Abstract
Background
The fibroblast growth factor (FGF) and FGF receptor (FGFR) axis plays a critical role in tumor-igenesis, but little is known of its influence in ovarian cancer. We sought to determine the association of genetic variants in the FGF pathway with risk, therapeutic response, and survival of patients with ovarian cancer.
Methods
We matched 339 non-Hispanic white ovarian cancer cases with 349 healthy controls and geno-typed them for 183 single-nucleotide polymorphisms (SNPs) from 24 FGF (fibroblast growth factor) and FGFR (fibroblast growth factor receptor) genes. Genetic associations for the main effect, gene– gene interactions, and the cumulative effect were determined.
Results
Multiple SNPs in the FGF–FGFR axis were associated with an increased risk of ovarian cancer. In particular, FGF1 [fibroblast growth factor 1 (acidic)] SNP rs7727832 showed the most significant association with ovarian cancer (odds ratio, 2.27; 95% CI, 1.31–3.95). Ten SNPs were associated with a reduced risk of ovarian cancer. FGF18 (fibroblast growth factor 18) SNP rs3806929, FGF7 (fibroblast growth factor 7) SNP rs9920722, FGF23 (fibroblast growth factor 23) SNP rs12812339, and FGF5 (fibroblast growth factor 5) SNP rs3733336 were significantly associated with a favorable treatment response, with a reduction of risk of nonresponse of 40% to 60%. Eleven SNPs were significantly associated with overall survival. Of these SNPs, FGF23 rs7961824 was the most significantly associated with improved prognosis (hazard ratio, 0.55; 95% CI, 0.39 – 0.78) and was associated with significantly longer survival durations, compared with individuals with the common genotype at this locus (58.1 months vs. 38.0 months, P = 0.005). Survival tree analysis revealed FGF2 rs167428 as the primary factor contributing to overall survival.
Conclusions
Significant associations of genetic variants in the FGF pathway were associated with ovarian cancer risk, therapeutic response, and survival. The discovery of multiple SNPs in the FGF–FGFR pathway provides a molecular approach for risk assessment, monitoring therapeutic response, and prognosis.
Ovarian cancer is the leading cause of death from gynecologic cancers and the fifth most lethal malignancy in women in the US. An estimated 22 240 new cases and 14 030 deaths from ovarian cancer will occur in the US in 2013 (1). The overall dismal 46% 5-year survival rate for ovarian cancer has remained unchanged for several decades (1). The main reason for this poor outcome is the lack of success in diagnosing ovarian cancer at an early stage, owing to an absence of obvious symptoms, clinical indications, and effective screening tests. A majority of women are diagnosed with a high-grade invasive cancer that is difficult to treat. In contrast, women have a 90% to 95% probability of survival when their ovarian cancer is detected at an early stage (2). The results obtained with current screening strategies to reduce mortality in women with ovarian cancer— which use the serum biomarkers cancer antigen 125 (CA125)5 and human epididymis protein 4 (HE4) along with the Risk of Ovarian Malignancy Algorithm and transvaginal ultrasonography— have not been encouraging (3). Currently, these serum biomarkers (CA125 and HE4) are being used mainly to monitor chemotherapeutic response and to detect recurrence after therapy, but none of the current biochemical markers are sufficient to guide the prediction, screening, and prognosis of ovarian cancer (4, 5). Therefore, the search for ovarian cancer biomarkers—in particular genetic markers—for risk assessment, monitoring of therapeutic response, and outcome prediction of ovarian carcinoma is of profound importance.
A number of common germline genetic alterations, including those identified via genome-wide association studies, have been associated with ovarian cancers (6–9). Candidate-gene and pathway-based approaches have also successfully identified ovarian cancer–susceptibility loci and loci associated with clinical outcomes (10). Our group previously demonstrated that nucleotide-excision repair polymorphisms are associated with recurrence and survival in ovarian cancer patients (11). Transforming growth factor β (TGF-β) enhances ovarian tumor metastasis, and we recently demonstrated that genetic variants in the TGF-β signaling pathway are associated with variation in the risk of developing ovarian cancer (12). In addition, we have identified several microRNA-related genetic polymorphisms that are associated with ovarian cancer risk and clinical outcomes (13); however, the full spectrum of the genetic loci contributing to ovarian cancer susceptibility and outcome remains to be revealed.
Ovarian cancer is a multifactorial and polygenic malignancy; therefore, any variation in a single gene will not be sufficient to provide comprehensive disease information. Fibroblast growth factors (FGFs) are a large family (24 members) of growth and differentiation factors. FGFs mediate their effects by binding to FGF receptors (FGFRs) on the cell surface. The signaling axis of FGFs and their receptors plays important roles in regulating cellular proliferation, migration, angiogenesis, wound repair, and differentiation (14). The FGF–FGFR axis has been demonstrated to modulate tumor stroma and cancer progression (15). On the other hand, FGF signaling may have tumor-suppressive functions in certain contexts (16). Compelling evidence has shown that FGF signaling pathways are implicated in cancer progression by inducing mitogenesis, cell migration, and tumor angiogenesis (16, 17). Therefore, aberrant FGF signaling can promote cancer development.
Several studies have demonstrated altered expression of genes encoding FGF receptors to be associated with ovarian cancer (18, 19). Furthermore, serum FGF2 (fibroblast growth factor 2) concentrations are increased in patients with ovarian cancer (20, 21), and amplification of FGF1 is correlated with poor survival in patients with advanced-stage serous ovarian cancer (22, 23). Similarly, altered expression of genes encoding FGFs have been reported for other human cancers (24, 25). This growing evidence for the role of FGF signaling in tumorigenesis has led to proposals for therapeutic strategies that target ovarian cancer via the FGF signaling pathway axis (14, 26, 27) The problem with developing such strategies, however, is that reports on the relationships between germline alterations in genes encoding FGFs or FGFRs and ovarian cancer are limited. Johnatty et al. investigated single-nucleotide polymorphisms (SNPs) in the FGF26 [fibroblast growth factor 2 (basic)] gene for ovarian cancer risk and observed no statistically significant associations (28); however, no studies have been conducted for other members of the FGF–FGFR axis or for associations with response to therapy or clinical outcome. Therefore, we investigated genetic variants within 24 FGF and FGFR genes for any associations with risk of ovarian cancer, therapeutic response to chemotherapy, and overall survival of patients with ovarian cancer.
Materials and Methods
Study Design
We recruited 417 ovarian cancer cases newly diagnosed and histologically confirmed at The University of Texas MD Anderson Cancer Center between August 1991 to January 2009. There were no age, ethnicity, and clinical-stage restrictions on recruitment. In parallel, we recruited a group of healthy women (n = 417) without prior history of cancer (except nonmelanoma skin cancer) from a large pool of individuals seeing a physician for routine health checkups or addressing health concerns at the Kelsey–Seybold Clinics, a large private multispecialty physician group in the Houston metropolitan area. Cases and controls were matched by age (±5 years) and ethnicity. To minimize population admixture, we included only non-Hispanic white individuals in the current analysis (339 cases and 349 controls).
Data Collection
Epidemiology, demographic, clinical, and follow-up data were obtained from medical records. Overall survival was calculated from the date of diagnosis to the date of death or the end of patient follow-up. Response to platinum-based chemotherapy was defined as evidence of residual disease, as indicated by various clinical measures, such as positron emission tomography and computed tomography scans, second-look surgery, and postchemotherapy CA125 concentration. Each patient and control individual signed a written informed-consent form. The study was approved by the Institutional Review Boards of The University of Texas MD Anderson Cancer Center.
SNP Selection and Genotyping
A peripheral blood sample was obtained from each study participant. Genomic DNA was extracted from peripheral blood with the QIAamp DNA Blood Maxi Kit (Qiagen) according to the manufacturer's protocol and stored for future use. For each gene encoding an FGF or FGFR, we extracted tag SNPs within 10 kb upstream of the transcriptional start site and 10 kb downstream of the transcriptional stop site. Selected tag SNPs had r2 values ≥0.80 and minor-allele frequencies (MAFs) ≥0.05. In addition, we identified potentially functional SNPs with MAF values ≥0.01, including coding SNPs and SNPs located in potential regulatory regions (promoter, splicing site, 5′ untranslated region, and 3′ untranslated region). We identified 183 SNPs in 24 genes encoding FGFs or FGFRs and sent a set of SNPs to Illumina technical support for custom iSelect Infinium II BeadChip design with Illumina's proprietary program. Genotyping followed the standard protocol for Illumina's Infinium iSelect HD Custom Genotyping BeadChip. For quality control, we randomly selected at least 2% to 3% of the samples for replicates. The concordance for all replicates was 100%. The call rate for all SNPs was 99.86%, and genotypes were autocalled with BeadStudio software (Illumina). All laboratory personnel were blinded to the case/control and outcome status of the study participants.
Statistical Analysis
The distributions of categorical variables and continuous variables between cases and controls were evaluated by Pearson χ2 tests and Student t-tests, respectively. The χ2 test was used to evaluate each SNP for the Hardy–Weinberg equilibrium in the population of control individuals; SNPs with P values <0.01 were removed from further analysis. Multivariate logistic regression analysis was performed to estimate odds ratios (ORs) and 95% CIs for each SNP's main effect while adjusting for age. The test with the highest level of statistical significance among the 3 genetic models of inheritance (dominant, recessive, and additive) was used to determine the statistical significance of each SNP. If the frequencies of the homozygous variant genotypes were <5% in the cases or controls, however, only the dominant model with the highest statistical power was considered. The effect of each SNP on survival was assessed with a multivariable Cox proportional hazards regression analysis adjusted for age, clinical stage, histology, and treatment regimen. We used Kaplan–Meier plots and log-rank tests to assess differences in overall survival for each SNP by genotype. For response to therapy, we carried out unconditional multivariate logistic regression analysis for each SNP while adjusting for age, clinical stage, histology, and treatment regimen. The cumulative effects of multiple unfavorable genotypes were evaluated for the SNPs that showed statistical significance in the main analysis (i.e., P < 0.05). As an alternative to external validation, we performed bootstrap resampling to internally validate the results. For single-SNP analysis, we conducted bootstrap re-sampling of 1000 runs. In each run, we performed bootstrap resampling 50 times to calculate the bootstrap P value. We then counted the number of times bootstrap P values were <0.05. For classification and regression tree (CART) analysis, survival tree analysis, and unfavorable-genotype analysis, we reported the bias-corrected bootstrap confidence intervals on the basis of performing bootstrap resampling 10 000 times. We used STATA software (version 10; StataCorp) for the statistical analyses described above, and we used HelixTree software (Golden Helix) for CART analysis to explore higher-order gene– gene interactions and to classify the study participants into distinct risk groups. Survival tree analysis was conducted with the STREE program (http://c2s2.yale.edu/software/stree) to build a decision tree via the recursive-partitioning method. In brief, the root nodes contained all the patients, and we defined the measure for goodness of split with the log-rank P value to select the optimal initial split and subsequent splits of the data set until no statistically significant split was identified (29). Owing to the small number of events for terminal node 1, we used terminal node 2 as the reference group to provide more-reliable estimates of the effect and the SE, thereby providing smaller 95% CIs. The terminal nodes were classified into low-risk, medium-risk, and high-risk groups according to their relative risk compared with terminal node 2. All P values reported were the results of 2-sided tests. Multiple hypothesis testing was conducted with the “q-value” package in R by controlling the false-discovery rate to <10% (30).
eQTL Analysis
To identify functional relevance in our findings, we checked for the potential functional effect of identified SNPs on gene expression by analyzing gene–SNP association in expression quantitative trait loci (eQTL) studies with the Genevar (GENe Expression VARiation) database (http://www.sanger.ac.uk/resources/software/genevar/) (31) in the HapMap3 data set. All analyses were performed for the CEU population with MAF values ≥0.05.
Results
Population Characteristics
The characteristics of the study population have been described (13). In brief, 417 case individuals and 417 control individuals were included in this study; 339 of the cases (81.3%) and 349 of the controls (83.7%) were for non-Hispanic white individuals. The mean age of the case and control individuals was 60.73 and 60.30 years, respectively. The difference in age between the cases and the controls was not significant (P = 0.554). To minimize the effects of treatment type on survival in clinical-outcome analyses, we focused the analysis on the non-Hispanic white patients who had received surgery and platinum-based chemotherapy (n = 319). For this group, 88% of the patients had a diagnosis of advanced-stage (stages III, IV) ovarian cancer. The majority (62%) of the tumors were of the serous sub-type. The median survival time was 48.26 months (median age, 60.73 years; range, 26 – 88 years). Slightly less than half of the patients (46%) had died by the end of the follow-up period, with 48% showing cancer recurrence and 33% not responding to treatment.
Association of Individual SNPs with Ovarian Cancer Risk
Of the 183 SNPs we analyzed from 24 genes encoding FGFs or FGFRs, 22 SNPs from 7 genes showed a significant association with ovarian cancer risk (i.e., P < 0.05, and q < 0.10; Table 1). The SNP with highest statistical significance was FGF1 [fibroblast growth factor 1 (acidic)] rs7727832. Individuals carrying at least 1 variant allele exhibited a 2.27-fold (95% CI, 1.31-fold to 3.95-fold; P = 0.0035) increased risk of ovarian cancer, compared with individuals with the wild-type genotype. The association with this SNP remained significant in >1000 bootstrap resamplings, providing strong support for the validity of this result. Carriers of at least 1 variant allele for rs3809495 in the FGF7 (fibroblast growth factor 7) gene had the highest risk of ovarian cancer (OR, 2.99; 95% CI, 1.07– 8.42; P = 0.038). Approximately half of the significant associations were associated with increased risk, whereas the other half conferred a protective effect. The protective SNP with the highest statistical significance was rs2288696 in the FGFR1 (fibroblast growth factor receptor 1) gene. Under the dominant model, this locus was associated with a 30% reduction in risk (95% CI, 0.51– 0.93; P = 0.017); this association remained significant in >90% of the bootstrap resamplings.
Table 1. Association of SNPs in FGF and FGFR genes with ovarian cancer risk.
| SNP | Gene | Position | Case (ww/wv/vv),a n | Control (ww/wv/vv), n | OR (95% CI)b | P | Modelc |
|---|---|---|---|---|---|---|---|
| rs7727832 | FGF1 | Intron | 297/42/0 | 328/20/1 | 2.27 (1.31–3.95) | 0.0035 | Domd |
| rs4752566 | FGFR2 | Intron | 91/183/65 | 123/151/75 | 1.49 (1.08–2.07) | 0.016 | Domd |
| rs2288696 | FGFR1 | Intron | 219/98/22 | 194/141/14 | 0.69 (0.51–0.93) | 0.017 | Domd |
| rs2981582 | FGFR2 | Intron | 134/141/63 | 136/171/42 | 1.67 (1.10–2.55) | 0.017 | Recd |
| rs4480740 | FGF7 | Intron | 155/150/34 | 135/161/53 | 0.77 (0.61–0.96) | 0.019 | Add |
| rs1823251 | FGF10 | 3′ Flanking region | 212/107/20 | 244/94/11 | 1.36 (1.05–1.78) | 0.022 | Add |
| rs8025433 | FGF7 | Intron | 326/13/0 | 321/28/0 | 0.46 (0.23–0.9) | 0.024 | Domd |
| rs15608 | FGF14 | 3′ UTR | 182/126/28 | 207/123/14 | 2.14 (1.10–4.14) | 0.025 | Recd |
| rs11639111 | FGF7 | Intron | 128/164/44 | 121/156/67 | 0.62 (0.41–0.94) | 0.026 | Recd |
| rs3135830 | FGFR2 | 3′ UTR | 323/16/0 | 343/6/0 | 2.82 (1.09–7.29) | 0.033 | Dom |
| rs2052006 | FGF1 | Intron | 175/145/19 | 174/140/35 | 0.53 (0.30–0.95) | 0.034 | Rec |
| rs2978073 | FGFR1 | Intron | 283/52/4 | 269/77/3 | 0.66 (0.45–0.97) | 0.035 | Dom |
| rs10070885 | FGF1 | Intron | 206/119/14 | 205/116/28 | 0.49 (0.25–0.95) | 0.036 | Rec |
| rs3809495 | FGF7 | 5′ Flanking region | 325/14/0 | 344/5/0 | 2.99 (1.07–8.42) | 0.038 | Dom |
| rs3925 | FGFR1 | Intron | 187/134/18 | 220/112/17 | 1.38 (1.02–1.88) | 0.038 | Dom |
| rs7317625 | FGF9 | 3′ UTR | 239/93/7 | 270/74/5 | 1.43 (1.02–2.02) | 0.041 | Dom |
| rs2981451 | FGFR2 | Intron | 84/172/83 | 103/182/64 | 1.25 (1.01–1.56) | 0.042 | Add |
| rs4338740 | FGF7 | Intron | 164/158/17 | 195/135/18 | 1.37 (1.01–1.85) | 0.042 | Dom |
| rs3135826 | FGFR2 | 3′ UTR | 325/13/0 | 321/26/0 | 0.49 (0.25–0.97) | 0.042 | Dom |
| rs1904316 | FGF7 | Intron | 107/165/66 | 86/183/79 | 0.71 (0.51–0.99) | 0.045 | Dom |
| rs2912759 | FGFR2 | Intron | 190/129/20 | 180/134/35 | 0.56 (0.32–0.99) | 0.047 | Rec |
| rs3715 | FGF9 | 3′ UTR | 240/92/7 | 270/74/5 | 1.41 (1.00–1.99) | 0.050 | Dom |
ww, homozygous for wild-type allele; wv, heterozygous for variant allele; vv, homozygous for variant allele; OR, odds ratio; UTR, untranslated region.
Adjusted for age.
Models of inheritance: Add, additive; Dom, dominant; Rec, recessive.
Remained significant for >75% of the 1000 bootstrap resamplings.
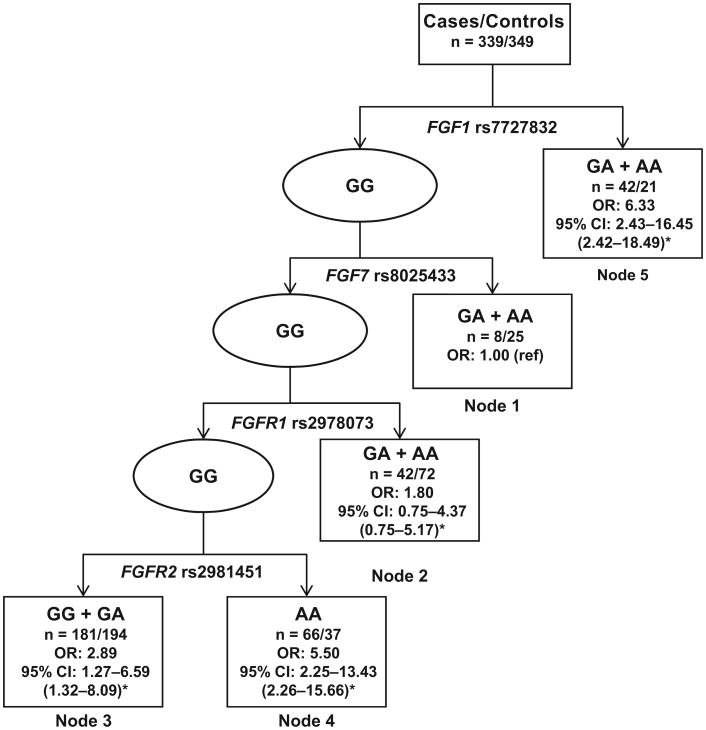
Because of the large number of variants in the FGF–FGFR axis that were significantly associated with ovarian cancer risk, we conducted a CART analysis to explore higher-order gene– gene interactions among these 22 significant SNPs. The final tree structure contained 5 terminal nodes with dramatically different risks for ovarian cancer (Fig. 1). The first split on the decision tree was FGF1 rs7727832, indicating that this SNP was the primary risk factor for ovarian cancer in the study population. With individuals of terminal node 1 used as the reference, the HRs for the other 4 terminal nodes ranged from 1.80 to 6.33.
Fig. 1. CART analysis of genetic variants in the FGF–FGFR axis and ovarian cancer risk.
*Bias-corrected CI from bootstrap analysis. n = Number of cases/number of controls; ref, reference.
Association of FGF and FGFR SNPs with Chemotherapeutic Response
Four SNPs were significantly associated with treatment response after platinum-based chemotherapy and surgery (Table 2). Interestingly, each of these loci was associated with a favorable response (a 35% to 56% reduction in risk of a poor response). These SNPs were FGF18 (fibroblast growth factor 18) rs3806929 (OR, 0.64; 95% CI, 0.44 – 0.94), FGF7 rs9920722 (OR, 0.65; 95% CI, 0.44 – 0.98), and FGF23 (fibroblast growth factor 23) rs12812339 (OR, 0.65; 95% CI, 0.43– 0.98) under the additive model, and FGF5 (fibroblast growth factor 5) rs3733336 (OR, 0.044; 95% CI, 0.19 – 0.98) under the recessive model. None of the SNPs remained significant, however, after adjustment for multiple comparisons (q > 0.1), and only 1 variant, rs3806929, reached significance in >50% of the bootstrap resamplings.
Table 2. Association of SNPs in FGF and FGFR genes with chemotherapeutic response in patients with ovarian cancer.
| SNP | Gene | Position | Nonresponders (ww/wv/vv)a | Responders (ww/wv/vv) | OR (95%CI)b | P | Modelc |
|---|---|---|---|---|---|---|---|
| rs3806929 | FGF18 | 5′ Flanking region | 42/45/9 | 66/92/41 | 0.64 (0.44–0.94) | 0.022 | Addd |
| rs9920722 | FGF7 | Intron | 47/42/7 | 71/99/29 | 0.65 (0.44–0.98) | 0.039 | Add |
| rs12812339 | FGF23 | 5′ Flanking region | 55/34/7 | 92/83/22 | 0.65 (0.43–0.98) | 0.039 | Add |
| rs3733336 | FGF5 | 3′ UTR | 37/49/10 | 71/92/36 | 0.44 (0.19–0.98) | 0.045 | Rec |
ww, homozygous for wild-type allele; wv, heterozygous for variant allele; vv, homozygous for variant allele; OR, odds ratio; UTR, untranslated region.
Adjusted for age, stage, histology, and treatment
Models of inheritance: Add, additive; Dom, dominant; Rec, recessive.
Remained significant for >50% of the 1000 bootstrap resamplings.
Overall Survival for FGF and FGFR Variants
Eleven SNPs from 7 genes were significantly associated with overall survival of ovarian cancer patients, with the results remaining highly significant in the bootstrap resampling (Table 3). FGF23 SNP rs7961824 had the highest significance. This association remained significant after multiple comparisons (q = 0.058) and was significant in 100% of the bootstraps. Individuals with at least one rs7961824 allele had a 45% reduction in risk of dying during the follow-up period (95% CI, 0.39 – 0.78), compared with women with the wild-type genotype. This favorable prognosis dramatically improved the median survival time (by 20 months), from 38.0 months for patients with the wild-type genotype to 58.1 months for those with the variant allele.
Table 3. Association of SNPs in FGF and FGFR genes with overall survival in ovarian cancer patients.
| SNP | Gene | Position | Event (ww/wv/vv)a | No event (ww/wv/vv) | HR (95% CI)b | P | Modelc |
|---|---|---|---|---|---|---|---|
| rs7961824 | FGF23 | 3′ Flanking region | 90/47/9 | 80/76/17 | 0.55 (0.39–0.78) | 0.0008 | Domd |
| rs7708357 | FGFR4 | 3′ Flanking region | 64/65/17 | 56/86/31 | 0.62 (0.44–0.87) | 0.0065 | Domd |
| rs17102287 | FGFR2 | Intron | 96/45/5 | 130/39/4 | 1.61 (1.12–2.31) | 0.010 | Domd |
| rs641101 | FGFR4 | 5′ Flanking region | 71/65/10 | 72/83/18 | 0.7 (0.53–0.93) | 0.013 | Addd |
| rs308441 | FGF2 | Intron | 102/39/5 | 97/66/10 | 0.63 (0.43–0.91) | 0.013 | Domd |
| rs167428 | FGF2 | Intron | 82/55/9 | 77/82/14 | 0.67 (0.48–0.95) | 0.023 | Domd |
| rs2981427 | FGFR2 | Intron | 47/75/24 | 54/81/38 | 0.59 (0.37–0.93) | 0.023 | Recd |
| rs17099029 | FGF1 | 3′ UTR | 120/23/3 | 148/25/0 | 1.71 (1.07–2.72) | 0.024 | Domd |
| rs7658439 | FGF5 | 5′ Flanking region | 131/13/1 | 164/9/0 | 1.88 (1.05–3.35) | 0.033 | Domd |
| rs376618 | FGFR4 | Exon | 82/57/6 | 93/64/16 | 0.74 (0.56–0.98) | 0.033 | Add |
| rs15608 | FGF14 | 3′ UTR | 73/57/14 | 102/58/12 | 1.86 (1.05–3.29) | 0.033 | Rec |
ww, homozygous for wild-type allele; wv, heterozygous for variant allele; vv, homozygous for variant allele; OR, odds ratio; UTR, untranslated region.
Adjusted for age, stage, histology, and treatment
Models of inheritance: Add, additive; Dom, dominant; Rec, recessive.
Remained significant for >75% of the 1000 bootstrap resamplings.
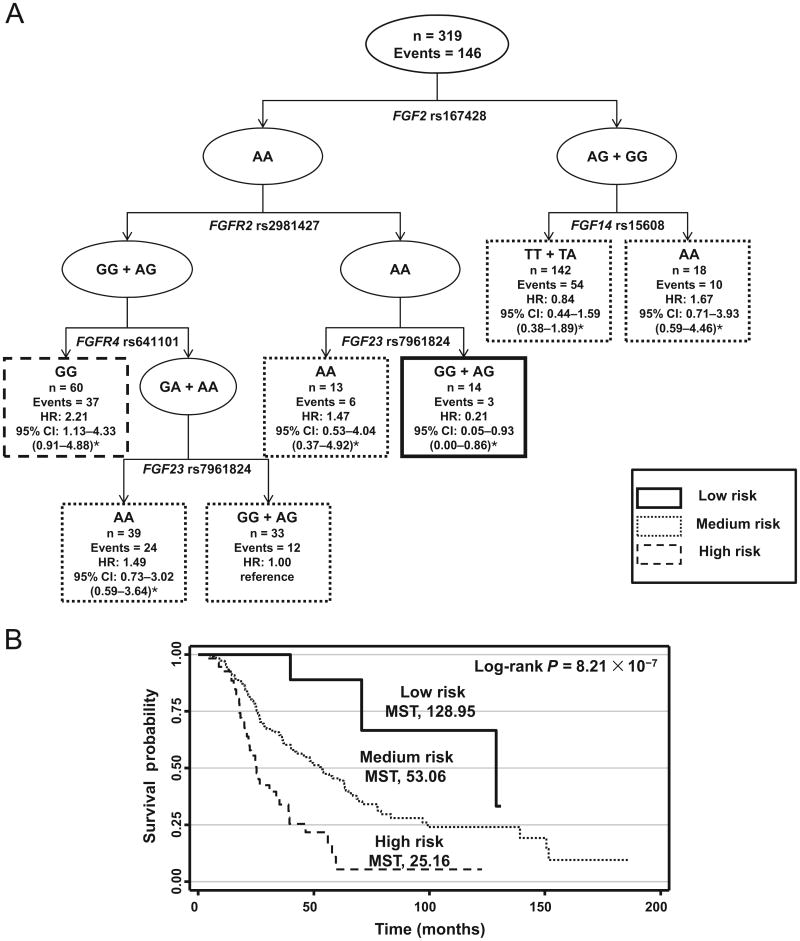
The remaining variants were borderline significant after adjustment for multiple comparisons (q values, 0.19 – 0.22), suggesting that they could have an effect on overall survival in the appropriate context. To assess this possibility, we performed a survival tree analysis for these 11 variants. The resulting tree structure comprised SNPs from 5 genes: FGF2, FGFR2 (fibroblast growth factor receptor 2), FGFR4 (fibroblast growth factor receptor 4), FGF14 (fibroblast growth factor 14), and FGF23 (Fig. 2A). The first split on the survival tree was FGF2 rs167428, indicating that this SNP is the primary factor contributing to overall survival. When we used individuals of terminal node 2 as a reference, the HRs for the other 6 terminal nodes ranged from 0.21 to 2.21. Grouping these terminal nodes into 3 risk groups—low, medium, and high— identified dramatic differences in survival durations. The median survival times for the patients in the low-risk, medium-risk, and high-risk groups were 128.95, 53.06, and 25.16 months, respectively (P = 8.21 × 10−7, log-rank test; Fig. 2B).
Fig. 2. Higher-order gene–gene interactions among genetic variants in the FGF–FGFR axis for overall survival of ovarian cancer.
(A), Survival tree structure. n, total population; Events, number of deaths; HR, hazard ratio. (B), Survival curves by the risk groups identified by the survival tree. *Bias-corrected CI from bootstrap analysis. MST, median survival time in months.
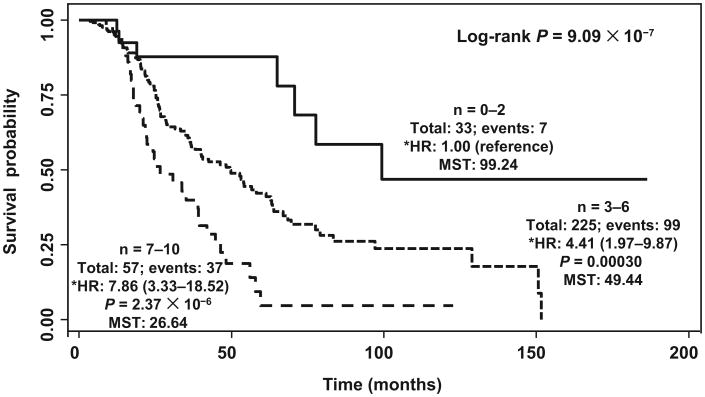
Next, we determined the cumulative effects of multiple unfavorable genotypes on ovarian cancer survival. Compared with patients carrying ≤2 unfavorable genotypes (low risk), patients carrying 3– 6 unfavorable genotypes (medium risk) and 7–10 unfavorable genotypes (high risk) exhibited a progressively increased risk of death, with HRs of 4.41 (95% CI, 1.97–9.87) and 7.86 (95% CI, 3.34 –18.52), respectively (P for trend = 2.49 × 10−7). This increase in risk produced highly significant differences in the median survival time. Patients in the medium-risk and high-risk groups had survival times of only 49.44 and 26.64 months, respectively, compared with 99.24 months for those in the low-risk group (P = 9.09 × 10−7, log-rank test; Fig. 3).
Fig. 3. Cumulative effect of genetic variants in the FGF–FGFR axis on overall survival.
n, number of unfavorable genotypes; Total, total number of patients; Events, number of deaths during study period; MST, median survival time in months; *HR, hazard ratio adjusted for age, stage, histology, and treatment.
SNP–Gene Association in cis eQTL
To check for possible functional effects of our identified SNPs on gene expression, we investigated whether any of the significant variants showed eQTL with gene expression with the Genevar database tool. One SNP (FGF1 rs17099029) associated with survival was in cis eQTL with FGF1 expression (P = 0.03; see Fig. 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol60/issue1). Individuals carrying a genotype with the variant had decreased FGF1 expression compared with those with the wild-type homozygous genotype. Interestingly, this SNP was also in linkage with the FGF1 SNP rs7727832 (r2 = 0.5) that was highly associated with ovarian cancer risk.
Discussion
Ovarian cancer is one of the most lethal gynecologic malignancies (1). The high mortality is often attributable to late diagnosis. Thus, considerable efforts have been made to identify high-risk populations to look not only for genetic markers associated with ovarian cancer risk that could improve early detection and screening approaches, but also for predictors of clinical outcomes. Several studies have been conducted to identify variants associated with ovarian cancer risk and clinical outcomes, but many of the findings of these studies are inconsistent (6 –10). Because of the increasing knowledge regarding the role of the FGF– FGFR axis in tumorigenesis in general and ovarian cancer specifically, we performed a comprehensive analysis of genetic variation in this pathway with respect to its association with ovarian cancer risk and outcomes.
One of the most important findings is that multiple SNPs in the FGF–FGFR pathway are associated with increased risk of ovarian cancer. In particular, FGF1 rs7727832 showed the most significant association with ovarian cancer in the main-effect analysis, and this result was supported by the CART gene– gene interactions analysis. Also showing significant associations with ovarian cancer were variants in 6 other genes, including FGFR1, FGFR2, FGF7, FGF9 (fibro-blast growth factor 9), FGF10 (fibroblast growth factor 10), and FGF14. Studies have shown that FGF10 over-expression leads to epithelial hyperproliferation (32) and that the FGF10 –FGFR1 or FGF10 –FGFR2 signaling axis plays a potential role in oncogenic transformation (33). Furthermore, evidence suggests that FGF1 overexpression may lead to increased angiogenesis and autocrine stimulation of cancer cells (23, 34). Specifically for ovarian cancer, there is strong evidence that these specific ligands and receptors are important in ovarian cancer tumorigenesis. For example, FGF1 overexpression has been observed in ovarian cancer and been associated with clinical prognosis (23, 24). Likewise, FGF7 is expressed in the majority of ovarian tumors and is detectable in malignant ovarian cancer– associated ascites (35). Cole et al. showed that FGFR2 and FGF7 stimulate ovarian cancer proliferation (36). An early study observed that the FGF2 and FGFR1 genes are highly expressed in human ovarian tumor endothelium (37). FGFR2 has been suggested to be involved in ovarian cancer pathogenesis (18). Indeed, increased FGFR2 levels have been found in ovarian cancer (36). The interaction of FGF7 with FGFR2 ligands and with FGFR2-IIIb induces proliferation, motility, and protection from cell death in epithelial ovarian cancer cell lines (38). Interestingly, approximately half of the significant associations observed were with a reduced risk of ovarian cancer, suggesting that FGF– FGFR signaling can exhibit tumor-suppressive functions in certain contexts (16).
We also identified that 4 SNPs (FGF18 rs3806929, FGF7 rs9920722, FGF23 rs12812339, and FGF5 rs3733336) were significantly associated with improved response to treatment. Experimental data have indicated that inhibition of FGFR along with the use of standard chemotherapeutic agents, including those used in ovarian cancer treatment, interacted synergistically to improve the therapeutic effect on endometrial cancer cells (39). Even more intriguing is that Cole et al. showed that the inhibition of FGFR1 and FGFR2 increased cisplatin sensitivity in ovarian cancer (36). Studies have demonstrated that FGFR4 is a potential therapeutic target and associated with a better therapeutic response, although we observed no association of variants within FGFR4 with any treatment response in our study (19, 20). Nevertheless, our results support targeting of the FGF–FGFR signaling axis as a potential therapeutic opportunity in ovarian cancer. Such targeting may provide a future approach to stratifying the patient population to improve the response to these agents.
Another major finding is that 11 SNPs from 7 genes were significantly associated with overall survival in ovarian cancer patients. Of these SNPs, FGF23 rs7961824 showed the most significant association with survival. Survival tree analysis revealed FGF2, FGFR2, FGFR4, FGF14, and FGF23 gene– gene interactions, indicating FGF2 rs167428 to be the primary factor contributing to overall survival. Patients in the low-risk group carrying these alleles have much longer survival times (>40 months) than those in the medium- and high-risk groups. In addition, the highly significant dose–response effect evident for an increasing number of unfavorable genotypes provides support for the global effect of variation within these important signaling genes. Previous studies have indicated that FGF1 overexpression may lead to increased angiogenesis, leading to poorer overall patient survival (23), and a recent study demonstrated that FGF1 expression adversely influenced survival in patients with ovarian tumors (24).
Interestingly, we found that one of the FGF1 SNPs associated with ovarian cancer survival (rs17099029) demonstrated a significant eQTL with FGF1 expression (Genevar analysis) (40). This SNP shows a degree of linkage with FGF1 rs7727832 (r2 = 0.5), the SNP with the highest risk, suggesting that changes in FGF1 expression may modify risk for both cancer susceptibility and outcome. rs17099029 is located in the 3′ untranslated region of FGF1, thus hinting at a possible role in affecting gene expression. Taken together, the novel findings of these genetic variants associated with ovarian cancer survival suggest, in the context of established tumor biology, that they have potential for use as prognostic markers for ovarian cancer.
While recognizing the original and significant findings of this current study, we understand that our study has some limitations. It is a hospital-based case control study, and thus selection bias may be present. The effects of these variants observed for non-Hispanic whites may not be generalizable to populations of other ethnicities. In addition, the SNPs we genotyped in this study are primarily tagging SNPs and most likely are not the true causal or functional variants. Therefore, it is difficult to ascertain the underlying biological mechanisms for these significant associations. Additional functional characterizations are needed. Nevertheless, our genetics-driven study used a well-characterized patient population with detailed clinical, treatment, and follow-up information. Although internal validation was conducted with bootstrap methods to add a level of confidence to the findings, further independent or external validation is necessary to verify the findings in our study.
In conclusion, we have conducted the first comprehensive study to identify significant associations of genetic variants in the FGF–FGFR pathway with ovarian cancer risk, therapeutic response, and survival. The association of multiple SNPs in the FGF pathway could provide a molecular approach for the development of new ovarian cancer biomarkers for these key end points, and the use of such markers could have a major impact on the survival of patients with this devastating disease. Furthermore, these findings lend support to the development of the FGF–FGFR axis as potential therapeutic targets in ovarian cancer.
Supplementary Material
Acknowledgments
Research Funding: Department of Defense Ovarian Cancer Research Program (award no. W81XWH-07-1-0449).
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: CA125, cancer antigen 125; HE4, human epididymis protein 4; TGF-β, transforming growth factor β; FGF, fibroblast growth factor; FGFR, FGF receptor; FGF2, fibroblast growth factor 2; SNP, single-nucleotide polymorphism; MAF, minor-allele frequency; OR, odds ratio; HR, hazard ratio; CART, classification and regression tree (analysis); eQTL, expression quantitative trait loci; Genevar, Gene Expression Variation (database).
Human genes: FGF2, fibroblast growth factor 2 (basic); FGF1, fibroblast growth factor 1 (acidic); FGF7, fibroblast growth factor 7; FGFR1, fibroblast growth factor receptor 1; FGF18, fibroblast growth factor 18; FGF23, fibroblast growth factor 23; FGF5, fibroblast growth factor 5; FGFR2, fibroblast growth factor receptor 2; FGFR4, fibroblast growth factor receptor 4; FGF14, fibroblast growth factor 14; FGF9, fibroblast growth factor 9; FGF10, fibroblast growth factor 10.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors' Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
Patents: None declared.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.American College of Obstetricians and Gynecologists Committee on Gynecologic Practice. Committee opinion no. 477: the role of the obstetrician-gynecologist in the early detection of epithelial ovarian cancer. Obstet Gynecol. 2011;117:742–6. doi: 10.1097/AOG.0b013e31821477db. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin LM, Trivers KF, Matthews B, Andrilla CH, Miller JW, Berry DL, et al. Vignette-based study of ovarian cancer screening: Do U.S. physicians report adhering to evidence-based recommendations? Ann Intern Med. 2012;156:182–94. doi: 10.7326/0003-4819-156-3-201202070-00006. [DOI] [PubMed] [Google Scholar]
- 4.Sturgeon CM, Duffy MJ, Stenman UH, Lilja H, Brünner N, Chan DW, et al. National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers. Clin Chem. 2008;54:e11–79. doi: 10.1373/clinchem.2008.105601. [DOI] [PubMed] [Google Scholar]
- 5.Buys SS, Partridge E, Black A, Johnson CC, Lam-erato L, Isaacs C, et al. for the PLCO Project Team. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA. 2011;305:2295–303. doi: 10.1001/jama.2011.766. [DOI] [PubMed] [Google Scholar]
- 6.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–9. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–70. 70e1–2. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolton KL, Ganda C, Berchuck A, Pharaoh PD, Gayther SA. Role of common genetic variants in ovarian cancer susceptibility and outcome: progress to date from the Ovarian Cancer Association Consortium (OCAC) J Intern Med. 2012;271:366–78. doi: 10.1111/j.1365-2796.2011.02509.x. [DOI] [PubMed] [Google Scholar]
- 11.Saldivar JS, Lu KH, Liang D, Gu J, Huang M, Vlastos AT, et al. Moving toward individualized therapy based on NER polymorphisms that predict platinum sensitivity in ovarian cancer patients. Gynecol Oncol. 2007;107:S223–9. doi: 10.1016/j.ygyno.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Yin J, Lu K, Lin J, Wu L, Hildebrandt MA, Chang DW, et al. Genetic variants in TGF-β pathway are associated with ovarian cancer risk. PLoS One. 2011;6:e25559. doi: 10.1371/journal.pone.0025559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye Y, et al. Genetic variants in microRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765–76. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–62. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 15.Czubayko F, Liaudet-Coopman ED, Aigner A, Tuveson AT, Berchem GJ, Wellstein A. A secreted FGF-binding protein can serve as the angiogenic switch in human cancer. Nat Med. 1997;3:1137–40. doi: 10.1038/nm1097-1137. [DOI] [PubMed] [Google Scholar]
- 16.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 17.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 18.Byron SA, Gartside MG, Wellens CL, Goodfellow PJ, Birrer MJ, Campbell IG, Pollock PM. FGFR2 mutations are rare across histologic subtypes of ovarian cancer. Gynecol Oncol. 2010;117:125–9. doi: 10.1016/j.ygyno.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Zaid TM, Yeung TL, Thompson MS, Leung CS, Harding T, Co NN, et al. Identification of FGFR4 as a potential therapeutic target for advanced-stage, high-grade serous ovarian cancer. Clin Cancer Res. 2013;19:809–20. doi: 10.1158/1078-0432.CCR-12-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen CV, Steffensen KD, Olsen DA, Waldstrøm M, Søgaard CH, Brandslund I, Jakobsen A. Serum platelet-derived growth factor and fibroblast growth factor in patients with benign and malignant ovarian tumors. Anticancer Res. 2012;32:3817–25. [PubMed] [Google Scholar]
- 21.Le Page C, Ouellet V, Madore J, Hudson TJ, Tonin PN, Provencher DM, Mes-Masson AM. From gene profiling to diagnostic markers: IL-18 and FGF-2 complement CA125 as serum-based markers in epithelial ovarian cancer. Int J Cancer. 2006;118:1750–8. doi: 10.1002/ijc.21521. [DOI] [PubMed] [Google Scholar]
- 22.Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, Bell A, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. Clin Oncol. 2007;25:2281–7. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 23.Smith G, Ng MT, Shepherd L, Herrington CS, Gourley C, Ferguson MJ, Wolf CR. Individuality in FGF1 expression significantly influences platinum resistance and progression-free survival in ovarian cancer. Br J Cancer. 2012;107:1327–36. doi: 10.1038/bjc.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy T, Darby S, Mathers ME, Gnanapragasam VJ. Evidence for distinct alterations in the FGF axis in prostate cancer progression to an aggressive clinical phenotype. J Pathol. 2010;220:452–60. doi: 10.1002/path.2657. [DOI] [PubMed] [Google Scholar]
- 25.Abdel-Rahman WM, Kalinina J, Shoman S, Eissa S, Ollikainen M, Elomaa O, et al. Somatic FGF9 mutations in colorectal and endometrial carcinomas associated with membranous β-catenin. Hum Mutat. 2008;29:390–7. doi: 10.1002/humu.20653. [DOI] [PubMed] [Google Scholar]
- 26.Burger RA. Overview of anti-angiogenic agents in development for ovarian cancer. Gynecol Oncol. 2011;121:230–8. doi: 10.1016/j.ygyno.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 27.Lieu C, Heymach J, Overman M, Tran H, Kopetz S. Beyond VEGF: inhibition of the fibroblast growth factor pathway and antiangiogenesis. Clin Cancer Res. 2011;17:6130–9. doi: 10.1158/1078-0432.CCR-11-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnatty SE, Beesley J, Chen X, Spurdle AB, Defazio A, Webb PM, et al. Polymorphisms in the FGF2 gene and risk of serous ovarian cancer: results from the Ovarian Cancer Association Consortium. Twin Res Hum Genet. 2009;12:269–75. doi: 10.1375/twin.12.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Singer BH. Recursive partitioning and applications. 2nd. New York: Springer; 2010. [Google Scholar]
- 30.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–6. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abate-Shen C, Shen MM. FGF signaling in prostate tumorigenesis—new insights into epithelialstromal interactions. Cancer Cell. 2007;12:495–7. doi: 10.1016/j.ccr.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Memarzadeh S, Xin L, Mulholland DJ, Mansukhani A, Wu H, Teitell MA, Witte ON. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12:572–85. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–53. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steele IA, Edmondson RJ, Bulmer JN, Bolger BS, Leung HY, Davies BR. Induction of FGF receptor 2-IIIb expression and response to its ligands in epithelial ovarian cancer. Oncogene. 2001;20:5878–87. doi: 10.1038/sj.onc.1204755. [DOI] [PubMed] [Google Scholar]
- 36.Cole C, Lau S, Backen A, Clamp A, Rushton G, Dive C, et al. Inhibition of FGFR2 and FGFR1 increases cisplatin sensitivity in ovarian cancer. Cancer Biol Ther. 2010;10:495–504. doi: 10.4161/cbt.10.5.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitworth MK, Backen AC, Clamp AR, Wilson G, McVey R, Friedl A, et al. Regulation of fibroblast growth factor-2 activity by human ovarian cancer tumor endothelium. Clin Cancer Res. 2005;11:4282–8. doi: 10.1158/1078-0432.CCR-04-1386. [DOI] [PubMed] [Google Scholar]
- 38.Steele IA, Edmondson RJ, Leung HY, Davies BR. Ligands to FGF receptor 2-IIIb induce proliferation, motility, protection from cell death and cytoskeletal rearrangements in epithelial ovarian cancer cell lines. Growth Factors. 2006;24:45–53. doi: 10.1080/08977190500361697. [DOI] [PubMed] [Google Scholar]
- 39.Byron SA, Loch DC, Pollock PM. Fibroblast growth factor receptor inhibition synergizes with paclitaxel and doxorubicin in endometrial cancer cells. Int J Gynecol Cancer. 2012;22:1517–26. doi: 10.1097/IGC.0b013e31826f6806. [DOI] [PubMed] [Google Scholar]
- 40.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.