Abstract
Heat shock protein 40 (Hsp40) co-chaperones assist in cellular protein folding and degradation through the binding and delivery of non-native proteins to heat shock protein 70 (Hsp70). The mechanism for substrate transfer from Hsp40s to Hsp70 is unknown. Two recent studies provide new details that shed light on novel mechanisms for substrate recognition by Hsp40s and a common mechanism for polypeptide transfer to Hsp70.
Hsp70s and protein folding
Heat shock protein 70 family members (hereafter referred to as Hsp70) interact with different combinations of heat shock protein 40s (Hsp40s) and other folding and degradatory co-chaperones to facilitate multiple processes required for protein homeostasis [1,2]. Defects in protein homeostasis can trigger aberrant protein aggregation, a hallmark of a broad class of diseases known collectively as ‘conformational disorders’. Hsp70 molecular chaperones protect cells against the accumulation of proteotoxic species by maintaining a delicate balance between protein synthesis, folding and degradation [3]. Yet the mechanism(s) by which functionally distinct Hsp40s (also known as J-proteins) bind and deliver a diverse array of proteins to the polypeptide-binding site of Hsp70 remains a major unanswered question.
Polypeptide binding and release by Hsp70 is coordinated via an array of co-chaperones that tightly regulate the Hsp70 ATP hydrolytic cycle [4–6] (Figure 1). This process is initiated when a non-native polypeptide is bound by an Hsp40 co-chaperone [1]. This large and structurally diverse family is defined by a highly conserved region called a J-domain that interacts with the Hsp70 nucleotide-binding domain (NBD) and stimulates intrinsic Hsp70 ATPase activity [1,7,8]. Hydrolysis of ATP to ADP in the Hsp70 NBD induces a conformational change in the Hsp70 substrate-binding domain (SBD), thus increasing the affinity of Hsp70 for the substrate [9,10]. In most cases, polypeptides are released from Hsp70 when ADP is exchanged for ATP; this reaction is facilitated by structurally diverse nucleotide exchange factors (NEFs) [11,12]. Once released, the polypeptide can fold to the native conformation, or remain misfolded and reenter the cycle for subsequent rounds of refolding. However, in some instances Hsp70-bound clients are recognized by the E3 ubiquitin ligase CHIP (carboxyl terminus of Hsc70-interacting protein) and targeted for degradation by the ubiquitin-proteasome system [13]. Interestingly, Hsp70 and Hsp40 proteins have recently been identified as partners for additional E3 ubiquitin ligases [14]. Thus, the fate of non-native polypeptides is determined via the association of Hsp70 with co-factors that facilitate protein folding or degradation via the ubiquitin-proteasome system.
Figure 1.

(a) Hsp70 polypeptide binding and release is regulated through cycles of ATP hydrolysis and exchange. Protein misfolding is managed, in part, through the action of Hsp70 molecular chaperones in coordination with various cellular cofactors. Initially, a non-native polypeptide is bound by an Hsp40 co-chaperone (light blue). (b) The Hsp40 J-domain then binds Hsp70 (blue-green) in an ATP-bound state. (c) J-domain-stimulated ATP hydrolysis in the nucleotide-binding domain (NBD) induces a conformational shift in the Hsp70 substrate-binding domain (SBD), increasing affinity for the non-native polypeptide that is released from the Hsp40. Additional cofactors such as the E3 ubiquitin ligase CHIP (brown) with a ubiquitin-conjugating E2 (blue) can bind to Hsp70 and the non-native polypeptide, targeting the substrate for degradation by the ubiquitin-proteasome system (Poly-Ub). (d) Nucleotide exchange factors (NEFs) such as the Hsp110s (green) replace the ADP with ATP, releasing the polypeptide from Hsp70. (e) If the polypeptide remains in a non-native conformation, the cycle can be repeated until folding is complete.
Hsp40s: diverse Hsp70-binding partners
Through direct binding to non-native substrates, Hsp40s serve as conduits that funnel non-native clients to Hsp70. Despite sharing a conserved J-domain, the Hsp40 family is extensive and members possess unique domains that bind to diverse clients and target Hsp70 to distinct cellular machineries. Hsp40s are categorized based on homology to the founding member in Escherichia coli, DnaJ [7,8]. Type I Hsp40s possess a J-domain, a glycine- and phenylalanine-rich (G/F-rich) region and a cysteine-rich, zinc-finger-like region (ZFLR). Type II Hsp40s contain the J-domain and G/F-rich region, whereas Type III Hsp40s retain only the J-domain. Action of the Hsp40 J-domain alone is sufficient to enable Hsp70 to perform its essential cellular functions [15]. However, the aforementioned specialized Hsp40 polypeptide-binding domain imparts considerable influence over Hsp40 quaternary structure and has crucial roles in directing Hsp70 to carry out a variety of specialized functions [16,17].
Importantly, evolutionary diversification of the Hsp40 family has expanded the repertoire of the Hsp40 co-chaperone class (Figure 2). Recent studies suggest that this structural diversification has generated an array of co-chaperones with unique polypeptide-binding domains that all interact with Hsp70 via the J-domain. This specialization is exemplified by new studies on the function of three different endoplasmic reticulum (ER)-localized Hsp40s [18–20]. For example, the Hsp40s P58 and ERdj3 specifically bind misfolded substrates in the ER lumen [19,20]. However, P58 is unique because this Hsp40 contains nine tetratricopeptide repeats that seem to participate in recognition and binding of misfolded proteins. Thus, P58 and ERdj3 might discriminate between distinct nonnative substrates within the ER. By contrast, the Type III Hsp40 ERdj5 is associated with a supramolecular complex that targets misfolded ER lumenal proteins for degradation via the ER-associated degradation pathways. ERdj5 contains three cysteine-rich thioredoxin domains that could be involved in substrate recognition and facilitate retrotranslocation by cleaving disulfide bonds in non-native proteins within the ER lumen. Thus, highly specialized ER-localized Hsp40s have evolved different mechanisms for substrate binding to meet the protein quality control requirements of this cellular sub-compartment.
Figure 2.
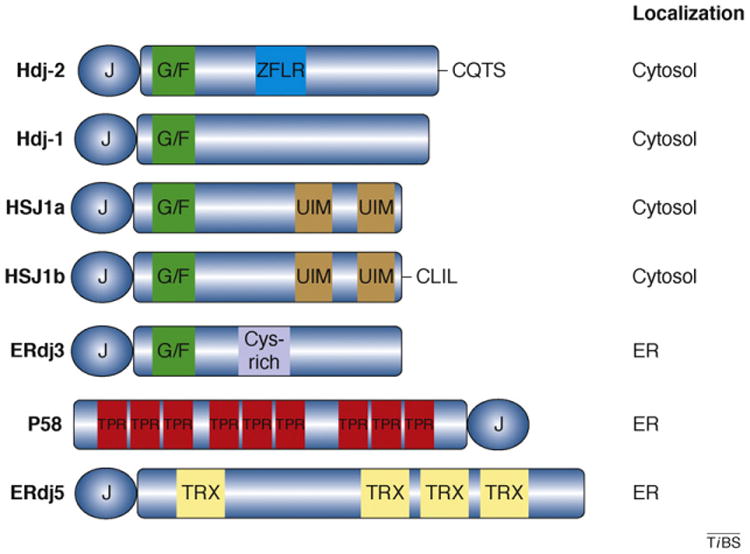
Domain structures of several human Hsp40s. Type I Hsp40s such as Hdj-2 (also called DnaJA1) possess a J-domain, G/F-rich region (green) and a ZFLR (blue). Hdj-2 is distinct from many Hsp40s because it is farnesylated at a C-terminal CAAX motif (CQTS). Type II Hsp40s, such as Hdj-1 (also called DnaJB1), lack the ZFLR. HSJ1a and HSJ1b (also called DnaJB2) are neuron-specific type II Hsp40 isoforms. Both isoforms possess two ubiquitin interaction motifs (UIMs; brown); however, Hsj1b also contains a CAAX motif that is geranylgeranylated (CLIL). ER-localized Hsp40s have also evolved specialized domains. For example, ERdj3 (also called DnaJB11) contains a cysteine-rich motif (purple) in the C terminus [21]. P58 (also called DnaJC3) contains nine tetratricopeptide repeats (TPR; red) that might bind non-native polypeptides, whereas ERdj5 contains three thioredoxin (TRX; yellow) domains, which are responsible for disulfide reductase activity. The length of each Hsp40 represents its relative size. The intracellular location of each Hsp40 is noted.
A unique feature of cytosolic Hsp40s is the modification of Type I and Type II family members by prenylation [1,7,8]. For example, the yeast Type I Hsp40 Ydj1p and its human homolog Hdj-2 are farnesylated at a C-terminal CAAX motif [21]. Whereas this lipid modification helps to localize a pool of Hsp40 to the cytosolic surface of the ER membrane, one recent study suggests that farnesylation is required for Type I Hsp40s to bind specific substrates [22,23]. In addition, the neuron-specific Type II Hsp40 HSJ1b is geranylgeranylated and localized to the ER surface. Similar to Type I Hsp40s, binding between HSJ1b and its client protein rhodopsin relies on this lipid modification [24]. Thus, lipid modification not only affects localization, but might also stabilize Hsp40–polypeptide complexes.
Polypeptide transfer and release from Hsp70
Once Hsp40s bind to non-native polypeptides, bound clients must be transferred to Hsp70 for subsequent processing. To begin to address how this occurs, given the extensive structural diversity within the Hsp40s, Petrova et al. [19] and Jin et al. [20] recently examined how two different ER-localized Hsp40s release substrates upon interaction with the Hsp70 BiP (immunoglobulin heavy-chain-binding protein). In these studies, the Hsp40s P58 and ERdj3 specifically bound misfolded or denatured substrates. Efficient substrate release from either Hsp40 relied upon interaction between BiP and the J-domain, specifically when BiP was ATP bound. Mutational analysis of BiP revealed that substrate release was functionally coupled to the ATPase activity of BiP, suggesting that a J-domain–BiP complex was not sufficient to dissociate the substrate. How does Hsp70 binding and ATP hydrolysis provoke Hsp40 to release its substrate? One hypothesis is that binding to Hsp70 induces an allosteric shift in the Hsp40 that reduces substrate affinity in the polypeptide-binding domain. However, Petrova et al. [16] found that the location of the J-domain within the Hsp40 was inconsequential to BiP-induced substrate release. Furthermore, the extensive variation present in the Hsp40 family argues against a unified allosteric mechanism. However, J-domain–Hsp70 binding might bring the substrate in close proximity to the Hsp70 SBD. After ATP hydrolysis, the increase in substrate affinity by Hsp70 could then out-compete the Hsp40 for binding.
This new model explains how divergent Hsp40s can uniformly release substrates for transfer to Hsp70 through interactions between the conserved J-domain and the Hsp70 NBD. However, some Hsp40s interact with regions within the Hsp70 SBD [25], suggesting that specific Hsp40s have evolved individualized mechanisms for cooperating with Hsp70. Ydj1p, in particular, requires its ZFLR for substrate transfer and chaperone-dependent refolding [26]. The Ydj1p ZFLR (located immediately adjacent to the J-domain) might help to orient substrates in a more amenable position for transfer to the Hsp70 SBD [16]. Therefore, distinct Hsp40 modules might confer not only unique binding preferences but also specialized interactions with Hsp70 that impart substantial regulatory oversight in this relationship.
Polypeptide release from Hsp70 is mediated by exchange of ADP for ATP. The NEFs that regulate this step are also highly diverse; they do not share a common domain responsible for this activity. Several recent structures suggest that individual NEFs might function by twisting the lobes of the Hsp70 ATPase domain in a manner that decreases affinity for ADP [12]. Interestingly, the NEF Sse1p and its human homologs are non-canonical Hsp70s that possesses polypeptide-binding activity and might interact with Hsp70-bound clients [27]. Thus, sequential interactions between non-native polypeptides with Hsp40, Hsp70 and other Hsp70 co-chaperones seem to have crucial roles in cellular folding and degradation pathways.
Concluding remarks
Hsp40 co-chaperones have emerged as complex and essential players in regulating protein homeostasis. This diverse family utilizes a variety of mechanisms to bind and funnel clients to Hsp70. Yet, recent studies suggest a uniform mechanism for substrate release and transfer from Hsp40 to Hsp70 [19,20]. Future studies are needed to dissect how specialized Hsp40s selectively recognize different protein conformers and specify Hsp70s cellular functions. In addition, we need to understand how interactions between Hsp70-bound protein clients and folding and degradatory co-chaperones mediate protein triage events [13,27]. Further knowledge of this process will lend crucial insight into the underlying causes and potential treatments for the expansive list of protein conformational disorders [2].
Acknowledgments
D.W.S. is funded by a predoctoral training grant from the National Institutes of Health (NIH; 5T32GM008581–09; www.nih.gov). P.M.D. is supported by a predoctoral fellowship from the American Heart Association (www.americanheart.org). C.H.I.R. is supported by grants from Fundação de Amparo a Pesquisa do Estado de São Paulo, Ministério da Ciência e Tecnologia/Conselho Nacional de Pesquisa e Desenvolvimento and NIH-R03TW007437 funded by the Fogarty International Center (www.fic.nih.gov). D.M.C. is supported by funds from the NIH (5R01GM067785-06).
References
- 1.Cyr DM, et al. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 2.Balch WE, et al. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 3.Cyr DM, et al. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 4.Langer T, et al. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 5.Cyr DM, et al. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- 6.Liberek K, et al. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the sigma 32 transcription factor. Proc Natl Acad Sci U S A. 1992;89:3516–3520. doi: 10.1073/pnas.89.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh P, et al. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu XB, et al. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel M, et al. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006;21:359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabo A, et al. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cyr DM. Swapping nucleotides, tuning Hsp70. Cell. 2008;133:945–947. doi: 10.1016/j.cell.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosser MF, et al. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 14.Han S, et al. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- 15.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci U S A. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos CH, et al. Conserved central domains control the quaternary structure of type I and type II Hsp40 molecular chaperones. J Mol Biol. 2008;383:155–166. doi: 10.1016/j.jmb.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan CY, et al. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ushioda R, et al. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 19.Petrova K, et al. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Y, et al. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 2008;27:2873–2882. doi: 10.1038/emboj.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caplan AJ, et al. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J Biol Chem. 1992;267:18890–18895. [PubMed] [Google Scholar]
- 22.Summers DW, et al. The type I HSP40 YDJ1 utilizes a farnesyl moiety and zinc finger like-region to suppress prion toxicity. J Biol Chem. 2008;284:3628–3639. doi: 10.1074/jbc.M807369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flom GA, et al. Farnesylation of ydj1 is required for in vivo interaction with hsp90 client proteins. Mol Biol Cell. 2008;19:5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapple JP, Cheetham ME. The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J Biol Chem. 2003;278:19087–19094. doi: 10.1074/jbc.M212349200. [DOI] [PubMed] [Google Scholar]
- 25.Freeman BC, et al. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan CY, et al. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J Biol Chem. 2005;280:695–702. doi: 10.1074/jbc.M410645200. [DOI] [PubMed] [Google Scholar]
- 27.Polier S, et al. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]


