Abstract
The incidence of Cryptococcus gattii infections in both Canada and the United States (US) is provided in this literature review beyond the British Columbia (BC) outbreak (1999–2013). Based on a search of the literature, case reports of C. gattii human infections including the prevalent molecular genotypes causing these infections in both Canada and the US have been documented since the C. gattii outbreak in BC. The literature reveals that: i) although C. gattii infections continue to be reported in both countries, the preliminary overall number of confirmed C. gattii infections may be decreasing in both Canada and the US (~23 cases each in 2012 versus ~17 and 20 cases, respectively in 2013); ii) C. gattii genotype distribution is region-dependent; iii) C. gattii is more frequently isolated from infections in the immunocompromised host (including acquired immune deficiency syndrome [AIDS] infection) than previously expected; iv) although pulmonary disease is higher than in C. neoformans infections, central nervous system disease is also reported among patients infected with C. gattii.
Keywords: C. gattii, human infections, incidence, molecular epidemiology, United States, Canada
Introduction
As with Cryptococcus neoformans and other fungal pathogens, Cryptococcus gattii is an environmental organism that can be isolated from the soil and decaying organic materials. Infections caused by cryptococcal species are acquired by the inhalation of basidiospores. Consequently, both C. neoformans and C. gattii commonly cause pneumonia that can disseminate and evolve into meningitis after 2–11 months of incubation, but the incubation period could be longer, especially among untreated immunocompromised patients.1 In contrast to infections caused by C. neoformans that are seen worldwide among immunocompromised hosts, C. gattii has been considered more geographically restricted. C. gattii can infect both immunocompromised and non-immunocompromised patients, and the infections are more difficult to treat.2,3 This review attempts to provide an update on the incidence of C. gattii infections in both Canada and the United States (US) beyond the Canadian and the Pacific Northwest outbreaks of the last decade. We searched the literature to trace the case reports of C. gattii human infections as well as the prevalent genotypes causing the infections from 1999–2013; case reporting after 2012 is limited in the literature. Although animal cases and environmental data are provided in several papers, the focus of this review is human disease.
Historical background
The historical summary provided here is found in more detail elsewhere.4 Between 1894 and 1895, Otto Busse and A Buschke reported the first known case of C. neoformans (as Saccharomyces hominis) in Zurich; the isolate was recovered from a woman who had an abscess of the tibia and had been diagnosed with sarcoma. Busse was able to produce similar lesions in mice as those seen in humans and obtained a pure culture of the organism. The patient died after developing similar lesions on the face and other areas of the body. In 1894, Francesco Sanfelice named the organism Saccharomyces neoformans and this name stayed until the organism was transferred to the genus Cryptococcus by Jean Paul Vuillemin in 1901 as C. hominis. The first case of cryptococcal meningitis was reported by D von Hansemann in 1905, but the first well-documented case of this infection was not reported in the US until 1923 in a patient diagnosed previously with Hodgkin’s disease. By 1926, 13 cryptococcosis cases had been documented (two in Germany and eleven in the US). In 1935, Rhoda W Benham concluded that the genus Cryptococcus could be separated into four groups based on cultural, serological, and animal pathogenicity studies of >40 isolates (including the two isolates from the original European cases), although she found that the isolates evaluated belonged to the same species, C. neoformans. In 1951, John F Kessel reported that C. neoformans could be divided into three serotypes, A, B, and C; serotype D (as R) was identified 17 years later by Ralph Vogel.
The introduction of the latex method to detect cryptococcal antigens in serum and cerebrospinal fluid (CSF) greatly improved the diagnosis of cryptococcal diseases as previous tests were negative for about 60% of serum specimens. The diagnosis of cryptococcosis frequently relies on direct microscopy, culture of clinical samples, and/or detection of cryptococcal antigen in body fluids. However, the organism is not identified to species level in many clinical microbiology laboratories. A key biochemical characteristic that distinguishes C. gattii from C. neoformans is its ability to produce blue coloration on L-canavanine-glycine-bromothymol blue (CGB) medium; occasionally, some isolates identified as C. gattii by molecular typing may be CGB negative.5
In 1970, the C. neoformans var. gattii (as an atypical strain of C. neoformans) was isolated from a leukemic patient in Congo, Africa (strain ATCC 32269) followed by the discovery of the sexual stage of C. neoformans and the establishment of the new genus Filobasidiella.6 Based on physiological assays, it was demonstrated that C. neoformans serotype B and C strains differed from C. neoformans serotype A and D, and C. bacillisporus was proposed for serotype B and C isolates.7 However, by 1982, further studies led to the recognition of these groups as two varieties of a single species, namely C. neoformans var. neoformans (serotypes A and D) and C. neoformans var. gattii (serotypes B and C).8 Further work including molecular studies has led to C. gattii being recognized as a separate species since 2002.9
C. gattii: general information
Molecular typing studies have been at the forefront of tracking C. gattii epidemiology. The techniques that have been used for this purpose are the following: polymerase chain reaction (PCR) fingerprinting, restriction fragment length polymorphism (RFLP) analysis, intergenic spacer sequencing, amplified fragment length polymorphism analysis, and more recently, multilocus sequence typing (MLST). Importantly, all of these methods support distinct and congruent molecular types, and a comprehensive overview of the molecular types and their nomenclature using different molecular typing methods may be found in Chen et al, 2014.5 As of now, C. gattii comprises four molecular types (VGI, VGII, VGIII, and VGIV) with varying distribution globally; in North America, at least three VGII subtypes (VGIIa, VGIIb, and VGIIc) and two VGIII subtypes (VGIIIa and VGIIIb), are present, among others.5,10,11 It is difficult to clearly identify the actual incidence of C. gattii unless molecular studies are performed or isolates are subtyped to the serotype or species level. The incidence of C. gattii infections is always lower than that attributed to C. neoformans. For example, in a US population-based surveillance study reported in 1996, only three of 248 cryptococcal isolates were C. gattii (as serotype B) that were obtained from human immunodeficiency virus (HIV)-negative individuals; one isolate originated in Georgia and two in San Francisco.12 In general, molecular types VGI and VGII are the cause of disease in otherwise healthy patients, whereas VGIII and VGIV are frequently isolated among immunocompromised patients, including those with HIV/AIDS infections. In addition, it has been postulated that patients considered “otherwise healthy” could have some unknown immunological defects that are not detected by the current methodologies employed in clinical laboratories.13
The polysaccharide capsule of C. neoformans and C. gattii is a major virulence determinant and both species also have the ability to grow at physiological temperatures, which contribute to their survival in the host. The enzyme phospholipase B, a potential determinant of tissue invasion, is produced in vitro and in vivo by C. gattii and plays a role in the virulence of C. neoformans.2,14 C. gattii produces an increased incidence of cryptococcomas in the lung and the brain, increased neurological morbidity, and a slower response to antifungal therapy as compared to the infection caused by C. neoformans.3,5,15 The Infectious Diseases Society of America (IDSA) recommendations for infections caused by C. gattii are based mostly on case reports and small case series; for central nervous system (CNS) disease IDSA recommendations are essentially the same as those for infections caused by C. neoformans.3,15
A variation in modal minimal inhibitory concentrations (MICs) among VGII genotypes has been noted, with flucytosine 2–8 μg/mL (VGIIa < VGIIc < VGIIb); fluconazole 4–6 μg/mL (VGIIa < VGIIb < VGIIc); voriconazole 0.12–0.25 μg/mL (VGIIa < VGIIb = VGIIc); posaconazole 0.12–0.5 μg/mL (VGIIb < VGIIa < VGIIc); and itraconazole 0.25–0.5 μg/mL (VGIIa = VGIIb < VGIIc).16,17 Another variation between C. gattii and C. neoformans and between molecular types was determined in epidemiological cut off values (ECVs); overall, ECVs for molecular types of C. neoformans were lower than those of C. gattii. Taken together, treatment guidelines may need to be tailored to species and/or molecular type.
Prior to the Canadian outbreak, C. gattii appeared to have a restricted global distribution, predominantly isolated in tropical and subtropical geographical areas.14 It was considered endemic in Australia, New Zealand, Papua New Guinea, South and Southeast Asia, parts of Latin America, Southern California, Mexico, Hawaii, Central and South Africa, and certain European areas.5,14,18 Climate change has been postulated as one of the reasons for the proliferation of C. gattii in Canada and outside Californian areas in the US.
In the 1990s, cryptococcal meningitis was the leading cause of adult meningitis in many African countries, including Malawi (27% of cases), Zimbabwe (45% of cases), and South Africa (31% of cases),19 but data are not available regarding the infecting cryptococcal species. In contrast to Europe and North America, a delayed and/or incomplete access to antiretroviral therapy (ART) led to a continuous high incidence of cryptococcal meningitis in geographical areas where access to healthcare is limited.20 By 2009, the Center for Disease Control and Prevention (CDC) (Atlanta, GA, USA) reported that throughout much of Sub-Saharan Africa (the global region most heavily affected by HIV), there were approximately 720,000 cryptococcal meningitis cases/year versus 70,000 in the Americas and the Caribbean among HIV-infected individuals. Cryptococcal meningitis is one of the leading causes of death (~504,000 deaths/year) in HIV/AIDS patients in Africa, higher than those caused by tuberculosis.20
Incidence of C. gattii in Canada
A total of 158 cryptococcal infections were diagnosed among HIV-negative patients between 1995 and 2004 in Vancouver Island Hospitals located at the Eastern edge of Vancouver Island with a marked incidental increase since 1999.21 In addition, by the end of March 2002, a total of 45 laboratory-confirmed animal cases were reported.22 All these cases were substantiated by cytological, histopathological, or culture methodologies. Cultures were not available from most of the 158 human infections, but serotyping of the 36 isolates that were available resulted in the following breakdown: 27 (75%) serotype A, six (17%) serotype D, and only three (8%) serotype B.21 The clinical presentation of the C. gattii (as serotype B) human cases was different, primarily a pulmonary presentation instead of CNS involvement. Three individuals (among 26 HIV-positive and negative patients with cryptococcosis), who lived on the British Columbia (BC) mainland but had traveled to Vancouver Island, were infected with the outbreak C. gattii strain between 1999 and 2002; again, cultures were not available from all 26 patients.23 However, spread from the Island to Vancouver and its surrounding communities became apparent when three C. gattii cases were reported between 2003 and 2005 in individuals from the BC mainland, who had not traveled to the Island and were HIV-negative.24 Before 1999, only three HIV-negative patients in the area were diagnosed with this infection. Remarkably, the majority of human cases occurred in people without recognized immunologic defects, thus highlighting the unusual pathogenicity of this species or the presence of an unrecognized defect with the current laboratory methodology.
In general, from 1999 to 2007, C. gattii veterinary cases (mostly companion animals) were diagnosed two to three times more frequently than human cases. For example, the report of the three human C. gattii infections on the mainland included infections in eight animals (dogs, cats, and a ferret).24 However, the incidence of human infections could have been higher since cultures were not available from all reported cases. The total number of deaths attributed to C. gattii infections was 19 during the period of 1999–2007; in 2003, C. gattii and other cryptococcal infections became reportable in BC, leading to improved surveillance efforts and providing the annual incidence in the area.25,26
In 2006 and 2007, BC reported the highest incidence of C. gattii infections in the world (nine and eight cases per million residents per year, respectively),26 with an average annual incidence of C. gattii infections of 5.8 per million persons on the island, versus 0.61 per million persons in Australia.5,25 A retrospective analysis of 124 confirmed C. gattii cases (of 218 total cases) reported in BC from 1999 to 2007 provided further understanding of this infection.25 Although the majority of the cases (73.9%) were from Vancouver Island, by 2007 the number of cases from the mainland was almost as high as those from the Island.26 Approximately 76% of the patients were treated for respiratory disease and of those 75% for lung cryptococcoma and only 7.8% for CNS disease; 10.1% were treated for both respiratory and CNS symptoms. Most hospitalized patients were HIV negative (59.3%); the mortality rate was 9.7% and occurred among the older population with CNS disease and/or if their infecting isolate was genotype VGIIb. By 2011, some predisposing risk factors for C. gattii infection had been reported: age (>50 years), smoking, corticosteroid use in the three months prior to onset, HIV infection, and a history of cancer and/or chronic lung disease.27
Since the release of the initial report in 1999 and following a plateau in the incidence of C. gattii infections between 2002 and 2005, two incidental increases have been reported from the first six cases in 1999, one in 2006 (38 cases: nine cases per million) and another in 2012 (24 confirmed and ten probable: seven cases per million). These higher incidences were mainly due to an increase in cases from the BC mainland and among adults.25,26 Although the incidence of C. gattii infections for 2014 has not yet been reported by the BC Ministry of Health, the increase in 2012 was followed by a 29% decrease in 2013 to 17 cases (4 per million population; culture confirmed cases) (Figure 1). The cause of these increases, especially the one in 2012, and the decrease in 2013, are unclear, but could be attributable to fluctuations in low numbers and the fact that only confirmed cases have been reported (E Galanis, personal communication, 2014).
Figure 1.
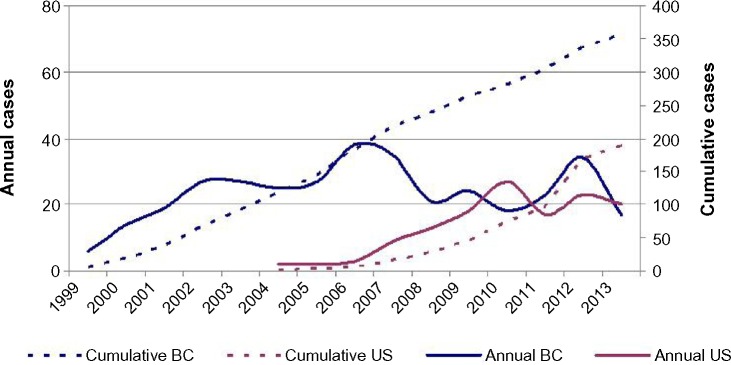
Approximate annual and cumulative incidence of reported human Cryptococcus gattii cases in Canada and the United States since 1999.
Abbreviation: BC, British Columbia.
Preliminary results indicated that C. gattii VGIIa (designated as the major outbreak strain) was the predominant genotype (90%) isolated from the Vancouver Island outbreak, followed by VGIIb in the Southern areas with occasional VGI cases.10,28–30 Further analyses determined that the VGII outbreak strain (C. gattii strain R265) was identical to two historic strains (C. gattii type strain NIH444 and C. gattii type strain CBS7750) recovered from clinical and environmental sources in the US as discussed in the Incidence of C. gattii in the US section, according to two independent MLST studies.28,29 Therefore, the major Vancouver outbreak genotype had been present in the US Pacific Northwest for at least 30 years prior to the outbreak. On the other hand, the VGIIb genotype was similar to an Australian recombinant population.28,29 However, a correlation with geographical groups was not possible with the C. gattii VGI isolates. The spread from Vancouver Island to the BC mainland became evident in 2006, with clinical/veterinary (without travel history) and environmental C. gattii isolates also belonging to the VGIIa and VGIIb genotypes.24 Figure 2 indicates a comparative genotype frequency between Canada and the US, and Figure 3 illustrates the distribution of genotypes from human isolates across Canada and the US.
Figure 2.
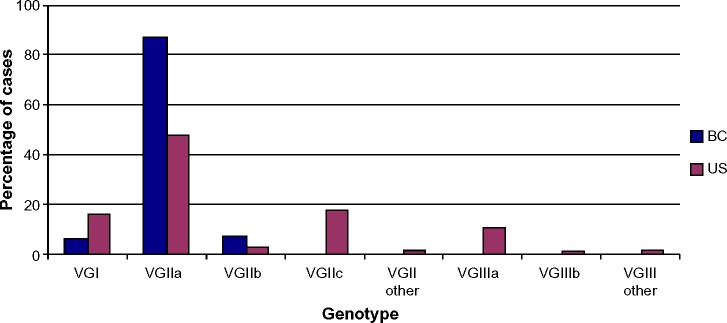
Cryptococcus gattii genotype frequency in Canada versus the United States, based upon 124 and 169 human culture confirmed cases diagnosed to 2007 and 2012, respectively.
Note: Data from Galanis et al25 and Lockhart et al.39
Abbreviation: BC, British Columbia.
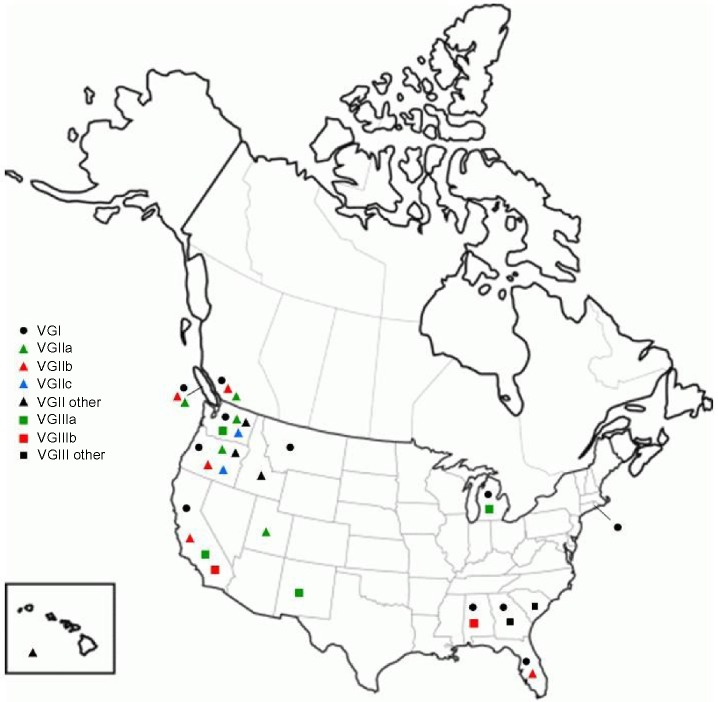
Figure 3.
Distribution of genotypes according to Province/State of residence of culture confirmed Cryptococcus gattii cases in Canada and the United States since 1999.
Incidence of C. gattii in the US
Sporadic C. gattii human infections were infrequently reported in various areas of the US before the BC outbreak and C. gattii was also prevalent among cryptococcal infected patients in Southern California. In 1977, it was noted that despite the absence of C. gattii (reported as serotypes B and C) in the soil and pigeon droppings, 25 of 49 isolates from cryptococcal infections were identified as C. gattii (reported as serotypes B and C).31 An additional 30 of 73 cryptococcal isolates were reported as C. gattii (serotype B, 19 isolates and serotype C, eleven isolates) from this region in 1984;32 some of the isolates were recovered from animals. In contrast, only five of 78 and two from Hawaii. In a subsequent surveillance study, where cryptococcal isolates were collected from four US regions (Atlanta, San Francisco, Alabama, and Houston: 1992 through 1994), C. gattii was isolated in just three of the 248 cases (1% of all cases).12 One of these three isolates (reported as serotype B) was recovered from one patient in Atlanta and the other two in San Francisco (one of the patients was HIV positive). Among other C. gattii strains isolated in the US prior to the outbreak in the Pacific Northwest, two of them are considered Type strains. The C. gattii strain NIH444 (ATCC 32609) recovered from a patient’s sputum in 1971 (Seattle) and the environmental C. gattii Type strain CBS7750 from a Eucalyptus camaldulensis tree (San Francisco) in 1990;33 it was later determined that both strains belong to the VGIIa genotype.10,28,29
An incidental increase of C. gattii infections was documented in the US since the Canadian outbreak, and this could have been due to the increased awareness of this infection. Two VGII cases from Oregon in 2005 and 2006 were reported by the BC cryptococcal group.24 The MLST genotypes of these isolates varied slightly from those of the predominant VGIIa and VGIIb genotypes; a recent re-analysis of these MLST sequences (by comparison to those in reference 11) indicated that one of those strains appears to be the VGIIc genotype, which had not been defined at that time (SE Kidd, unpublished data, 2014). In 2006, a C. gattii (VGIIa) case was diagnosed in Orcas, Washington State,34 and by 2008 the number of culture positive cases was 19 (nine from Oregon and ten from Washington State).24,35 While all but one isolate from Washington State were genotype VGIIa, three distinct VGII subtypes were detected in Oregon, including the VGIIc subtype. On the other hand, the genotype VGIII was isolated from Washington State.35 Some of these individuals had not traveled to BC outbreak areas. Since then, C. gattii infections became reportable in both states, but still are considered rare infections. C. gattii VGIII isolates (28 of 276 cryptococcal strains) were diagnosed earlier from HIV/AIDS patients in Los Angeles County, California; genotypes VGI and VGII were also present.11,36 The VGIII isolates were divided into molecular subgroups, VGIIIa (17 isolates) and VGIIIb (eleven isolates).11,36
In 2009, the count of confirmed cases had increased from nine to 19 in Oregon, most of them with a pulmonary clinical presentation.37 By July 2011, 96 C. gattii cases were reported to the CDC (Atlanta) by their respective state health department or patients’ physicians.38 Most of the cases were from Oregon (61 cases) and Washington State (20 cases) and the patients’ onset of illness was from December 2004 to June 2011. In addition to those two states, case reports also originated from Alaska, Hawaii, Idaho, Michigan, Montana, North Carolina, New Mexico (one case each), Georgia (two cases), and California (five cases); culture identification was confirmed at the CDC.38 Three patients reported travel to California (patient from North Carolina), Washington State, or Oregon (patients from Alaska and Idaho). Most of the strains were VGIIa (52%), VGIIc (23%), and VGIIb (6%). The CDC reported169 human cases isolated from January 2005 to January 2013. However, this second report was the cumulative count of confirmed culture positive C. gattii cases reported to the CDC up to that date.38–40 The report included additional states where C. gattii infections were diagnosed, one isolate each from Alabama, Colorado, Rhode Island, South Carolina, and Utah and two isolates from Florida. However, the highest number of isolates originated in Georgia (eight isolates), California (28 isolates), Washington (31 isolates), and Oregon (88 isolates).39 This information revealed an incidental increase of this pathogen in the US up to January 2013 (Figure 1), as well as an expansion to areas outside the Pacific Northwest and California. There were sufficient cases/isolates including the 104 strains from veterinary sources to provide the actual distribution of the genotypes. As expected, 214 (78%) isolates that originated in the Pacific Northwest (human and veterinary origin) were mostly VGIIa (64%) and VGIIc (21.6%), while the 59 isolates from outside this area were mostly VGIII (44%) and VGI (41%) (Figures 2 and 3).25,39 The third CDC report provides the following data for 96 of the 196 patients: 64% had been previously healthy; 84% were male and 76% had CNS disease; and ten patients had not traveled to endemic areas.40 A few of these CDC cases were previously reported (Georgia [three cases], Florida, North Carolina, and New Mexico).41–44 In addition, since the early 1980s, C. gattii infections (reported as B or C serotypes) were diagnosed from Alabama, Georgia, Louisiana, Tennessee, New York City, and, more recently, New England (an adolescent VGI infection).45–47
Preliminary data from the CDC indicates that the incidence of cases in the US from 2012 through 2013 is as follows: an increase in Washington State from approximately five to eight cases and a decrease in Oregon from 18 to 12 cases, followed by lower numbers thus far for 2014. In summary, US cases continue to be reported at a lower rate since 2012, following a similar trend to BC (S Lockhart, personal communication, December 2014).
Clinical importance of C. gattii infections
An epidemiological study of the BC cases revealed that while the majority (86.7% of 124 culture confirmed cases) were caused by the VGIIa strain, this strain did not appear to cause greater illness or death than either the VGIIb (7.2%) or VGI (6.4%) cases. However, the patients who died were more likely to be older, to have CNS disease, and have infection caused by the VGIIb strain. The patients >50 years of age were more likely to be infected with VGIIa (P=0.002) or VGIIb (P=0.006) than with VGI.15,25 These observations are supported by the findings from US patients,40 but are at odds with experimental murine virulence studies, where VGIIa isolates were more virulent than the VGIIb strains.29,48 The reasons for this observation are not clear. In contrast, a recent Galleria mellonella virulence study revealed a range of virulence within all molecular types/subtypes that was associated with melanin production (P<0.05) rather than genotype.49
A global epidemiological study of C. gattii infections was reported by the Mycoses Study Group during Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC), 2014.50 This retrospective cohort study evaluated patients with confirmed cryptococcosis from six institutions (Australia, Alabama, Oregon, South Carolina, and both CDC organizations from BC and the US) from 1995 to 2012 (some of them included in the studies discussed so far). The characteristics of 724 of 850 patients with C. neoformans or C. gattii infection were compared. The important differences between the infections produced by both species were: the incidence of C. gattii infections was lower (20.2%); clinical findings were more common among C. neoformans than C. gattii patients; and pulmonary disease was higher in C. gattii than in C. neoformans infections (59% versus 35%; P<0.001). Overall, 1-year mortality was lower among C. gattii patients (15.1% versus 34.7%; P<0.001); successful (complete or partial) clinical response to therapy at 6 months was higher for C. gattii (93.8% versus 83.7%; P=0.023); and clinical response to therapy at 6 months for patients with meningitis was greater among C. gattii patients (95% versus 75%; P=0.0014). Some of these factors are different from what has been so far investigated or reported. However, the most frequent clinical presentation among these patients with cryptococcosis caused by both C. neoformans and C. gattii was CNS disease.
Origin and significance of genotypes
The introduction of C. gattii strains into the Pacific Northwest and the factors that led to the emergence of C. gattii as an important etiologic agent of disease have not been completely elucidated. Overall, the VGII group appears to have originated in South America, and the VGIIa and VGIIb genotypes within this lineage can also be traced to populations in Australia and the US Pacific Northwest, probably as a result of multiple dispersal events.2,28,29,51,52 To our knowledge, the VGIIc genotype has not been identified outside the US (mainland states). VGIII isolates have been identified among isolates from Central and South America, South Africa, Australia, New Zealand, and India.5 The first reported environmental isolations of C. gattii in 1990 (San Diego, CA) included two VGIII isolates from a Eucalyptus citriodora tree and another from eucalypt debris;33,53 thus VGIII parallels VGII in its longstanding presence in North America prior to the emergence of infections. VGI is overall the most commonly isolated of the C. gattii lineages globally,5 but is comparatively less prevalent in North America.
The overall molecular type distribution inside the Pacific Northwest (US) since 1997 has been VGIIa (≥50%), VGIIc (≤32%), and VGIIb (≤10%).25,39 VGIII and VGI are more predominant outside the Pacific Northwest (44% and 41%, respectively of total isolates outside the Pacific Northwest area).25,39 The latter two genotypes have been found in Southeastern US areas as well as in California, but the Californian VGIIIa subgroup also appears to originate from Australia and the VGIIIb subgroup from North America, South America, and Asia.11 A study of potential dispersal mechanisms in BC showed an association of C. gattii environmental isolations with high-traffic/visitor locations demonstrated by carriage on car wheels and footwear, and increased airborne C. gattii detection around forestry activities,54 which may in part account for the global distribution of particular genotypes.
A model of patient travel and the spread of these infections from one area to another are supported, among others, by a report of a C. gattii infection in a patient who had traveled the prior year to San Francisco, CA.41 The patient’s C. gattii VGI isolate, recovered from cultures of both leg and brain biopsies, was genetically identical to two isolates recovered from patients living in California and Australia. It was suggested that this patient acquired the infection while traveling.41
Conclusion
It is evident that by 2013 in the US and Canada, i) the distribution and incidence of C. gattii genotypes and subtypes were geographically dependent and attributable infections can be caused by a number of C. gattii molecular types and their subtypes; ii) C. gattii was more frequently found in HIV/AIDS patients than previously estimated; iii) of the four molecular types of C. gattii, VGI and VGII produce disease in otherwise healthy hosts in the Pacific Northwest, and VGIII and VGIV tend to produce disease among AIDS patients; iv) VGII isolates tend to cause primarily pulmonary disease, whereas VGI and VGIII isolates tend to cause CNS infections; v) infections caused by C. gattii continue to be reported in Canada and the US, but no obvious increase in the incidence of C. gattii infections has been recently noted in either country; vi) instead, preliminary information suggests either another plateau or a slight incidental decrease of these infections in both countries.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.MacDougall L, Fyfe M. Emergence of Cryptococcus gattii in a novel environment provides clues to its incubation period. J Clin Microbiol. 2006;44(5):1851–1852. doi: 10.1128/JCM.44.5.1851-1852.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 3.Perfect JR, Dismukes WE, Dromer F, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of America. Clin Infect Dis. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinel-Ingroff A. Medical Mycology in the United States (1894–1996) Dordrecht, the Netherlands: Kluger Academic Publishers; 2003. [Google Scholar]
- 5.Chen SC, Meyer W, Sorrell TC. Cryptococcus gattii infections. Clin Microbiol Rev. 2014;27:980–1024. doi: 10.1128/CMR.00126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanbreuseghem R, Takashio M. An atypical strain of C. neoformans (San Felice) Vuillemin. Part II. Cryptococcus neoformans var. gattii. var. nov. Ann Soc Belges Med Trop Parasitol Mycol. 1970;50:695–702. [PubMed] [Google Scholar]
- 7.Kwon-Chung KJ, Bennett JE, Theodore TS. Cryptococcus bacillisporus sp. nov.: serotype B-C of Cryptococcus neoformans. Int J Syst Bacteriol. 1978;28:616–620. [Google Scholar]
- 8.Kwon-Chung KJ, Bennett JE, Rhodes JC. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Van Leeuwenhoek. 1982;48:25–38. doi: 10.1007/BF00399484. [DOI] [PubMed] [Google Scholar]
- 9.Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae) Taxon. 2002;51:804–806. [Google Scholar]
- 10.Kidd SE, Hagen F, Tscharke RL, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci U S A. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrnes EJ, 3rd, Li W, Ren P, et al. A diverse population of Cryptococcus gattii molecular type VGIII in Southern Californian HIV/AIDS patients. PLoS Pathog. 2011;7:e1002205. doi: 10.1371/journal.ppat.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt ME, Hutwagner LC, Klug LA, et al. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. Cryptococcal Disease Active Surveillance Group. J Clin Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon-Chung KJ, Fraser JA, Doering TL, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4:a019760. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorrell TC. Cryptococcus neoformans variety gattii. Med Mycol. 2001;39:155–168. [PubMed] [Google Scholar]
- 15.La Hoz RM, Pappas PG. Cryptococcal infections: changing epidemiology and implications for therapy. Drugs. 2013;73(6):495–504. doi: 10.1007/s40265-013-0037-z. [DOI] [PubMed] [Google Scholar]
- 16.Espinel-Ingroff A, Chowdhary A, Cuenca-Estrella M, et al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for amphotericin B and flucytosine. Antimicrob Agents Chemother. 2012;56:3107–3113. doi: 10.1128/AAC.06252-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinel-Ingroff A, Aller AI, Canton E, et al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole and voriconazole. Antimicrob Agents Chemother. 2012;56:5898–5906. doi: 10.1128/AAC.01115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viviani MA, Cogliati M, Esposto MC, et al. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res. 2006;6:614–619. doi: 10.1111/j.1567-1364.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- 19.Sloan DJ, Parris V. Cryptococcal meningitis: epidemiology and therapeutic options. Clin Epidemiol. 2014;6:169–182. doi: 10.2147/CLEP.S38850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 21.Fyfe M, MacDougall L, Romney M, et al. Cryptococcus gattii infections on Vancouver Island, British Columbia, Canada: emergence of a tropical fungus in a temperate environment. Can Commun Dis Rep. 2008;34:1–12. [PubMed] [Google Scholar]
- 22.Stephen C, Lester S, Black W, Fyfe M, Raverty S. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can Vet J. 2002;43:792–794. [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang LM, Maguire JA, Doyle P, Fyfe M, Roscoe DL. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997–2002): epidemiology, microbiology and histopathology. J Med Microbiol. 2004;53:935–940. doi: 10.1099/jmm.0.05427-0. [DOI] [PubMed] [Google Scholar]
- 24.MacDougall L, Kidd SE, Galanis E, et al. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis. 2007;13:42–50. doi: 10.3201/eid1301.060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galanis E, MacDougall L, Kidd S, Morshed M, The British Columbia Cryptococcus gattii Working Group Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999–2007. Emerg Infect Dis. 2010;16:251–257. doi: 10.3201/eid1602.090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.BC Center for Disease Control . British Columbia Annual Summary of Reportable Diseases. Vancouver, BC, Canada: BCCDC; 2014. [Google Scholar]
- 27.MacDougall L, Fyfe M, Romney M, Starr M, Galanis E. Risk factors for Cryptococcus gattii infection, British Columbia, Canada. Emerg Infect Dis. 2011;17:193–199. doi: 10.3201/eid1702.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kidd SE, Guo H, Bartlett KH, Xu J, Kronstad JW. Comparative Gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryot Cell. 2005;4(10):1629–1638. doi: 10.1128/EC.4.10.1629-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser JA, Giles SS, Wenink EC, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 30.Bartlett KH, Kidd SE, Kronstad JW. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr Infect Dis Rep. 2008;10:58–65. doi: 10.1007/s11908-008-0011-1. [DOI] [PubMed] [Google Scholar]
- 31.Bennett JE, Kwon-Chung KJ, Howard DH. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 32.Kwon-Chung KJ, Bennett JE. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984;120:123–130. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 33.Pfeiffer T, Ellis D. Environmental isolation of Cryptococcus neoformans-gattii from California. J Infect Dis. 1991;163(4):929–930. doi: 10.1093/infdis/163.4.929. [DOI] [PubMed] [Google Scholar]
- 34.Upton A, Fraser JA, Kidd SE, et al. First contemporary case of human infection with Cryptococcus gattii in Puget Sound: evidence for spread of the Vancouver Island outbreak. J Clin Microbiol. 2007;45:3086–3088. doi: 10.1128/JCM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byrnes EJ, Bildfell R, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the Vancouver Island Cryptococcus gattii outbreak has expanded into the United States Pacific Northwest. J Infect Dis. 2009;199:1081–1086. doi: 10.1086/597306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaturvedi S, Dyavaiah M, Larsen RA, Chaturvedi V. Cryptococcus gattii in AIDS patients, Southern California. Emerg Infect Dis. 2005;11:1686–1692. doi: 10.3201/eid1111.040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datta K, Bartlett KH, Baer R, et al. Spread of Cryptococcus gattii-into Pacific Northwest region of the United States. Emerg Infect Dis. 2009;15:1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris JR, Lockhart SR, Debess E, et al. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin Infect Dis. 2011;53:1188–1195. doi: 10.1093/cid/cir723. [DOI] [PubMed] [Google Scholar]
- 39.Lockhart SR, Iqbal N, Harris JR, et al. Cryptococcus gattii in the United States: genotypic diversity of human and veterinary isolates. PLoS One. 2013;8:e74737. doi: 10.1371/journal.pone.0074737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris JR, Lockhart SR, Sondermeyer G, et al. Cryptococcus gattii infections in multiple states outside the US Pacific Northwest. Emerg Infect Dis. 2013;10:1620–1626. doi: 10.3201/eid1910.130441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byrnes EJ, 3rd, Li W, Lewit Y, et al. First reported case of Cryptococcus gattii in the Southeastern USA: implications for travel associated acquisition of an emerging pathogen. PLoS One. 2009;4:e5851. doi: 10.1371/journal.pone.0005851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walraven CJ, Gerstein W, Hardison SE, et al. Fatal disseminated Cryptococcus gattii infection in New Mexico. PLoS One. 2011;6(12):e28625. doi: 10.1371/journal.pone.0028625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sellers B, Hall P, Cine-Gowdie S. Cryptococcus gattii: an emerging fungal pathogen in the Southeastern United States. Am J Med Sci. 2012;343:1547–1559. doi: 10.1097/MAJ.0b013e3182464bc7. [DOI] [PubMed] [Google Scholar]
- 44.Kunadharaju R, Choe U, Harris JR, Lockhart SR, Greene JN. Cryptococcus gattii, Florida, USA, 2011. Emerg Infect Dis. 2013;19:519–521. doi: 10.3201/eid1903.121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fromtling RA, Shadomy S, Shadomy HJ, Dismukes WE. Serotype B/C Cryptococcus neoformans isolated from patients in nonendemic areas. J Clin Microbiol. 1982;16:408–410. doi: 10.1128/jcm.16.2.408-410.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bottone EJ, Kirschner PA, Salkin IF. Isolation of highly encapsulated Cryptococcus neoformans serotype B from a patient in New York City. J Clin Microbiol. 1986;23:186–188. doi: 10.1128/jcm.23.1.186-188.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCulloh RJ, Phillips R, Perfect JR, Byrnes EJ, 3rd, Heitman J, Dufort E. Cryptococcus gattii genotype VGI Infection in New England. Pediatr Infect Dis J. 2011;30:1111–1114. doi: 10.1097/INF.0b013e31822d14fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngamskulrungroj P, Serena C, Gilgado F, Malik R, Meyer W. Global VGIIa isolates are of comparable virulence to the major fatal Cryptococcus gattii Vancouver Island outbreak genotype. Clin Microbiol Infect. 2011;17:251–258. doi: 10.1111/j.1469-0691.2010.03222.x. [DOI] [PubMed] [Google Scholar]
- 49.Firacative C, Duan S, Meyer W. Galleria mellonella model identifies highly virulent strains among all major molecular types of Cryptococcus gattii. PLoS One. 2014;9(8):e105076. doi: 10.1371/journal.pone.0105076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baddley JW, Perfect JR, Chen S, et al. Epidemiology and outcomes of cryptococcosis: description of an international, multi-center cohort; Abstract M1787 presented at: Interscience Conference on Antimicrobial Agents and Chemotherapy; September 5–9, 2014; Washington, DC. [Google Scholar]
- 51.Hagen F, Ceresini PC, Polacheck I, et al. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One. 2013;8(8):e71148. doi: 10.1371/journal.pone.0071148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engelthaler DM, Hicks ND, Gillece JD, et al. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio. 2014;5(4):e1414–e1464. doi: 10.1128/mBio.01464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sorrell TC, Chen SC, Ruma P, et al. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kidd SE, Bach PJ, Hingston AO, et al. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg Infect Dis. 2007;13(1):51–57. doi: 10.3201/eid1301.060823. [DOI] [PMC free article] [PubMed] [Google Scholar]