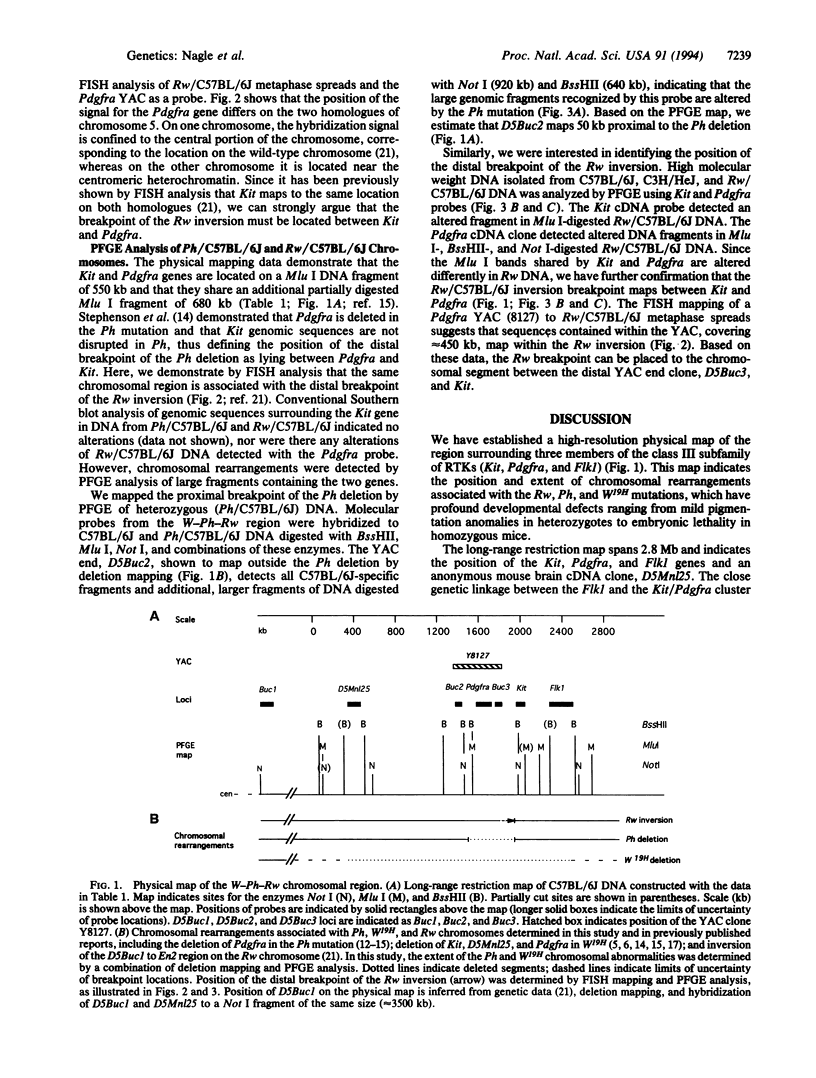
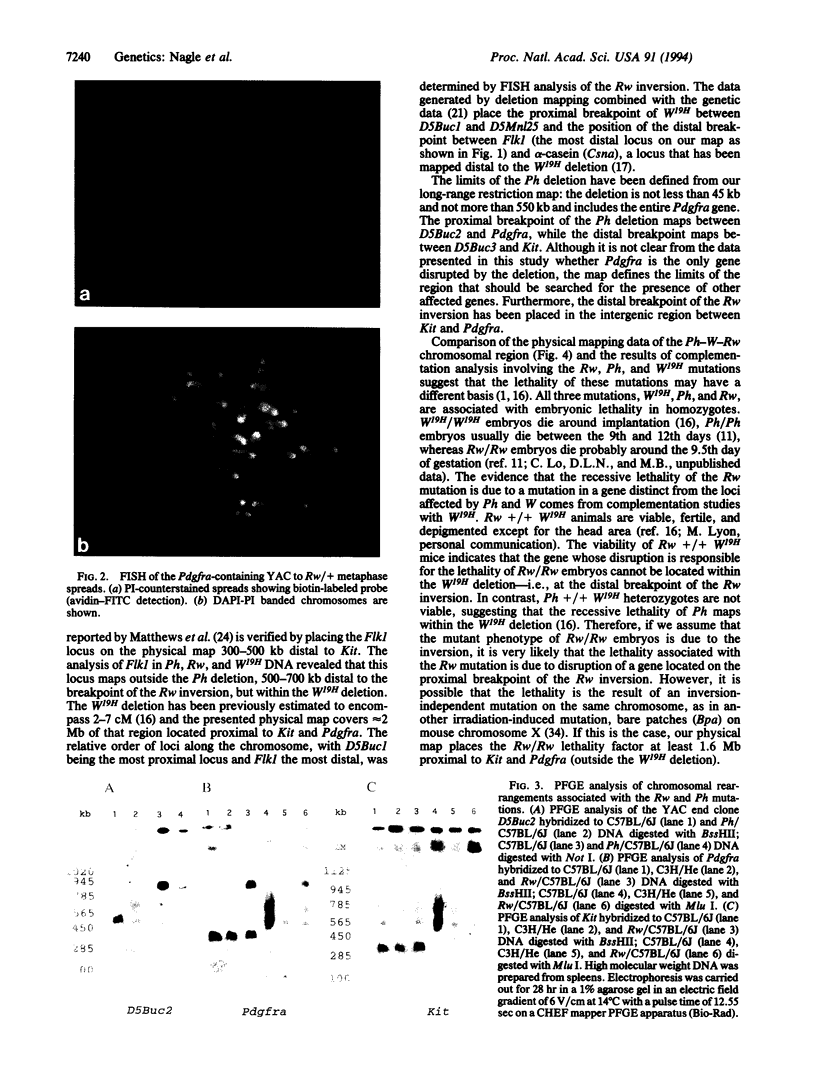
Abstract
We are studying the chromosomal structure of three developmental mutations, dominant spotting (W), patch (Ph), and rump white (Rw) on mouse chromosome 5. These mutations are clustered in a region containing three genes encoding tyrosine kinase receptors (Kit, Pdgfra, and Flk1). Using probes for these genes and for a closely linked locus, D5Mn125, we established a high-resolution physical map covering approximately 2.8 Mb. The entire chromosomal segment mapped in this study is deleted in the W19H mutation. The map indicates the position of the Ph deletion, which encompasses not more than 400 kb around and including the Pdgfra gene. The map also places the distal breakpoint of the Rw inversion to a limited chromosomal segment between Kit and Pdgfra. In light of the structure of the Ph-W-Rw region, we interpret the previously published complementation analyses as indicating that the pigmentation defect in Rw/+ heterozygotes could be due to the disruption of Kit and/or Pdgfra regulatory sequences, whereas the gene(s) responsible for the recessive lethality of Rw/Rw embryos is not closely linked to the Ph and W loci and maps proximally to the W19H deletion. The structural analysis of chromosomal rearrangements associated with W19H, Ph, and Rw combined with the high-resolution physical mapping points the way toward the definition of these mutations in molecular terms and isolation of homologous genes on human chromosome 4.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Besmer P., Murphy J. E., George P. C., Qiu F. H., Bergold P. J., Lederman L., Snyder H. W., Jr, Brodeur D., Zuckerman E. E., Hardy W. D. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature. 1986 Apr 3;320(6061):415–421. doi: 10.1038/320415a0. [DOI] [PubMed] [Google Scholar]
- Chabot B., Stephenson D. A., Chapman V. M., Besmer P., Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988 Sep 1;335(6185):88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- Duttlinger R., Manova K., Chu T. Y., Gyssler C., Zelenetz A. D., Bachvarova R. F., Besmer P. W-sash affects positive and negative elements controlling c-kit expression: ectopic c-kit expression at sites of kit-ligand expression affects melanogenesis. Development. 1993 Jul;118(3):705–717. doi: 10.1242/dev.118.3.705. [DOI] [PubMed] [Google Scholar]
- Evans E. P., Phillips R. J. Inversion heterozygosity and the origin of XO daughters of Bpa/+female mice. Nature. 1975 Jul 3;256(5512):40–41. doi: 10.1038/256040a0. [DOI] [PubMed] [Google Scholar]
- Fleischman R. A. From white spots to stem cells: the role of the Kit receptor in mammalian development. Trends Genet. 1993 Aug;9(8):285–290. doi: 10.1016/0168-9525(93)90015-a. [DOI] [PubMed] [Google Scholar]
- Fleischman R. A., Saltman D. L., Stastny V., Zneimer S. Deletion of the c-kit protooncogene in the human developmental defect piebald trait. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10885–10889. doi: 10.1073/pnas.88.23.10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler E. N., Cheng S. V., Gusella J. F., Housman D. E. Genetic analysis of the dominant white-spotting (W) region on mouse chromosome 5: identification of cloned DNA markers near W. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9635–9639. doi: 10.1073/pnas.85.24.9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler E. N., Ryan M. A., Housman D. E. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell. 1988 Oct 7;55(1):185–192. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- Green E. D., Olson M. V. Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1213–1217. doi: 10.1073/pnas.87.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann B. G., Barlow D. P., Lehrach H. A large inverted duplication allows homologous recombination between chromosomes heterozygous for the proximal t complex inversion. Cell. 1987 Mar 13;48(5):813–825. doi: 10.1016/0092-8674(87)90078-x. [DOI] [PubMed] [Google Scholar]
- Herrmann B., Bućan M., Mains P. E., Frischauf A. M., Silver L. M., Lehrach H. Genetic analysis of the proximal portion of the mouse t complex: evidence for a second inversion within t haplotypes. Cell. 1986 Feb 14;44(3):469–476. doi: 10.1016/0092-8674(86)90468-x. [DOI] [PubMed] [Google Scholar]
- Hsieh C. L., Navankasattusas S., Escobedo J. A., Williams L. T., Francke U. Chromosomal localization of the gene for AA-type platelet-derived growth factor receptor (PDGFRA) in humans and mice. Cytogenet Cell Genet. 1991;56(3-4):160–163. doi: 10.1159/000133076. [DOI] [PubMed] [Google Scholar]
- Larin Z., Monaco A. P., Lehrach H. Yeast artificial chromosome libraries containing large inserts from mouse and human DNA. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4123–4127. doi: 10.1073/pnas.88.10.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F., Glenister P. H., Loutit J. F., Evans E. P., Peters J. A presumed deletion covering the W and Ph loci of the mouse. Genet Res. 1984 Oct;44(2):161–168. doi: 10.1017/s0016672300026367. [DOI] [PubMed] [Google Scholar]
- Matsui T., Heidaran M., Miki T., Popescu N., La Rochelle W., Kraus M., Pierce J., Aaronson S. Isolation of a novel receptor cDNA establishes the existence of two PDGF receptor genes. Science. 1989 Feb 10;243(4892):800–804. doi: 10.1126/science.2536956. [DOI] [PubMed] [Google Scholar]
- Matthews W., Jordan C. T., Gavin M., Jenkins N. A., Copeland N. G., Lemischka I. R. A receptor tyrosine kinase cDNA isolated from a population of enriched primitive hematopoietic cells and exhibiting close genetic linkage to c-kit. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9026–9030. doi: 10.1073/pnas.88.20.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H., Davisson M. T., Doolittle D. P., Grant P., Hillyard A. L., Kosowsky M. R., Roderick T. H. Comparative map for mice and humans. Mamm Genome. 1992;3(9):480–536. doi: 10.1007/BF00778825. [DOI] [PubMed] [Google Scholar]
- Nocka K., Majumder S., Chabot B., Ray P., Cervone M., Bernstein A., Besmer P. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice--evidence for an impaired c-kit kinase in mutant mice. Genes Dev. 1989 Jun;3(6):816–826. doi: 10.1101/gad.3.6.816. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A., Bedford M. T., Do M. S., Eisenbach L., Lonai P. Developmental expression of the alpha receptor for platelet-derived growth factor, which is deleted in the embryonic lethal Patch mutation. Development. 1992 May;115(1):289–303. doi: 10.1242/dev.115.1.289. [DOI] [PubMed] [Google Scholar]
- Pawson T., Bernstein A. Receptor tyrosine kinases: genetic evidence for their role in Drosophila and mouse development. Trends Genet. 1990 Nov;6(11):350–356. doi: 10.1016/0168-9525(90)90276-c. [DOI] [PubMed] [Google Scholar]
- Reith A. D., Rottapel R., Giddens E., Brady C., Forrester L., Bernstein A. W mutant mice with mild or severe developmental defects contain distinct point mutations in the kinase domain of the c-kit receptor. Genes Dev. 1990 Mar;4(3):390–400. doi: 10.1101/gad.4.3.390. [DOI] [PubMed] [Google Scholar]
- Sawyer J. R., Moore M. M., Hozier J. C. High resolution G-banded chromosomes of the mouse. Chromosoma. 1987;95(5):350–358. doi: 10.1007/BF00293182. [DOI] [PubMed] [Google Scholar]
- Schatteman G. C., Morrison-Graham K., van Koppen A., Weston J. A., Bowen-Pope D. F. Regulation and role of PDGF receptor alpha-subunit expression during embryogenesis. Development. 1992 May;115(1):123–131. doi: 10.1242/dev.115.1.123. [DOI] [PubMed] [Google Scholar]
- Searle A. G., Truslove G. M. A gene triplet in the mouse. Genet Res. 1970 Apr;15(2):227–235. doi: 10.1017/s0016672300001555. [DOI] [PubMed] [Google Scholar]
- Smith E. A., Seldin M. F., Martinez L., Watson M. L., Choudhury G. G., Lalley P. A., Pierce J., Aaronson S., Barker J., Naylor S. L. Mouse platelet-derived growth factor receptor alpha gene is deleted in W19H and patch mutations on chromosome 5. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4811–4815. doi: 10.1073/pnas.88.11.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz R. A., Droetto S., Fukushima Y. Deletion of the KIT and PDGFRA genes in a patient with piebaldism. Am J Med Genet. 1992 Nov 1;44(4):492–495. doi: 10.1002/ajmg.1320440422. [DOI] [PubMed] [Google Scholar]
- Stephenson D. A., Mercola M., Anderson E., Wang C. Y., Stiles C. D., Bowen-Pope D. F., Chapman V. M. Platelet-derived growth factor receptor alpha-subunit gene (Pdgfra) is deleted in the mouse patch (Ph) mutation. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):6–10. doi: 10.1073/pnas.88.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. C., Nocka K., Ray P., Traktman P., Besmer P. The dominant W42 spotting phenotype results from a missense mutation in the c-kit receptor kinase. Science. 1990 Jan 12;247(4939):209–212. doi: 10.1126/science.1688471. [DOI] [PubMed] [Google Scholar]
- Wang C., Kelly J., Bowen-Pope D. F., Stiles C. D. Retinoic acid promotes transcription of the platelet-derived growth factor alpha-receptor gene. Mol Cell Biol. 1990 Dec;10(12):6781–6784. doi: 10.1128/mcb.10.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]