Abstract
Sinking particles mediate the transport of carbon and energy to the deep-sea, yet the specific microbes associated with sedimenting particles in the ocean's interior remain largely uncharacterized. In this study, we used particle interceptor traps (PITs) to assess the nature of particle-associated microbial communities collected at a variety of depths in the North Pacific Subtropical Gyre. Comparative metagenomics was used to assess differences in microbial taxa and functional gene repertoires in PITs containing a preservative (poisoned traps) compared to preservative-free traps where growth was allowed to continue in situ (live traps). Live trap microbial communities shared taxonomic and functional similarities with bacteria previously reported to be enriched in dissolved organic matter (DOM) microcosms (e.g., Alteromonas and Methylophaga), in addition to other particle and eukaryote-associated bacteria (e.g., Flavobacteriales and Pseudoalteromonas). Poisoned trap microbial assemblages were enriched in Vibrio and Campylobacterales likely associated with eukaryotic surfaces and intestinal tracts as symbionts, pathogens, or saprophytes. The functional gene content of microbial assemblages in poisoned traps included a variety of genes involved in virulence, anaerobic metabolism, attachment to chitinaceaous surfaces, and chitin degradation. The presence of chitinaceaous surfaces was also accompanied by the co-existence of bacteria which encoded the capacity to attach to, transport and metabolize chitin and its derivatives. Distinctly different microbial assemblages predominated in live traps, which were largely represented by copiotrophs and eukaryote-associated bacterial communities. Predominant sediment trap-assocaited eukaryotic phyla included Dinoflagellata, Metazoa (mostly copepods), Protalveolata, Retaria, and Stramenopiles. These data indicate the central role of eukaryotic taxa in structuring sinking particle microbial assemblages, as well as the rapid responses of indigenous microbial species in the degradation of marine particulate organic matter (POM) in situ in the ocean's interior.
Keywords: metagenomics, marine particles, sediment trap, biological pump, microbiology
Introduction
Particulate organic matter (POM) generated in the euphotic zone is the major conduit of matter and energy transport to the deep sea and also represents the primary mechanism of carbon removal from surface waters via the biological pump (McCave, 1975; Volk and Hoffert, 1985). POM is operationally defined as particles ranging from 0.1 μm to centimeters in size, and is further qualitatively subcategorized into macroaggregates (marine snow; centimeters to 500 μm in diameter), microaggregates (500–1 μm), and submicron particles (1–0.1 μm) (Simon et al., 2002). Sinking POM can be collected in situ using sediment traps that contain saline solutions slightly denser than seawater that retain sinking particles (Knauer et al., 1979). This broad size spectrum of POM harbors a diverse and complex variety of inorganic as well as living and non-living organic materials (Volkman and Tanoue, 2002; Nebbioso and Piccolo, 2013).
Much of the current knowledge of POM-degrading microbial communities is derived from studies of suspended POM. Analysis of whole seawater segregated into particle-associated (>1 μm) and free-living size fractions has revealed taxonomically and functionally distinct microbial communities in marine anoxic zones (Ganesh et al., 2014), coastal ecosystems (Allen et al., 2012; Smith et al., 2013), estuarine environments (Crump et al., 1999; Waidner and Kirchman, 2007), inland seas (Moeseneder et al., 2001; Fuchsman et al., 2011, 2012; Crespo et al., 2013), phytoplankton blooms (Riemann et al., 2000; Fandino et al., 2005; Teeling et al., 2012), ocean trenches (Eloe et al., 2011), and the open ocean (Kellogg and Deming, 2009; Allen et al., 2012). These studies have shown that in particular, members of the Bacteroidetes, Planctomycetes, and Deltaproteobacteria are often enriched in larger particle size fractions. Studies of microbial community composition on sinking particles are less extensive than those on suspended particles. Research programs such as the Vertical Transport and Exchange (VERTEX)(Martin et al., 1987) and VERtical Transport In the Global Ocean (VERTIGO) (Buesseler et al., 2008) supported diverse process-oriented studies that revealed the importance of chemolithotrophs like nitrifiers (Karl et al., 1984), organotrophs (Boyd et al., 1999), and exoenzyme-driven degradation on sinking particles (Smith et al., 1992). These findings laid the foundation for phylogentically-oriented studies that suggested that Bacteroidetes, Planctomycetes, and Roseobacter can act as sinking particle colonizers in the upper water column (DeLong et al., 1993; LeCleir et al., 2013). While sediment traps have proven useful for over 30 years in studies of sinking POM (Karl and Knauer, 1984), to date there exists only one report of the phylogenetic diversity of sediment-trap collected microbes, which grew over 24 h in sediment-trap captured particles from 100 to 120 m (LeCleir et al., 2013).
Given the diverse sources and sinks of sinking particles in the ocean's interior (Honjo et al., 2008), much remains to be learned about the microbes and processes that regulate the degradation of sinking POM. In this study, we sought to examine the nature of sinking particles collected in poisoned traps, which we hypothesized would help preserve sinking materials and allow us to identify (using metagenomics) the sources of larger sinking particulates including larger eukaryotes that are known to aggregate and sink to the deep-sea. We also included paired, un-poisoned traps (live) in our experiments, postulating that these might reveal the nature and identity of microorganisms capable of growth on the collected organic material at the in situ temperatures and pressures of trap deployment. We reasoned that the phylogenetic identity of poisoned vs. live traps would reveal the identity and genomic potential of microbes capable of growth on sinking particulate organic materials in situ in the ocean's interior.
Materials and methods
Sample collection
A free-drifting sediment trap array identical to those used in the VERTEX and HOT field programs (Knauer et al., 1979) was deployed at station ALOHA (22.75°N, 158°W) in the North Pacific Subtropical Gyre on July 14, 2012. Each trap tube (cross sectional area of 0.0039 m2) was filled with approximately 1.8 liters of either an 0.2 μm-filtered brine solution (Knauer et al., 1984) (“live”) or an 0.2 μm-filtered RNAlater solution (“poisoned”) adjusted to a density of 1.05 g/cc (see Supplementary Material, for further methodological details on trap solutions). Both sets of traps were fitted with a 335 um Nitex screen below the topmost baffle in order to exclude larger zooplankton. The array drifted north-west for 75 nautical miles before recovery on July 26, 2012. Prior to filtration, the 335 μm Nitex screen was removed along with approximately 500 mL of seawater overlying the higher density hypersaline trap solution. Following recovery, particles in the 0.2–335 μm fraction were collected on Sterivex filters (EMD Millipore, Billerica, MA, USA) and preserved with 1.5 mL RNAlater (Ambion, Carlsbad, CA, USA). Filters were stored at −80°C prior to nucleic acid isolation.
Microscopy
Epifluorescence and optical microscopy
For each depth, 10 mL of fixed sediment trap sample (2% formaldehyde final concentration) was filtered onto black 0.2 μm pore size polycarbonate filters and allowed to dry completely. Ethanol cleansed surgical scissors were used to cut 1/8 pieces from each filter and four pieces were then positioned on a microscope slide (Fisherbrand Superfrost precleaned microscope slides #12-550-143). A 24 × 50 mm coverslip with #1.5 thickness was placed below the slide. Antifade mounting medium (Patel et al., 2007) containing 1 μg mL−1 of nucleic acid stain 4′,6-diamidino-2-phenylindole (DAPI) was spotted on the coverslip (15 μL) to align with the filter pieces. The coverslip was inverted onto the slide and the filters were stained for 10 min. Filters were visualized at 1000× total magnification on a Nikon 90i epifluorescence microscope with excitation/emission settings for DAPI, chlorophyll, and phycoerythrin. Images were acquired with a QImaging Retiga EXi camera using optimized exposure times and analyzed with Nikon NIS-Elements software.
For optical microscopy, unmounted filters were visualized through a Nikon AZ100 Multizoom microscope with a 10× objective at 20× and 40× magnification (zoom settings 4 and 8) using a Nikon NI-150 illuminator. Images were captured via NIS-Elements software using a Nikon DS-Fi1 camera.
Library preparation and sequencing
Approximately one-half of each Sterivex filter was used for extraction of total community DNA using the Powerwater DNA isolation kit (Mobio, Carlsbad, CA, USA), with modifications (see Supplementary Material for details). Library preparation followed the Nextera XT DNA sample preparation protocol (Illumina, San Diego, CA, USA). Samples were dual indexed and 10 samples pooled per sequencing run on a MiSeq using MiSeq reagent kit v3 (Illumina, San Diego, CA, USA). Sequencing and quality control followed the manufacturer's recommendations.
Sequence analysis and annotation
Sequencing and annotation statistics for sediment trap and seawater samples are summarized in Table A1 in Supplementary Material. Metagenomic sequences were filtered with Trimmomatic (Bolger et al., 2014), PandaSeq (Masella et al., 2012), and SortMeRNA to identify rRNA-containing reads (Kopylova et al., 2012) as previously described (Lincoln et al., 2014) with one modification. Unjoined read pairs output from PandaSeq were joined with 6 N's and tracked along with paired reads. The taxonomic origins of rRNA and non-rRNA reads were determined by comparison against SILVA release 115 and the NCBI RefSeq release 61 databases, respectively, using lastal (Kiełbasa et al., 2011). Function of non-rRNA reads was determined by comparison with the September 2013 version of the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) database using lastal and the March 22, 2013 version of the Carbohydrate Active Enzyme (CAZy) database (Cantarel et al., 2009) using HMMER3.0 (Eddy, 2011) and hidden Markov models of CAZy signature domains (Yin et al., 2012). See Supplementary Material for further details.
Statistical analyses
All metagenomic sequence counts were normalized and variance stabilized using the regularized log transformation in DESeq2 (Love et al., 2014). Ordination of normalized sequences used principal coordinate analysis with Bray-Curtis distance in the phyloseq R package (McMurdie and Holmes, 2013). Significance (p < 0.05) of clusters was determined using non-parametric analysis of variance based on dissimilarities in the vegan R package (Dixon, 2003). A negative binomial Wald test in DESeq2 was used to identify statistically significant differences in taxonomic and functional non-normalized gene counts among live traps, poisoned traps, and seawater (data not shown). The presence of copepods in live and poisoned traps was also confirmed by optical microscopy (Figures 1G,H). As replicates of sediment traps at each depth were not available, all four depths belonging to a treatment were modeled as biological replicates. A false discovery rate threshold of 0.01 was used for detecting differentially abundant taxa or functions. For statistical validation of depth-specific taxonomic differences, Fisher's exact test as implemented in the STAMP v2.01 program (Parks and Beiko, 2010) was used for pairwise comparisons of 150 m vs. 500 m RefSeq-identified non-normalized taxa within treatment types. A false discovery rate threshold of 0.05 and a difference between proportions cutoff of 1 were used to assess statistical and biological significance, respectively.
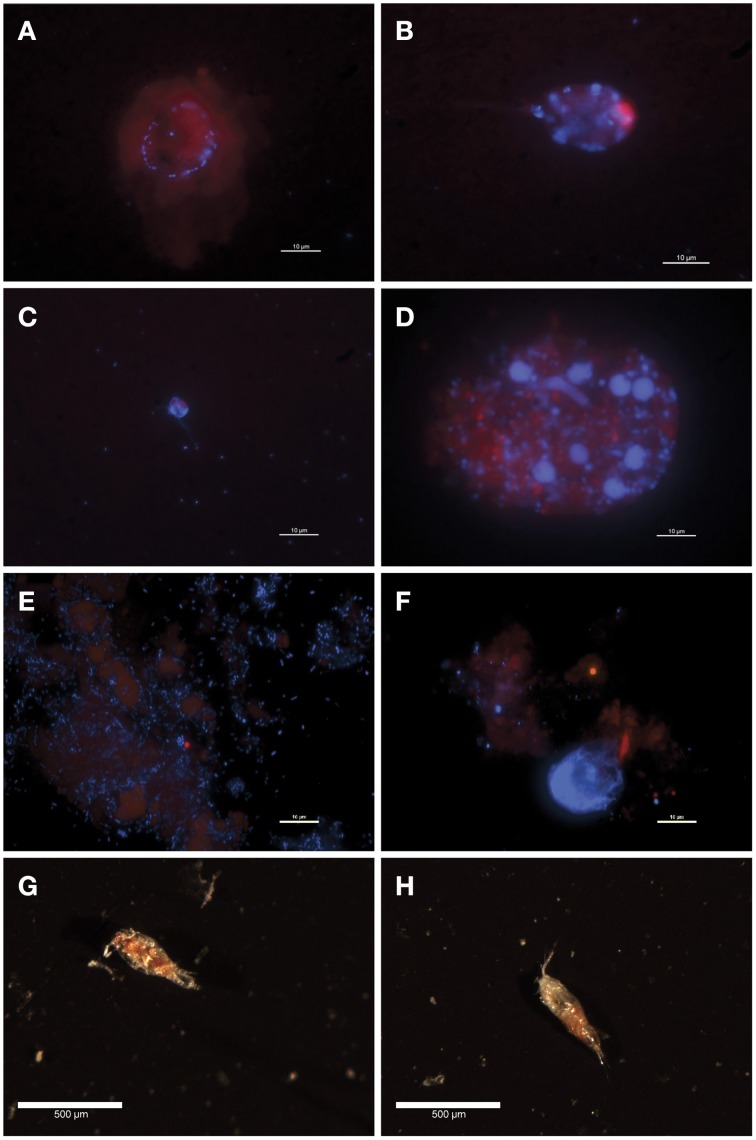
Figure 1.
Fluorescence (A–F) and optical (G,H) microscopy images of sediment trap POM. (A) 150m, poisoned trap; A circle of bacteria associated with a chlorophyll-containing particle (B) 200 m, poisoned trap; Unattached, ovoid pigmented cell in class Dinophyceae (C) 300 m, poisoned trap; A dually flagellated cell in class Chrysophyceae loosely surrounded by bacteria (D) 500 m, poisoned trap; A chlorophyll-containing particle covered with non-pigmented cells of varying sizes including flagellates in class Chrysophyceae and large bacteria (E) 150 m, live trap; Abundant bacteria, some associated with diffuse chlorophyll-containing particles (F) 150 m, poisoned trap; Sparsely distributed bacteria associated with a chlorophyll-containing particle (G) 150 m, live trap; Partially degraded copepod (H) 150 m, poisoned trap; Slightly degraded copepod.
Sequence data
The sequences reported in this paper have been deposited in the Genbank Short Read Archive (Bioproject PRJNA270248).
Results and discussion
Sinking particles were collected using a free-drifting sediment trap array deployed in the North Pacific Subtropical Gyre, from the base of the photic zone and into the mesopelagic [150, 200, 300, and 500 m (Figure A1 in Supplementary Material)]. The sediment traps included two treatments: a “live” trap which contained a solution of sterile seawater adjusted to a density of 1.05 g/cc with NaCl and a “poisoned” trap which contained a preservative to prevent in situ growth and preserve DNA. Fluorescence microscopy of trap-collected POM revealed bacteria associated with chlorophyll-containing particles in both live and poisoned sediment traps (Figures 1A,D–F). Bacteria in the sediment traps were either unattached, loosely surrounding protists (Figure 1C), or directly associating with amorphous particles (Figure 1D). Unattached bacteria were most abundant in 150 m live traps likely due to the opportunity for enhanced growth during deployment (Figure 1E).
Domain level taxonomic composition in live vs. poisoned sediment traps
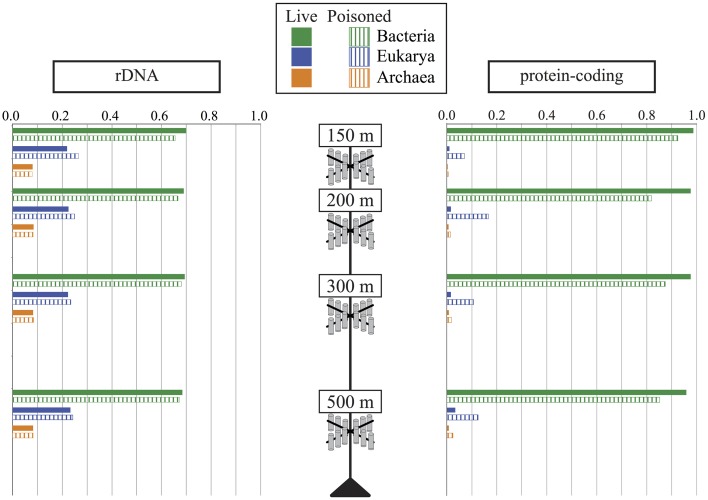
DNA was extracted and shotgun sequenced from both live and poisoned sediment trap particulates collected at the base of the photic zone and into the mesopelagic (Figure A1 in Supplementary Material) to determine the taxonomic and functional diversity associated with sinking particles. The diversity of the particle-associated microbes at the domain-level indicated the dominance of Bacteria and Eukarya in all traps at all depths sampled (Figure 2). Archaea represented less than 10% of total in both live and poisoned sediment trap microbial assemblages at all depths (Table 1).
Figure 2.
The relative abundance of Archaea, Bacteria, and Eukarya in live (filled bars) and poisoned (striped bars) sediment traps at the indicated depths as determined by the taxonomic identifications of small subunit ribosomal RNA genes (rDNA) and protein coding genes.
Table 1.
The relative abundance of Bacteria, Eukaryota, and Archaea in live (L) and dead (D) sediment traps.
| rDNAa | ||||||||
|---|---|---|---|---|---|---|---|---|
| 150Lb(%) | 150D (%) | 200L (%) | 200D (%) | 300L (%) | 300D (%) | 500L (%) | 500D (%) | |
| Bacteria | 70.0 | 65.6 | 69.1 | 66.7 | 69.4 | 68.1 | 68.4 | 67.3 |
| Eukaryota | 22.0 | 26.6 | 22.5 | 25.0 | 22.3 | 23.5 | 23.3 | 24.3 |
| Archaea | 8.0 | 7.8 | 8.4 | 8.2 | 8.3 | 8.5 | 8.3 | 8.3 |
| Protein-codingc | ||||||||
| 150L (%) | 150D (%) | 200L (%) | 200D (%) | 300L (%) | 300D (%) | 500L (%) | 500D (%) | |
| Bacteria | 98.7 | 92.4 | 97.6 | 81.9 | 97.6 | 87.4 | 95.9 | 85.1 |
| Eukaryota | 1.0 | 7.0 | 1.6 | 16.6 | 1.7 | 10.7 | 3.4 | 12.5 |
| Archaea | 0.3 | 0.6 | 0.7 | 1.5 | 0.8 | 1.9 | 0.8 | 2.4 |
Normalized relative abundance determined by rDNA, small subunit ribosomal RNA genes.
Numbers preceding L and D refer to sediment trap depths in meters (150, 200, 300, and 500 m).
Normalized relative abundance determined by protein-coding genes.
Eukaryotic rRNA genes were more highly represented than identifiable protein-encoding sequence reads, likely due to the inherently lower gene density in eukaryotes vs. bacteria, as well as the lack of closely related eukaryotic reference genomes for a number of highly represented taxa. Notably, the percentage of total annotated genes in poisoned traps was consistently lower than that of live traps (Table A1 in Supplementary Material). This result most likely reflects the higher relative abundance of eukaryotic DNA in the poisoned traps compared to live traps. In total, these results are consistent with previous observations that particles are more enriched in eukaryote-associated and unclassified protein-coding genes (Allen et al., 2013; Smith et al., 2013; Ganesh et al., 2014). The decreased ratio of eukaryote-to-bacteria DNA in the live traps was apparently due to in situ microbial growth in the trap during the deployment as POM underwent decomposition, and possibly concomitant degradation of the particle-associated eukaryotic nucleic acids.
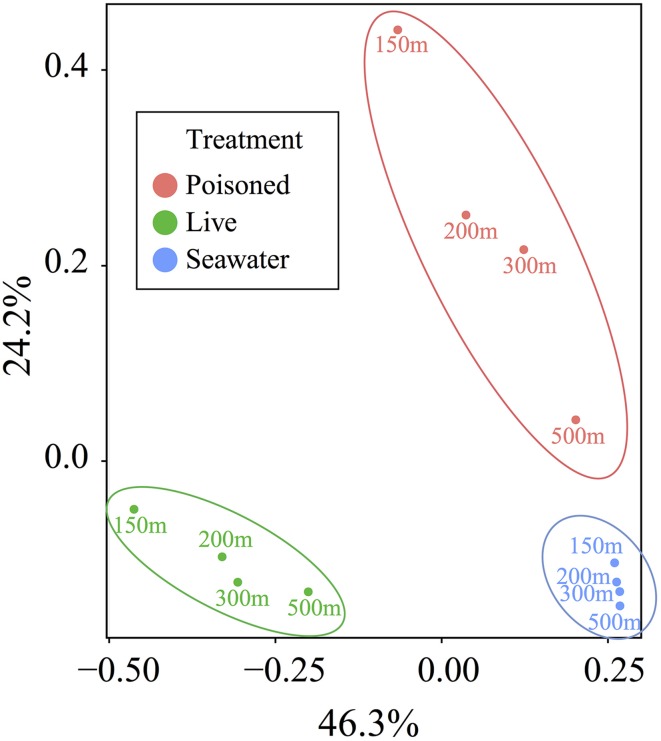
Principal coordinate analysis of protein-coding genes indicated the presence of distinctive communities in live traps, poisoned traps, and surrounding seawater (p < 0.001) and a much lesser effect of depth on community composition (Figure 3). We therefore focused subsequent analyses on the taxonomic and functional differences between live and poisoned sediment traps with minor attention given to depth-related differences.
Figure 3.
Principal coordinate analysis of the relative abundance of taxa, as determined by protein-coding genes, in live particle, poisoned particle, and seawater samples. Samples were grouped into significant clusters according to treatment, as assessed by non-parametric analysis of variance based on dissimilarities (p < 0.001).
Live vs. poisoned sediment trap bacterial assemblages
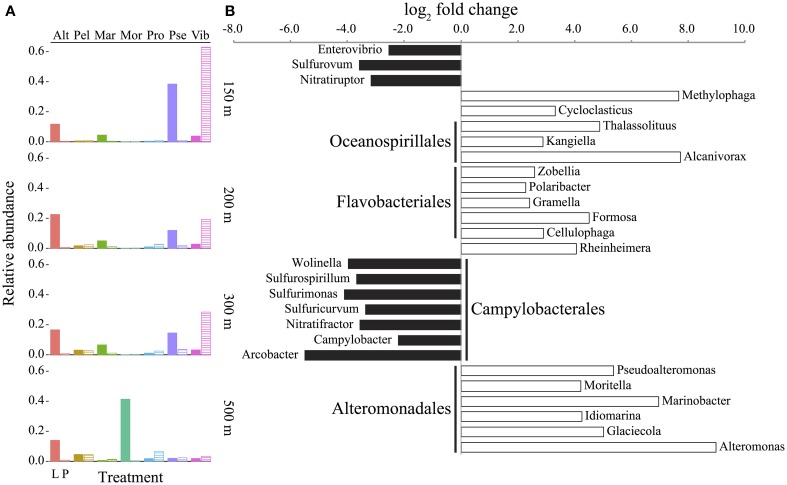
The most abundant bacterial genera in the live traps were affiliated with the order Alteromonadales (Alteromonas, Marinobacter, Moritella, and Pseudoalteromonas), in contrast to poisoned sediment traps where Vibrio was the most highly represented genus (Figure 4A). These trends were consistent across all sampled depths except 500 m, where in the poisoned traps there was a lower more even distribution of different bacterial groups, that included Pelagibacter which is typically more abundant in seawater and much less so on particles. The detection of Prochlorococcus DNA at 500 m in the poisoned traps may reflect their entrainment on particles or in fecal pellets and subsequent transport into deeper waters, perhaps reflecting the positive correlation between picophytoplankton productivity and their export to the deep sea (Richardson and Jackson, 2007).
Figure 4.
Abundance and enrichment of bacteria in sediment traps assessed by the taxonomic identification of protein-coding genes. (A) Genera with at least 2% mean abundance in traps relative to all sequences (including Archaea and Eukarya) in live (L, filled columns) and poisoned (P, striped columns) sediment traps at the indicated depths. Alt, Alteromonas; Pel, Candidatus Pelagibacter; Mar, Marinobacter; Mor, Moritella; Pro, Prochlorococcus; Pse, Pseudoalteromonas; Vib, Vibrio. (B) Representative bacterial genera (for full figure and dataset see Figure A3 and Dataset A1 in Supplementary Material) that are significantly enriched (FDR < 1%) in live (positive, white) or poisoned (negative, black) sediment traps. Depths were treated as biological replicates to identify statistically significant differences between live and poisoned traps (see Materials and Methods). Order-level identifications are listed in bold for those genera with at least three taxa represented in the comparison.
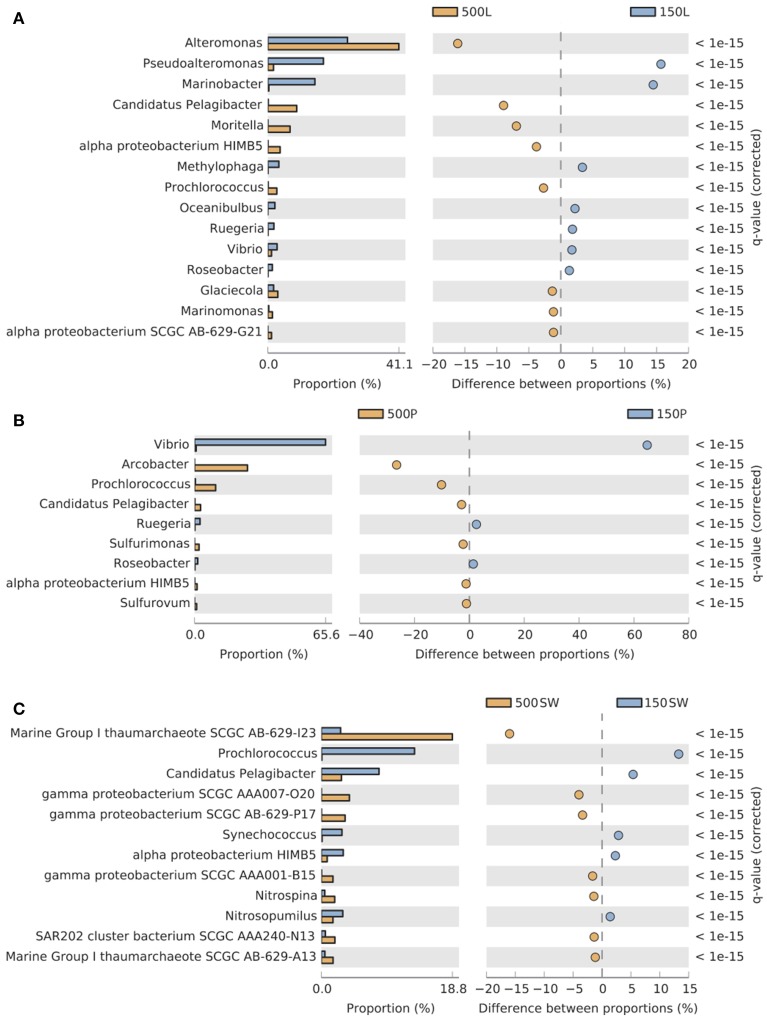
Significant depth-related partitioning in bacterial abundance in the live traps was detected for Alteromonadales, with Alteromonas, Moritella, and Glaciecola significantly enriched at 500 m and Pseudoalteromonas and Marinobacter enriched at 150 m (Figure 5A). In the poisoned traps, Vibrio was also enriched at 150 m (Figure 5B). Depth-related partitioning, also observed in seawater (Figure 5C), may be linked to different lifestyles of the bacteria. Marinobacter and Pseudoalteromonas are known to associate with eukaryotes (Thomas et al., 2008; Gärdes et al., 2010) and their enrichment at 150 m could be due to their attachment to eukaryotic biomass originating from the euphotic zone. Many alteromonads are typical r-strategists, capable of multiplying rapidly in response to nutrient-rich particles and Alteromonas species have been found in suspended particle fractions in a variety of ocean basins (Garcia-Martinez et al., 2002; López-Pérez et al., 2012). This may explain their overall high abundance in live traps and enrichment on 500 m particles (Figures 4A, 5A) where they outcompete k-strategists like Pelagibacter which are generally more abundant in seawater at this depth at Station ALOHA (DeLong et al., 2006).
Figure 5.
Pairwise comparisons of selected Bacterial and Archaeal taxa in 150 m vs. 500 m (A) live sediment traps, (B) poisoned sediment traps, and (C) seawater. Only taxa significantly enriched (FDR < 0.05) at shallow (blue, 150 m) and deep (orange, 500 m) depths with a difference between proportions >1 are shown. The bar plot displays the mean proportion of sequences assigned to each taxon in each sample.
At all depths, the live traps were significantly enriched in bacterial taxa previously implicated with particle-association, hydrocarbon and dissolved organic matter (DOM) degradation, and eukaryote associations. There was a significant enrichment of Oceanospirillales, Flavobacteriales, and Alteromonadales in live traps (Figure 4B and Figure A3 in Supplementary Material). Taxa within the Bacteroidetes, including Cytophaga and Flavobacteria, are often found associated with marine snow and phytoplankton blooms in marine environments (DeLong et al., 1993; Teeling et al., 2012). Many Flavobacteriales such as Zobellia, Gramella, Formosa, and Cellulophaga specialize in algal-derived organic matter degradation (Bauer et al., 2006; Bowman, 2006; Mann et al., 2013), which may be an ancestral trait of this group (Thomas et al., 2012). Methylophaga, Alteromonas, Idiomarina, Glaciecola, Rheinheimera, Polaribacter, and Formosa were also enriched in the live traps, and have also been detected in DOM enrichment events such as phytoplankton blooms and experimental microcosms (Brettar et al., 2006; McCarren et al., 2010; Teeling et al., 2012; von Scheibner et al., 2013). These bacterial types were either growing directly on POM, or on DOM generated in situ from POM degradation. Cycloclasticus, Thalassolituus, Alcanivorax, and Marinobacter were also enriched in live traps (Figure 4B). Species within these genera are often found in high abundance oil-contaminated marine environments, and include some obligately hydrocarbonoclastic species (Yakimov et al., 2007). Enrichment of hydrocarbons on marine POM has long been postulated due their hydrophobicity which may lead to their adsorption on marine POM (Lee et al., 1978; Evans et al., 1990). Recent studies have reported that obligately hydrocarbonoclastic bacteria may associate specifically with phytoplankton as well (Gutierrez et al., 2012). Since hydrocarbons comprise a measurable fraction of carbon in POM (Wakeham and Volkman, 1991) and lipids are the second largest identifiable POM compound class (Lee et al., 2004), these hydrocarbonoclastic species may be participating in the degradation of adsorbed hydrocarbons or those derived directly from eukaryotic plankton (Yoshimura and Hama, 2012; Wei et al., 2013). Several species within Marinobacter and Pseudoalteromonas are reportedly eukaryote-associated (Thomas et al., 2008; Gärdes et al., 2010) and might be expected to be well-represented in live and poisoned sediment traps. Their enrichment in live traps (Figure 4B), however, suggests that they may be actively growing and degrading their deceased eukaryotic hosts as has been suggested for a several marine symbionts (Grossart, 1999).
The poisoned traps were significantly enriched in chemoautotrophic bacterial types and those with eukaryote-associated lifestyles (Figure 4B and Figures A3, A4 in Supplementary Material). These included epsilon-proteobacteria, particularly Campylobacterales, in the poisoned traps (Figure 4B, Figure A3 in Supplementary Material). Presumptive chemoautotrophic sulfur-oxidizing bacteria including Sulfurimonas, Sulfurovum, and Sulfuricurvum (Campbell et al., 2006) were also enriched in the poisoned traps (Figure 4B, Figure A3 in Supplementary Material). Sulfurospirillum, which contains sulfur- and nitrate-reducing heterotrophic species (Stolz et al., 1999), was also enriched in poisoned traps. The presence of sulfur-oxidizing and -reducing taxa in poisoned traps is consistent with previous reports of these metabolic pathways on suspended particles (Fuchsman et al., 2011; Swan et al., 2011). Sulfurovum and other epsilon-proteobacterial species have also been found as ectosymbionts of marine invertebrates in both hydrothermal vent and coastal environments (Goffredi, 2010; Ruehland and Dubilier, 2010) suggesting a diverse habitat range for these bacteria on metazoan surfaces in niches where both reduced sulfur compounds and oxygen are readily available. Nitratiruptor and Nitratifractor are chemolithotrophic hydrogen-oxidizing denitrifiers (Nakagawa et al., 2005) that were also enriched in the poisoned traps (Figure 4B, Figure A3 in Supplementary Material). Denitrification is not considered to be a significant process in well-oxygenated seawater; however it may occur in anoxic microniches within large particles (Karl et al., 1984) or more likely within the intestinal tracts of decaying zooplankton carcasses that have been shown to provide anoxic niches for marine bacteria (Tang et al., 2011; Bickel and Tang, 2014).
Several particle-associated taxa enriched in the poisoned traps were closely related to well-described eukaryote-associated groups, that included Enterovibrio, Arcobacter, Wolinella, and Campylobacter (Figure 4B). Their presence in poisoned traps is consistent with the detection of diverse eukaryote-associated microbes in seawater and on marine metazoan surfaces (Gugliandolo et al., 2008; Preheim et al., 2011; Turner et al., 2014). Notably, Vibrionales genera were similarly enriched in both live and poisoned traps, as compared to seawater (Figure A4 in Supplementary Material). Many Vibrionales associate with diverse eukaryotes and are capable of degrading a wide variety of abundant marine biopolymers (Thompson et al., 2004; Takemura et al., 2014). Taken together, these data indicate that bacteria enriched in poisoned traps were most likely associated with eukaryotic surfaces or digestive tracts which is consistent with phyto- and zooplankton detritus constituting the major fraction of marine POM (Simon et al., 2002).
Live vs. poisoned sediment trap eukaryotic assemblages
The most abundant eukaryotic phyla across all depths in both live and poisoned sediment traps were Dinoflagellata, Protalveolata, Retaria, Metazoa, and unclassified Stramenopiles, which are likely representative of the composition of sinking particles at Station ALOHA (Figure A5 in Supplementary Material). Protists in class Dinophyceae were also identified microscopically in the 200-m poisoned trap (Figure 1B). Dinoflagellates, radiolarians, and foraminiferan protists, algae, metazoans, and heterotrophic protists were expected to be captured in live and poisoned sediment traps at similar rates.
Differences between live and poisoned traps were most pronounced for Metazoa and Retaria. The most abundant metazoa across live and poisoned traps were unclassified Maxillopoda and a variety of copepod genera (data not shown). Copepod taxa in live traps included Corycaeus, Clausocalanus, and Oithona while Scolecithrix and the ostracod Conchoecia were most abundant in poisoned traps (data not shown).
Retaria species were slightly more abundant in the poisoned vs. live traps with the most abundant taxa classified as Acantharia (poisoned vs. live mean abundance; 1.8% vs. 1.6%) and Polycystinea (2.8% vs. 2.4%). A colonial polycystine protist, Sphaerozoum, appeared significantly enriched in the poisoned trap (Figure A6 in Supplementary Material). The silica skeleton of polycystines may be solubilized rapidly in the upper water column at Station ALOHA, with approximately 40% lost within the mesopelagic (Lamborg et al., 2008). Again, in situ microbial degradation in the live traps are likely responsible for the differences in eukaryotic taxon abundance between live and poisoned traps. The detection of Acantharia, which have strontium sulfate skeletons, in both live and poisoned traps is notable because they are not typically detected in traps without the addition of strontium to the capture solution to inhibit their dissolution (Michaels et al., 1995). This suggests that acantharians detected in the poisoned trap were intact just prior to capture, and that remnants of their DNA in the live trap endured during the 12-day deployment.
Functional gene categories associated with live and poisoned sediment trap assemblages
KEGG pathways and genes associated with sediment trap DNA were surveyed to functionally profile live vs. poisoned sediment trap metagenome content. A large variety of pathways were significantly enriched in the live vs. poisoned traps, including those associated with motility, amino acid, carbohydrate, and energy metabolism, signal transduction, and cofactors and vitamin biosynthesis (Figure A7 in Supplementary Material). Notably, only the siderophore biosynthesis pathway was enriched in poisoned vs. live sediment trap microbial assemblages. Similarly, siderophore biosynthesis was significantly enriched in poisoned sediment traps compared to seawater (Figure A7 in Supplementary Material). In a recent study, a high frequency of strains containing siderophore biosynthetic genes were linked to eukaryote-associated lifestyles (Cordero et al., 2012), which were enriched in our poisoned sediment traps (Figure A4 in Supplementary Material). These siderophore biosynthesis pathways are likely affiliated with eukaryote-associated taxa since iron acquisition is important for microbial colonization of eukaryotes (Miethke and Marahiel, 2007). Pathways for membrane transport and cell motility were significantly enriched in both live and poisoned sediment traps, compared to seawater (Figure A7 in Supplementary Material).
Bacterial genes and pathways enriched in the live sediment traps
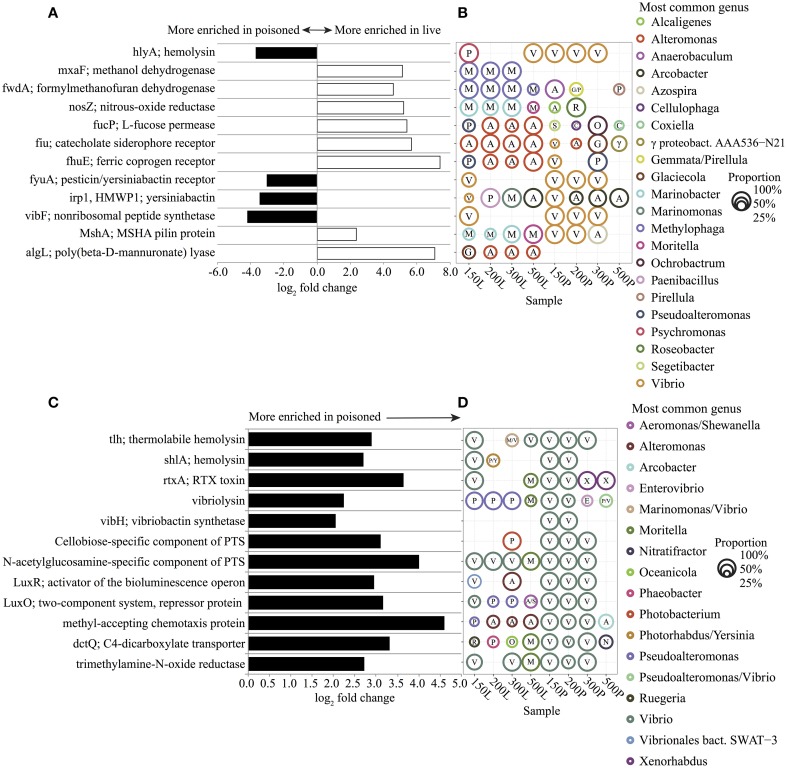
To explore the potential functions and metabolism associated with in situ particle degradation, we surveyed genes and metabolic pathways that were enriched in sediment trap bacterial DNA. Live sediment traps were found to be significantly enriched in genes for TonB-dependent iron transporters (TBDTs), assimilatory and dissimilatory single-carbon compound utilization, and polysaccharide utilization (Figures 6A,B). The majority of sequences matching TBDTs (fiu, fhuE) were associated with Alteromonas spp. (Figures 6A,B). Alteromonas-like gene sequences matching algae- (alginate) and diatom-derived polysaccharide (fucose) utilization were also prevalent (Figures 6A,B). Alginate and fucose utilization genes have been linked to phytoplankton decomposition and assimilation of diatom exopolysaccharides in previous studies (Teeling et al., 2012; Smith et al., 2013). Methylophaga-like gene sequences matching key genes of the ribulose monophosphate (RuMP) assimilatory pathway for formaldehyde fixation and detoxification (hxlA/hxlB) and the tetrahydromethanopterin (THMPT)-dependent dissimilatory formaldehyde oxidation pathway (FwdABC, ftr, mch) were also prevalent (Figures 6A,B, Dataset A2 in Supplementary Material). These data are consistent with previous microcosm experiments that showed the enrichment of Alteromonadaceae and Methylophaga phylotypes, as well as their expressed genes, including Alteromonadaceae TBDTs and Methylophaga-like key enzymes of the RuMP pathway and the THMPT-dependent pathway (Pinhassi et al., 2004; Neufeld et al., 2008; McCarren et al., 2010). Potential growth of these groups on DOM generated in the live trap by POM breakdown is also consistent with previous zooplankton and phytoplankton degradation studies that have demonstrated rapid accumulation of DOM over the course of just a few days during POM diagenesis (Yoshimura et al., 2009; Yoshimura and Hama, 2012).
Figure 6.
Representative significantly enriched KEGG genes (FDR < 1%) in live (positive, white) and poisoned (negative, black) sediment traps for comparisons between (A) live and poisoned and (C) poisoned and seawater. For full gene list see Dataset A2 in Supplementary Material. The most common genus of sequences matching the selected genes and its proportional representation within live (L) and poisoned (P) sediment traps at 150, 200, 300 and 500 meter depths (B,D). The first letter of each genus name is listed within each circle. The circle area represents the proportional representation of each genus within the specified sample. In cases where two genera each represented 50% of the sequences, both are listed.
Other bacterial genes enriched in the live sediment traps included those associated with denitrification from nitrate to N2 (nitrate reductase; narG, nitrate reductase; nirS, nitric oxide reductase; norB, and nitrous oxide reductase; nosZ). (Figures 6A,B, Dataset A2 in Supplementary Material) Genes encoding a mannose-sensitive hemagglutinin (MSHA) pilus were found associated with Marinobacter at depths between 150–300 m, and Moritella at 500 m (Figures 6A,B). The MSHA pilus has been implicated in attachment to animal surfaces in Vibrio and Pseudoalteromonas (Chiavelli et al., 2001; Dalisay, 2006) and it may also play a role in attachment by Moritella and Marinobacter, which have previously been shown to associate with eukaryotes (Gärdes et al., 2011; Tunsjø et al., 2011). The pilA gene encoding the pilin protein of a novel chitin-regulated pilus (ChRP;K02650) along with a chitin-binding protein gene (CBP; K03933) were also highly enriched in live traps (Dataset A2 in Supplementary Material). In addition, a variety of heavy-metal resistance genes associated with Alteromonadales were enriched in the live traps (Figures A8A,B and Dataset A2 in Supplementary Material). The czcABCD genes encode a heavy metal efflux pump involved in resistance to cobalt, zinc, and cadmium that were affiliated with Alteromonas. CusAB and cusRS encode copper efflux proteins and copper two-component sensor systems, respectively, that were affiliated with Alteromonas, Glaciecola, and Marinobacter. The genes involved in mercury resistance (merABR) and transport (merTP) were affiliated with Alteromonas and Marinobacter. The enrichment of metal resistance genes in the poisoned traps may be linked to their growth on particles, which are known to concentrate heavy metals (Puig et al., 1999).
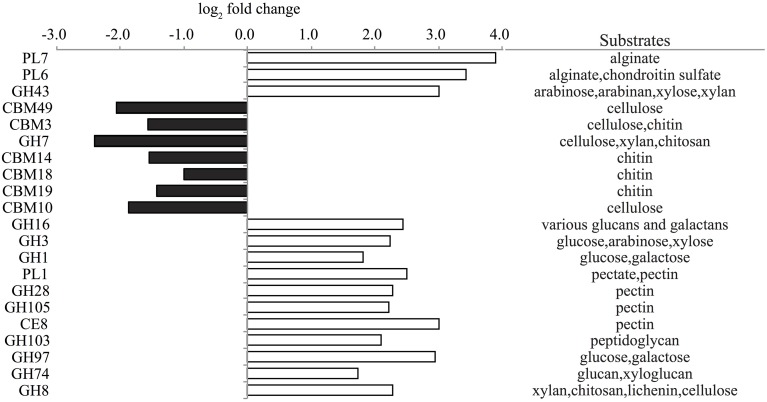
To further evaluate the potential for carbohydrate degradation capabilities of trap associated microbial assemblages, peptide encoding sequences were compared to a carbohydrate-active enzymes database (CAZymes) (Yin et al., 2012). A large number of glycoside hydrolase (GH) families associated with polysaccharide degradation found in algae and bacterial cell walls including arabinose, pectin, cellulose, and peptidoglycan were significantly enriched in the live traps (Figure 7). Polysaccharide lysase (PL) families 6,7 and 1 and carbohydrate esterase family 8 complement the degradation potential of GHs (Cantarel et al., 2009) and potentially may enhance the degradation of the algal substrates alginate and pectin in live traps (Figure 7). Several carbohydrate-binding module (CBM) families targeting chitin (e.g., CBM1, CBM14, and CBM18) were also enriched in live traps (Dataset A4 in Supplementary Material).
Figure 7.
Representative significantly enriched CAZy families (FDR < 1%) in live (positive, white) and poisoned (negative, black) sediment traps along with their potential substrates. For full family list, see Dataset A4 in Supplementary Material. PL, polysaccharide lyase; GH, glycoside hydrolase; CBM, carbohydrate-binding modules; CE, carbohydrate esterase.
Bacterial genes and pathways enriched in the poisoned sediment traps
Poisoned sediment trap particles were significantly enriched in a variety of iron-scavenging genes and virulence factors, primarily associated with Vibrio spp. (Figures 6A–D). Vibrio spp. are known to engage in pathogenic, symbiotic, and saprophytic associations with a wide variety of eukaryotes in the marine environment (Takemura et al., 2014). Vibrio-like genes for carbohydrate uptake and chemotaxis, supporting eukaryote-associated lifestyles, were significantly enriched in poisoned sediment traps (Figures 6C,D). Vibrio genes for chitin utilization, including those associated with sensing, attachment, degradation, and uptake of chitin derivatives (Keyhani and Roseman, 1999; Beier and Bertilsson, 2013) were also enriched in the poisoned sediment traps. These included a methyl-accepting chemotaxis protein that mediates a chemotactic response to N-acetylglucosamine (GlcNAc) (Meibom et al., 2004) and a CBP (K03933; Cazy AA10) that mediates Vibrio spp. attachment to chitin surfaces and enzymatically cleaves chitin, which were both highly enriched in poisoned traps (Figures 6C,D and Dataset A2 in Supplementary Material) (Vaaje-Kolstad et al., 2010; Frederiksen et al., 2013). Transporters mediating uptake of cellobiose and GlcNAc were also highly enriched in poisoned traps (Figures 6C,D). Together, these data support the association of Vibrio in poisoned traps with chitin substrates, and are consistent with the presence of copepods detected in DNA analyses and in optical microscopy (Figure 1H) in the same traps.
A variety of genes involved in quorum sensing and anaerobic metabolism, also associated with Vibrio spp., were significantly enriched in poisoned traps (Figures 6C,D). They included the luxS-luxP/Q quorum-sensing system, the luxOR bioluminescence regulators, trimethylamine N-oxide (TMAO) reductase, and the TMAO two-component regulatory sensors (torR/S) (Figures 6C,D and Dataset A2 in Supplementary Material). TMAO is an abundant osmolyte found in the tissues of marine eukaryotes that can be utilized aerobically and anaerobically by diverse marine bacteria (Barrett and Kwan, 1985; Proctor and Gunsalus, 2000; Sun et al., 2011). Enrichment of TMAO genes is thus consistent with eukaryotic association in live animals or sedimenting particles that entered the poisoned traps.
Comparisons with the CAZyme database revealed that a variety of CBM families targeting cellulose and chitin were enriched in the poisoned traps (Figure 7). CBMs complement the activity of other enzymes by promoting extended interactions with substrates (Cantarel et al., 2009). Further, GH families 7 and 19 catalyze the degradation of cellulose and chitin and were highly enriched in poisoned traps as compared to live traps and seawater, respectively (Figure 7 and Dataset A4 in Supplementary Material). Auxiliary activity (AA) family 10, formerly classified as CBM family 33, capable of cleaving chitin and cellulose (Aachmann et al., 2012) was enriched in both live and poisoned traps, as compared to seawater (Dataset A4 in Supplementary Material). These data support KEGG functional profiles indicating the potential for chitin degradation in both live and poisoned sediment traps.
Conclusion
Sinking particles represent the primary vehicles of organic carbon flux from surface waters to the deep ocean (Volk and Hoffert, 1985), yet to date, few data are available on the specific microbes and metabolic pathways responsible for POM degradation throughout the water column. There is a general consensus that particles represent hotspots of microbial activity in the ocean (Karl and Knauer, 1984; Turley and Mackie, 1994; Crump et al., 1999; Bochdansky et al., 2010; Smith et al., 2013), but the nature of those processes and microorganisms responsible still need to be better described.
In this study, sediment trap metagenomic analyses revealed dramatic differences in the taxonomic diversity and functional potential of microbes associated with sinking particles in poisoned sediment traps, compared to those that grew in situ in live traps. Both live and poisoned sediment trap microbial assemblages were distinctly different from those found in seawater, which is consistent with the conclusions of several recent studies (LeCleir et al., 2013; Smith et al., 2013). Live particle-trap assemblages shared many similarities with communities found in microcosm DOM enrichments, with the added dimension of known particle-associated bacteria (e.g., Flavobacteriales) and potentially eukaryote-associated bacteria (e.g., Pseudoalteromonas and Marinobacter). The functional gene content in live traps pointed to the potential for growth by alteromonads on labile DOM produced in situ from sinking POM. Apparently, the contained environment within the sediment trap acted similarly to microcosm enrichment experiments, where fast-growing copiotrophic bacteria out-competed the particle- and eukaryote-associated bacteria for the nutrients available in the trap. The poisoned sediment trap-associated metagenomic analyses provided a clear contrast to live traps and presumably reflected the biological material and microbial assemblages associated with sinking particles. The differences in composition between live and poisoned traps were much greater than depth associated differences, consistent with previous studies of suspended particulates found in oxygen minimum zones (Ganesh et al., 2014).
In total, these findings are consistent with a previous study that suggested initial particle-colonizers are surface-colonizing (or eukaryote-associated) specialists (LeCleir et al., 2013). Our metagenomic data further indicated that microbes in poisoned sediment traps were often associated with eukaryotic surfaces and intestinal tracts as symbionts, pathogens, or saprophytes. Some of these eukaryote-associated bacteria may alternate between symbiotic to pathogenic or saprophytic lifestyles, as has been shown for some phytoplankton symbionts (Grossart, 1999; Seyedsayamdost et al., 2011). The functional gene content in poisoned traps, which included a variety of genes involved in virulence, anaerobic metabolism, attachment to chitinaceous surfaces, and chitin degradation were consistent with this conclusion. Notably, genes for attachment to chitinaceous surfaces and anaerobic metabolism were also detected in live traps, though they were associated with a different set of microbial taxa. Thus, eukaryote-associated communities captured in live and poisoned traps differed, most likely due to bacterial growth in the live trap.
Our data also provide new perspective on the taxonomic identity of the particulate matter itself, namely the eukaryotic taxa that contribute to the complex mixture of detritus and minerals that make up marine particles. While previous studies have reported marine particles as consisting of eukaryote-derived detritus, the analyses we report here suggests that specific interactions between eukaryotes and bacteria may be centrally important in the transport and degradation processes associated with sinking POM. The presence of chitinaceous surfaces provides a habitat for a specialized bacterial community adapted to sense, attach, degrade, and take up chitin derivatives. These same habitats appeared to coincide with the development of copiotrophs known to respond rapidly to labile DOM inputs. The probable sources of labile DOM include turnover of phytoplankton captured in the traps, the excretions of swimming zooplankton, and substrates from the degradation of captured chitinaceous detritus.
This study provides a baseline for understanding microbial community assemblages and metabolic activities associated with the transport and degradation of sinking POM. To date, gene expression associated with POM degradation has not yet been reported, partly due to the technical difficulties associated with preserving RNA in situ. Future metatranscriptomic analyses have the potential to identify those metabolic pathways that are expressed in situ on sinking particles and help to define the processes that actively drive particle degradation in the ocean's interior. Finer scale studies of particle transport and degradation should also help to define hypothetical successional cascades that reflect sequential processing of POM to DOM during its transport to the deep-sea.
Author contributions
KF, ED, DK designed research. JE contributed bioinformatics tools. KF performed research. TS performed microscopy. KF, JE, and ED analyzed data. KF, ED, and DK wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the C-MORE team, the captain and crew of the R/V Kilo Moana for their expert assistance at sea, Tara Clemente for assistance deploying and processing sediment traps, HOE-DYLAN chief scientist Sam Wilson, and Oscar Sosa for collecting the sediment trap samples. This work was supported by grants #3777 and #3794 from the Gordon and Betty Moore Foundation (ED, DK, respectively), a gift from the Agouron Institute (AI-MO9.12.1) to ED, and NSF Science and Technology Center grant, Center for Microbial Oceanography, Research and Education EF0424599 (DK, ED), and grants from the Simons Foundation (Award number 329108 to DK, ED). KF was supported by a NSF Postdoctoral Research Fellowship in Biology DBI-1202684. This work is a contribution of the Center for Microbial Oceanography: Research and Education (C-MORE) and the Simons Collaboration on Ocean Processes and Ecology (SCOPE).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00469/abstract
References
- Aachmann F. L., Sørlie M., Skjåk-Bræk G., Eijsink V. G. H., Vaaje-Kolstad G. (2012). NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc. Natl. Acad. Sci. U.S.A. 109, 18779–18784. 10.1073/pnas.1208822109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A. E., Allen L. Z., McCrow J. P. (2013). Lineage specific gene family enrichment at the microscale in marine systems. Curr. Opin. Microbiol. 16, 605–617. 10.1016/j.mib.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Allen L. Z., Allen E. E., Badger J. H., McCrow J. P., Paulsen I. T., Elbourne L. D., et al. (2012). Influence of nutrients and currents on the genomic composition of microbes across an upwelling mosaic. ISME J. 6, 1403–1414. 10.1038/ismej.2011.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E. L., Kwan H. S. (1985). Bacterial reduction of trimethylamine oxide. Annu. Rev. Microbiol. 39, 131–149. 10.1146/annurev.mi.39.100185.001023 [DOI] [PubMed] [Google Scholar]
- Bauer M., Kube M., Teeling H., Richter M., Lombardot T., Allers E., et al. (2006). Whole genome analysis of the marine Bacteroidetes “Gramella forsetii” reveals adaptations to degradation of polymeric organic matter. Environ. Microbiol. 8, 2201–2213. 10.1111/j.1462-2920.2006.01152.x [DOI] [PubMed] [Google Scholar]
- Beier S., Bertilsson S. (2013). Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front. Microbiol. 4:149. 10.3389/fmicb.2013.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel S. L., Tang K. W. (2014). Carbon substrate usage by zooplankton-associated bacteria, phytoplankton-associated bacteria, and free-living bacteria under aerobic and anaerobic conditions. Mar. Biol. 161, 2233–2242. 10.1007/s00227-014-2501-z [DOI] [Google Scholar]
- Bochdansky A. B., van Aken H. M., Herndl G. J. (2010). Role of macroscopic particles in deep-sea oxygen consumption. Proc. Natl. Acad. Sci. U.S.A. 107, 8287. 10.1073/pnas.0913744107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. P. (2006). The marine clade of the family flavobacteriaceae: the genera Aequorivita, Arenibacter, Cellulophaga, Croceibacter, Formosa, Gelidibacter, Gillisia, Maribacter, Mesonia, Muricauda, Polaribacter, Psychroflexus, Psychroserpens, Robiginitalea, Salegentibacter, Tenacibaculum, Ulvibacter, Vitellibacter and Zobellia, in The Prokaryotes, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York, NY: Springer; ), 677–694. [Google Scholar]
- Boyd P. W., Sherry N. D., Berges J. A., Bishop J. K. B., Calvert S. E., Charette M. A., et al. (1999). Transformations of biogenic particulates from the pelagic to the deep ocean realm. Deep Sea Res. II Top. Stud. Oceanogr. 46, 2761–2792. 10.1016/S0967-0645(99)00083-1 [DOI] [Google Scholar]
- Brettar I., Christen R., Höfle M. G. (2006). Rheinheimera perlucida sp. nov., a marine bacterium of the Gammaproteobacteria isolated from surface water of the central Baltic Sea. Int. J. Syst. Evol. Microbiol. 56, 2177–2183. 10.1099/ijs.0.64172-0 [DOI] [PubMed] [Google Scholar]
- Buesseler K. O., Trull T. W., Steinberg D. K., Silver M. W., Siegel D. A., Saitoh S. I., et al. (2008). VERTIGO (VERtical Transport In the Global Ocean): a study of particle sources and flux attenuation in the North Pacific. Deep Sea Res. II Top. Stud. Oceanogr. 55, 1522–1539. 10.1016/j.dsr2.2008.04.024 [DOI] [Google Scholar]
- Campbell B. J., Engel A. S., Porter M. L., Takai K. (2006). The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468. 10.1038/nrmicro1414 [DOI] [PubMed] [Google Scholar]
- Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiavelli D. A., Marsh J. W., Taylor R. K. (2001). The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67, 3220–3225. 10.1128/AEM.67.7.3220-3225.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero O. X., Ventouras L.-A., DeLong E. F., Polz M. F. (2012). Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. U.S.A. 109, 20059–20064. 10.1073/pnas.1213344109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo B. G., Pommier T., Fernández Gómez B., Pedrós Alió C. (2013). Taxonomic composition of the particle-attached and free-living bacterial assemblages in the Northwest Mediterranean Sea analyzed by pyrosequencing of the 16S rRNA. Microbiologyopen 2, 541–552. 10.1002/mbo3.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump B. C., Armbrust E. V., Baross J. A. (1999). Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65, 3192–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalisay D. S. (2006). A mannose-sensitive haemagglutinin (MSHA)-like pilus promotes attachment of Pseudoalteromonas tunicata cells to the surface of the green alga Ulva australis. Microbiology 152, 2875–2883. 10.1099/mic.0.29158-0 [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Franks D. G., Alldredge A. L. (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934. 10.4319/lo.1993.38.5.0924 [DOI] [Google Scholar]
- DeLong E. F., Preston C. M., Mincer T., Rich V., Hallam S. J., Frigaard N. U., et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311, 496–503. 10.1126/science.1120250 [DOI] [PubMed] [Google Scholar]
- Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. 10.1111/j.1654-1103.2003.tb02228.x [DOI] [Google Scholar]
- Eddy S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloe E. A., Shulse C. N., Fadrosh D. W., Williamson S. J., Allen E. E., Bartlett D. H. (2011). Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ. Microbiol. Rep. 3, 449–458. 10.1111/j.1758-2229.2010.00223.x [DOI] [PubMed] [Google Scholar]
- Evans K. M., Gill R. A., Robotham P. W. J. (1990). The PAH and organic content of sediment particle size fractions. Water Air Soil Pollut. 51, 13–31. 10.1007/BF00211500 [DOI] [Google Scholar]
- Fandino L. B., Riemann L., Steward G. F., Azam F. (2005). Population dynamics of Cytophaga-Flavobacteria during marine phytoplankton blooms analyzed by real-time quantitative PCR. Aquat. Microb. Ecol. 40, 251–257. 10.3354/ame040251 [DOI] [Google Scholar]
- Frederiksen R. F., Paspaliari D. K., Larsen T., Storgaard B. G., Larsen M. H., Ingmer H., et al. (2013). Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology (Reading, Engl.) 159, 833–847. 10.1099/mic.0.051839-0 [DOI] [PubMed] [Google Scholar]
- Fuchsman C. A., Kirkpatrick J. B., Brazelton W. J., Murray J. W., Staley J. T. (2011). Metabolic strategies of free-living and aggregate-associated bacterial communities inferred from biologic and chemical profiles in the Black Sea suboxic zone. FEMS Microbiol. Ecol. 78, 586–603. 10.1111/j.1574-6941.2011.01189.x [DOI] [PubMed] [Google Scholar]
- Fuchsman C. A., Staley J. T., Oakley B. B., Kirkpatrick J. B., Murray J. W. (2012). Free-living and aggregate-associated Planctomycetes in the Black Sea. FEMS Microbiol. Ecol. 80, 402–416. 10.1111/j.1574-6941.2012.01306.x [DOI] [PubMed] [Google Scholar]
- Ganesh S., Parris D. J., DeLong E. F., Stewart F. J. (2014). Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 8, 187–211. 10.1038/ismej.2013.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez J., Acinas S. G., Massana R., Rodriguez-Valera F. (2002). Prevalence and microdiversity of Alteromonas macleodii-like microorganisms in different oceanic regions. Environ. Microbiol. 4, 42–50. 10.1046/j.1462-2920.2002.00255.x [DOI] [PubMed] [Google Scholar]
- Gärdes A., Iversen M. H., Grossart H.-P., Passow U., Ullrich M. S. (2011). Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 5, 436–445. 10.1038/ismej.2010.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärdes A., Kaeppel E., Shehzad A., Seebah S., Teeling H., Yarza P., et al. (2010). Complete genome sequence of Marinobacter adhaerens type strain (HP15), a diatom-interacting marine microorganism. Stand. Genomic Sci. 3, 97–107. 10.4056/sigs.922139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffredi S. K. (2010). Indigenous ectosymbiotic bacteria associated with diverse hydrothermal vent invertebrates. Environ. Microbiol. Rep. 2, 479–488. 10.1111/j.1758-2229.2010.00136.x [DOI] [PubMed] [Google Scholar]
- Grossart H. P. (1999). Interactions between marine bacteria and axenic diatoms (Cylindrotheca fusiformis, Nitzschia laevis, and Thalassiosira weissflogii) incubated under various conditions in the lab. Aquat. Microb. Ecol. 19, 1–11. 10.3354/ame019001 [DOI] [Google Scholar]
- Gugliandolo C., Irrera G. P., Lentini V., Maugeri T. L. (2008). Pathogenic Vibrio, Aeromonas and Arcobacter spp. associated with copepods in the Straits of Messina (Italy). Mar. Pollut. Bull. 56, 600–606. 10.1016/j.marpolbul.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Gutierrez T., Nichols P. D., Whitman W. B., Aitken M. D. (2012). Porticoccus hydrocarbonoclasticus sp. nov., an aromatic hydrocarbon-degrading bacterium identified in laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 78, 628–637. 10.1128/AEM.06398-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo S., Manganini S. J., Krishfield R. A., Francois R. (2008). Particulate organic carbon fluxes to the ocean interior and factors controlling the biological pump: a synthesis of global sediment trap programs since 1983. Prog. Oceanogr. 76, 217–285. 10.1016/j.pocean.2007.11.003 [DOI] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D. M., Knauer G. A. (1984). Detritus-microbe interactions in the marine pelagic environment - selected results from the VERTEX experiment. Bull. Mar. Sci. 35, 550–565. [Google Scholar]
- Karl D. M., Knauer G. A., Martin J. H., Ward B. B. (1984). Bacterial chemolithotrophy in the ocean is associated with sinking particles. Nature 309, 54–56. 10.1038/309054a0 [DOI] [Google Scholar]
- Kellogg C. T., Deming J. W. (2009). Comparison of free-living, suspended particle, and aggregate-associated bacterial and archaeal communities in the Laptev Sea. Aquat. Microb. Ecol. 57, 1–18. 10.3354/ame01317 [DOI] [Google Scholar]
- Keyhani N. O., Roseman S. (1999). Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473, 108–122. 10.1016/S0304-4165(99)00172-5 [DOI] [PubMed] [Google Scholar]
- Kiełbasa S. M., Wan R., Sato K., Horton P., Frith M. C. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493. 10.1101/gr.113985.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer G. A., Karl D. M., Martin J. H., Hunter C. N. (1984). In situ effects of selected preservatives on total carbon, nitrogen and metals collected in sediment traps. J. Mar. Res. 42, 445–462. 10.1357/002224084788502710 [DOI] [Google Scholar]
- Knauer G. A., Martin J. H., Bruland K. W. (1979). Fluxes of particulate carbon, nitrogen, and phosphorus in the upper water column of the northeast Pacific. Deep Sea Res. 26, 97–108. 10.1016/0198-0149(79)90089-X [DOI] [Google Scholar]
- Kopylova E., Noé L., Touzet H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. 10.1093/bioinformatics/bts611 [DOI] [PubMed] [Google Scholar]
- Lamborg C. H., Buesseler K. O., Valdes J., Bertrand C. H., Bidigare R., Manganini S., et al. (2008). The flux of bio- and lithogenic material associated with sinking particles in the mesopelagic “twilight zone” of the northwest and North Central Pacific Ocean. Deep Sea Res. II Top. Stud. Oceanogr. 55, 1540–1563. 10.1016/j.dsr2.2008.04.011 [DOI] [Google Scholar]
- LeCleir G. R., DeBruyn J. M., Maas E. W., Boyd P. W., Wilhelm S. W. (2013). Temporal changes in particle-associated microbial communities after interception by nonlethal sediment traps. FEMS Microbiol. Ecol. 87, 153–163. 10.1111/1574-6941.12213 [DOI] [PubMed] [Google Scholar]
- Lee C., Wakeham S., Arnosti C. (2004). Particulate organic matter in the sea: the composition conundrum. Ambio 33, 565–575. 10.1639/0044-7447(2004)033[0565:POMITS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lee R. F., Gardner W. S., Anderson J. W., Blaylock J. W., Barwell-Clarke J. (1978). Fate of polycyclic aromatic hydrocarbons in controlled ecosystem enclosures. Environ. Sci. Technol. 12, 832–838. 10.1021/es60143a007 [DOI] [Google Scholar]
- Lincoln S. A., Wai B., Eppley J. M., Church M. J., Summons R. E., DeLong E. F. (2014). Planktonic Euryarchaeota are a significant source of archaeal tetraether lipids in the ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 9858–9863. 10.1073/pnas.1409439111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Pérez M., Gonzaga A., Martin-Cuadrado A.-B., Onyshchenko O., Ghavidel A., Ghai R., et al. (2012). Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci. Rep. 2, 696. 10.1038/srep00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann A. J., Hahnke R. L., Huang S., Werner J., Xing P., Barbeyron T., et al. (2013). The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl. Environ. Microbiol. 79, 6813–6822. 10.1128/AEM.01937-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. H., Knauer G. A., Karl D. M., Broenkow W. W. (1987). VERTEX: carbon cycling in the northeast Pacific. Deep Sea Res. 34, 267–285. 10.1016/0198-0149(87)90086-0 [DOI] [Google Scholar]
- Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., Neufeld J. D. (2012). PANDAseq: PAired-eND Assembler for Illumina sequences. BMC Bioinformatics 13:31. 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarren J., Becker J. W., Repeta D. J., Shi Y., Young C. R., Malmstrom R. R., et al. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U.S.A. 107, 16420–16427. 10.1073/pnas.1010732107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCave I. N. (1975). Vertical flux of particles in the ocean. Deep Sea Res. 22, 491–502. 10.1016/0011-7471(75)90022-4 [DOI] [Google Scholar]
- McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom K. L., Li X. B., Nielsen A. T., Wu C.-Y., Roseman S., Schoolnik G. K. (2004). The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101, 2524–2529. 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels A. F., Caron D. A., Swanberg N. R., Howse F. A., Michaels C. M. (1995). Planktonic sarcodines (Acantharia, Radiolaria, Foraminifera) in surface waters near Bermuda: abundance, biomass and vertical flux. J. Plankton Res. 17, 131–163. 10.1093/plankt/17.1.131 [DOI] [Google Scholar]
- Miethke M., Marahiel M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeseneder M. M., Winter C., Herndl G. J. (2001). Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol. Oceanogr. 46, 95–107. 10.4319/lo.2001.46.1.0095 [DOI] [Google Scholar]
- Nakagawa S., Takai K., Inagaki F., Horikoshi K., Sako Y. (2005). Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the epsilon-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55, 925–933. 10.1099/ijs.0.63480-0 [DOI] [PubMed] [Google Scholar]
- Nebbioso A., Piccolo A. (2013). Molecular characterization of dissolved organic matter (DOM): a critical review. Anal. Bioanal. Chem. 405, 109–124. 10.1007/s00216-012-6363-2 [DOI] [PubMed] [Google Scholar]
- Neufeld J. D., Boden R., Moussard H., Schafer H., Murrell J. C. (2008). Substrate-specific clades of active marine methylotrophs associated with a phytoplankton bloom in a temperate coastal environment. Appl. Environ. Microbiol. 74, 7321–7328. 10.1128/AEM.01266-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D. H., Beiko R. G. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26, 715–721. 10.1093/bioinformatics/btq041 [DOI] [PubMed] [Google Scholar]
- Patel A., Noble R. T., Steele J. A., Schwalbach M. S., Hewson I., Fuhrman J. A. (2007). Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2, 269–276. 10.1038/nprot.2007.6 [DOI] [PubMed] [Google Scholar]
- Pinhassi J., Sala M. M., Havskum H., Peters F., Guadayol Ò., Malits A., et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766. 10.1128/AEM.70.11.6753-6766.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preheim S. P., Boucher Y., Wildschutte H., David L. A., Veneziano D., Alm E. J., et al. (2011). Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ. Microbiol. 13, 265–275. 10.1111/j.1462-2920.2010.02328.x [DOI] [PubMed] [Google Scholar]
- Proctor L. M., Gunsalus R. P. (2000). Anaerobic respiratory growth of Vibrio harveyi, Vibrio fischeri and Photobacterium leiognathi with trimethylamine N−oxide, nitrate and fumarate: ecological implications. Environ. Microbiol. 2, 399–406. 10.1046/j.1462-2920.2000.00121.x [DOI] [PubMed] [Google Scholar]
- Puig P., Palanques A., Sanchez-Cabeza J. A., Masqué P. (1999). Heavy metals in particulate matter and sediments in the southern Barcelona sedimentation system (North-western Mediterranean). Mar. Chem. 63, 311–329. 10.1016/S0304-4203(98)00069-3 [DOI] [Google Scholar]
- Richardson T. L., Jackson G. A. (2007). Small phytoplankton and carbon export from the surface ocean. Science 315, 838–840. 10.1126/science.1133471 [DOI] [PubMed] [Google Scholar]
- Riemann L., Steward G. F., Azam F. (2000). Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66, 578–587. 10.1128/AEM.66.2.578-587.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruehland C., Dubilier N. (2010). Gamma−and epsilonproteobacterial ectosymbionts of a shallow−water marine worm are related to deep−sea hydrothermal vent ectosymbionts. Environ. Microbiol. 12, 2312–2326. 10.1111/j.1462-2920.2010.02256.x [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost M. R., Carr G., Kolter R., Clardy J. (2011). Roseobacticides: small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 133, 18343–18349. 10.1021/ja207172s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Grossart H. P., Schweitzer B., Ploug H. (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211. 10.3354/ame028175 [DOI] [Google Scholar]
- Smith D. C., Simon M., Alldredge A. L., Azam F. (1992). Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359, 139–142. 10.1038/359139a0 [DOI] [Google Scholar]
- Smith M. W., Zeigler Allen L., Allen A. E., Herfort L., Simon H. M. (2013). Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front. Microbiol. 4:120. 10.3389/fmicb.2013.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz J. F., Ellis D. J., Blum J. S., Ahmann D., Lovley D. R., Oremland R. S. (1999). Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the epsilon Proteobacteria. Int. J. Syst. Bacteriol. 49, 1177–1180. 10.1099/00207713-49-3-1177 [DOI] [PubMed] [Google Scholar]
- Sun J., Steindler L., Thrash J. C., Halsey K. H., Smith D. P., Carter A. E., et al. (2011). One carbon metabolism in SAR11 pelagic marine bacteria. PLoS ONE 6:e23973. 10.1371/journal.pone.0023973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan B. K., Martinez-Garcia M., Preston C. M., Sczyrba A., Woyke T., Lamy D., et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1300. 10.1126/science.1203690 [DOI] [PubMed] [Google Scholar]
- Takemura A. F., Chien D. M., Polz M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5:38. 10.3389/fmicb.2014.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K. W., Glud R. N., Glud A., Rysgaard S., Nielsen T. G. (2011). Copepod guts as biogeochemical hotspots in the sea: evidence from microelectrode profiling of Calanus spp. Limnol. Oceanogr. 56, 666–672. 10.4319/lo.2011.56.2.0666 [DOI] [Google Scholar]
- Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Thomas F., Barbeyron T., Tonon T., Génicot S., Czjzek M., Michel G. (2012). Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ. Microbiol. 14, 2379–2394. 10.1111/j.1462-2920.2012.02751.x [DOI] [PubMed] [Google Scholar]
- Thomas T., Evans F. F., Schleheck D., Mai-Prochnow A., Burke C., Penesyan A., et al. (2008). Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS ONE 3:e3252. 10.1371/journal.pone.0003252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson F. L., Iida T., Swings J. (2004). Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunsjø H. S., Wiik-Nielsen C. R., Grove S., Skjerve E., Sørum H., L'Abée-Lund T. M. (2011). Putative virulence genes in Moritella viscosa: activity during in vitro inoculation and in vivo infection. Microb. Pathog. 50, 286–292. 10.1016/j.micpath.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Turley C. M., Mackie P. J. (1994). Biogeochemical significance of attached and free-living bacteria and the flux of particles in the NE Atlantic Ocean. Mar. Ecol. Prog. Ser. 115, 191–203. 10.3354/meps115191 [DOI] [Google Scholar]
- Turner J. W., Malayil L., Guadagnoli D., Cole D., Lipp E. K. (2014). Detection of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae with respect to seasonal fluctuations in temperature and plankton abundance. Environ. Microbiol. 16, 1019–1028. 10.1111/1462-2920.12246 [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G., Westereng B., Horn S. J., Liu Z., Zhai H., Sorlie M., et al. (2010). An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222. 10.1126/science.1192231 [DOI] [PubMed] [Google Scholar]
- Volk T., Hoffert M. I. (1985). Ocean carbon pumps: Analysis of relative strengths and efficiencies in ocean−driven atmospheric CO2 changes, in The Carbon Cycle and Atmospheric CO2: Natural variations Archean to Present, eds Sundquist E. T., Broecker W. S. (Washington, DC: American Geophysical Union; ), 99–110. 10.1029/GM032p0099 [DOI] [Google Scholar]
- Volkman J. K., Tanoue E. (2002). Chemical and biological studies of particulate organic matter in the ocean. J. Oceanogr. 58, 265–279. 10.1023/A:1015809708632 [DOI] [Google Scholar]
- von Scheibner M., Dörge P., Biermann A., Sommer U., Hoppe H.-G., Jürgens K. (2013). Impact of warming on phyto-bacterioplankton coupling and bacterial community composition in experimental mesocosms. Environ. Microbiol. 16, 718–733. 10.1111/1462-2920.12195 [DOI] [PubMed] [Google Scholar]
- Waidner L. A., Kirchman D. L. (2007). Aerobic anoxygenic phototrophic bacteria attached to particles in turbid waters of the Delaware and Chesapeake estuaries. Appl. Environ. Microbiol. 73, 3936–3944. 10.1128/AEM.00592-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeham S. G., Volkman J. K. (1991). Sampling and analysis of lipids in marine particulate matter, in Marine Particles: Analysis and Characterization, eds Hurd D. C., Spencer D. W. (Washington, DC: American Geophysical Union; ), 171–179. [Google Scholar]
- Wei N., Quarterman J., Jin Y.-S. (2013). Marine macroalgae: an untapped resource for producing fuels and chemicals. Trends Biotechnol. 31, 70–77. 10.1016/j.tibtech.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Yakimov M. M., Timmis K. N., Golyshin P. N. (2007). Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18, 257–266. 10.1016/j.copbio.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451. 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K., Hama T. (2012). Degradation and dissolution of zooplanktonic organic matter and lipids in early diagenesis. Earth Planet. Sci. Lett. 68, 205–214. 10.1007/s10872-011-0091-7 [DOI] [Google Scholar]
- Yoshimura K., Ogawa T., Hama T. (2009). Degradation and dissolution properties of photosynthetically-produced phytoplankton lipid materials in early diagenesis. Mar. Chem. 114, 11–18. 10.1016/j.marchem.2009.03.002 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.