Introduction
The increase in drug resistant bacteria continues to pose a significant threat to human health worldwide, and new antibiotics are required to combat resistant strains (Center for disease Control and Prevention, 2013). Drug discovery typically begins with identification of an appropriate target pathway, isolation of “hit” compounds that inhibit (or dysregulate) the pathway, validation, and optimization of the hit into a lead compound for development. trans-Translation has been an intriguing target for new antibiotics because the pathway is required for virulence or viability in most bacterial pathogens that have been examined, but it is not present in metazoans, so specific inhibitors might have few side effects (Keiler and Alumasa, 2013; Ramadoss et al., 2013; Giudice et al., 2014). Recent publication of a high-throughput screen capable of identifying inhibitors of trans-translation with antibiotic activity validated the pathway as a potential drug target and provided a key tool for isolating lead compounds (Ramadoss et al., 2013). We argue that this high-throughput assay, coupled to click-chemistry techniques to identify the molecular targets of active compounds, should enable widespread efforts to develop drugs targeted to trans-translation.
Reasons for targeting trans-translation
trans-Translation is a pathway for rescuing ribosomes trapped at the 3' end of an mRNA (Keiler et al., 1996). Bacteria do not use the same mRNA quality control mechanisms employed by eukaryotes to ensure that mRNAs are intact before translation begins (Keiler, 2008). As a result, translation can initiate on truncated or damaged mRNAs. In addition, mRNA damage can occur after ribosomes have initiated translation. When the ribosome reaches the 3′ end of such an mRNA, there is no stop codon to terminate translation. Estimates in E. coli indicate that ~2–4% of all translation initiations terminate in stalled complexes (Ito et al., 2011). Accumulation of these stalled complexes is lethal, and bacteria require at least one mechanism to rescue stalled ribosomes (Feaga et al., 2014; Keiler and Feaga, 2014). The primary rescue pathway in bacteria is trans-translation, in which the protein SmpB and the specialized RNA, tmRNA, recognize stalled ribosomes. During trans-translation, the ribosome resumes translation on an open reading frame within tmRNA, adding a peptide tag to the C terminus of the nascent polypeptide and releasing the previously stalled ribosome. The peptide tag is ultimately recognized by multiple proteases, which degrade the tagged protein (Keiler, 2008). Genes encoding tmRNA and SmpB have been identified in almost all bacterial genomes (Hudson et al., 2014). Some species also have backup rescue systems, the ArfA or ArfB proteins (Chadani et al., 2011, 2012) that can release ribosomes when trans-translation activity is not available.
A number of key attributes make trans-translation an attractive antibiotic target. First, it is essential for viability or virulence in many pathogenic species, including Neisseria gonorrhoeae, Mycobacterium tuberculosis, and Staphylococcus aureus (Huang et al., 2000; Zhang et al., 2012b; Fey et al., 2013). Disruption of trans-translation also results in hyper-sensitivity to antibiotics in some species (Abo et al., 2002; Li et al., 2013). Second, tmRNA and SmpB are universally conserved in bacteria, but are absent in metazoans, so specific inhibition of trans-translation should not cause toxicity in the host (Hudson et al., 2014). Third, the pathway includes several molecules that are not targeted by existing antibiotics, including tmRNA, SmpB, and stalled ribosomes, therefore cross-resistance with existing antibiotics should be minimal.
An HTS assay to identify inhibitors
To validate trans-translation as a target and to begin the drug discovery process, an assay for trans-translation inhibitors was developed and used in a high-throughput screen (Ramadoss et al., 2013). The assay uses an E. coli strain engineered to express luciferase from a truncated mRNA. When there is no inhibition, all the luciferase is tagged by trans-translation and degraded, but when trans-translation is inhibited active luciferase is produced. The Z' score under high-throughput conditions is 0.75, and the assay is appropriate for a wide range of screening platforms. Screening of a library of small molecules identified specific inhibitors that have promising antibacterial properties against both Gram-positive and Gram-negative bacteria and exhibit low toxicity against human cells (Ramadoss et al., 2013).
This assay is cell-based, which ensures that hit compounds will be able to function in the context of the bacterial cell. However, inhibitors could target one of several potential molecules including both nucleic acids and proteins (Keiler and Alumasa, 2013; Ramadoss et al., 2013; Giudice et al., 2014), and the molecular target of each inhibitor must be identified to facilitate optimization.
Molecular target identification strategies
The main genetic approach to molecular target identification typically involves isolating and sequencing resistant mutants to identify candidate targets (Lerner et al., 2005; Wacker et al., 2012; Schenone et al., 2013). This method is generally successful when resistant mutants can be obtained (Wacker et al., 2012). However, there are several reasons why target-based resistance will be rare or nonexistent: (i) mutations in the target that eliminate inhibitor activity may themselves be lethal; (ii) multiple mutations may be required for resistance, for example if the target is encoded in redundant genes; (iii) there may be multiple targets for the compound. No target-based resistant mutants have been obtained for any of the trans-translation inhibitors, although it is not known why resistance is rare in this case (Ramadoss et al., 2013).
Alternative approaches to identify molecular targets use chemical biology, computational, or biochemical techniques (Schenone et al., 2013). Chemical biology techniques typically employ the use of small molecule probes that mimic the structure of the inhibitor. Probe-aided pull down assays, which use an affinity tag (biotin) fused to the inhibitor, fall into this category. Although this approach has been shown to work independently, greater success occurs when it is coupled with orthogonal methods such as photo-affinity labeling (Li et al., 2004; Kotake et al., 2007; Yamazaki et al., 2010). A significant drawback of this approach is that structural modifications performed on the pharmacophore to incorporate the affinity tag and a linker usually result in altered physicochemical properties of the probe that may affect target binding (Kashiwayama et al., 2010; Zhang et al., 2012a). Click chemistry could provide a solution to this problem through a two step process incorporating the use of a minimally-modified pharmacophore for target binding, and a secondary probe for isolation of the target molecule.
Click chemistry
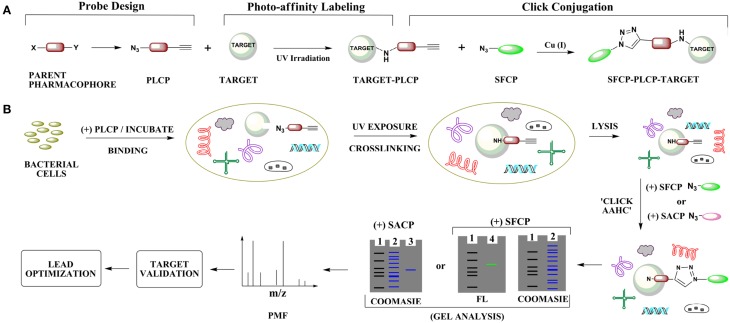
The concept of click chemistry broadly encompasses groups of chemical reactions that are fast, versatile, high yielding and simple to use (Kolb et al., 2001). The copper catalyzed Azide-Alkyne Huisgen Cycloaddition (AAHC) is among the most common click chemistry reactions (Spiteri and Moses, 2010). AAHC requires both azide and alkyne functional groups, which react almost exclusively with each other under a defined set of conditions, minimizing unwanted side reactions (Thirumurugan et al., 2013). Notable advantages of this method include its specificity, mild reaction conditions, irreversibility and selectivity (Kolb and Sharpless, 2003). When applied to molecular target identification, AAHC can be coupled with methods such as photo-affinity labeling, making the identification process more efficient (Sumranjit and Chung, 2013). In the example we consider here, photo-affinity labeling can be used to covalently attach a probe to the target, and a subsequent AAHC reaction can link the probe to a fluorescent moiety or affinity tag to enable isolation (Figure 1). This strategy circumvents addition of bulky substituents to the probe that may otherwise affect probe-target binding.
Figure 1.
Identification of molecular targets by photo-affinity labeling and click chemistry. (A) Overview of photo-labile click probe (PLCP) design, photo-affinity labeling and click bioconjugation using a fluorescent reporter probe. (B) Cell-based strategy for click-based molecular target identification for trans-translation inhibitors. The process involves intracellular probe-target binding, intracellular probe-target covalent cross-linking, cell lysis, click bioconjugation using either a secondary fluorescent click probe (SFCP) or secondary affinity click probe (SACP), target isolation, identification by peptide mass fingerprinting (PMF), and validation all leading to lead optimization. Abbreviations: AAHC, Azide-alkyne Huisgen Cycloaddition; FL, fluorescence-exposed gel; Gel lanes, one ladder; two cell lysate; three target isolated from a click-based affinity pull-down assay; 4 target isolated from a click-based fluorescence assay.
Compatibility of this reaction with physiological conditions provides myriad avenues for target identification studies, and this procedure has been successfully employed to identify molecular targets for anticancer and antibacterial agents. For example, the click chemistry approach was used to identify the target for LW6 an inhibitor targeting the regulation of tumor angiogenesis and metastasis in colon cancer cell lines (Lee et al., 2013). Similarly, this procedure was successfully used to identify inhibitors of the biotin protein ligase possessing antibacterial activity (Tieu et al., 2013). Many other examples have been described, demonstrating the efficiency of this method in identifying molecular targets within biological environments (Colca et al., 2003; Thirumurugan et al., 2013). It is noteworthy that the click-based method can be used in live cells, in contrast to pull-down assays that are normally performed in vitro (Colca et al., 2003; Speers and Cravatt, 2004). This is an important property that would allow probe-target interaction to occur in cells, prior to target isolation. For all such techniques, however, success is largely dependent upon nature of the probe.
Click probe design
Probe design is perhaps the most challenging task requiring careful assessment in the initial steps of target identification. An efficient click probe must retain key properties of the parent molecule, including biological activity and cell permeability, while accommodating the azide and alkyne moieties that enable target cross-linking and subsequent click coupling. Placement of these groups can be guided by structure-activity relationship (SAR) studies aimed at identifying sites within the basic pharmacophore that allow modification without eliminating activity or increasing off-target binding interactions. A generalized design for a photo-labile click probe (PLCP), harboring a photo-labile terminal azide group and a terminal alkyne, is shown in Figure 1A. The azide group in this PLCP can be activated by UV irradiation to cross-link the PLCP to the target. The alkyne can then be used for click conjugation to a secondary probe containing a fluorescent or biotinylated group to facilitate purification.
Molecular target identification
To identify the molecular target, the PLCP can be cross-linked to interacting molecules in vivo, in a cell extract, or in a purified system. If the probe is cell permeable, it can be added to bacterial cells to allow target binding (Figure 1B) (Colca et al., 2003). UV irradiation (typically 254–366 nm) is then used to activate the azide and cross-link the PCLP to binding partners. The cells are lysed under denaturing conditions, excess PCLP is removed, and the secondary probe is attached via click chemistry. Denaturation ensures that the click reaction can proceed even if the PCLP binds in a site that is inaccessible under native conditions. If a fluorescent secondary probe is attached, gel electrophoresis can be used to isolate fluorescent bands (Figure 1B). Conversely, if the secondary probe contains biotin or any other affinity tag, the target can be isolated on an appropriate affinity resin (Pozdnyalov et al., 2013). In either case, the target molecule is then identified by mass spectrometry. If the PLCP is not cell permeable, it can be added to a cell extract or to purified candidate molecules prior to UV irradiation, but the rest of the identification procedure would be unchanged. Once the target is identified, target validation and lead optimization can proceed.
Summary
trans-Translation is an attractive target for new antibiotic development because the reaction is required for virulence or viability in many pathogens, and several small molecules that inhibit the pathway have been identified (Ramadoss et al., 2013). A cell-based assay for high-throughput screening has been engineered, enabling screening of diverse libraries for inhibitors that are active in vivo (Ramadoss et al., 2013). The click-based molecular target identification strategy described here should enable rapid advancement through the hit validation process. With these tools available, trans-translation should be pursued as a drug target on many fronts.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors were supported by NIH grants AI113993 and AI111692.
References
- Abo T., Ueda K., Sunohara T., Ogawa K., Aiba H. (2002). SsrA mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells 7, 629–638. 10.1046/j.1365-2443.2002.00549.x [DOI] [PubMed] [Google Scholar]
- Center for disease Control and Prevention. (2013). Threat Report: Antibiotic Resistance Threats in the United States. Available online at: http://www.cdc.gov/drugresistance/threat-report-2013/index.html
- Chadani Y., Ito K., Kutsukake K., Abo T. (2012). ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl–tRNA hydrolysis in Escherichia coli. Mol. Microbiol. 86, 37–50. 10.1111/j.1365-2958.2012.08190.x [DOI] [PubMed] [Google Scholar]
- Chadani Y., Ono K., Kutsukake K., Abo T. (2011). Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA and ArfA-mediated pathways. Mol. Microbiol. 80, 772–785. 10.1111/j.1365-2958.2011.07607.x [DOI] [PubMed] [Google Scholar]
- Colca J. R., McDonald W. G., Waldon D. J., Thomasco L. M., Gadwood R. C., Lund E. T., et al. (2003). Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J. Biol. Chem. 278, 21972–21979. 10.1074/jbc.M302109200 [DOI] [PubMed] [Google Scholar]
- Feaga H. A., Viollier P. H., Keiler K. C. (2014). Release of nonstop ribosomes is essential. MBio 5:e01916. 10.1128/mBio.01916-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P. D., Endres J. L., Yajjala V. K., Widhelm T. J., Boissy R. J., Bose J. L., et al. (2013). A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537–e00512. 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice E., Macé K., Gillet R. (2014). trans-Translation exposed: understanding the structures and functions of tmRNA-SmpB. Front. Microbiol. 5:113 10.3389/fmicb.2014.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wolfgang M. C., Withey J., Koomey M., Friedman D. I. (2000). Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 19, 1098–1107. 10.1093/emboj/19.5.1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C. M., Lau B. Y., Williams K. P. (2014). Ends of the line for tmRNA-SmpB. Front. Microbiol. 5:421. 10.3389/fmicb.2014.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Chadani Y., Nakamori K., Chiba S., Akiyama Y., Abo T. (2011). Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS ONE 6:e28413. 10.1371/journal.pone.0028413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwayama Y., Tomohiro T., Narita K., Suzumura M., Glumoff T., Hiltunen J. K., et al. (2010). Identification of a substrate-binding site in a peroxisomal β-oxidation enzyme by photoaffinity labeling with a novel palmitoyl derivative. J. Biol. Chem. 285, 26315–26325. 10.1074/jbc.M110.104547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C. (2008). Biology of trans-translation. Annu. Rev. Microbiol. 62, 133–151. 10.1146/annurev.micro.62.081307.162948 [DOI] [PubMed] [Google Scholar]
- Keiler K. C., Alumasa J. N. (2013). The potential of trans-translation inhibitors as antibiotics. Future Microbiol. 8, 1235–1237. 10.2217/fmb.13.110 [DOI] [PubMed] [Google Scholar]
- Keiler K. C., Feaga H. A. (2014). Resolving nonstop translation complexes is a matter of life or death. J. Bacteriol. 196, 2123–2130. 10.1128/JB.01490-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C., Waller P. R., Sauer R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 27, 990–993. 10.1126/science.271.5251.990 [DOI] [PubMed] [Google Scholar]
- Kolb H. C., Finn M. G., Sharpless K. B. (2001). Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- Kolb H. C., Sharpless B. K. (2003). The growing impact of click chemistry on drug discovery. Drug Discov. Today 8, 1128–1137. 10.1016/S1359-6446(03)02933-7 [DOI] [PubMed] [Google Scholar]
- Kotake Y., Sagane K., Owa T., Mimori−Kiyosue Y., Shimizu H., Uesugi M., et al. (2007). Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3, 570–575. 10.1038/nchembio.2007.16 [DOI] [PubMed] [Google Scholar]
- Lee K., Ban H. S., Naik R., Hong Y. S., Son S., Kim B., et al. (2013). Identification of malate dehydrogenase 2 as a target protein of the HIF-1 inhibitor LW6 using chemical probes. Angew. Chem. 125, 10476–10479. 10.1002/ange.201304987 [DOI] [PubMed] [Google Scholar]
- Lerner C. G., Kakavas S. J., Wagner C., Chang R. T., Merta P. J., Ruan X., et al. (2005). Novel approach to mapping of resistance mutations in whole genomes by using restriction enzyme modulation of transformation efficiency. Antimicrob. Agents Chemother. 49, 2767–2777. 10.1128/AAC.49.7.2767-2777.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ji L., Shi W., Xie J., Zhang Y. (2013). trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 68, 2477–2481. 10.1093/jac/dkt231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. M., Xu M., Lai M. T., Hunag Q., Castro J. L., DiMuzio−Mower J., et al. (2004). Photoactivated-γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature 405, 689–694. 10.1038/35015085 [DOI] [PubMed] [Google Scholar]
- Pozdnyalov N., Murrey H. E., Crump C. J., Petterson M., Ballard T. E., Ende C. W., et al. (2013). γ-Secretasemodulator (GSM) photoaffinity probes reveal distinct allosteric binding sites on presenilin. J. Biol. Chem. 288, 9710–9720. 10.1074/jbc.M112.398602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadoss N. S., Alumasa J. N., Cheng L., Wang Y., Li S., Chambers B. S., et al. (2013). Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proc. Natl. Acad. Sci. U.S.A. 110, 10282–10287. 10.1073/pnas.1302816110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenone M., Danèík V., Wagner B. K., Clemons P. A. (2013). Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 9, 232–240. 10.1038/nchembio.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers A. E., Cravatt B. F. (2004). Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 11, 535–546. 10.1016/j.chembiol.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Spiteri C., Moses J. E. (2010). Copper-catalyzed azide–alkyne cycloaddition: regioselective synthesis of 1,4,5-trisubstituted 1,2,3-triazoles. Angew. Chem. Int. Ed. Engl. 49, 31–33. 10.1002/anie.200905322 [DOI] [PubMed] [Google Scholar]
- Sumranjit J., Chung S. J. (2013). Recent advances in target characterization and identification by photoaffinity probes. Molecules 18, 10425–10451. 10.3390/molecules180910425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumurugan P., Matosiuk D., Jozwiak K. (2013). Click chemistry for drug development and diverse chemical-biology applications. Chem. Rev. 113, 4905–4979. 10.1021/cr200409f [DOI] [PubMed] [Google Scholar]
- Tieu W., Soares da Costa T. P., Yap M. N., Keeling K. L., Wilce M. C. J., Wallace J. C., et al. (2013). Optimising in situ click chemistry: the screening and identification of biotin protein ligase inhibitors. Chem. Sci. 4, 3533–3537. 10.1039/c3sc51127h [DOI] [Google Scholar]
- Wacker S. A., Houghtaling B. R., Elemento O., Kapoor T. M. (2012). Using transcriptome sequencing to identify mechanism of drug action and resistance. Nat. Chem. Biol. 8, 235–237. 10.1038/nchembio.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y., Sumikura M., Hidaka K., Yasui H., Kiso Y., Yakushiji F., et al. (2010). Antimicrotubule ‘plinabulin’ chemical probe KPU-244 labeled both α- and β- tubulin. Bioorg. Med. Chem. 18, 3169–3174. 10.1016/j.bmc.2010.03.037 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang Y., Dong J., Liu J., Zhang L., Sun S. (2012a). Design and synthesis of novel photoaffinity probes for study of the target proteins of oleanolic acid. Bioorg. Med. Chem. Lett. 22, 1036–1039. 10.1016/j.bmcl.2011.11.123 [DOI] [PubMed] [Google Scholar]
- Zhang Y. J., Ioerger T. R., Huttenhower C., Long J. E., Sassetti C. M., Sacchettini J. C., et al. (2012b). Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 8:e1002946. 10.1371/journal.ppat.1002946 [DOI] [PMC free article] [PubMed] [Google Scholar]