB2 genes. Interestingly, Ls-VLRA has a small ORF, which is located within the TGB2 gene in a different reading frame, encoding a small (42 aa in length) hydrophobic TGB3-like protein (Figure 1). These findings strongly support our previous suggestion (Morozov and Solovyev, 2012) that the second step of the TGB module formation in plant virus evolution could be the acquisition of TGB2-like protein in addition to an autonomized helicase domain. Existence of this stage in the TGB evolution, previously merely hypothetical, is strongly supported by finding Ls-VLRA, which encodes no TGB3 protein. On the other hand, Lc-VLRA TGB3-like ORF might illustrate the TGB3 gene origination by overprinting of TGB2.
Virus Genomes Encoding Both SF-I and SF-II Helicases
An initial step of the TGB origination could take place as duplication or horizontal gene transfer (HGT) of the replicative helicase domain (Morozov and Solovyev, 2012). If we consider the acquisition of an additional helicase gene per se (not in the context of TGB evolution), available examples of viral genomes with non-TGB “accessory” helicases suggest that this process could occur through HGT. The genome of Chara australis virus (CAV; Gibbs et al., 2011) encodes a large protein, which shows the relationship with RNA polymerases of benyviruses, and the CP related to the CP of tobamoviruses. The helicase domain of this replicative protein belongs to SF-I. Two additional CAV ORFs code for a non-replicative RNA helicase and a protein of unknown function. Importantly, this CAV helicase is related to CI helicase (SF-II) of Ipomoviruses (family Potyviridae). RNA virus with a similar genome organization was recently found in charcoal rot (NCBI accession NC_025674). This fungal virus (Macrophomina phaseolina tobamo-like virus) encodes a large ORF with replicative MET/SF-I HEL/POL domains related to tobamoviruses and a SF-II helicase ORF showing highest sequence similarity to CI helicase of Potato virus Y (family Potyviridae; data not shown).
One additional example of HGT resulting in a combination of SF-I and SF-II helicases in single virus genome is provided by endornavirus Gremmeniella abietina type B RNA virus XL (GaBRV-XL; Tuomivirta et al., 2009). Members of the genus Endornavirus (the family Endornaviridae) have linear double-stranded RNA genomes of 9.8–17.6 kb in length containing one ORF coding for a single polypeptide, which is thought to be processed by a proteinase. The encoded polyprotein comprises conserved POL domain and, in most cases, SF-I RNA helicase domain (Song et al., 2013). The second SF-II DExH box helicase of GaBRV-XL is most similar to the HEL domain from Classical swine fever virus (the family Flaviviridae, genus Pestivirus; Tuomivirta et al., 2009). Endornaviruses seem not to form true virions and found in plants, fungi, and protists (Song et al., 2013). Emergence of a second SF-II helicase in addition to unrelated replicative SF-I helicase in the CAV, Macrophomina virus, and GaBRV-XL genomes is intriguing assuming that no definite function can be attributed so far to these accessory helicases.
Posible Scenarios of TGB Evolution
The transition to multicellularity could be beneficial for life forms, which had been earlier evolved to acquire mechanisms of antiviral silencing at a single-cell evolutionary stage. Indeed, small RNA, rapidly moving between the cells of a primordial multicellular organism ahead of the advancing infection, could increase the immunity of distant cell and, consequently, the viability of the multicellular organism as it was described for present-day plants (Hyun et al., 2011; Melnyk et al., 2011; Amari et al., 2012). Along with PCD (Iranzo et al., 2014), this defense mechanism could provide evolutionary advantages to multicellular organisms over communities of unicellular organisms, serving therefore as a driving force in the evolution of multicellularity. We have speculated that plus-RNA viruses of unicellular algae in the course of transition of their hosts to multicellularity may have evolved additional RNA helicase genes by shuffling with distantly related viruses or by duplication of helicase domain in their own replicase. These novel genes combined VSR and MP functions for efficient spread over the multicellular plant organism (Morozov and Solovyev, 2012). Further evolution of virus genomes could have resulted in the origination of the separated MP and VSR proteins.
Considering the possible mechanisms of evolving the genes coding for such proteins, one can propose a combination of horizontal transfer (HGT) and de novo origin by duplication and/or overprinting (Rancurel et al., 2009; Pavesi et al., 2013). Particularly, the MPs of Alphaflexiviridae and Betaflexiviridae are located in the same genomic position (downstream of the replicase gene) but encoded by either the TGB type or the single MP gene (Martelli et al., 2007). Somewhat different situation was found in Tombusviridae and Virgaviridae, where the transport module is positioned differently among virus genera and consists of the single MP gene or double gene block/TGB, respectively, (Morozov and Solovyev, 2003; Stuart et al., 2004; Adams et al., 2009; Schoelz et al., 2011). These facts strongly indicate that at least one type of MPs have been acquired by viruses of these families by HGT. HGT may be also considered as the most probable evolutionary event in acquisition of SF-II helicase gene by plant virus CAV, fungal Macrophomina virus, or GaBRV-XL endornavirus.
New observations reported here allowed us to extend the understanding of possible scenarios for TGB evolution and its successive steps. First, accessory helicase gene was acquired in the process of HGT or gene duplication that might occur independently in different virus groups. Second, the TGB2 gene could have been evolved by HGT or autonomization of the C-terminal transmembrane domain, found in at least one TGB1 helicase, by a frame-shift mutation bringing the future TGB2 sequence into another reading frame (Morozov and Solovyev, 2012).
The TGB3 gene has most likely evolved in the genomic block consisting of TGB1 and TGB2 genes at the latest step of TGB formation. The TGB3 gene, which often overlaps the TGB2 gene, was predicted to appear by overprinting, in which an existing coding sequence is becoming to be translated in two reading frames (Rancurel et al., 2009; Morozov and Solovyev, 2012). A possible loss of virus fitness due to its reduced ability to tolerate mutations in gene overlap regions can be compensated by an evolutionary advantage, as it was found in studies of the paramyxovirus overlapping P/C genes coding for identical functional motifs (Lo et al., 2014). In the case of TGB2 and TGB3 it may be advantageous to encode essential functional hydrophobic motifs in overlapping frames (provided that they are rather short). Finally, an important feature of overlapping genes is that they provide a regulatory advantage that may recompense the increased constraints they impose on the virus, by encoding two proteins that are co-regulated and have coordinated functions (Pavesi et al., 2013). Indeed, the expression of the TGB2 and TGB3 proteins is co-regulated, since they are transcribed from the same messenger RNA (Verchot-Lubicz et al., 2010; Solovyev et al., 2012).
In the evolution of different TGB types, the TGB3 gene could originate by the process of overprinting, the TGB2 gene duplication and subsequent divergence of the emerged TGB3 gene, or an acquisition of an TGB2-independent sequence encoding a progenitor of the TGB3 gene. Possible independent origin of small TGB3 genes in some virus families explains the structural and functional diversity of TGB3 proteins identified among viruses sequenced so far (Morozov and Solovyev, 2012). Impressive example of this evolutionary diversity could present HGSV-related TGBs. Indeed, HGSV TGB3 gene is encoded just downstream TGB2 gene; potential TGB3 ORF in Lc-VLRA is located completely within the TGB2 gene (Figure 1), and Ls-VLRA possesses no identifiable TGB3 ORF. Therefore, the presented analysis of new virus and virus-like sequences confirms the previously suggested scenario for the TGB evolution and provides new models as well as a framework for further functional studies of non-canonical TGB modules.
In general, analysis of plant VLRAs appeared to be a powerful tool, which can help to shed a new light on the details of diversity and evolution of RNA viruses, and, particularly, TGB-containing genomes. Additionally, new TGB-possessing VLRAs can provide new experimental models for exploration of TGB protein functions. Deep sequencing technologies have enabled detection of both known and novel viruses with unprecedented sensitivity. However, the large numbers of reads generated by these methods necessitate new approaches for filtering raw data and discriminating putative viral transcripts for further detailed analysis. In should be mentioned that, in addition to TGB helicases, we used viral RNA-dependent RNA polymerase signatures to select plant virus sequence homologs in the TSA sequence data. The identified transcripts carrying potential viral signatures have enabled the reconstruction of dozens of previously unknown, nearly complete viral genomes from overlapping reads (to be published elsewhere). Therefore, we further expand our analysis to plant TSA libraries in a search for unknown ssRNA viruses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the researchers who contributed samples used in this study to the 1KP initiative. This work was supported by the Russian Foundation for Basic Research (grant 15-04-03922-a).
References
- Adams M. J., Antoniw J. F., Kreuze J. (2009). Virgaviridae: a new family of rod-shaped plant viruses. Arch. Virol. 154 1967–1972. 10.1007/s00705-009-0506-6 [DOI] [PubMed] [Google Scholar]
- Amari K., Vazquez F., Heinlein M. (2012). Manipulation of plant host susceptibility: an emerging role for viral movement proteins? Front. Plant Sci. 3:10 10.3389/fpls.2012.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne E. H., Rakitina D. V., Morozov S. Y., Baulcombe D. C. (1995). Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 44 471–482. 10.1111/j.1365-313X.2005.02539.x [DOI] [PubMed] [Google Scholar]
- Burgyán J., Havelda Z. (2011). Viral suppressors of RNA silencing. Trends Plant Sci. 16 265–272. 10.1016/j.tplants.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Byrd A. K., Raney K. D. (2012). Superfamily 2 helicases. Front. Biosci. (Landmark Ed.) 17:2070–2088. 10.2741/4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S., Chung B. Y., Bass D., Moureau G., Tang S., McAlister E., et al. (2013). Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts. PLoS ONE 8:e80720 10.1371/journal.pone.0080720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T., Bovi A., Dalmay T., Burgyán J. (2007). The p122 subunit of Tobaccomosaic virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and MicroRNA-mediated pathways. J. Virol. 81 11768–11780. 10.1128/JVI.01230-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X. S., Liu J., Cheng N. H., Folimonov A., Hou Y. M., Bao Y., et al. (2004). The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol. Plant Microbe Interact. 17 583–592. 10.1094/MPMI.2004.17.6.583 [DOI] [PubMed] [Google Scholar]
- Gibbs A. J., Torronen M., Mackenzie A. M., Wood J. T., II, Armstrong J. S., Kondo H., et al. (2011). The enigmatic genome of Chara australis virus. J. Gen. Virol. 92 2679–2690. 10.1099/vir.0.033852-0 [DOI] [PubMed] [Google Scholar]
- Gilhooly N. S., Gwynn E. J., Dillingham M. S. (2013). Superfamily 1 helicases. Front. Biosci. (Schol. Ed.) 5x:206–216. [DOI] [PubMed] [Google Scholar]
- Hickman A. B., Dyda F. (2005). Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15 77–85. 10.1016/j.sbi.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Hyun T. K., Uddin M. N., Rim Y., Kim J. Y. (2011). Cell-to-cell trafficking of RNA and RNA silencing through plasmodesmata. Protoplasma 248 101–116. 10.1007/s00709-010-0225-6 [DOI] [PubMed] [Google Scholar]
- Iranzo J., Lobkovsky A. E., Wolf Y. I., Koonin E. V. (2014). Virus-host arms race at the joint origin of multicellularity and programmed cell death. Cell Cycle 13 3083–3088. 10.4161/15384101.2014.949496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L. M., Leipe D. D., Koonin E. V., Aravind L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146 11–31. 10.1016/j.jsb.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Jankowsky E., Fairman M. E. (2007). RNA helicases - one fold for many functions. Curr. Opin. Struct. Biol. 17 316–324. 10.1016/j.sbi.2007.05.007 [DOI] [PubMed] [Google Scholar]
- Johnson M. T., Carpenter E. J., Tian Z., Bruskiewich R., Burris J. N., Carrigan C. T., et al. (2012). Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE 7:e50226 10.1371/journal.pone.0050226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junglen S., Drosten C. (2013). Virus discovery and recent insights into virus diversity in arthropods. Curr. Opin. Microbiol. 16 507–513. 10.1016/j.mib.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Hirano S., Chiba S., Andika I. B., Hirai M., Maeda T., et al. (2013). Characterization of burdock mottle virus, a novel member of the genus Benyvirus, and the identification of benyvirus-related sequences in the plant and insect genomes. Virus Res. 177 75–86. 10.1016/j.virusres.2013.07.015 [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Dolja V. V. (1993). Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28 375–430. 10.3109/10409239309078440 [DOI] [PubMed] [Google Scholar]
- Kubota K., Tsuda S., Tamai A., Meshi T. (2003). Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 77 11016–11026. 10.1128/JVI.77.20.11016-11026.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. K., Søgaard T. M., Karlin D. G. (2014). Evolution and structural organization of the C proteins of paramyxovirinae. PLoS ONE 9:e90003 10.1371/journal.pone.0090003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli G. P., Adams M. J., Kreuze J. F., Dolja V. V. (2007). Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45 73–100. 10.1146/annurev.phyto.45.062806.094401 [DOI] [PubMed] [Google Scholar]
- Matasci N., Hung L. H., Yan Z., Carpenter E. J., Wickett N. J., Mirarab S., et al. (2014). Data access for the 1,000 Plants (1KP) project. Gigascience 3:17 10.1186/2047-217X-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk C., Molnar A., Baulcombe D. C. (2011). Intercellular and systemic movement of RNA silencing signals. EMBO J. 30 3553–3563. 10.1038/emboj.2011.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer M. J., Sether D. M., Borth W. B., Hu J. S. (2012). Characterization of a virus infecting citrus volkameriana with Citrus Leprosis-Like Symptoms. Phytopathology 102 122–127. 10.1094/PHYTO-01-11-0013 [DOI] [PubMed] [Google Scholar]
- Morozov S. Y., Solovyev A. G. (2003). Triple gene block: modular design of a multi-functional machine for plant virus movement. J. Gen. Virol. 84 1351–1366. 10.1099/vir.0.18922-0 [DOI] [PubMed] [Google Scholar]
- Morozov S. Y., Solovyev A. G. (2012). Did silencing suppression counter-defensive strategy contribute to origin and evolution of the triple gene block coding for plant virus movement proteins? Front. Plant Sci. 3:136 10.3389/fpls.2012.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A., Magiorkinis G., Karlin D. G. (2013). Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of Deltaretroviruses. PLoS Comput. Biol. 9:e1003162 10.1371/journal.pcbi.1003162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Voinnet O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 11 745–760. 10.1038/nrmicro3120 [DOI] [PubMed] [Google Scholar]
- Rancurel C., Khosravi M., Dunker A. K., Romero P. R., Karlin D. (2009). Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 83 10719–10736. 10.1128/JVI.00595-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles J. W., Rohde W. (1990). Nicotiana velutina mosaic virus: evidence for a bipartite genome comprising 3 kb and 8 kb RNAs. J. Gen. Virol. 71 1019–1027. 10.1099/0022-1317-71-5-1019 [DOI] [PubMed] [Google Scholar]
- Roossinck M. J. (2015). Metagenomics of plant and fungal viruses reveals an abundance of persistent life styles. Front. Microbe Sci. 5:767 10.3389/fmicb.2014.00767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K., Breitbart M. (2011). Exploring the viral world through metagenomics. Curr. Opin. Virol. 1 289–297. 10.1016/j.coviro.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Schoelz J. E., Harries P. A., Nelson R. S. (2011). Intracellular transport of plant viruses: finding the door out of the cell. Mol. Plant 4 813–831. 10.1093/mp/ssr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senshu H., Yamaji Y., Minato N., Shiraishi T., Maejima K., Hashimoto M., et al. (2011). A dual strategy for the suppression of host antiviral silencing: two distinct suppressors for viral replication and viral movement encoded by Potato virus M. J. Virol. 85 10269–10278. 10.1128/JVI.05273-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev A. G., Kalinina N. O., Morozov S. Y. (2012). Recent advances in research of plant virus movement mediated by triple gene block. Front. Plant Sci. 3:276 10.3389/fpls.2012.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Cho W. K., Park S. H., Jo Y., Kim K. H. (2013). Evolution of and horizontal gene transfer in the Endornavirus genus. PLoS ONE 8:e64270 10.1371/journal.pone.0064270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorel M., Garcia J. A., German-Retana S. (2014). The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein. Mol. Plant Microbe Interact. 27 215–226. 10.1094/MPMI-11-13-0333-CR [DOI] [PubMed] [Google Scholar]
- Stuart G., Moffett K., Bozarth R. F. (2004). A whole genome perspective on the phylogeny of the plant virus family Tombusviridae. Arch. Virol. 149 1595–1610. 10.1007/s00705-004-0298-7 [DOI] [PubMed] [Google Scholar]
- Tuomivirta T. T., Kaitera J., Hantula J. (2009). A novel putative virus of Gremmeniella abietina type B (Ascomycota: Helotiaceae) has a composite genome with endornavirus affinities. J. Gen. Virol. 90 2299–2305. 10.1099/vir.0.011973-0 [DOI] [PubMed] [Google Scholar]
- Vasilakis N., Forrester N. L., Palacios G., Nasar F., Savji N., Rossi S. L., et al. (2013). Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J. Virol. 87 2475–2488. 10.1128/JVI.00776-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot-Lubicz J., Torrance L., Solovyev A. G., Morozov S. Y., Jackson A. O., Gilmer D. (2010). Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe Interact. 23 1231–1247. 10.1094/MPMI-04-10-0086 [DOI] [PubMed] [Google Scholar]
- Wang L.-Y., Lin S.-S., Hung T.-H., Li T.-K., Lin N.-C., Shen T.-L. (2012). Multiple domains of the Tobacco mosaic virus p126 protein can independently suppress local and systemic RNA silencing. Mol. Plant Microbe Interact. 25 648–657. 10.1094/MPMI-06-11-0155 [DOI] [PubMed] [Google Scholar]
- Wang Y., Dang M., Hou H., Mei Y., Qian Y., Zhou X. (2014). Identification of an RNA silencing suppressor encoded by a mastrevirus. J. Gen. Virol. 95 2082–2088. 10.1099/vir.0.064246-0 [DOI] [PubMed] [Google Scholar]
- Xie Y., Wu G., Tang J., Luo R., Patterson J., Liu S., et al. (2014). SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30 1660–1666. 10.1093/bioinformatics/btu077 [DOI] [PubMed] [Google Scholar]