Abstract
Purpose
The global spread of blaCTX-M-I extended-spectrum beta-lactamase (ESBL)-producing Salmonella spp. remains a major threat to treatment and control. Evidence of emergence and spread of this marker are lacking in Nigeria. This study investigated blaCTX-M-I ESBL production among Salmonella isolates from hospitalized patients.
Methods
Patients (158 total) made up of two groups were evaluated. Group A was composed of 135 patients with persistent pyrexia and group B was composed of 23 gastroenteritis patients and their stool samples. Samples were cultured, and isolates were identified and were subjected to antibiotic susceptibility testing by standard methods. Isolates were further screened for ESBL production, blaCTX-M-I genes and transferability by double disk synergy test, plasmid extraction, polymerase chain reaction, and conjugation experiment.
Results
Thirty-five (25.9%) Salmonella isolates were identified from group A, of which 74.3% were S. typhi, 22.9% were S. paratyphi and two (5.7%) were invasive non-typhoidal S. enteritidis. Nine Plasmodium falciparum infections were recorded, four of which were identified as co-infections with typhoidal Salmonella. Only two (8.7%) S. enteritidis samples were obtained from group B (P>0.05). A total of 24 isolates were ESBL-positive, eliciting resistance to five to seven antibiotics, and were multiple-drug resistant. ESBL production due to the blaCTX-M-I gene cluster was detected in eleven (45.8%) Salmonella isolates. Nine (81.8%) of the eleven blaCTX-M-I ESBL producers were S. typhi and two (18.2%) isolates were S. enteritidis. Four of nine S. typhi blaCTX-M-I ESBL-producing strains harbored 23 kb self-transmissible plasmid that was co-transferred with cefotaxime and augmentin resistance to Escherichia coli j53-2 transconjugants.
Conclusion
This study revealed the emergence of blaCTX-M-I S. typhi as an agent of persistent pyrexia with potential to spread to other Enterobacteriaceae in Lagos, Nigeria. Cautionary prescription and judicious use of third-generation cephalosporins, particularly cefotaxime, for the treatment of typhoid fever and routine screening for P. falciparum co-infection with ESBL-producing Salmonella in the laboratories during diagnosis of persistent pyrexia conditions in patients are recommended.
Keywords: ESBL, emergence, plasmid, Salmonella, cephalosporin
Video abstract
Introduction
In developing countries, diseases such as enteric fever and diarrhea are on the increase due to poor sanitation and inadequate potable water supply.1 An estimated annual incidence of 540 per 100,000 for typhoid fever in developing countries and about 21 million cases worldwide had been reported.2 The number of strains of Salmonella enterica serovars that have developed resistance to one or more antibacterial agent has steadily increased, probably due to continuous antibiotic pressure.3 Resistance to third-generation cephalosporins (3GCs) due to acquisition and expression of bla-CTX-M-mediated, extended-spectrum β-lactamase (ESBL) enzymes among Gram-negative bacteria in the family Enterobacteriaceae is also on the increase. These enzymes are produced exclusively by Gram-negative bacteria and are active against 3GCs, especially cefotaxime, compared to bla-TEM and bla-SHV ESBLs.3 These ESBL enzymes are usually plasmid-mediated, but are susceptible to cephamycins such as cefoxitin, beta-lactamase inhibitors such as clavulanate, sulbactam, tazobactam, and carbapenems.4
ESBL-producing organisms are reported to account for a significant proportion of intensive care infections and mortality in children and immunocompromised patients.5 Problems of ESBLs have led to limited as well as expensive treatment options, and have impacted negatively on clinical outcomes.6 Growing incidences of ESBLs in Salmonella species have been identified in numerous countries of Latin America, Africa, Europe, and Asia.3,7,8 In Nigeria, blaCTX-M-I ESBL-producing Escherichia coli, Enterobacter spp., and Klebsiella spp. have been documented.9–11 However, data on blaCTX-M-I-mediated ESBL-producing S. enterica serovars are currently not available in Nigeria. In a recent work carried out by our research team, we provided evidence of ESBL-bla-CTX-M-producing S. enterica serovars in Lagos.12 Unfortunately, these strains were not characterized for carriage of the blaCTX-M-I gene cluster, which is associated with community-associated infections in many countries including Nigeria.11,13 This family of blaCTX-M-I, reported to have replaced the bla-CTX-M-2 gene cluster in the late 1990s, have now spread worldwide among many members of the Enterobacteriaceae, including Salmonella.14
In recent times, increasing episodes of persistent fever among patients affected by Salmonella typhi and Plasmodium falciparum infections has been a major concern in hospitals and clinics in Lagos and other parts of Nigeria.15,16 This is because of the perceived clinical failure associated with the use of 3GCs, particularly cefotaxime, ceftriaxone, and cefuroxime, and increasing drug pressure with arthemisin combination therapies.15,16 The current World Health Organization WHO) recommendation for parasite-based diagnosis of malaria in pyrexia cases before treatment in the era of artemisinin-based combination therapy (ACT) has further made it imperative to screen for malaria parasites by microscopic or rapid diagnostic method in countries like Nigeria, where ACT has replaced monotherapies as first- and second-line treatments for uncomplicated malaria.17,18 In the current study, we investigated the carriage of blaCTX-M-I ESBL among Salmonella strains isolated from patients, and the possibility of Plasmodium spp. co-infection.
Materials and methods
Patient population and study design
A total of 158 patients who sought treatment at referral centers including Ikeja General Hospital, Lagos; Infectious Diseases Hospital Mainland, Lagos; Central Bank of Nigeria Clinics Satellite, Lagos; and Central Medical Laboratory Health Centre, Lagos from October 2010 to July 2011 were recruited for the study. Important biodata, history of vaccination, antimicrobial therapy, time of onset of illness, etc, of these patients were recorded. Ethics approval from the ethics committee of each institution was obtained prior to patients’ enrollment.
Case definition, sample processing, Plasmodium and Salmonella detection
Two categories of patients were demarcated. A total of 135 patients were assigned to group A, and had been diagnosed by a physician for persistent fever (≥37.5°C) in the previous 72 hours with or without one or more of the following symptoms: diarrhea, headache, abdominal pains, loss of appetite, vomiting, and/or nausea for 5 consecutive days. In detail, 4 mL of blood was collected from each of the patients at the early onset of symptoms. Three of the 4 mL blood samples in each case were inoculated into 27 mL of brain-heart infusion (BHI) broth (Oxoid, Basingstoke, UK) for bacteriological culture, while the remaining blood sample was used for the preparation of thick and thin blood films on grease-free slides (two per sample) for the detection and speciation of Plasmodium parasites by light microscopy. Group B was made up of 23 patients who had presented with frequent stools for 2 or more days (diarrhea). A fresh stool sample from each patient was inoculated into Cary-Blair transport medium (10 mL/tube) and was brought to the laboratory for bacteriological culture.
Bacterial agent isolation
Blood samples in the inoculated BHI broth culture bottles were incubated overnight aerobically at 37°C. Similarly, stool samples from Cary-Blair medium were inoculated into enrichment Selenite F broth (Oxoid) and were incubated at 37°C for 18–24 hours aerobically. Thereafter, sub-cultures were made onto deoxycholate citrate agar, Salmonella-Shigella agar, and MacConkey agar plates. All the culture agar plates were incubated at 37°C aerobically for 18–24 hours. In negative blood samples, sub-cultures were repeated daily from the BHI broth cultures for 7 consecutive days, after which the samples were disposed of. After overnight incubation, culture plates were examined for colonial morphology and Gram stained. Colonies were first identified by standard methods, as described in Cowan and Steel’s Manual.19 The Analytical Profile Index 20E identification system (Institut Mérieux, Marcy l’Etoile, France) was used for the confirmation of the Salmonella isolates. Further identification of Salmonella species based on their somatic (O) and flagella (H) antigen characteristics was done using polyvalent antisera (Wellcome Diagnostic, London, UK).
Antimicrobial susceptibility testing
All Salmonella isolates were investigated for their in vitro susceptibilities to 13 antibiotics by disk diffusion, as described by Clinical and Laboratory Standard Institute (CLSI) guidelines.20 Disks with the following preparations were used for susceptibility testing: ampicillin (25 μg), chloramphenicol (30 μg), co-trimoxazole (25 μg), tetracycline (25 μg), nalidixic acid (30 μg), ciprofloxacin (20 μg), ofloxacin (20 μg), gentamicin (10 μg), cefotaxime (30 μg), augmentin (30 μg; amoxicillin 20 μg/clavulanic acid 10 μg combination), ceftriaxone (30 μg), ceftazidime (30 μg), imipenem (30 μg), levofloxacin (10 μg), and azithromycin (15 μg) (Oxoid). The plates were incubated aerobically at 37°C for 18–24 hours. The diameter of the zones of inhibition were measured with a ruler and compared with a zone interpretation chart.14 E. coli American Type Culture Collection (ATCC) 25922 was used as a control. Multidrug resistance phenotype was defined as resistance to three or more classes of antibiotics.
ESBL assay
All the isolates that exhibited reduced susceptibility and/or resistance to 3GCs were screened for ESBL production, using the double disk synergy test method. This was done by placing the 3GC antibiotics, ie, ceftazidime (30 μg) and ceftriaxone (30 μg) at a distance of 15 mm (center to center) from 30 μg augmentin (20 μg amoxicillin combined with 10 μg clavulanic acid), using CLSI interpretative guidelines as the standard.21 An aliquot of a 0.5 μl Klebsiella pneumoniae ATCC 700603 was used as the positive control and E. coli ATCC 25922 was used as the negative control in each test batch.
Plasmid DNA extraction
Plasmid extraction was performed by a simplified alkaline lysis method described by Cheng et al.22 Briefly, overnight culture (1.5 mL) was centrifuged at 5,000 rpm for 1 minute to pellet the cells. The supernatant was gently decanted. After washing, the cell pellet was re-suspended in 300 μL Tris (tris(hydroxymethyl)aminomethane)-EDTA(ethylenediaminetetraacetic acid)-NaOH-SDS (sodium dodecyl sulfate) (TENS) buffer solution and mixed by gentle inversion (five times) within 3–5 minutes of incubation on ice to obtain a straw-like, sticky lysate. This was followed by neutralization by adding 200 μL of 5 M potassium acetate buffer (pH 5.2), followed by incubation on ice for 10 minutes. Plasmid DNA solution was recovered as supernatant after centrifugation at 10,000 rpm for 10 minutes. Plasmid pellets isolated with ice-cold absolute ethanol were washed in 70% ethanol before re-suspension in 20–40 μL Tris (10 mM)-EDTA (1 mM) buffer (pH 8.0). The plasmid DNAs were separated by electrophoresis on 0.8% agarose pre-stained with ethidium bromide (0.5 μg/mL).
Detection of blaCTX-M-I gene cluster
Genomic DNA was extracted from the Salmonella isolates by boiling method and was used as a template for the detection of ESBL blaCTX-M-I gene cluster by polymerase chain reaction (PCR). The 25 μL PCR reaction mixture contained 10 pmol of each forward and reverse primer pair for the blaCTX-M-I gene cluster, 200 μM of deoxynucleotide triphosphate, 1 μL of Taq DNA polymerase, and 1 μL of DNA solution. The PCR program consisted of an initial denaturation step of 5 minutes at 94°C, a 30 cycle period (each cycle consisting of 30 seconds at 94°C, 40 seconds at 52°C, and 50 seconds at 72°C), and then a final extension step of 5 minutes at 72°C. The primer nucleotide sequences used for the 415 bp blaCTX-M-I group gene were 5′-AAA AAT CAC TGC GCC AGT TC-3′ and 5′-AGC TTA TTC ATC GCC ACG TT-3′ for Enterobacteriaceae species, as previously reported by Woodford et al.23 The 415 bp blaCTX-M-I gene was resolved on 1.5% agarose gel pre-stained with ethidium bromide (0.5 μg/mL) by electrophoresis.
Conjugation experiment
Salmonella isolates harboring the blaCTX-M-I group of ESBLs were selected for the conjugation experiment using the broth mating technique described by Chen et al24 with rifampicin-resistant E. coli j53-2 as the recipient. Transconjugants were selected on a BHI agar plate containing cefotaxime (4 μg/mL) and rifampicin (200 μg/mL). Transfer of blaCTX-M-I gene, plasmids, and co-dissemination of antibiotic resistance was confirmed by PCR, plasmid isolation, and antibiotic susceptibility testing for the transconjugants and recipient, as previously done for the donor.
Statistical analyses
Data were expressed as frequencies or percentages. Fischer’s exact test was performed on 2×2 and 2×3 contingency tables of data for assessing disparity in Salmonella isolation rates between blood and stool samples, and ESBL patterns among the three S. enterica serovars isolated. Outcomes of P<0.05 were taken to be significant.
Results
Out of the 135 samples screened in group A, 35 Salmonella isolates (25.9%) were made up of three serotypes; S. typhi (71.4%), S. paratyphi (22.9%), and two (5.7%) of Salmonella enteritidis. Nine Plasmodium spp. were identified. Three strains of S. typhi and a strain of S. paratyphi were isolated from patients with Plasmodium sp. and complications. Thirteen of the 23 stool samples from group B were confirmed positive for bacterial pathogens, made up of eleven strains of E. coli and two strains of S. enteritidis (Table 1). Further analysis revealed non-significant difference (P>0.05) in the observed disparity in the isolation rate of S. enterica isolates or S. enteritidis isolates between culture-positive stool and blood samples from the two groups of patients studied.
Table 1.
Percentage distribution of pathogens isolated according to clinical samples
| Source (sample size) | No of positive sample (%) |
Salmonella enterica serotypes isolated
|
Other pathogens obtained (% from total positive sample)
|
||||
|---|---|---|---|---|---|---|---|
| No of isolates (%) | S. typhi | S. paratyphi | S. enteritidis | E. coli | Plasmodium sp. | ||
| Blood (135) | 46 (78.0) | 35 (94.6)a | 25 (71.4) | 8 (22.9) | 2 (5.7) | 0 (0) | 9 (19.6) |
| Stool (23) | 13 (22.0) | 2 (5.4) | 0 (0.0) | 0 (0.0) | 2 (100)a | 11 (84.6) | Not applicable |
| Total | 59 (100.0) | 37 (100) | 26 (70.3) | 8 (21.6) | 3 (8.1) | 11 (100) | 9 (100) |
Notes:
P>0.05 (blood vs stool), Fischer’s exact test.
Abbreviations: No, number; vs, versus.
Antibiotic resistance patterns of the isolated 37 S. enterica serovars are presented in Table 2. A total of 24 isolates were ESBL producers, elicited resistance to 5–7 antibiotics, and accounted for eleven (78.6%) of the 14 antibiotic resistance patterns observed. On the whole, 75.7% of the recovered S. enterica serovars isolated were multidrug resistant. PCR analysis revealed ESBL production due to blaCTX-M-I gene cluster in eleven (45.8%) S. enterica serovars. These strains also accounted for 76.9% (ten of 13 cases) of augmentin resistance observed. Carriage of blaCTX-M-I was also significantly associated with cefotaxime resistance (Table 2). However, all the Salmonella isolates were sensitive to imipenem, levofloxacin, and azithromycin. Of the eleven blaCTX-M-I ESBL producers detected, nine (81.8%) were S. typhi and two (18.2%) were S. enteritidis. Non-production of ESBL by 24%, 75%, and 25% of S. typhi, S. paratyphi, and S. enteritidis (P=0.03), respectively, was also observed (Figure 1). Conjugation experiments revealed a 23 kb self-transmissible plasmid and co-transfer of cefotaxime and augmentin resistance to E. coli j53-2 transconjugants by four of the eleven blaCTX-M-I-producing donor serovars, all of which were S. typhi strains (Figure 2 and Table 3).
Table 2.
Antibiotic resistance patterns of Salmonella enterica serovars producing bla-CTX-M-1 extended-spectrum beta lactamase
| S/N | Resistance pattern | ESBL− | ESBL+ | blaCTX-M-1 | Non-blaCTX-M-1 | P-value |
|---|---|---|---|---|---|---|
| 1 | AMPTETNALAUGCROCAZ | 0 | 2 | 0 | 2 | 0.19 |
| 2 | AMPTETCOTCOLAUGCROCTX | 0 | 2 | 2 | 0 | 0.08 |
| 3 | AMPTETNALCROCAZ | 0 | 1 | 0 | 1 | 0.95 |
| 4 | AMPTETCOTAUGCAZCROCTX | 0 | 6 | 5 | 1 | 0.004 |
| 5 | AMPTETCOTNALCAZ | 0 | 2 | 0 | 2 | 0.2 |
| 6 | AMPTETCOTGENCAZ | 0 | 2 | 0 | 2 | 0.2 |
| 7 | AMPCOTTETNALCRO | 0 | 2 | 0 | 2 | 0.2 |
| 8 | AMPTETNALCROCAZ | 0 | 3 | 1 | 2 | 0.39 |
| 9 | AMPTETNALCROCAZ | 0 | 1 | 0 | 1 | 0.53 |
| 10 | AMPNALAUGCROCTX | 0 | 1 | 1 | 0 | 0.3 |
| 11 | AMPCOTAUGCAZCTX | 0 | 2 | 2 | 0 | 0.08 |
| 12 | AMPCOTTETNAL | 4 | 0 | 0 | 0 | 0.02 |
| 13 | AMPTET | 5 | 0 | 0 | 0 | 0.004 |
| 14 | AMP | 4 | 0 | 0 | 0 | 0.02 |
| TOTAL | 13 | 24 | 11 | 13 |
Abbreviations: AMP, ampicillin; TET, tetracycline; COT, cotrimoxazole; NAL, nalidixic acid; GEN, gentamicin; COL, colistin; STR, streptomycin; AUG, augmentin; CRO, ceftriaxone; CAZ, ceftazidime; CTX, cefotaxime; S/N, sample number; ESBL, extended-spectrum beta lactamase.
Figure 1.
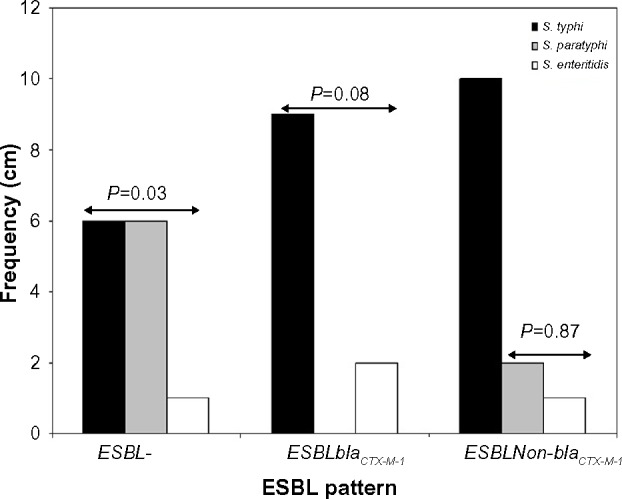
Distribution of Salmonella enterica serovars producing extended spectrum beta lactamases.
Notes: P<0.05 was significant. S. enterica serovars as per ESBL patterns 2×3 contingency table, Fischer’s exact test.
Abbreviation: ESBL, extended spectrum beta lactamase.
Figure 2.
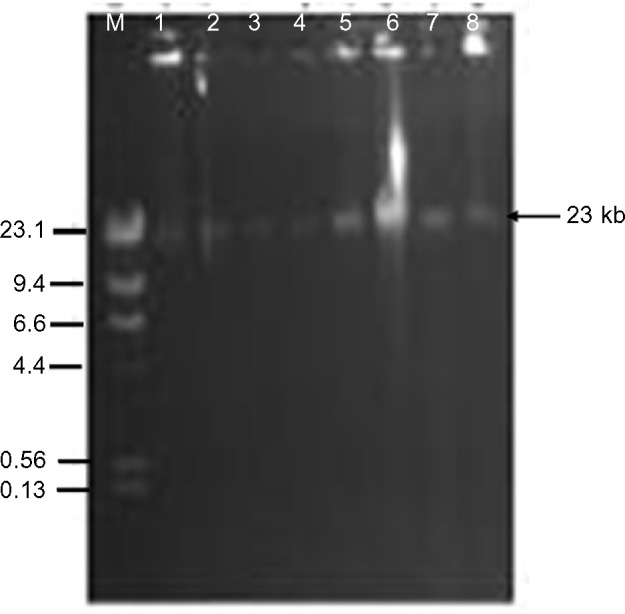
Plasmid transfer by conjugation from donor Salmonella typhi strains to recipient Escherichia coli j53-2. Lanes 1, 3, 5, and 7 are S. typhi donors Lag-003, -004, -007, and -010. Lanes 2, 4, 6, and 8 are E. coli j53-2 transconjugants. Lane M represents Lamda DNA Hind III markers.
Abbreviation: DNA, deoxyribonucleic acid.
Table 3.
Antibiotic resistance and plasmid transfer by conjugation from the donor Salmonella typhi isolates to the Escherichia coli j53-2 recipient
| Donor S. typhi isolate
|
E. coli j53-2 transconjugant
|
|||
|---|---|---|---|---|
| Strain ID | Plasmid profile, kb | Antibiotic resistance profile | Plasmid transferred, kb | Transferred antibiotic resistant profile |
| Lag-001 | 30.5, 23, 9.0, 2.1 | AMPTETCOTCOLAUGCROCTX | 23 | AMPCOTAUGCTX |
| Lag-004 | 23, 9.0 | AMPTETCOTAUGCAZCROCTX | 23 | AMPAUGCAZCTX |
| Lag-007 | 23, 17, 2.1 | AMPNALAUGCROCTX | 23 | AMPAUGCTX |
| Lag-010 | 23, 9.0, 5.1 | AMPCOTAUGCAZCTX | 23 | AMPCTX |
Abbreviations: ID, identification; AMP, ampicillin; TET, tetracycline; COT, cotrimoxazole; NAL, nalidixic acid; COL, colistin; AUG, augmentin; CRO, ceftriaxone; CAZ, ceftazidime; CTX, cefotaxime.
Discussion
3GCs remain the most commonly prescribed class of antibiotics for case management of typhoidal and non-typhoidal salmonellosis in many countries of the world, including Nigeria12,25 In this study, 37 Salmonella isolates, which included 35 (25.9%) from blood of patients with persistent pyrexia and two (8.7%) S. enteritidis from stool samples of patients with gastroenteritis were isolated to yield a prevalence of 20.8% salmonellosis. S. typhi was further found to account for 71.4% of persistent pyrexia cases due to Salmonella isolates producing blaCTX-M-I ESBLs that were mediated by a 23 kb plasmid. These phenotypic and molecular characteristics were observed in 75.6% of Salmonella isolates that were multidrug resistant.
The findings from the current study indicated that the epidemiology of Salmonella as a public health burden in Lagos has not changed from the previous reports from Lagos12,15,26 and from other parts of Nigeria.16 Interestingly, in this study, some of the bacteremic strains of S. typhi that produced blaCTX-M-I ESBL can be used to explain why persistent pyrexia occurs in affected patients despite them receiving treatment with 3GCs. The implication of this is the potential for spread of emerging blaCTX-M-I-producing S. typhi in Lagos, which will add to the prevailing public health burdens in the state. In our previous study in the same environment, we found 53.1% of Salmonella isolates, composed of Salmonella typhimurium, S. enteritidis, S. paratyphi, and Salmonella choleraesuis from gastroenteritis cases to be blaCTX-M-I positive.12 The present result found eleven (nine S. typhi and two S. enteritidis) of 37 Salmonella isolates recovered to be blaCTX-M-I-positive, with four S. typhi isolates transferring this genetic marker to E. coli j53-2. This finding indicates that other groups of bla-CTX-M exist among clinical Salmonella isolates in Lagos and that blaCTX-M-I carriage may be an emerging trend with pathogenic advantage to cause persistent pyrexia in patients with typhoid fever. The blaCTX-M-I rate of 45.6% observed in this study is higher than the 7.7% detected among other clinical enteric pathogens from Abeokuta, Nigeria by Akinduti et al.27 It is also higher than the 6% rate recently reported among clinical Salmonella isolates in France.28 In previous studies on blaCTX-M-I gene variants in Lagos, blaCTX-M-15 was detected in 17 urinary pathogens of K. pneumoniae by Soge et al29 and in Enterobacter cloacae and Pantoea agglomerans isolates by Aibinu et al.30 Also in south-eastern Nigeria, Iroha et al11 reported the carriage of blaCTX-M-I ESBL among 44 clinical isolates of E. coli from two hospitals in 2012. These findings indicate increasing spread of blaCTX-M-I among Enterobacteriaceae members in Nigeria.
Continuous surveillance of blaCTX-M-I-producing pathogens is needed to guide preventive interventions in Nigeria. BlaCTX-M-I gene cluster carriage is associated with increased resistance to cefotaxime. Therefore, the clinical practice of switching Salmonella bacteremic and febrile patients who failed treatment with ceftriaxone, cefuroxime, or ceftazidime empirically to cefotaxime also has the risk of treatment failure in this environment. For such patients, this study recommends the use of imipenem or levofloxacin, or azithromycin for case management. The clinical effectiveness of these antibiotics for treating multidrug-resistant infections has been reported in African countries such as Tanzania,31 in North and South America,32 Nepal,33 and India.34 These antibiotics are also included in the drug formulary of many hospitals in Lagos and other states in Nigeria, where they are used for case management of inpatients and outpatients affected by other bacterial infections.9–12 The results of the conjugation experiment not only showed the blaCTX-M-I gene carried by S. typhi was plasmid-mediated, but also revealed the potential for rapid spread of this genetic marker to other members of Enterobacteriaceae such as E. coli, which are often encountered in polymicrobial infections in Lagos and other regions of Nigeria.35
Similar plasmid-mediated transfer of blaCTX-M-I and resistance to antibiotics such as cefotaxime, aminoglycosides, and beta-lactamase inhibitor-containing antibiotics such as augmentin, as demonstrated in the present study, have been documented by previous investigators from other countries of the world.31,36–38 However, disparity can be seen in the molecular size of the plasmid transferred. Jin and Ling36 reported the presence of bla-CTX-M genes on 62 kb, 70 kb, and 92 kb harbored by S. enteritidis and S. typhimurium from Hong Kong, while Mshana et al31 found a 231 kb plasmid to be responsible for the carriage of bla-CTX-M-15 gene among S. enterica isolates from Tanzania. In a study carried out by Bado et al37 in Uruguay, where the first description of blaCTX-M-I gene cluster in S. enteritidis was reported, a 195 kb plasmid was incriminated as a vector. In Germany, Fischer et al38 reported the transferability of blaCTX-M-15 among Salmonella isolates from horse and swine through 95 kb IncF and Inc1I plasmids. In Lagos, an earlier study by Soge et al29 revealed the presence of the blaCTX-M-I gene on plasmids of sizes 58–320 kb in K. pneumoniae.
Nevertheless, it is sufficient to suggest that multiple self-transmissible plasmids are involved in the spread of blaCTX-M-I across the different regions and countries of the world. Their active roles for dissemination of cefotaxime resistance appear to vary according to geographical region, reservoir of Salmonella isolates (ie, food animals or pets of humans), time of study, and their genetic environment. The latter can be used to explain why blaCTX-M-I carriage accounted for 76.9% resistance to augmentin, as seen in the current study. This was further confirmed by co-transfer of resistance to antibiotics to E. coli j53-2 by conjugation. Studies conducted in other countries have reported carriage of augmentin resistance markers such as ampC, aminoglycoside modification genes, and metallo beta lactamase genes on blaCTX-M-I plasmids.29–31,36–39
Furthermore, additional antibiotic resistance conferred by this genetic marker may also set the stage for the emergence of pan-resistant S. typhi in Nigeria, as recently documented in some Asian countries.40 The emergence of multidrug-resistant typhoidal and non-typhoidal Salmonella have caused life-threatening invasive disease outbreaks in children and adults in many African countries, including Zaire,41 Malawi,42 and Kenya.43 Similarly, multidrug-resistant Salmonella serotypes have been widely prevalent in Kuwait and India.44,45
It is worthy to note in this study that four Salmonella isolates with P. falciparum-associated co-infection were detected, and that S. typhi mono-infection was characterized by ceftriaxone and cefotaxime resistance; the current study has further revealed the relevance of blaCTX-M-I ESBL testing for rational use of both ACT and antibiotics for good outcomes in patients with persistent pyrexia-associated Salmonella or Plasmodium, or both. In this context, the reported risk of poor treatment outcomes in patients admitted in hospitals with laboratories that do not perform tests for detection of ESBLs and do not report ESBL producers as resistant to cephalosporins is now apparent in Lagos.46 The potential spread of blaCTX-M-I-producing S. typhi to other states of the country and other neighboring countries is also a possibility.
The inability to confirm the variants of blaCTX-M-I gene cluster carried by the positive S. typhi strains, which is essential to infer clonal relatedness of these strains, is one of the limitations of this study. Molecular typing techniques such as multilocus sequence typing and pulse-field gel electrophoresis to reveal information about the upstream and downstream genetic environment of the recovered blaCTX-M-I plasmid of the positive S. typhi isolates is essential to assess their levels of homogeneity or heterogeneity. Such information will also be needed to further understand mechanisms of dissemination and extra-antibiotic resistance mechanisms of blaCTX-M-I plasmids among S. typhi isolates in our environment.
Despite these limitations, this study has revealed the emergence of resistance to 3GC antibiotics due to acquisition and expression of plasmid-borne blaCTX-M-I gene cluster among S. typhi strains, and this emergence calls for cautionary parenteral use of cefotaxime and other 3GCs for the treatment of typhoid fever in Lagos, Nigeria. A need for routine screening for P. falciparum co-infection with ESBL-producing Salmonella in the laboratories during diagnosis of persistent pyrexia conditions in patients is recommended.
Acknowledgments
We are grateful to all the staff of the public hospitals used for this work. We are equally grateful to the Staff of the Department of Microbiology of Lagos State University and the Nigerian Institute of Medical Research, Yaba for their technical support.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Akinyemi KO, Iwalokun BA, Foli F, Oshodi K, Coker AO. Prevalence of multiple drug resistance and screening of enterotoxin gene (stn) in Salmonella enterica serovars from water sources in Lagos, Nigeria. Public Health. 2011;125:65–71. doi: 10.1016/j.puhe.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 2.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. B World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 3.Goque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008;13(47):19051. [PubMed] [Google Scholar]
- 4.Jacoby AG, Munoz-Price SL. The new β-lactamases. N Engl J Med. 2005;352:380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 5.Menashe G, Borer A, Yagupsky P, et al. Clinical significance and impact on mortality of extended-spectrum beta lactamase-producing Enterobacteriaceae isolates in nosocomial bacteremia. Scand J Infect Dis. 2001;33:188–193. doi: 10.1080/00365540151060806. [DOI] [PubMed] [Google Scholar]
- 6.Ndugulile F, Jureen R, Harthug S, Urassa W, Langeland N. Extended spectrum β-lactamases among gram-negative bacteria of nosocomial origin from an intensive care unit of a tertiary health facility in Tanzania. BMC Infect Dis. 2005;5:1–6. doi: 10.1186/1471-2334-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tassios PT, Gazouli M, Tzelepi E, et al. Spread of a Salmonella typhimurium clone resistant to expanded-spectrum cephalosporins in three European countries. J Clin Microbiol. 1999;37(11):3774–3777. doi: 10.1128/jcm.37.11.3774-3777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winokur PL, Canton R, Casellas JM, Legakis N. Variations in the prevalence of strains expressing an extended-spectrum beta-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin Infect Dis. 2001;32:S94–S103. doi: 10.1086/320182. [DOI] [PubMed] [Google Scholar]
- 9.Aibinu IE, Ohaegbulam VC, Adenipekun EA, Ogunsola FT, Odugbemi TO, Mee BJ. Extended-spectrum beta-lactamase enzyme in clinical isolates of Enterobacter species from Lagos, Nigeria. J Clin Microbiol. 2003;41:2197–2200. doi: 10.1128/JCM.41.5.2197-2200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iroha IR, Amadi ES, Agabus AC, Oji AE. Susceptibility pattern of extended spectrum beta-lactamase producing Klebsiella pneumoniae from clinical isolates. Int J Microbiol. 2008;5:34–37. [Google Scholar]
- 11.Iroha IR, Esimone CO, Neumann S, et al. First description of Escherichia coli producing CTX-M-15-extended spectrum beta lactamase (ESBL) in out-patients from south eastern Nigeria. Ann Clin Microbiol Antimicrob. 2012;11:19. doi: 10.1186/1476-0711-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akinyemi KO, Iwalokun BA, Agboyinu JA, Ogunyemi O, Fasure AK. Emergence of third generation cephalosporins and typing by rapid amplified polymorphic DNA among clinical Salmonella isolates from Lagos, Nigeria. Brit Microbiol Res J. 2014;6:668–677. [Google Scholar]
- 13.Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev. 2013;26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radice M, Power P, Di Conza J, Gutkind G. Early dissemination of CTX-M-derived enzymes in South America. Antimicrob Agents Chemother. 2012;46:602–604. doi: 10.1128/AAC.46.2.602-604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akinyemi KO, Oshundare YO, Oyeyinka OG, Coker AO. A retrospective study of community-acquired Salmonella infections in patients attending public hospitals in Lagos, Nigeria. J Infect Dev Ctries. 2012;6(5):387–395. doi: 10.3855/jidc.2120. [DOI] [PubMed] [Google Scholar]
- 16.Fashae K, Ogunshola F, Aerestrup FM, Hendriksen RS. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J Infect Dev Ctries. 2010;4:484–494. doi: 10.3855/jidc.909. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization . Guidelines for the Treatment of Malaria. 2nd ed. Geneva: World Health Organization; 2010. [Google Scholar]
- 18.World Health Organization . The Implementation of the Global Malaria Control Strategy: WHO Technical Report Series No 839. Geneva: World Health Organization; 1993. [PubMed] [Google Scholar]
- 19.Barrow GI, Feltham RKA, editors. Cowan and Steel’s Manual for the identification of Medical Bacteria. 3rd edn. Cambridge University Press; 1993. p. 331. [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards . Performance Standards for Antimicrobial Susceptibility Testing: Ninth Informational Supplement, Version 19. Wayne, PA: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 21.Clinical Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing: 19th Informational Supplement M100S19. Wayne, PA: Clinical Laboratory Standards Institute; 2009. [Google Scholar]
- 22.Cheng S, Zhao S, White DG, Meng J. Characterization of multiple antimicrobial resistant Salmonella serovars isolated from retail meats. Appl Environ Microbiol. 2004;70:1–7. doi: 10.1128/AEM.70.1.1-7.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodford N, Fagan EJ, Ellington LJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2006;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Chen ZL, Liu JH, Zeng ZL, Ma JY, Jiang HX. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J Antimicrob Chemother. 2007;59:880–885. doi: 10.1093/jac/dkm065. [DOI] [PubMed] [Google Scholar]
- 25.Arlet G, Barrett TJ, Butaye P, Cloeckaert A, Mulvey MR, White DG. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 2006;8:1945–1954. doi: 10.1016/j.micinf.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 26.Akinyemi KO, Smith SI, Oyefolu AO, Coker AO. Multidrug resistance in Salmonella enterica serovar typhi isolated from patients with typhoid fever complications in Lagos, Nigeria. Public Health. 2005;119:321–327. doi: 10.1016/j.puhe.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Akinduti PA, Oluwadun A, Iwalokun BA, Onagbesan OM, Ejilude O. Community-acquired CTX-M beta lactamase isolates from Abeokuta, Nigeria. Brit Microbiol Res J. 2014;5:351–358. [Google Scholar]
- 28.Haenni M, Saras E, Métayer V, Médaille C, Madec JY. High prevalence of blaCTX-M-1/IncI1/ST3 and blaCMY-2/IncI1/ST2 plasmids in healthy urban dogs in France. Antimicrob Agents Chemother. 2014;58:5358–5362. doi: 10.1128/AAC.02545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soge OO, Adeniyi BA, Roberts MC. New antibiotic resistance genes associated with CTX-M plasmids from uropathogenic Nigerian Klebsiella pneumoniae. J Antimicrob Chemother. 2006;58(5):1048–1053. doi: 10.1093/jac/dkl370. [DOI] [PubMed] [Google Scholar]
- 30.Aibinu I, Pfeifer Y, Peters F, et al. Emergence of bla(CTX-M-15), qnrB1 and aac(6′)-Ib-cr resistance genes in Pantoea agglomerans and Enterobacter cloacae from Nigeria (sub-Saharan Africa) J Med Microbiol. 2012;61:165–167. doi: 10.1099/jmm.0.035238-0. [DOI] [PubMed] [Google Scholar]
- 31.Mshana SE, Gerwing L, Minde M, et al. Outbreak of a novel Enterobacter sp. carrying blaCTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int J Antimicrob Agents. 2011;38(3):265–269. doi: 10.1016/j.ijantimicag.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Biedenbach DJ, Toleman M, Walsh TR, Jones RN. Analysis of Salmonella spp. with resistance to extended-spectrum cephalosporins and fluoroquinolones isolated in North America and Latin America: report from the SENTRY Antimicrobial Surveillance Program (1997–2004) Diagn Microbiol Infect Dis. 2006;54(1):13–21. doi: 10.1016/j.diagmicrobio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Pathak A1, Marothi Y, Kekre V, Mahadik K, Macaden R, Lundborg C. High prevalence of extended-spectrum β-lactamase-producing pathogens: results of a surveillance study in two hospitals in Ujjain, India. Infect Drug Resist. 2012;5:65–73. doi: 10.2147/IDR.S30043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pokharel BM, Koirala J, Dahal RK, Mishra SK, Khadga PK, Tuladhar NR. Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. Int J Infect Dis. 2006;10:434–468. doi: 10.1016/j.ijid.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Iwalokun BA, Gbenle GO, Smith SI, Ogunledun A, Akinsinde KA, Omonigbehin EA. Epidemiology of shigellosis in Lagos, Nigeria: trends in antimicrobial resistance. J Health Popul Nutr. 2001;19:183–190. [PubMed] [Google Scholar]
- 36.Jin Y, Ling JM. CTX-M-producing Salmonella spp. in Hong Kong: an emerging problem. J Med Microbiol. 2006;55(Pt 9):1245–1250. doi: 10.1099/jmm.0.46637-0. [DOI] [PubMed] [Google Scholar]
- 37.Bado I, García-Fulgueiras V, Cordeiro NF, et al. First human isolate of Salmonella enterica serotype Enteritidis harboring bla-CTX-M-14 in South America. Antimicrob Agents Chemother. 2012;56:2132–2134. doi: 10.1128/AAC.05530-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer J, Rodríguez I, Baumann B, et al. bla-CTX-M-15-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J Antimicrob Chemother. 2014;69:2951–2958. doi: 10.1093/jac/dku270. [DOI] [PubMed] [Google Scholar]
- 39.Baba Ahmed-Kazi Tani Z, Arlet G. News of antibiotic resistance among Gram-negative bacilli in Algeria. Pathol Biol (Paris) 2014;62:169–178. doi: 10.1016/j.patbio.2014.01.005. French. [DOI] [PubMed] [Google Scholar]
- 40.Tadesse G. A meta-analysis of the proportion of antimicrobial resistant human Salmonella isolates in Ethiopia. BMC Pharmacol Toxicol. 2014;15:51. doi: 10.1186/2050-6511-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milledge J, Calis JC, Graham SM. The effect of HIV infection or neonate bacterial meningitis in Blantyre, Malawi. Arch Dis Child. 2003;88:1112–1118. doi: 10.1136/adc.88.12.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karuiki S, Revathi G, Kariuki N, et al. Multidrug-resistant non-typhoidal Salmonella infection in Africa: zoonotic or anthroponotic transmission. J Med Microbiol. 2006;55:585–591. doi: 10.1099/jmm.0.46375-0. [DOI] [PubMed] [Google Scholar]
- 43.Amatya NM, Shrastha B, Lekhak B. Etiological agents of bacteraemia and antibiotic susceptibility pattern in Kathmandu Model hospital. JNMA J Nepal Med Assoc. 2007;46:112–118. [PubMed] [Google Scholar]
- 44.Dimitrov T, Udo EE, Albaksami O, et al. Clinical and microbiological investigations of typhoid fever in an infectious disease hospital in Kuwait. J Med Microbiol. 2007;56:538–544. doi: 10.1099/jmm.0.46814-0. [DOI] [PubMed] [Google Scholar]
- 45.Shukla I, Tiwan R, Agarwal M. Prevalence of Extended spectrum beta-lactamase producing Klebsiella pneumoniae in a tertiary care hospital. Indian J Med Microbiol. 2004;22:87–91. [PubMed] [Google Scholar]
- 46.Kruger T, Szabo D, Keddy KH, et al. Infections with Non-typhoidal Salmonella species producing TEM-63 or a novel TEM enzyme, TEM-131, in South Africa. Antimicrob Agents Chemother. 2004;48:4263–4270. doi: 10.1128/AAC.48.11.4263-4270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


