Abstract
Endophytic nitrogen fixing bacteria were isolated from the leaves, stems and roots of industrial variety (cv. U-Thong 3; UT3), wild and chewing sugarcane plants grown for 6 weeks in nitrogen (N)-free sand. Eighty nine isolates of endophytic bacteria were obtained on N-free agar. An acetylene reduction assay (ARA) detected nitrogenase activity in all 89 isolates. Three isolates from the chewing (C2HL2, C7HL1 and C34MR1) sugarcane and one isolate from the industrial sugarcane (UT3R1) varieties were characterised, and their responses to different yeast extract concentrations were investigated. Three different responses in nitrogenase activity were observed. Isolates C2HL2 and C7HL1 exhibited major increases with the addition of 0.005% yeast extract, C34MR1 exhibited no response, and UT3R1 exhibited a significant decrease in nitrogenase activity with 0.005% yeast extract. In all the isolates, nitrogenase activity decreased with further increase of the yeast extract to 0.05%. The highest nitrogenase activity was observed in isolates C2HL2 and C7HL1, which had 16S rRNA gene sequences that were closely related to Novosphingobium sediminicola and Ochrobactrum intermedium, respectively.
Keywords: Endophyte, Nitrogen Fixing Bacteria, Sugarcane
Abstract
Bakteria pengikat nitrogen endofit telah diasingkan daripada daun, batang dan akar pokok tebu jenis industri (cv. U-Thong 3; UT3), liar dan kunyah yang telah bertumbuh selama 6 minggu dalam pasir bebas nitrogen (N). Lapan puluh sembilan pencilan bakteria endofit telah didapati daripada agar bebas N. Cerakin penurunan asetilena (ARA) telah mengesan aktiviti nitrogenase dalam kesemua 89 pencilan. Ciri-ciri tiga pencilan daripada tebu jenis kunyah (C2HL2, C7HL1 and C34MR1) dan satu daripada jenis industri (UT3R1) dan respons terhadap extrak yis yang berbeza kepekatan telah dikaji. Tiga respons aktiviti nitrogenase berlainan telah diperhatikan. Pencilan C2HL2 dan C7HL1 menunjukkan peningkatan major dengan penambahan ekstrak yis sebanyak 0.005%, C34MR1 tidak menunjukkan sebarang respons, dan C7HL1 menunjukkan pengurangan yang signifikan dalam aktiviti nitrogenase dengan penambahan ekstrak yis sebanyak 0.005%. Dalam kesemua pencilan, aktiviti nitrogenase berkurangan dengan peningkatan yis ekstrak hingga 0.05%. Aktiviti nitrogenase tertinggi diperhatikan dalam pencilan C2HL2 dan C7HL1, yang mempunyai jujukan gen 16S rRNA yang menyerupai Novosphingobium sediminicola dan Ochrobactrum intermedium, masing-masing.
Keywords: Endofit, Bakteria Pengikat Nitrogen, Tebu
INTRODUCTION
Sugarcane (Saccharum officinarum L.) is a major crop in the tropics, where it is grown for the production of biofuel as well as food. In Brazil, ethanol produced from sugarcane has been used extensively to replace petroleum fuel in cars. It was previously suggested that sugarcane could benefit from nitrogen fixing bacteria (Döbereiner et al. 1972; Boddey et al. 2003), and Acetobacter diazotrophicus (now renamed Gluconacetobacter diazotrophicus) was isolated from sugarcane (Boddey et al. 2003). Many species of endophytic nitrogen fixing bacteria have since been isolated from sugarcane (Baldani et al. 1992; Dong et al. 1994; Loiret et al. 2004) and other plants, e.g., rice, kallar grass and maize, and these bacteria supply fixed nitrogen (N) to their hosts (Hallmann et al. 1997; James 2000; Baldani et al. 2002). Other nitrogen fixing bacteria, including Azospirillum spp., Herbaspirillum spp., Burkholderia spp., Enterobacter cloacae, Klebsiella oxytoca, Klebsiella pneumoniae and Pantoea sp., were reported to have been isolated from the roots, stems and leaves of sugarcane (Govindarajan et al. 2007, 2008; Mendes et al. 2007). Symbiotic associations between sugarcane and its endophytic nitrogen fixing bacteria provide mutual benefits such as a combined N (NH3) supply to the plant and photosynthesis, refuge and transmission to the bacteria. Co-evolution of these partners to engage in symbiosis would be expected in advantageous environments. As Thailand lies within the Vavilov Center of Diversity for sugarcane (Ladizinsky 1998), diversity in terms of endophytic nitrogen fixing bacteria might be expected. The aim of this study was to isolate and characterise endophytic nitrogen fixing bacteria from the tissues of industrial variety, wild and chewing sugarcane grown in Thailand.
MATERIALS AND METHODS
Sugarcane Samples
The sugarcane plants used in this study consisted of an industrial variety (U-Thong 3; UT3), two varieties of wild sugarcane (W1 and W2) obtained from Suphanburi Field Crops Research Center and five local varieties of chewing sugarcane with either low plant N levels of 0.31%–0.33% N (C13L and C15L), medium levels of 0.66%–0.70% N (C34M) or high levels of 0.81%–0.95% N (C2H and C7H) obtained from local sugarcane plantations in Chiang Mai province. Sugarcane cuttings that were 20 cm long with two buds for each variety were planted in drainable pots (30 cm diameter, 30 cm deep), with one plant per pot filled with washed river quartz sand (collected from Chiang Mai province). The sugarcane plants were watered twice daily with N-free nutrient solution containing 1000 µM calcium chloride (CaCl2), 50 µM potassium dihydrogen phosphate (KH2PO4), 10 µM ferric citrate (C6H5O7Fe), 250 µM magnesium sulphate (MgSO4), 250 µM potassium sulphate (K2SO4), 1 µM manganese sulphate (MnSO4), 0.5 µM zinc sulphate (ZnSO4), 0.2 µM copper sulphate (CuSO4), 0.1 µM cobalt sulphate (CoSO4), 0.1 µM sodium molybdate (Na2MoO4) and 2 µM boric acid (H3BO3).
Bacterial Isolation and Culture
A one-gram sample each of the fresh roots (R), stems (S) and leaves (L) of the sugarcane plants was taken for bacterial isolation at 6 weeks after planting. The samples were surface sterilised by sequential washing in 50% ethanol for 1 min, 2% sodium hypochlorite for 3 min, and 50% ethanol for 30 sec and then rinsed twice with sterile distilled water. Each sample was ground in a sterile mortar, suspended in a N-free broth (0.1 g of dipotassium hydrogen phosphate [K2HPO4], 0.4 g of KH2PO4, 0.2 g of MgSO4, 0.1 g of sodium chloride [NaCl], 0.02 g of CaCl2, 0.01 g of ferric chloride [FeCl3], 0.002 g of sodium molybdate [NaMoO4], and 10 g of glucose in 1000 ml of distilled water) (Döbereiner et al. 1972) for 24 h at 30°C. Ten-fold serial dilutions of the suspensions ranging from 10−1 to 10−4 were spread on N-free agar (NFA) in triplicate. After 72 h of incubation, individual bacterial colonies on NFA were isolated and purified on new NFA. The bacterial cultures were stored in 25% glycerol at −20°C.
Morphological and Biochemical Characterisation
Bacterial isolates were characterised according to Bergey’s Manual of Determinative Bacteriology (Holt et al. 1994). All cultures and reactions were incubated at 30°C. Colony morphologies were examined on NFA after 24 to 48 h of incubation. Cell morphology observations and Gram typing of bacteria were determined by Gram staining with a 3% potassium hydroxide (KOH) confirmation test within 24 h of incubation. Bacterial isolates grown on nutrient agar (NA) were tested for the presence of catalase and oxidase within 24 h. Indole production was examined for each culture grown in tryptone broth within 24 h via a reaction with Kovac’s reagent. Methyl red (MR) and Voges-Proskauer (VP) tests were performed on each culture grown in MR-VP medium after 48 h of incubation via reaction with MR solution for the MR test and 5% α–naphthol solution and 40% KOH for the VP test. Motility was examined on cultures grown in semi-solid motility test medium. Starch hydrolysis was tested by flooding cultures grown on starch agar containing 2% soluble starch after 24 h of cultivation with Lugol’s iodine. A urease test was performed on each bacterial isolate grown in urea broth after 15, 30, and 60 min and up to 4 h of incubation. A citrate test was performed on point inocula grown on Simmon’s citrate agar after 24 h. Utilisation of glucose, arabinose, rhamnose and mannitol as carbon sources were assayed on oxidative fermentative medium after 24 h of cultivation.
Determination of Nitrogenase Activity
Nitrogenase activity was determined with an acetylene reduction assay (ARA). Each bacterial isolate was grown in a 20 ml test tube containing 10 ml of N-free semi-solid medium for 72 h at 30°C. Each test tube was sealed with a rubber stopper, and 1 ml of acetylene gas was added to the air in the headspace (10 ml). The test tubes were incubated at 30°C for 24 h. One ml of each gas sample from the headspace was assayed for ethylene (C2H4) production by gas chromatography (GOW MAC series 750, New Jersey, USA) equipped with a hydrogen flame ionisation detector (FID) and a Porapack N column. Nitrogenase activity was calculated as nmol of ethylene per tube per h. To examine the effect of yeast extract on nitrogenase activity, ARA was performed as mentioned above, but N-free semi-solid medium was added with yeast extract at 0%, 0.005%, 0.01% and 0.05%.
Isolation of Total DNA
Bacterial isolates with the highest nitrogenase activity were selected and grown in nutrient broth (NB) at 30°C for 48 h and centrifuged at 12000 rpm. Bacterial DNA was extracted according to Prakamhang et al. (2009). The DNA pellets were resuspended in 720 µl of extraction buffer (100 mM Tris-HCl, 50 mM ethylene diamine tetra acetic acid [EDTA], 500 mM NaCl and 1.25% sodium dodecyl sulfate [SDS]) and incubated in a water bath at 65°C for 20 min. Proteins were precipitated by adding 225 ml of 5 M potassium acetate and incubated on ice for 20 min before decanting supernatant into a new microcentrifuge tube. The cleaned DNA lysates were extracted with 500 µl of phenol:chloroform:isoamyl alcohol (25:24:1). The DNA extracts were precipitated by adding 2/3 volume cold isopropanol, precipitated again with 300 ml of 70% cold ethanol then suspended in 20 ml of Tris-EDTA (TE) buffer and stored at −20°C.
PCR Amplification and Sequencing of the 16S rRNA Gene
The 16S rRNA gene sequences of the isolates were determined by PCR amplification with the universal primers 27F (5′ - AGA GTT TGA TCM TGG CTCAG - 3′) and 1525R (5′- AAG GAG GTG WTC CAR CC - 3′) (Lane 1991). Each amplification was performed in a total volume of 50 µl of reaction mixture. Each reaction mixture contained 5.0 µl of 10× Tris-acetate-EDTA (TAE) buffer, 2.0 µl of 50 mM magnesium chloride (MgCl2), 1.0 µl of 10 mM dNTP mix, 1.0 µl each of 1 nm primers, 0.4 µl of Taq DNA polymerase, 1.0 µl of template DNA, 2.5 µl of 5% dimethyl sulfoxide (DMSO) and 36.1 µl of deionised water. PCR amplifications were carried out with the following temperature profile: initial denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 30 sec, with a final extension at 72°C for 7 min. PCR products were electrophoresed on a 0.8% agarose gel in 1× TAE buffer. The PCR products of the 16S rRNA gene were sequenced and aligned with the corresponding sequences of representatives within the related genera Ancylobacter, Novosphingobium and Ochrobactrum, which were retrieved from the GenBank databases, using Bioedit software version 7.0.9.1 (Hall 1999). Phylogenetic trees were inferred with Neighbour-Joining (NJ) trees using MEGA 4 (Saitou & Nei 1987).
RESULTS AND DISCUSSION
Eighty nine isolates of endophytic bacteria were obtained from the leaves, stems and roots of industrial variety, wild and chewing sugarcane grown in N-free sand (Table 1). All the isolates grew in N-free media and had nitrogenase activity that was detectable with ARA, confirming that they were nitrogen fixing bacteria. Colonies of most isolates on NFA were slimy, slightly viscous, white or pale yellow, smooth, and convex with intact margins. Sixty three isolates (71%) were Gram-negative, and 26 isolates (29%) were Gram-positive. Many genera of endophytic nitrogen fixing bacteria were actually Gram-negative (Iniquez et al. 2004; Loiret et al. 2004; Reis et al. 2004). Four Gram-negative isolates with the highest nitrogenase activity were selected for further investigation. These isolates were UT3R1, isolated from the root of industrial sugarcane, C7HL1 and C2HL2 from the leaf of chewing sugarcane, which both exhibited high N content while growing in N-free sand, and C34MR1 from the root of chewing sugarcane, which exhibited moderate N content in N-free sand. Isolates with the highest nitrogenase activity did not include those from the wild or chewing sugarcane plants with low N content in N-free sand.
Table 1:
Bacterial isolates from sugarcane.
| Varieties of sugarcane | Number of isolates | |||
|---|---|---|---|---|
|
| ||||
| Root | Stem | Leaf | Total | |
| U-Thong 3 (UT3) | 6 | 3 | 4 | 13 |
| Chewing (C13L) | 3 | 3 | 2 | 8 |
| Chewing (C15L) | 2 | 2 | 2 | 6 |
| Chewing (C34M) | 4 | 3 | 3 | 10 |
| Chewing (C2H) | 3 | 3 | 4 | 10 |
| Chewing (C7H) | 5 | 4 | 3 | 12 |
| Wild (W1) | 4 | 4 | 3 | 11 |
| Wild (W2) | 3 | 4 | 3 | 10 |
|
| ||||
| Total | 29 | 29 | 31 | 89 |
Nitrogenase activities of the bacterial isolates responded differently to increasing yeast extract concentrations (Table 2). The responses were divided into three groups: C2HL2 and C7HL1 exhibited increases in nitrogenase activity with the addition of 0.005% yeast extract, and C34MR1 demonstrated no response, whereas UT3R1 exhibited a significant decrease in nitrogenase activity at the lowest concentration of yeast extract. Nitrogenase activity generally declined in all isolates with further increase of the yeast extract concentration to 0.05%. Reis et al. (2004) reported that the growth and nitrogenase activity of Burkholderia tropica were stimulated by the addition of 0.01% yeast extract. Yeast extract is a source of vitamins that act as coenzymes as well as an alternative N source. Thus, the bacteria should be able to directly use organic N compounds such as amino acids for growth. For symbioses between legumes and nodule bacteria, it has been well established that there is a shift from N fixation to ammonium and nitrate as alternative sources of N with high levels of soil and N fertiliser (Evans 1982; Saxena et al. 1996). The suppression of nitrogenase activity at higher concentrations of yeast extract, particularly at 0.05% in this study, is indicative of the shift towards the yeast extract as a N source instead of N fixation by the endophytes. Variation in the responses of the isolates to 0.005% yeast extract in terms of nitrogenase activity ranged from a negative response (UT3R1) to no response (C34MR1) to positive responses (C2HL2 and C7HL1), indicating that differential adaptation may have practical implications for sugarcane cultivation as well as our understanding of the N fixing symbiosis between sugarcane and these endophytes.
Table 2:
Nitrogenase activities of four endophytic bacterial isolates from sugarcane with varying concentrations of yeast extract in the media.
| Isolates | Nitrogenase activity by ARA (nmol C2H4/mg protein/h) | |||
|---|---|---|---|---|
|
| ||||
| Yeast extract | ||||
|
| ||||
| 0% | 0.005% | 0.01% | 0.05% | |
| UT3R1 | 52.9f | 22.1g | 15.1g | 6.5g |
| C2HL2 | 354.1b | 403.8a | 320.9c | 23.6g |
| C7HL1 | 22.5g | 208.7d | 9.6g | 9.4g |
| C34MR1 | 23.0g | 24.1g | 15.4g | 5.6g |
Note: Different letters indicate a significant difference at the 5% level (according to Duncan’s multiple range test) with five replicates.
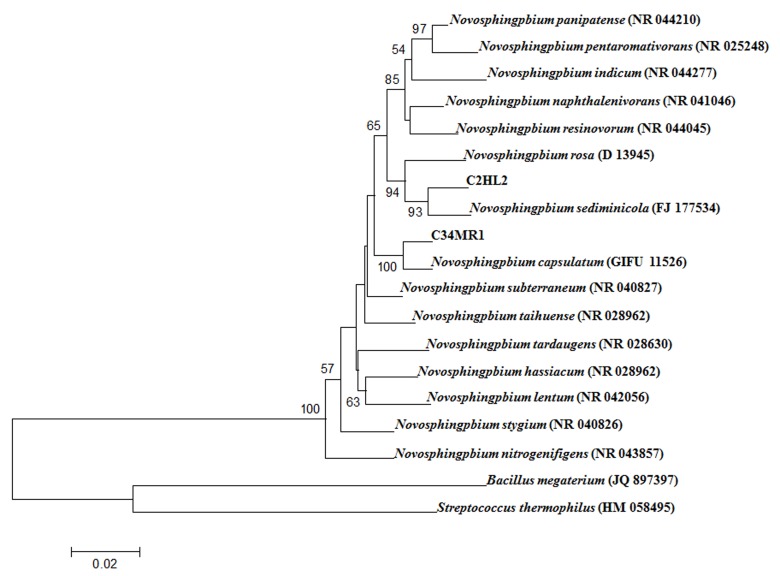
Characterisation of the bacterial isolates UT3R1, C7HL1, C2HL2 and C34MR1 via morphological and biochemical tests was carried out according to Bergey’s Manual of Determinative Bacteriology (Holt et al. 1994). The 16S rRNA gene sequences of the four bacterial isolates were analysed. All the isolates were Gram-negative, had short rods, and had colonies that were circular with intact margins and a smooth surface. They were catalase-positive and negative for indole production and starch hydrolysis (Tables 3 and 4). The phylogenetic tree of the 16S rRNA gene sequence from UT3R1 (1391 bp) demonstrated that this isolate belonged to the genus Ancylobacter (Fig. 1). Bacteria in this genus, such as A. rudongensis, A. vacuolatus sp. nov. and A. polymorphus sp. nov., have been reported to possess nitrogenase activity (Xin et al. 2004, 2006). C7HL1 exhibited some characteristics of the genus Ochrobactrum (Holt et al. 1994), and the phylogenetic tree of the 16S rRNA gene sequence (1487 bp) was 98.7% similar to O. intermedium (Fig. 2). Ochrobactrum oryzae sp. nov. and Ochrobactrum anthropi are endophytic bacteria that were isolated from rice (Tripathi et al. 2006; Mano & Morisaki 2008). C2HL2 and C34MR1 exhibited some characteristics of the genus Novosphingobium, such as yellow colonies, Gram-negative, short rods, non-motile, non-spore forming and catalase-positive (Takeuchi et al. 2001; Baek et al. 2011). The 16S rRNA gene sequence of C2HL2 (1426 bp) had the highest similarity to Novosphingobium sediminicola at 97.4%, and that of C34MR1 (1409 bp) was 98.29% similar to Novosphingobium capsulatum (Fig. 3). Islam et al. (2010) reported that N. capsulatum was isolated from rice paddy soil. Bacteria in the genus Novosphingobium are plant endophytes and include N. rosa, which was isolated from rose roots (Takeuchi et al. 1995), and N. tardaugens, which was isolated from Oryza sativa (Mano & Morisaki 2008). Addison et al. (2007) reported that N. nitrogenifigens sp. nov. is a polyhydroxy alkanoate-accumulating diazotroph isolated from pulp and paper waste water.
Table 3:
Characteristics of selected endophytic bacterial isolates.
| Characteristics | Isolates | |||
|---|---|---|---|---|
|
| ||||
| UT3R1 | C2HL2 | C7HL1 | C34MR1 | |
| Colony forming on N-free agar: | ||||
| Size (mm) | 2–3 | 3–5 | 1–3 | 3–5 |
| Pigmentation | White | White | White | Yellow |
| Shape | Circular | Circular | Circular | Circular |
| Margin | Entire | Entire | Entire | Entire |
| Elevation | Raised | Convex | Raised | Convex |
| Microscopic characteristics: | ||||
| Gram staining | Negative | Negative | Negative | Negative |
| Cell shape | Rod | Rod | Rod | Rod |
Table 4:
Biochemical tests of selected endophytic bacterial isolates.
| Biochemical tests | Isolates | |||
|---|---|---|---|---|
|
| ||||
| UT3R1 | C2HL2 | C7HL1 | C34MR1 | |
| Catalase | + | + | + | + |
| Indole production | − | − | − | − |
| MR | − | − | + | − |
| Motility | − | − | + | − |
| Oxidase | + | + | + | + |
| Starch hydrolysis | − | − | − | − |
| VP | − | − | + | − |
| Urease | + | − | + | − |
| Citrate | − | − | + | − |
| Carbon source utilisation: | ||||
| Mannitol | − | + | − | − |
| Rhamnose | − | + | + | + |
| Arabinose | − | + | + | + |
| Glucose | − | + | + | + |
Notes: For biochemical tests: (+) = positive reaction; (−) = negative reaction
For carbon source utilization: (+) = presence of growth; (−) = absence of growth
Figure 1:
Phylogenetic position of UT3R1 within the genus Ancylobacter.
Figure 2:
Phylogenetic positions of C7HL1 within the genus Ochrobactrum.
Figure 3:
Phylogenetic positions of C2HL2 and C34MR1 within the genus Novosphingobium.
CONCLUSION
Endophytic nitrogen fixing bacteria were found on the roots, leaves and stems of industrial, chewing and wild sugarcane plants. Although nitrogenase activity was detected in all 89 isolates, their response to 0.005% yeast extract varied from an increase in activity to no response or even a decrease. The highest nitrogenase activity was observed for C2HL2 and C7HL1, which had 16S rRNA gene sequences closely related to Novosphingobium sediminicola and Ochrobactrum intermedium, respectively. These bacteria were isolated from the leaves of chewing sugarcane plants and were able to achieve high N concentrations when grown in N-free sand.
Acknowledgments
The authors acknowledge the financial support from the National Research Universities Program of Thailand Commission on Higher Education. We thank the Multiple Cropping Centre and the Department of Biology, Faculty of Science, Chiang Mai University for the research facilities and the Suphanburi Field Crops Research Center for providing sugarcane samples.
REFERENCES
- Addison SL, Foote SM, Reid NM, Lloyd-Jones G. Novosphingobium nitrogenifigens sp. nov., a polyhydroxyalkanoate-accumulating diazotroph isolated from a New Zealand pulp and paper wastewater. International Journal of Systematic and Evolutionary Microbiology. 2007;57(11):2467–2471. doi: 10.1099/ijs.0.64627-0. [DOI] [PubMed] [Google Scholar]
- Baek SH, Lim JH, Jin L, Lee HG, Lee ST. Novosphingobium sediminicola sp. nov. isolated from freshwater sediment. International Journal of Systematic and Evolutionary Microbiology. 2011;61(10):2464–2468. doi: 10.1099/ijs.0.024307-0. [DOI] [PubMed] [Google Scholar]
- Baldani JI, Reis VM, Baldani VLD, Döbereiner J. A brief story of nitrogen fixation in sugarcane – reasons for success in Brazil. Functional Plant Biology. 2002;29(4):417–423. doi: 10.1071/PP01083. [DOI] [PubMed] [Google Scholar]
- Baldani VLD, Baldani JI, Olivares FL, Döbereiner J. Identification and ecology of Herbaspirillum seropedicae and closely related Pseudomonas rubrisubalbicans. Symbiosis. 1992;13(2):65–73. [Google Scholar]
- Boddey RM, Urquiaga S, Alves BJR, Reis V. Endophytic nitrogen fixation in sugarcane: Present knowledge and future application. Plant and Soil. 2003;252(1):139–149. [Google Scholar]
- Döbereiner J, Day JM, Dart PJ. Nitrogenase activity in the rhizosphere of sugarcane and other tropical grasses. Plant and Soil. 1972;37(1):191–196. [Google Scholar]
- Dong Z, Canny MJ, McCully ME, Roboredo MR, Cabadilla CF, Ortega E, Odes R. A nitrogen-fixing endophyte of sugarcane stems (a new role for the apoplast) Plant Physiology. 1994;105(4):1139–1147. doi: 10.1104/pp.105.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. Response of soybean-rhizobium symbioses to mineral nitrogen. Plant and Soil. 1982;66(3):439–442. [Google Scholar]
- Govindarajan M, Balandreau J, Kwon SW, Weon HY, Lakshminarasimhan C. Effects of the inoculation of Burkholderia vietnamensis and related endophytic diazotrophic bacteria on grain yield of rice. Microbial Ecology. 2008;55(1):32–37. doi: 10.1007/s00248-007-9247-9. [DOI] [PubMed] [Google Scholar]
- Govindarajan M, Kwon SW, Weon HY. Isolation, molecular characterization and growth-promoting activities of endophytic sugarcane diazotroph Klebsiella sp. GR9. World Journal of Microbiology and Biotechnology. 2007;23(7):997–1006. [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hallmann J, Mahaffee WF, Kloepper JW, Quadt-Hallmann A. Bacterial endophytes in agricultural crops. Canadian Journal of Microbiology. 1997;43(10):895–914. [Google Scholar]
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. Bergey's manual of determinative bacteriology. 7th ed. Baltimore, USA: William and Wilkin Company; 1994. [Google Scholar]
- Iniquez AL, Dong Y, Triplett EW. Nitrogen fixation in wheat provided by Klebsiella pneumonia 342. Molecular Plant-Microbe Interactions. 2004;17(10):1078–1085. doi: 10.1094/MPMI.2004.17.10.1078. [DOI] [PubMed] [Google Scholar]
- Islam R, Trivedi P, Madhaiyan M, Seshadri S, Lee G, Yang J, Kim Y, Kim M, Han G, Chauhan PS, et al. Isolation, enumeration, and characterization of diazotrophic bacteria from paddy soil sample under long-term fertilizer management experiment. Biology and Fertility of Soils. 2010;46(3):261–269. [Google Scholar]
- James EK. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Research. 2000;65(2–3):197–209. [Google Scholar]
- Ladizinsky G. Plant evolution under domestication. Dordrecht, Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- Loiret FG, Ortega E, Kleiner D, Ortega–Rodes P, Rodes R, Dong Z. A putative new endophytic nitrogen–fixing bacterium Pantoea sp. from sugarcane. Journal of Applied Microbiology. 2004;97(3):504–511. doi: 10.1111/j.1365-2672.2004.02329.x. [DOI] [PubMed] [Google Scholar]
- Mano H, Morisaki H. Endophytic bacteria in the rice plant. Microbes and Environments. 2008;23(2):109–117. doi: 10.1264/jsme2.23.109. [DOI] [PubMed] [Google Scholar]
- Mendes R, Pizzirani-Kleiner AA, Araujo WL, Raaijmakers JM. Diversity of cultivated endophytic bacteria from sugarcane: Genetic and biochemical characterization of Burkholderia cepacia complex isolates. Applied and Environmental Microbiology. 2007;73(22):7259–7267. doi: 10.1128/AEM.01222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakamhang J, Minamisawa K, Teamtaisong K, Boonkerd N, Teaumroong N. The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L.) Applied Soil Ecology. 2009;42(2):141–149. [Google Scholar]
- Reis VM, Estrada-de los Santos P, Tenorio-Salgado S, Vogel J, Stoffels M, Guyon S, Mavingui P, Baldani VLD, Schmid M, Baldani JI, et al. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. International Journal of Systematic and Evolutionary Microbiology. 2004;54(6):2155–2162. doi: 10.1099/ijs.0.02879-0. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saxena AK, Rathi SK, Tilak KVBR. Selection and evaluation of nitrate tolerant strains of Rhizobium leguminasarun biovar viceae specific to the lentil. Biology and Fertility of Soils. 1996;22(1–2):126–130. [Google Scholar]
- Takeuchi M, Hamana K, Hiraishi A. Hiraishi, proposal of the genus Sphingomonas sensustricto and three new genera, analyses. International Journal of Systematic and Evolutionary Microbiology. 2001;51(4):1405–1417. doi: 10.1099/00207713-51-4-1405. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Sakane T, Yanagi M, Yamasato K, Hamana K, Yokota A. Taxonomic study of bacteria isolated from plants: Proposal of Sphingomonas rosa sp. nov., Sphingomonas pruni sp. nov., Sphingomonas asaccharolytica sp. nov., and Sphingomonas mali sp. Nov. International Journal of Systematic and Evolutionary Microbiology. 1995;45(2):334–341. doi: 10.1099/00207713-45-2-334. [DOI] [PubMed] [Google Scholar]
- Tripathi AK, Verma SC, Chowdhury SP, Lebuhn M, Gattinger A, Schloter M. Ochrobactrum oryzae sp. nov., an endophytic bacterial species isolated from deep water rice in India. International Journal of Systematic and Evolutionary Microbiology. 2006;56(7):1677–1680. doi: 10.1099/ijs.0.63934-0. [DOI] [PubMed] [Google Scholar]
- Xin YH, Zhou YG, Chen WX. Ancylobacter polymorphus sp. nov. and Ancylobacter vacuolatus sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2006;56(6):1185–1188. doi: 10.1099/ijs.0.64118-0. [DOI] [PubMed] [Google Scholar]
- Xin YH, Zhou YG, Zhou HL, Chen WX. Ancylobacter rudongensis sp. nov., isolated from roots of Spartina anglica. International Journal of Systematic and Evolutionary Microbiology. 2004;54(2):385–388. doi: 10.1099/ijs.0.02466-0. [DOI] [PubMed] [Google Scholar]