Summary
The mechanisms regulating human embryonic stem (ES) cell self-renewal and differentiation are not well defined in part due to the lack of tools for forward genetic analysis. We present a piggyBac transposon gain of function screen in human ES cells that identifies DENND2C, which genetically cooperates with NANOG to maintain self-renewal in the presence of retinoic acid. We show that DENND2C negatively regulates RHOA activity, which cooperates with NANOG to block differentiation. It has been recently shown that RHOA exists in the nucleus and is activated by DNA damage; however, its nuclear function remains unknown. We discovered that RHOA associates with DNA and that DENND2C affects nuclear RHOA localization, activity, and DNA association. Our study illustrates the power of piggyBac as a cost-effective, efficient, and easy to use tool for forward genetic screens in human ES cells and provides insight into the role of RHOA in the nucleus.
Highlights
-
•
NANOG-sensitized piggyBac screen for regulators of stem cell differentiation
-
•
DENND2C is a negative regulator of RHOA
-
•
Genetic cooperation exists between RHOA and NANOG
-
•
Nuclear RHOA associates with DNA
In this article, Xu and colleagues demonstrate the utility of piggyBac insertional mutagenesis in human embryonic stem cells. In screening for genes cooperating with the expression of the self-renewal factor NANOG to resist differentiation, they identify the gene DENND2C as a mediator of retinoic acid resistance through repression of both nuclear and cytoplasmic RHOA.
Introduction
The molecular basis for the self-renewal and differentiation of human embryonic stem (ES) cells is not fully understood. In the mouse, self-renewal depends on the maintenance of a core regulatory network of three transcription factors, Nanog, Oct4, and Sox2, which function as a unit to block differentiation (reviewed in Jaenisch and Young, 2008). Mouse embryos null for any of these factors are incapable of maintaining a pluripotent inner cell mass, and cells destined to become epiblast instead develop into extraembryonic lineages (Avilion et al., 2003; Nichols et al., 1998; Chambers et al., 2003; Mitsui et al., 2003). The involvement of these transcription factors has been more recently extended to human ES cells, as they occupy the promoters of a number of genes shown to be differentially upregulated or repressed in human ES cells versus differentiated cells (Boyer et al., 2005). Unlike mouse ES cells, these three factors do not function as a unit to regulate self-renewal of human ES cells and each represses the differentiation of different cell fates (Wang et al., 2012).
Little is known about the factors working with either NANOG, OCT4, or SOX2 to block lineage specific differentiation in human ES cells. Co-immunoprecipitation experiments have been successfully utilized to detect proteins binding to and cooperating with Nanog, Oct4, and Sox2 in mouse ES cells (Wang et al., 2006; Liang et al., 2008; van den Berg et al., 2010; Mallanna et al., 2010; Pardo et al., 2010). We would like to develop a complementary forward genetic approach to identify genes that cooperate with a factor such as NANOG in regulating important biological processes in human ES cells. A forward genetic approach not only has the power to interrogate the genome in an unbiased fashion, but also has the potential to identify cooperating genes that are either not in the same protein complex or have low transcript or protein abundance.
We have previously shown that the piggyBac (PB) transposon modified from moth can efficiently transpose in the mouse and human genomes (Ding et al., 2005). Here we present a gain of function screen in human ES cells using PB transposon mutagenesis. The transposon is specially designed for identifying genes that cooperate with NANOG to block differentiation and support human ES cell self-renewal. As proof of principle, we show that the screen identified DENND2C, whose overexpression is capable of genetic cooperation with NANOG to block retinoic acid (RA)-induced differentiation. Further characterization revealed that DENND2C negatively regulates RHOA, affecting the localization, activity, and DNA association of nuclear RHOA.
Results
PB Insertional Mutagenesis Screen in Human ES Cells
The PB transposon has been demonstrated to be a useful tool for efficient transgenesis and insertional mutagenesis in both mouse and human immortalized cells (Ding et al., 2005). The transposon can efficiently mediate both loss- and gain-of-function insertional mutagenesis in mice (Ding et al., 2005; Rad et al., 2010; Landrette et al., 2011). Given that PB can also mediate efficient gene transfer in human ES cells (Chen et al., 2010), we decided to develop a PB vector for insertional mutagenesis screens in human ES cells.
To identify factors that cooperate with NANOG to block RA-induced differentiation, we have taken the advantage of PB’s large cargo capacity to develop an insertional mutagenesis vector that can simultaneously express the NANOG transgene (PB[IM,N]; Figure 1A; Ding et al., 2005; Li et al., 2011). PB[IM,N] insertion upstream of a gene results in constitutive overexpression of the downstream gene, while insertion within a transcription unit can result in overexpression of a truncated gene product downstream of the insertion site, leading to constitutive activation, dominant-negative effects, or heterozygous knockout of the gene (Figure 1A). In addition to mutagenesis, such insertions result in fluorescent labeling of mutated cells with Katushka marker. Upon co-transfection with a helper plasmid carrying the PB transposase transgene, PB[IM,N] stably integrates into the genome of about 30% of transfected human ES cells with an average of five copies of transposon per genome (Figure 1B; Experimental Procedures). Of 133 PB insertions mapped in human ES cells to date, 93% of insertions are within 200 kb of a gene, and 53% are located within an intron. This is consistent with data showing that PB frequently integrates near to or within coding units, making this transposon a useful tool for insertional mutagenesis of genes (Ding et al., 2005). The puromycin antibiotic resistance marker within the construct allows for the selection of cells with stable PB integration. Thus, this vector allows one to generate a library of individually mutagenized human ES cells with the condition of NANOG overexpression in a quick, easy, and cost-effective fashion.
Figure 1.
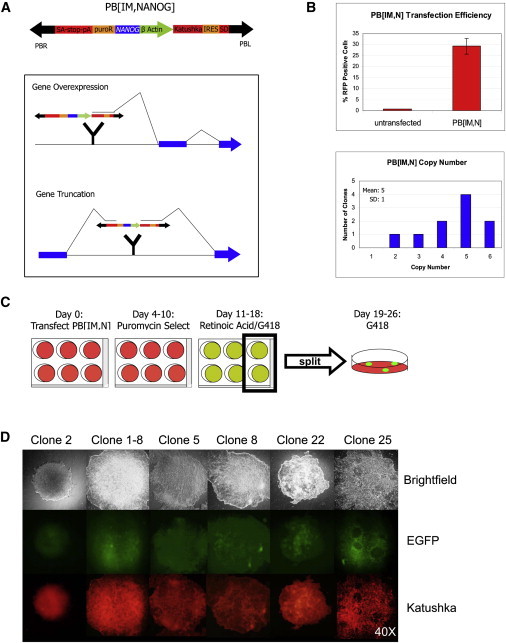
PB Mutagenesis in OCT4 Reporter Human ES Cells
(A) Splicing consequences of PB[Insertional Mutagenesis,NANOG] (PB[IM,N]) insertion either in front of or within the intron of a gene. PB[IM,N] insertion into any reading frame is capable of inducing both overexpression of the downstream transcript with the β actin promoter and triple reading frame start cassette (not shown) as well as simultaneous expression of Katushka fluorescent marker by the splicing of Katushka/IRES into the native transcript with the splice donor (SD). This construct is also able to create N-terminally truncated transcripts with the splice acceptor, triple reading frame stop cassette, and poly A tail (SA-stop-pA). PB[IM,N] can also constitutively overexpress NANOG transcript from the independent NANOG overexpression cassette.
(B) (Top) FACS analysis of Katushka expression from PB[IM,N] in human ES cells 48 hr after transfection in single-cell suspension using the RFP channel. Mean of triplicate independent experiments is shown. Error bars represent SD. (Bottom) PB copy number within ten individual PB[IM,N] transfected human ES cell clones as assessed by real time PCR. There is a mean of five PB[IM,N] insertions per clone.
(C) Protocol of screen performed in H1 OCT4-EGFP cells for resistance to RA-induced differentiation using PB[IM,N]. Cells in six-well plates were transfected with PB[IM]N, puromycin selected, and subsequently treated with RA and G418. Cells were then split from the six-well plates into 10-cm plates at a 2:1 ratio and maintained on G418 alone.
(D) Brightfield and fluorescent images of the six unique clones recovered from the screen. All clones are positive for EGFP and Katushka fluorescent reporters for OCT4 expression and PB[IM,N] insertion within an actively expressed gene.
For this screen, we have used an H1 human ES cell line containing an EGFP reporter and neomycin resistance cassette knocked into the OCT4 3′UTR (Zwaka and Thomson, 2003; H1 OCT4-EGFP or WT cells). Undifferentiated cells that continue to express the self-renewal factor OCT4 are both EGFP positive and G418 resistant. We utilized PB[IM,N] to screen for genes capable of genetically cooperating with NANOG to block differentiation of H1 OCT4-EGFP human ES cells. For the screen, approximately 9.6 × 107 cells were transfected in single-cell suspension with PB[IM,N] and the PB transposase transgene plasmid in 24 Matrigel-coated 6-well plates, allowing for the isolation of individual mutant clones after puromycin selection. Surviving adherent and puromycin selected cells were subsequently treated with a combination of RA and G418 in the same monolayer culture. Six surviving independent clones with undifferentiated morphology were isolated and confirmed in retesting (Figure 1C; see Experimental Procedures for details). All of these Katushka-positive clones express OCT4-EGFP, indicating that they remain undifferentiated (Figure 1D).
In human ES cells, the expression of a NANOG transgene confers inhibition of spontaneous differentiation and fibroblast growth factor (FGF) independence (Zhang et al., 2009; Darr et al., 2006). In mouse ES cells, expression of a Nanog transgene has been shown to confer several effects, including resistance to RA-induced differentiation, leukemia inhibitory factor (LIF) independence, and increased colony formation (Chambers et al., 2003; Mitsui et al., 2003; reviewed in Pan and Thomson, 2007). We confirmed that expression of NANOG by a PB vector in the H1 OCT4-EGFP cells stabilizes human ES cells to resist spontaneous differentiation (PB[NANOG] or NANOG; Figure 2A). Unlike in mouse ES cells, NANOG overexpression in human ES cells confers only weak resistance to RA-induced differentiation. All WT human ES cells appear morphologically differentiated in response to RA treatment (Figure 2B). This differentiation is reflected by induction of the GATA6 endoderm marker (Figures 2B and 2C). In contrast, NANOG expressing colonies after RA treatment contain small patches of cells that appear morphologically undifferentiated and continue to express NANOG (Figures 2B and 2C). This weak resistance to RA is also observed in cells transfected with the PB[IM,N] mutagenic construct (Figure 2D). Concordant with these results, only a few PB[NANOG] or PB[IM,N] colonies are able to survive when RA treatment is combined with G418, as differentiated cells no longer express the G418 resistance marker (Figure 2E). Thus, NANOG overexpression in human ES cells offers an ideal condition for forward genetics, as the small degree of resistance to differentiation allows for a sensitized screen without the drawback of excessive background. The clones identified from the screen display robust resistance to RA treatment in comparison to NANOG overexpressing escapers (Figure 2E). Indeed, only the identified clones can survive more than one round of RA treatment.
Figure 2.
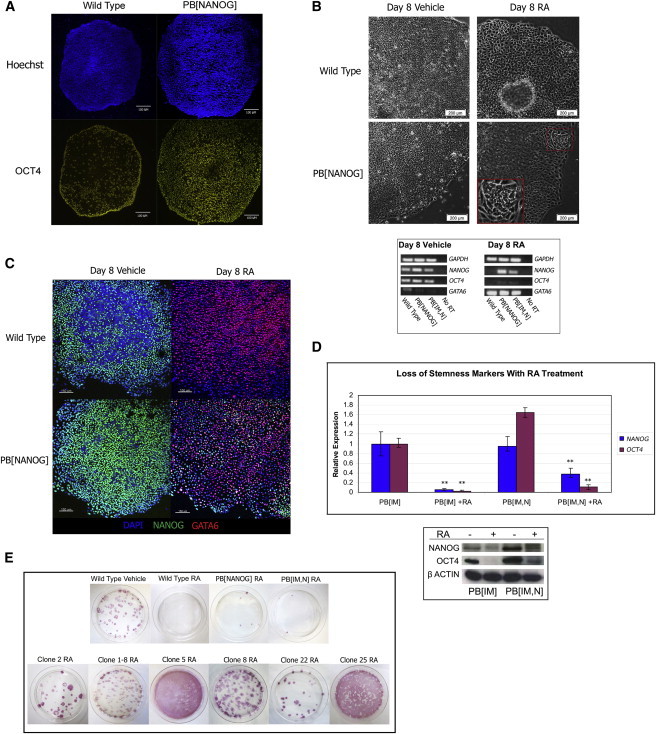
NANOG Expression from PB Constructs Induces Resistance to RA-Induced Differentiation
(A) OCT4 immunostaining in WT and NANOG expressing human ES cells. NANOG expressing cells resist spontaneous differentiation and maintain OCT4 signal in colony centers at larger diameters than WT cells. Scale bar represents 100 μm.
(B) (Top) Morphology of RA differentiated cells. The red square highlights a cluster of morphologically undifferentiated NANOG expressing cells after RA treatment, magnified in inset. Scale bar represents 200 μm. (Bottom) RT-PCR analysis of RA-differentiated cells.
(C) Immunostaining of RA-differentiated cells co-stained for NANOG and GATA6. RA induces differentiation of human ES cells as assessed by GATA6 endoderm marker. Scale bar represents 150 μm.
(D) Real-time PCR (top) and western blot (bottom) analysis of stemness marker expression in RA-treated cells. The addition of a NANOG overexpression cassette to PB[IM] induces protein overexpression of both NANOG and the NANOG target OCT4 and induces weak persistence of NANOG and OCT4 after RA treatment. Graph shows mean of triplicate independent experiments. Error bars represent SD. ∗∗p < 0.001 in Student’s t test.
(E) Alkaline phosphatase staining of combined RA/G418-treated cells after 16 days. Experiments are performed in duplicate. A representative plate is shown.
The PB insertion sites in the resistant clones were determined by PCR and sequencing (Experimental Procedures; Table S1). Detailed characterization of one of the isolated clones is presented below, while other information will be described elsewhere.
Identification of DENND2C as a NANOG Collaborator for Blocking Differentiation
The clones isolated from the screen exhibit varying degrees of blocking RA-induced differentiation and different morphologies, suggesting different underlying mechanisms of resistance (Figure 2E). Clone 2 was of particular interest to us because of its unique morphology. Cells from this clone resisted spontaneous differentiation and instead piled into dense, multilayered colonies, with some cells aggregating into balls. These cells exhibited a weak attachment to Matrigel as they are readily detached after collagenase IV treatment. Clone 2 had three PB insertions, the examination of which led to the identification of the causative gene, DENND2C. DENND2C belongs to a family of proteins containing DENN domains (Differentially Expressed in Normal versus Neoplastic; reviewed in Marat et al., 2011). A PB insertion is located within an intron of the DENND2C gene, resulting in the overexpression of the C-terminal DENN domain of the gene while not having significant effects on the expression of surrounding genes (Figure 3A). More importantly, overexpression of the DENND2C gene product alone, either truncated or full length, in the NANOG-expressing parental cells phenocopies clone 2 morphology and behavior (Figure 3B). This indicates that PB disruption of DENND2C is the causative alteration (Figures 3A and 3B).
Figure 3.
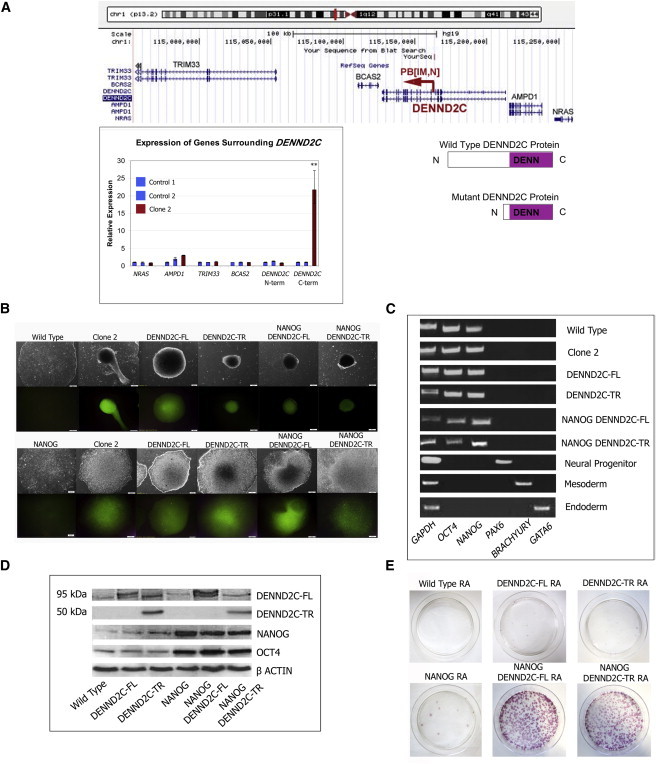
PB Mutagenesis of DENND2C Causes Morphological Changes and Resistance to RA
(A) (Top) Screenshot from the UCSC Genome Browser detailing the position and orientation of the PB[IM,N] insertion in DENND2C mapped in clone 2 (red arrow). (Bottom right) PB[IM,N] insertion results in overexpression of the C-terminal half of DENND2C, made up primarily of its DENN domain. (Bottom left) The C terminus of DENND2C is significantly upregulated in clone 2 by real-time PCR. Graph shows mean of triplicate technical replicates, and error bars represent SD. ∗∗p < 0.001 in Student’s t test.
(B) Brightfield and OCT4-EGFP reporter images of clone 2 and DENND2C expressing human ES cells. Clone 2 aggregates into either balls of cells or flat, extremely dense colonies that resist spontaneous differentiation. Cells stably transfected with full-length (FL) or truncated (TR) DENND2C also display this behavior. Scale bar represents 50 μm.
(C) RT-PCR showing maintenance of stemness in DENND2C expressing human ES cells, which do not display markers of differentiation.
(D) Western blot demonstrating that DENND2C overexpressing cells produce NANOG and OCT4 proteins at levels comparable to the parental WT and NANOG overexpressing cell lines. Overexpressed DENND2C-FL runs as a doublet, while only DENND2C-TR lysate exhibits a band running at the predicted 50 kDa of truncated DENND2C.
(E) Alkaline phosphatase-stained plates after RA/G418 treatment. Experiments were performed in triplicate. A representative plate for each set of experiments is shown.
See Figure S1 for additional information.
These cells maintain the expression of stemness markers OCT4, NANOG, and alkaline phosphatase and are able to form well-differentiated teratomas in mice, indicating that they are undifferentiated (Figures 3C, 3D, and S1A). DENND2C expressing cells are resistant to spontaneous differentiation as well as several passages in RA-treated media; however, resistance is not observed with other differentiating agents such as 12-O-Tetradecanoylphorbol-13-acetate (TPA). Furthermore, the effects of DENND2C on colony morphology were reversible in a doxycycline-inducible system (data not shown).
By comparing cells overexpressing either DENND2C alone or DENND2C expressed in the PB[NANOG] background, we discovered that DENND2C alone is responsible for the morphological properties of clone 2, while cooperation of DENND2C and NANOG is responsible for blocking RA-induced differentiation (Figures 3B and 3E). This genetic cooperation is also observed in induced pluripotent stem cells co-expressing DENND2C and NANOG (Figure S1B).
DENND2C Functions as a Negative Regulator of RHOA and RAC1
DENN domain proteins have been shown to function as RAB GEFs (GDP-GTP exchange factors) as well as interact with non-RAB proteins (Allaire et al., 2010; Yoshimura et al., 2010; reviewed in Marat et al., 2011). The DENND2 subfamily consists of four members, A through D. While all four DENND2 subfamily members could behave as RAB9 GEFs by in vitro GDP release activity, the in vivo functions for DENND2B, DENND2C, and DENND2D are unknown, as only DENND2A knockdown phenocopies the lysosomal defect caused by RAB9 inactivation in HeLa cells (Yoshimura et al., 2010). Consistent with the previous report, we detected a lysosomal defect for DENND2A, but not for DENND2C in knockdown experiments in human ES cells (Figure S1C). Furthermore, deregulation of RAB9A and B activities using dominant-negative and active mutants does not result in any of the ES cell phenotypes caused by DENND2C (Figure S1D). These data indicate that DENND2C does not function as a regulator for RAB9 in human ES cells.
DENND2 proteins are expressed within the nucleus and cytoplasm of both undifferentiated and differentiated human ES cells (Figure S1E). While DENND2C knockdown does not induce differentiation, it is possible that it shares redundant function with other DENND2 proteins. We were unable to induce sufficient simultaneous knockdown of DENND2 proteins to induce differentiation in human ES cells.
We thus further examined the colony morphology phenotype caused by DENND2C, as it could provide clues for underlying molecular mechanisms. DENND2C-expressing colonies have a strikingly disorganized cortical F-actin staining pattern, which differs from the organized cortical F-actin pattern observed in WT human ES cells (Figure 4A). In addition, these cells are smaller in cell and nuclear sizes, which is more dramatic toward the interior of colonies (Figure 4A). Despite these morphological changes, these cells remain actively proliferative with no changes in either DNA content or cell-cycle profile compared with WT human ES cells (Figure S1F).
Figure 4.
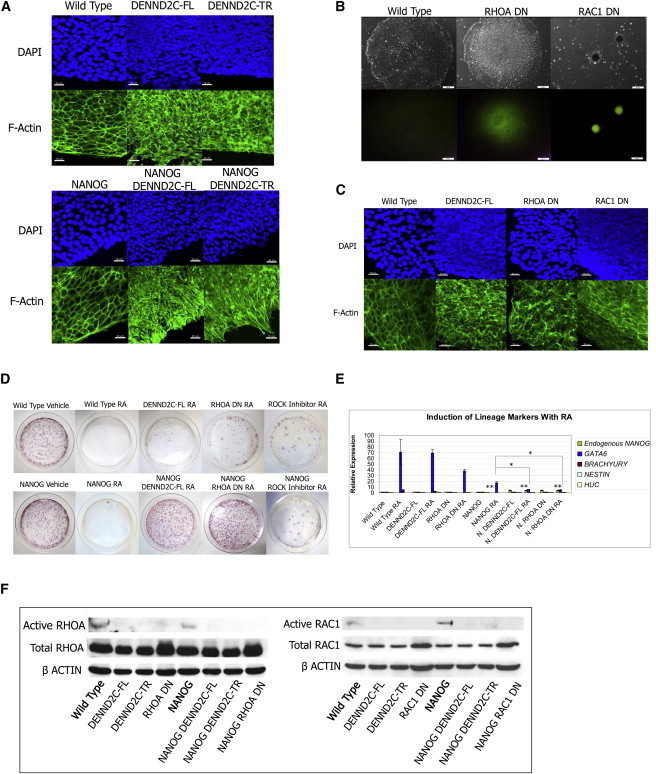
DENND2C Phenotype Is Induced by Loss of Active RHOA and RAC1
(A) DAPI and F-actin immunostaining of WT and DENND2C expressing human ES cells. Scale bar represents 30 μm.
(B) Brightfield and OCT4-EGFP reporter images of cells stably expressing dominant-negative RHOA T19N (RHOA DN) and dominant-negative RAC1 T17N (RAC1 DN). Scale bar represents 50 μm.
(C) DAPI and F-actin immunostaining of WT and RHOA DN and RAC1 DN mutant human ES cells. Scale bar represents 30 μm.
(D) Plates stained for alkaline phosphatase after RA/G418 treatment. Experiments were performed in triplicate. A representative plate for each set of experiments is shown.
(E) The induction of lineage markers with RA assessed by real-time PCR. RA treatment induces differentiation as reflected by the induction of the endoderm marker GATA6. NANOG expression from PB induces some repression of GATA6 and no upregulation of endogenous NANOG. NANOG/DENND2C and NANOG/RHOA DN cells display enhanced repression of GATA6 and upregulation of endogenous NANOG at the RNA level. Graph shows mean of triplicate independent experiments. Error bars represent SD. ∗∗p < 0.001, ∗p < 0.05 in Student’s t test.
(F) Immunoprecipitation of active RHOA and RAC1 in DENND2C expressing cells. DENND2C expressing cells have decreased active RHOA and RAC1 relative to WT cells.
See Figure S2 for additional information.
The striking cytoskeletal and morphological changes in clone 2 and DENND2C-expressing cells suggested that a cytoskeletal regulator could be the potential effector of DENND2C. Thus, we tested components of several pathways regulating the cytoskeleton, cell size, and cell adhesion by stably transfecting PB constructs containing WT, dominant-negative, and constitutively active forms of these genes into WT human ES cells. These candidate genes were examined for morphologies similar to DENND2C expressing cells as well as the ability to block RA-induced differentiation when transfected into the PB[NANOG] cell line. We discovered that reduction of RHOA and RAC1 activities by the expression of dominant-negative alleles, knockdown, or direct chemical inhibition could recapitulate the DENND2C-expressing phenotypes. Downregulation of either RHOA or RAC1 causes the cells to pile into dense, multilayer colonies (Figures 4B and S2A–S2F). However, only downregulation of RAC1 results in cells aggregating into balls (Figures 4B and S2A). Expression of dominant-negative RAC1 (T17N, RAC1 DN) results in the small cell and nuclei phenotypes observed in DENND2C-expressing cells (Figure 4C). On the other hand, expression of dominant-negative RHOA (T19N, RHOA DN) results in altered F-actin morphology phenotypes (Figure 4C). Expression of dominant-negative RHOB or RHOC (T19N) does not elicit these morphological changes, instead triggering differentiation (data not shown), indicating that the morphological changes are specific to downregulation of RHOA activity. Finally, similar to clone 2 and DENND2C-expressing clones, cells with reduction of either RHOA or RAC1 activity maintain the expression of stemness markers (Figure S2G).
Although we were unable to test the effects of RAC1 disruption in blocking RA due to its suppression of proliferation, disruption of RHOA exhibits genetic cooperation with NANOG expression in a manner similar to DENND2C to block RA-induced differentiation (Figures 4D and S2D). This genetic cooperation enhances NANOG-induced repression of the endoderm differentiation trigger GATA6 (Figure 4E; Capo-Chichi et al., 2005; Cai et al., 2008).
As it has been shown that chemical inhibition of the RHOA effector ROCK with the drug Y-27632 (ROCK inhibitor) results in resistance to RA, we wondered whether ROCK inhibitor treatment of NANOG expressing cells could block RA-induced differentiation (Krawetz et al., 2011). However, unlike Rho inhibitor treatment, ROCK inhibitor treatment does not cooperate with NANOG to block RA-induced differentiation (Figures 4D and S2D). Given that NANOG overexpression also results in the overexpression of its target OCT4, we further wondered whether OCT4 overexpression could cooperate with RHOA disruption to block RA-induced differentiation. However, this was not the case (Figure S2H). These results indicate that our approach of NANOG sensitized PB mutagenesis is useful in identifying relevant genetic interactions with NANOG capable of blocking RA-induced differentiation.
Inactivation of RHOA and RAC1 recapitulates DENND2C-expressing phenotypes and suggests that DENND2C could be a negative regulator of RHOA and RAC1. We next examined the possibility that DENND2C physically interacts with RHOA and RAC1; however, we did not detect such interactions by co-immunoprecipitation (data not shown). We thus directly examined the activities of RHOA and RAC1 in human ES cells expressing either full-length or truncated DENND2C by immunoprecipitation of the active forms of these GTPases. Indeed, in both cases, overexpression of DENND2C results in decreased levels of active RHOA and RAC1 (Figure 4F).
DENND2C Affects Nuclear RHOA Function
Given that DENND2C negatively regulates RHOA activity, we examined whether DENND2C expression affected RHOA localization. Interestingly, while WT cells displayed RHOA staining throughout the cell, in DENND2C expressing cells, RHOA was relocalized away from the interior of the nucleus (Figure 5A). Although RHOA plays an essential role in the regulation of cytoskeleton dynamics, many of the known functions of RHOA relate to its functions in the cytoplasm (reviewed in Burridge and Doughman, 2006). RHOA has been recently documented to reside in the nucleus (Dubash et al., 2011). In fact, actin and many actin nucleators have also been found in the nucleus, implicating functional significance for the nuclear cytoskeleton (reviewed in de Lanerolle, 2012; Weston et al., 2012; Hofmann, 2009). While it has been shown that nuclear RHOA is activated in response to DNA damage, its function within the nucleus remains unknown (Dubash et al., 2011).
Figure 5.
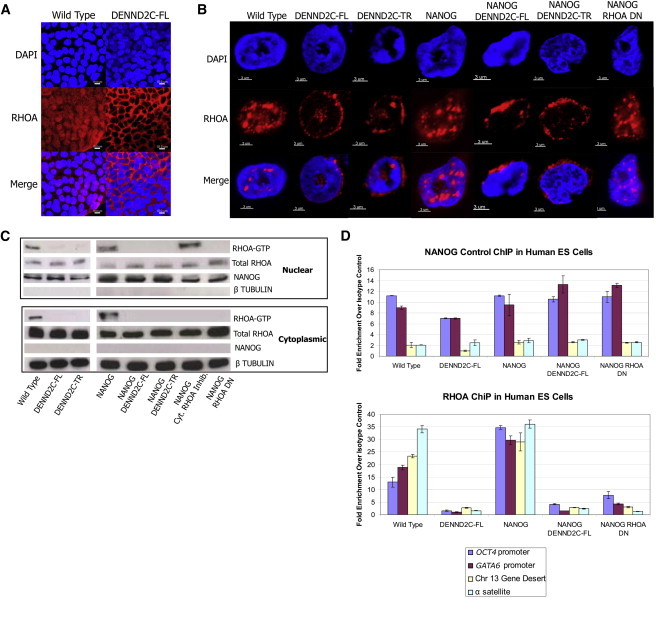
DENND2C Negatively Regulates Nuclear RHOA Activities
(A) DAPI and RHOA immunostaining of WT and DENND2C expressing ES cell colonies. Scale bar represents 10 μm.
(B) DAPI and RHOA immunostaining of isolated WT and DENND2C expressing nuclei. Scale bar represents 3 μm.
(C) Immunoprecipitation of active RHOA from nuclear and cytoplasmic fractions of WT and mutant cells. DENND2C and RHOA DN cells have reduced active RHOA in both nuclear and cytoplasmic fractions. Cytoplasmic RHOA inhibitor (RAB35) is included as a negative control.
(D) ChIPs performed in human ES cells with a RHOA monoclonal antibody and NANOG-positive control antibody. Presented is the average fold enrichment over IgG control of the OCT4 promoter, GATA6 promoter, chromosome 13 gene desert, and α satellite DNA. Error bars represent the mean percent fold enrichment over IgG control ± the SEM for triplicate technical replicates.
See Figure S3 for additional information.
We therefore examined whether DENND2C affected the localization and activity of nuclear RHOA. By immunostaining human ES cell nuclei isolated by density gradient centrifugation, it was evident that WT and NANOG overexpressing cells contained RHOA throughout the nucleus (Figure 5B). In contrast, RHOA in DENND2C overexpressing nuclei is relocalized to the nuclear periphery (Figure 5B). We next examined the activity of RHOA in nuclear and cytoplasmic fractions of WT and DENND2C expressing human ES cells (Guilluy et al., 2011). In addition to reduced cytoplasmic RHOA activation, DENND2C expressing cells have significant reduction of nuclear RHOA activation (Figure 5C). We observed similar reduction of active nuclear RHOA in the RHOA DN-expressing positive control (Figure 5C).
As a negative control, we expressed RAB35 in human ES cells, which has been implicated in the cytoplasmic transport and regulation of Rho GTPases (Chevallier et al., 2009; Shim et al., 2010). Indeed, expression of RAB35 inhibits cytoplasmic RHOA activity, but not nuclear RHOA activity, nor does RAB35 expression result in cooperation with NANOG in blocking RA-induced differentiation (Figures 5C and S3A–S3E). Furthermore, the expression of RHOA DN tagged with a nuclear exclusion signal does not cooperate with NANOG to block differentiation (Figure S3F). Together, our data suggest that DENND2C downregulates both nuclear and cytoplasmic RHOA activity, which cooperates with NANOG to block RA-induced differentiation. Indeed, nuclear RHOA is hyperactivated during RA-induced differentiation of WT human ES cells, supporting this hypothesis (Figure S3G).
Given that DENND2C negatively regulates nuclear RHOA, we wondered what function RHOA had within the nucleus. Other cytoskeletal effectors are known to associate with DNA either globally or with certain promoters when activated, and furthermore, nuclear RHOA is activated in response to DNA damage (Mazumdar et al., 2011; Buongiorno et al., 2008; Dubash et al., 2011). We therefore hypothesized that nuclear RHOA may associate with DNA. Using chromatin immunoprecipitation (ChIP) with an anti-RHOA monoclonal antibody that works well for immunoprecipitation (Figure S3H), we were able to pull down large amounts of DNA in undifferentiated WT and NANOG expressing human ES cells (Figure 5D). In contrast, the amount of DNA associated with RHOA in DENND2C overexpressing human ES cells is dramatically reduced (Figure 5D). The RHOA-precipitated DNA includes the OCT4 and GATA6 promoters, DNA from a gene desert located on chromosome 13, as well as centromeric α satellite DNA, indicating that RHOA does not exclusively associate with transcription units (Figure 5D). RHOA association with DNA was also confirmed in other human ES cell lines (H9 and parental H1; Figure S3I). Overall, these data show that nuclear RHOA associates with chromatin in ES cells and that this ability is affected upon downregulation of nuclear RHOA activity by DENND2C.
Discussion
To date, only one genome-wide genetic screen has been performed in human ES cells (Chia et al., 2010). The high cost associated with an siRNA library can deter investigators from utilizing this approach to decipher human stem cell biology. In this study, we show that the piggyBac transposon can be used to conduct genetic screens in human ES cells. The ability to rapidly and cost-effectively generate a large collection of cells in which each cell has different genes mutated by simple transfection enables one to perform phenotypic-based genetic screens in human cells in a manner similar to yeast genetics. Combined with high-throughput sequencing, the genes disrupted by the piggyBac transposon can be easily identified, allowing one to study the molecular mechanisms underlying any biological process.
The unique ability of the piggyBac transposon to carry large inserts offers the opportunity to conduct sophisticated insertional mutagenesis screens (Ding et al., 2005; Li et al., 2011). Here we were able to combine mutagenesis and simultaneous overexpression of a gene into a single transposon vector, performing a genetic screen in a sensitized background. Different from proteomic approaches, this sensitized screening strategy has the power to detect genes that cooperate in a biological process, e.g., blocking differentiation, regardless whether gene products physically interact with each other. With this strategy, we are able to probe the genome for genes capable of cooperating with NANOG, and by characterizing one of the clones, we were able to identify a poorly characterized gene, DENND2C, which is capable of cooperation with NANOG overexpression to block RA-induced differentiation. Although our piggyBac screen utilizes gain-of-function mutagenesis, such mutations can easily lead to the identification of loss-of-function alterations in genes in the same pathway or process, as every pathway has both positive and negative regulatory components. Indeed, we discovered that DENND2C is a negative regulator of RHOA. Concordantly, RHOA inactivation also cooperates with NANOG in blocking RA-induced differentiation. Although DENND2C was artificially overexpressed, we were able to phenocopy its effects with several different methods of RHOA inactivation, demonstrating that gain-of-function screens can be used to elucidate real biological mechanisms. Notably, both RHOA and DENND2C are expressed in human ES cells as well as differentiated cells and would have been missed as potential regulators of self-renewal and differentiation by more traditional approaches that focused on probing genes expressed uniquely in ES cells. Much of our understanding of RHOA comes from studies for its role in the cytoplasm as a key regulator of cytoskeleton dynamics (reviewed in Burridge and Doughman, 2006). Recently, RHOA has been found to be located within the nucleus, and its nuclear activity is stimulated by DNA damage (Dubash et al., 2011). We found that DENND2C negatively regulates the activity of nuclear RHOA in human ES cells. Furthermore, we discovered that nuclear RHOA is associated with DNA in both coding and non-coding regions and that this association is negatively regulated by DENND2C.
In summary, forward genetic analysis by piggyBac gain of function mutagenesis has multiple advantages complimenting genomic and biochemical methods and empowers individual investigators to functionally interrogate the human ES cell genome to elucidate mechanisms underlying biology, disease, and therapeutic strategy.
Experimental Procedures
Cell Culture
The H1 OCT4-EGFP human ES cell line was created in the James Thomson laboratory and obtained from the WiCell Research Institute under the appropriate material transfer agreement (MTA). Cells were maintained on Matrigel (BD Biosciences catalog number 354277) in a 37°C incubator at 5% CO2. Cells were passaged with manual dissociation with a 5-ml pipette after collagenase IV treatment (GIBCO catalog number 17104019) and maintained as originally described (Thomson et al., 1998). Additional information is in Supplemental Experimental Procedures.
Transfections and Drug Selection
For the screen, 9.6 × 107 human ES cells were pretreated with ROCK inhibitor as described (EMD/Calbiochem catalog number 688000; Watanabe et al., 2007) for 3 hr, dissociated with Accutase (Millipore catalog number SCR005), and split in single-cell suspension onto Matrigel coated six-well plates. Each well was treated with a cocktail consisting of 3-μg piggyBac, 1-μg PB transposase helper plasmid, 6-μl Fugene HD (Roche catalog number 04883560001), and 90-μl OptiMEM (Invitrogen catalog number 11058-021) for 48 hr. Cells containing stable piggyBac integration were selected with 1-μg/ml puromycin (Sigma catalog number P8833) starting 48 hr after transfection for a period of 2 weeks.
For transfections of all other piggyBac constructs, adherent human ES cells 2 days after passaging were treated with the same transfection cocktail and drug selection protocol.
RA Treatment
Human ES cells at approximately 30% confluency in six-well plates were treated with a combination of 1-μM all-trans RA (Sigma catalog number R2625) and 0.2-mg/ml G418 (Sigma catalog number A1720) for 8 days. Cells were then split into 10-cm plates at a ratio of two wells to one 10-cm plate. Cells were kept on G418 for another 8 days. For the screen, surviving undifferentiated colonies were individually isolated from the 10-cm plates, expanded, and retested for RA resistance. Of an initial 50 colonies isolated, only six unique clones survived this retest. For assaying resistance to differentiation, 10-cm plates were stained for the stemness marker alkaline phosphatase (Millipore catalog number SCR004).
Mapping of Insertions
Mapping was performed with a combination of linker-mediated PCR as described in Landrette et al. (2011), splinkerette PCR as described in Potter and Luo (2010) and Horn et al. (2007), and vectorette PCR as described by Carl Friddle (available at http://www.princeton.edu/genomics/botstein/protocols/vectorette.html). PCR products created with Taq polymerase (Denville catalog number CB4050) were TOPO-TA cloned (Invitrogen catalog number 450641), and competent cells were transformed with the resulting plasmid DNA. Individual bacterial colonies carrying an insert were picked, expanded, and DNA isolated and sequenced through the Yale Keck Sequencing Facility. True insertions were separated from PCR artifacts by the identification of PBL next to the TTAA transposition site within the sequence. Sequences identified by PCR were used to identify the location and orientation of the transposon within the genome through human BLAT search (http://genome.ucsc.edu/). Additional information is in Supplemental Experimental Procedures.
Reverse Transcription and PCR
Equal amounts of total RNA was reverse transcribed using Superscript III and oligo(dT)20 (Invitrogen catalog number 18080-051). On column DNase treatment was performed as described in the instruction manual. Semiquantitative PCR was carried out in accordance with the protocol used by Darr et al. (2006). Quantitative real-time PCR was carried out on the cDNA using SYBR Green Supermix (Bio-Rad catalog number 172-5100). Samples were normalized to GAPDH amplification. Primer sequences available in Supplemental Experimental Procedures.
Active GTPase Immunoprecipitation
Immunoprecipitation of total active RHOA from 350-μg whole-cell lysate was performed with the NewEast Biosciences RhoA Activation Assay (catalog number 80601). Immunoprecipitation of active RHOA from 350-μg fractionated nuclear and cytoplasmic lysates isolated as described in Guilluy et al. (2011) was performed with the Cell Biolabs RhoA Activation Assay (catalog number STA-403-A). Immunoprecipitation of active RAC1 from 500-μg whole-cell lysate was performed with the Cell Biolabs Rac1 Activation Assay (catalog number STA-401-1).
Nuclear Isolation
The protocol is as described in Guilluy et al. (2011).
Overexpression and Mutant Constructs
All genes overexpressed in human ES cells were amplified from either human ES cell or 293T cDNA created with the Superscript III kit (Invitrogen catalog number 18080-051) with the exceptions of NANOG, which was amplified from Addgene plasmid 16578, a gift from Dr. James Thomson, and RHOA, which was amplified off Addgene plasmid 23224, a gift from Dr. Channing Der. All cDNA was amplified with MluI restriction sites and cloned into a piggyBac construct designed to stably overexpress the cloned gene with a β actin promoter with the sole exception of NANOG, which was overexpressed with the ubiquitin C promoter in all PB constructs used in this study. Dominant-negative and constitutively active constructs were created by introducing the mutations on primers and subsequent subcloning of PCR products. For assessment of cooperation with NANOG, all constructs were transfected into the same parental NANOG overexpressing cell line. Primer sequences are available in Supplemental Experimental Procedures.
ChIP
ChIP was performed using reagents from the Pierce Agarose ChIP kit (Pierce catalog number 26156), MagnaChIP Protein AG Beads (Millipore catalog number 16-663), and 6 × 106 cells (approximately two to three 10-cm plates of human ES cells) per immunoprecipitation. For each chip, we used either 2-μg NANOG antibody or rabbit IgG control (Cell Signaling catalog numbers 5232S and 2729S) or 5-μg RHOA antibody (Abcam catalog number ab54835) or mouse IgG control (Cell Signaling catalog number 5415S). Fresh chromatin and fresh antibody were incubated with 20-μl magnetic beads overnight at 4°C. Eluted chromatin was treated with RNase for 30 min prior to decrosslinking and column purification. Real-time PCR was performed in triplicate on precipitated DNA with OCT4 promoter primers (Cell Signaling catalog number 4641S), GATA6 promoter primers, gene desert primers (Supplemental Experimental Procedures), and α satellite repeat primers (Cell Signaling catalog number 4486S). For fold enrichment calculations, the average percentage of input DNA pulled down with NANOG, or RHOA antibody was divided by the average percentage of input DNA pulled down with the relevant IgG isotype control.
Acknowledgments
We thank Drs. D. Krause, J. Lu, V. Horsley, and Y. Chabu for reading the manuscript and suggestions. This study was supported in part by funding from Connecticut Stem Cell Funds and is taken from a dissertation submitted to fulfill in part the requirements for the degree of Doctor of Philosophy, Yale University. S.G. is a predoctoral student supported by National Research Service Award T32-HG003198 from the NIH, and T.X. is an HHMI investigator.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information
References
- Allaire P.D., Marat A.L., Dall’Armi C., Di Paolo G., McPherson P.S., Ritter B. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell. 2010;37:370–382. doi: 10.1016/j.molcel.2009.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buongiorno P., Pethe V.V., Charames G.S., Esufali S., Bapat B. Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wnt-responsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Mol. Cancer. 2008;7:73. doi: 10.1186/1476-4598-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., Doughman R. Front and back by Rho and Rac. Nat. Cell Biol. 2006;8:781–782. doi: 10.1038/ncb0806-781. [DOI] [PubMed] [Google Scholar]
- Cai K.Q., Capo-Chichi C.D., Rula M.E., Yang D.H., Xu X.X. Dynamic GATA6 expression in primitive endoderm formation and maturation in early mouse embryogenesis. Dev. Dyn. 2008;237:2820–2829. doi: 10.1002/dvdy.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi C.D., Rula M.E., Smedberg J.L., Vanderveer L., Parmacek M.S., Morrisey E.E., Godwin A.K., Xu X.X. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev. Biol. 2005;286:574–586. doi: 10.1016/j.ydbio.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen Y.T., Furushima K., Hou P.S., Ku A.T., Deng J.M., Jang C.W., Fang H., Adams H.P., Kuo M.L., Ho H.N. PiggyBac transposon-mediated, reversible gene transfer in human embryonic stem cells. Stem Cells Dev. 2010;19:763–771. doi: 10.1089/scd.2009.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier J., Koop C., Srivastava A., Petrie R.J., Lamarche-Vane N., Presley J.F. Rab35 regulates neurite outgrowth and cell shape. FEBS Lett. 2009;583:1096–1101. doi: 10.1016/j.febslet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.S. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Darr H., Mayshar Y., Benvenisty N. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 2006;133:1193–1201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- de Lanerolle P. Nuclear actin and myosins at a glance. J. Cell Sci. 2012;125:4945–4949. doi: 10.1242/jcs.099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., Han M., Zhuang Y., Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Dubash A.D., Guilluy C., Srougi M.C., Boulter E., Burridge K., García-Mata R. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PLoS ONE. 2011;6:e17380. doi: 10.1371/journal.pone.0017380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C., Dubash A.D., García-Mata R. Analysis of RhoA and Rho GEF activity in whole cells and the cell nucleus. Nat. Protoc. 2011;6:2050–2060. doi: 10.1038/nprot.2011.411. [DOI] [PubMed] [Google Scholar]
- Hofmann W.A. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 2009;273:219–263. doi: 10.1016/S1937-6448(08)01806-6. [DOI] [PubMed] [Google Scholar]
- Horn C., Hansen J., Schnütgen F., Seisenberger C., Floss T., Irgang M., De-Zolt S., Wurst W., von Melchner H., Noppinger P.R. Splinkerette PCR for more efficient characterization of gene trap events. Nat. Genet. 2007;39:933–934. doi: 10.1038/ng0807-933. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz R.J., Taiani J., Greene A., Kelly G.M., Rancourt D.E. Inhibition of Rho kinase regulates specification of early differentiation events in P19 embryonal carcinoma stem cells. PLoS ONE. 2011;6:e26484. doi: 10.1371/journal.pone.0026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrette S.F., Cornett J.C., Ni T.K., Bosenberg M.W., Xu T. piggyBac transposon somatic mutagenesis with an activated reporter and tracker (PB-SMART) for genetic screens in mice. PLoS ONE. 2011;6:e26650. doi: 10.1371/journal.pone.0026650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.A., Turner D.J., Ning Z., Yusa K., Liang Q., Eckert S., Rad L., Fitzgerald T.W., Craig N.L., Bradley A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S.Y., Qin J., Wong J., Cooney A.J., Liu D., Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Mallanna S.K., Ormsbee B.D., Iacovino M., Gilmore J.M., Cox J.L., Kyba M., Washburn M.P., Rizzino A. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28:1715–1727. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat A.L., Dokainish H., McPherson P.S. DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M., Sung M.H., Misteli T. Chromatin maintenance by a molecular motor protein. Nucleus. 2011;2:591–600. doi: 10.4161/nucl.2.6.18044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Pan G., Thomson J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M.M., Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C.J., Luo L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS ONE. 2010;5:e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad R., Rad L., Wang W., Cadinanos J., Vassiliou G., Rice S., Campos L.S., Yusa K., Banerjee R., Li M.A. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J., Lee S.M., Lee M.S., Yoon J., Kweon H.S., Kim Y.J. Rab35 mediates transport of Cdc42 and Rac1 to the plasma membrane during phagocytosis. Mol. Cell. Biol. 2010;30:1421–1433. doi: 10.1128/MCB.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van den Berg D.L., Snoek T., Mullin N.P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R.A. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K., Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Weston L., Coutts A.S., La Thangue N.B. Actin nucleators in the nucleus: an emerging theme. J. Cell Sci. 2012;125:3519–3527. doi: 10.1242/jcs.099523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., Barr F.A. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Neganova I., Przyborski S., Yang C., Cooke M., Atkinson S.P., Anyfantis G., Fenyk S., Keith W.N., Hoare S.F. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J. Cell Biol. 2009;184:67–82. doi: 10.1083/jcb.200801009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka T.P., Thomson J.A. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


