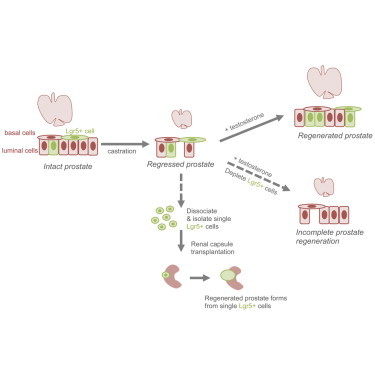
Summary
The adult prostate possesses a significant regenerative capacity that is of great interest for understanding adult stem cell biology. We demonstrate that leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) is expressed in a rare population of prostate epithelial progenitor cells, and a castration-resistant Lgr5+ population exists in regressed prostate tissue. Genetic lineage tracing revealed that Lgr5+ cells and their progeny are primarily luminal. Lgr5+ castration-resistant cells are long lived and upon regeneration, both luminal Lgr5+ cells and basal Lgr5+ cells expand. Moreover, single Lgr5+ cells can generate multilineage prostatic structures in renal transplantation assays. Additionally, Lgr5+ cell depletion revealed that the regenerative potential of the castrated adult prostate depends on Lgr5+ cells. Together, these data reveal insights into the cellular hierarchy of castration-resistant Lgr5+ cells, indicate a requirement for Lgr5+ cells during prostatic regeneration, and identify an Lgr5+ adult stem cell population in the prostate.
Graphical Abstract
Highlights
-
•
Castration-resistant Lgr5+ basal and luminal cells exist in regressed prostate
-
•
Lgr5+ castration-resistant cells are long lived
-
•
During regeneration, Lgr5+ basal and luminal cells expand
-
•
Prostate regeneration requires Lgr5+ cells
In this article, Junttila and colleagues show that a castration-resistant Lgr5+ cell population exists in the adult prostate. Luminal and basal Lgr5+ castration-resistant cells are found, and both expand during the regenerative process. Moreover, castration-resistant Lgr5+ cells are a long-lived adult stem cell population in the prostate that are required for complete tissue regeneration.
Introduction
The secretory prostate organ requires androgen signaling both for development and for proper maintenance of structure and function. Although the adult homeostatic prostate is a predominantly quiescent organ, it harbors a tremendous regenerative capacity that allows it to undergo multiple rounds of androgen-mediated involution and regeneration. Understanding this profound capacity for adult prostate tissue renewal is of great interest for elucidating stem cell biology and may also provide insight into disease.
Unlike the human prostate, which is organized into a single lobe with varying zonal layers, the murine prostate has four distinct lobes surrounding the urethra. The adult prostate contains three epithelial lineages, i.e., basal, luminal, and neuroendocrine cells (Karthaus et al., 2014; Marker et al., 2003; Wang et al., 2014b). Luminal cells are columnar androgen-receptor-positive cells that are identified by the cytokeratin markers CK8 and CK18. Basal cells reside adjacent to the luminal cells and express the markers CK5, P63, and CK14. Upon androgen deprivation via castration, 90% of luminal cells undergo apoptosis; however, the luminal cells can be rapidly regenerated following readministration of testosterone, suggesting the existence of stem cells and their important role in prostate regeneration (Choi et al., 2012; Goldstein et al., 2008; Tsujimura et al., 2002). Many hypotheses have been suggested to explain the regenerative process of the adult prostate. Previous work has established that stem cells within the basal compartment divide asymmetrically, generating one stem cell and one committed progenitor cell that differentiates to ultimately yield a luminal or neuroendocrine cell during prostate postnatal development (Ousset et al., 2012; Pignon et al., 2013). However, some studies have described multipotency for both luminal (Wang et al., 2009) and basal (Wang et al., 2014a) cells following castration-induced involution, while others have supported committed lineage-specified progenitors for luminal and basal cells during adult prostatic regeneration (Choi et al., 2012; Liu et al., 2011). In addition, an intermediate cell phenotype that is both CK5+ and CK18+ has been reported (Xue et al., 1998).
During development, prostate expansion is regulated by Wnt signaling (Isaacs and Coffey, 1989; Marker et al., 2003; Wang et al., 2008). The Wnt pathway is highly activated during early stages of prostate development, as well as during prostate regeneration. The Wnt-regulated gene Lgr5 has been shown to serve as a stem cell marker in several adult tissues, including the ovary, mammary gland, intestinal tract, and hair follicle (Barker et al., 2007; Jaks et al., 2008; Ng et al., 2014; Plaks et al., 2013; Rios et al., 2014).
Here, we investigated the possibility that Lgr5+ cells are a stem cell population within the adult prostate. Our results demonstrate that prostatic Lgr5+ cells comprise subpopulations within both the luminal and basal compartments. Using in vivo lineage tracing, we discovered that castration-resistant Lgr5+ cells are long-lived, multipotent progenitors. Moreover, depletion of Lgr5+ cells revealed that the regenerative potential of the castrated adult prostate requires the Lgr5+ population of cells. Together, these findings indicate that Lgr5 identifies an adult stem cell population in the prostate.
Results
Lgr5 Is Expressed in a Rare Subpopulation of Cells in the Adult Prostate
By Lgr5 in situ hybridization on intact adult prostate, we found that Lgr5 is expressed in a rare subpopulation of cells located in the anterior, dorsal, lateral, and ventral lobes. Lgr5+ cells were observable in both the basal and luminal compartments of all lobes, as well as in the stromal compartment of the ventral lobes (Figures 1A and 1B). To further determine the expression pattern of Lgr5 in the prostate, we pulse labeled Lgr5CreER2;Rosa26.LSL.tdTomato (LT) mice with tamoxifen for 24 hr (Figure 1C). Labeled cells were found in the dorsal, lateral, ventral, and anterior lobes, albeit infrequently, with no observable preference by lobe. The Lgr5+ (RFP+) cells expressed either basal or luminal markers (77.4% of the RFP+ cells were luminal cells and 22.6% were basal cells; n = 4 animals, 66 sections, and 97 RFP+ cells; Figures 1D–1F). Label-positive cells within the stroma were confined to the ventral lobes, consistent with our in situ hybridization data (data not shown). Immunostaining revealed that Lgr5+ cells did not colocalize with Ki67 (Figure 1G), providing evidence that these cells were not actively dividing in the adult quiescent prostate. Notably, we did not observe any Lgr5+ cells coexpressing synaptophysin, a neuroendocrine cell marker (data not shown). Lgr5+ cells were found in the basal and luminal compartments in all four pairs of lobes (dorsal, lateral, ventral, and anterior) and within the stroma of the ventral lobes of the mouse prostate.
Figure 1.
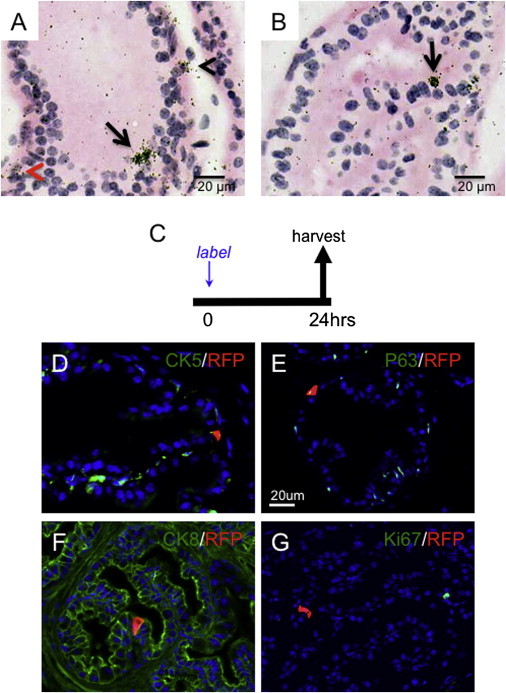
Lgr5 Is Expressed in the Intact Adult Mouse Prostate
(A and B) In situ hybridization of Lgr5 in adult mice (8–10 weeks old; n = 4) luminal (arrows), basal (black arrowhead), and stroma (red arrowhead) cells in ventral (A) and anterior (B) lobes.
(C) Experimental design.
(D–G) Immunofluorescent images of Lgr5+ cells pulse labeled with tamoxifen for 24 hr in Lgr5CreER;Rosa26.LSL.tdTomato (LT) mice, showing RFP positivity and CK5 (D), P63 (E), CK8 (F), and Ki67 (G) in anterior (D and G) and dorsolateral (E and F) lobes.
Lgr5+ Cells Are Castration Resistant
To determine the androgen dependency of Lgr5+ cells, we castrated adult LT mice, allowed them to regress for 4 weeks, and then pulse labeled them for 4 consecutive days prior to terminal tissue assessment (Figure 2A). We identified a castration-resistant subpopulation of Lgr5+ cells in the fully regressed prostate (Figures 2B–2E). Sporadic Lgr5+ cells comprised only 0.068% of total cells in the anterior lobes of the fully regressed mouse prostate (Figure 2B). Moreover, Lgr5 mRNA levels did not decrease in 6-week castrated, regressed prostates compared with intact prostates (Figure 2C). The rare castration-resistant Lgr5+ cells were in the basal and luminal epithelia in all lobes (Figures 2D and 2E). However, the labeling efficiency was most prominent in the anterior lobe, and poor in both the ventral and dorsal-lateral lobes. To directly compare the abundance of Lgr5+ cells in intact and regressed prostates, we utilized Lgr5DTR.EGFP mice, which express a knockin GFP fusion protein (Tian et al., 2011). Lin−/PI−/EpCAM+/GFP+ cell abundance was 0.023% ± 0.011% in intact prostates and 0.036% ± 0.001% in 6-week castrated prostates (Figures S1A and S1B), suggesting retention of Lgr5+ castration-resistant cells during regression. The majority (94%) of label-positive Lgr5+ cells were single cells after the first round of castration and regression, and further quantitation revealed that 75% of these cells were luminal and 25% were basal cells (Figure S2A) expressing the basal cell marker CK5. Taken together, these data demonstrate that a castration-resistant Lgr5+ population exists in the adult prostate.
Figure 2.
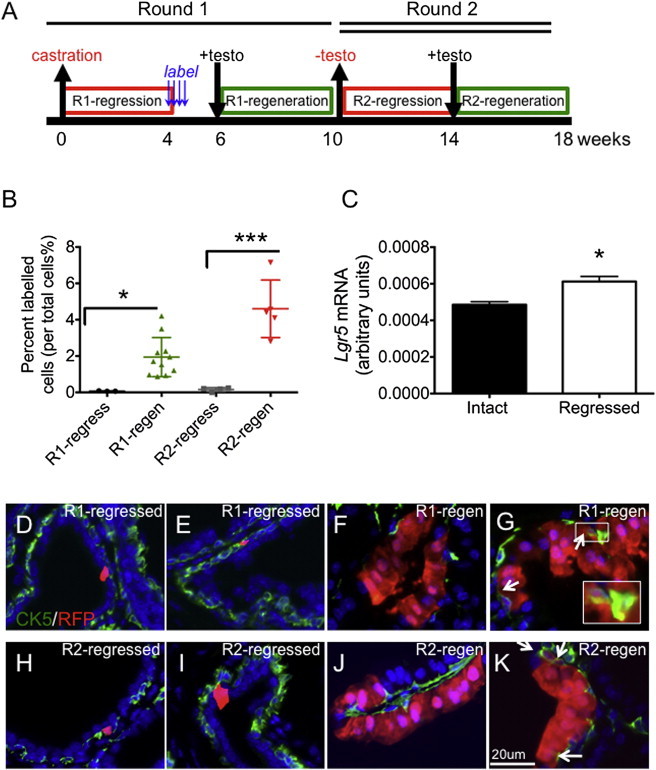
Lineage Tracing of Lgr5+ Cells in Two Rounds of Prostate Regression and Regeneration
(A) Experimental design of lineage-tracing studies.
(B) Percentage of label-positive cells per total cells at different stages of the two rounds of regression and regeneration: round 1 regressed, n = 3 mice, number of cells counted (cn) = 18,159; round 1 regenerated, n = 11 mice, cn = 31,375; round 2 regressed, n = 7 mice, cn = 11,756; round 2 regenerated, n = 5 mice, cn = 18,070.
(C) Quantification of relative Lgr5 expression in intact and castrated prostates; n = 4 mice per group.
(D and E) Immunofluorescent images of Lgr5+ (RFP+) cells (red) and CK5 (green) in anterior lobes of round 1 (R1) regressed prostates. Labeled cells were identified in the luminal (D) and basal (E) compartments.
(F and G) Immunofluorescent images of Lgr5+ (RFP+) progeny (red) and CK5 (green) in round 1 regenerated prostates. Labeled cells were in the luminal compartment only (F) or in both the luminal and basal compartments, arrows (G).
(H and I) Immunofluorescent images of Lgr5+ (RFP+) progeny (red) and CK5 (green) in anterior lobes of round 2 (R2) regressed prostates. Labeled cells were observed in both the basal (H) and luminal (I) compartments.
(J and K) Immunofluorescent images of Lgr5+ (RFP+) progeny (red) and CK5 (green) in anterior lobes of round 2 regenerated prostates. Labeled cells were exclusively in the luminal compartment (J) or in both the luminal and basal compartments, arrows (K).
All values are presented as mean and SEM. testo, testosterone; n, number of mice; cn, number of cells counted. ∗p < 0.05, ∗∗∗p < 0.001. See also Figure S1.
Prostatic Lgr5+ Cells Are Long-Lived Stem Cells
To assess the potential of the castration-resistant population of Lgr5+ cells to generate progeny during prostate regeneration, we performed in vivo lineage tracing from Lgr5+ cells. Tamoxifen-mediated labeling in fully regressed prostates and subsequent testosterone-mediated prostate regeneration revealed ribbons (defined as three or more cells) of labeled cells (Figures 2F and 2G). Quantification of labeled cells demonstrated a 28-fold expansion following the first round of regeneration (0.068% in first-round fully regressed prostates to 1.942% after the first round of regeneration; Figure 2B). Following a second round of regression (Figures 2H and 2I), the labeled castration-resistant cells were frequently clustered as doublets, and twice as many labeled cells were identified compared with the first round of regression (round 1: 0.068%; round 2: 0.156%; Figure 2B). When the cells were allowed to regenerate a second time (Figures 2J and 2K), a 29-fold expansion of labeled cells between fully regressed and regenerated prostates was observed (0.156% in round 2 regressed to 4.60% in round 2 regenerated prostates; Figure 2B). These findings show that Lgr5+ cells and their progeny participate in prostatic expansion during prostate regeneration, and that Lgr5+ cells not only survived >19 weeks of regression and regeneration but also maintained a comparable regenerative capacity during the two rounds of regeneration. This indicates that prostatic Lgr5+ cells are long lived and are capable of self-renewal.
Lgr5+ Luminal Cells Are Unipotent and Basal Cells Are Bipotent
Quantification of the label-positive cells revealed an average of one cell after the first round of regression and two cells after the second round of regression (Figure 3A). In the regenerated state, the average number of label-positive cells per unit increased to 6 and 10 cells per unit for first and second rounds of regeneration, respectively (Figure 3A). The labeled units were comprised of basal and/or luminal cells, and the numbers of these cells did not decrease on a per-unit basis during the two rounds of regression and regeneration (Figures 3B and 3C). In fact, the labeled luminal population doubled in the regressed setting between the first and the second round of regression, suggesting that a subset of Lgr5+ cell progeny may also be castration resistant (Figure 3B). Further analysis of the label-positive cells from the first round of regression and regeneration revealed that 70% of the labeled units were of the luminal cell type only (Figure 3D). In contrast, the average number of basal cells per unit was less than one, and no ribbons (defined as three or more cells) of purely basal cells were observed in either regressed or regenerated prostate (Figure 3C). These findings indicate that the majority of labeled basal Lgr5+ cells do not give rise exclusively to basal cell progeny during the first round of prostate regeneration, which is consistent with limited loss of basal cells and extensive loss of luminal cells upon androgen deprivation. Overall, the data show that cell expansion from Lgr5+ cells occurs mostly in the luminal cell compartment during regeneration.
Figure 3.
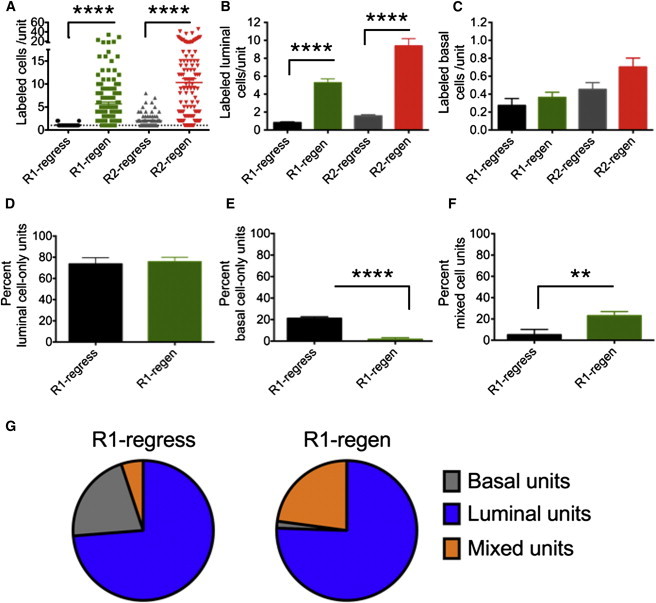
Unit Analysis of Lgr5+ Progeny Reveals Uni- and Bipotent Properties
(A) Quantitation of labeled cells at the indicated stages of regression and regeneration per unit.
(B and C) Quantitation of unit composition as the number of labeled luminal (B) or basal (C) cells per unit.
(D and E) Unit characterization as the percentage of luminal-only (D), basal-only (E), or mixed (F) units.
(G) Pie graph representation of unit content at the indicated stages of prostate regression and regeneration, showing the number of animals per stage (n) and total number of units counted (uc): R1 regressed, n = 3, uc = 33; R1 regenerated, n = 11, uc = 150; R2 regressed, n = 7, uc = 84; R2 regenerated, n = 5, uc = 121. Units counted per mouse = 9–43.
All values are represented as mean and SEM; n.s., not significant; ∗∗p < 0.01, ∗∗∗∗p < 0.0001. See also Figure S2.
Since we found Lgr5+ cells within both the basal and luminal compartments upon prostate regression, we sought to elucidate whether they were giving rise to progeny in a uni- or bipotent manner. Label-positive units were found to be composed of luminal cells only, basal cells only, or mixed units containing both luminal and basal cells. Seventy-four percent of the units contained only luminal cells, and the percentage of luminal-only units did not change significantly during regeneration (Figure 3D), indicating that the majority of luminal Lgr5+ cells at castrated stages were unipotent. However, the percentage of basal-cell-only units dropped following the first round of regeneration (round 1: 21.2% in the regressed prostate versus 1.6% in the regenerated prostate; Figure 3E). In contrast, the percentage of mixed units increased following regeneration (round 1: 5.1% in the regressed prostate versus 23% in the regenerated prostate; Figures 3F and 3G). Notably, in the second round of regression and regeneration, we observed a similar trend, i.e., the percentage of luminal-only units did not change. However, the basal-cell-only units significantly decreased (round 2: 12.6% in the regressed prostate versus 0% in the regenerated prostate) and the mixed units significantly increased (round 2: 17.2% in the regressed prostate versus 36.1% in the regenerated prostate) during the second round of regeneration (Figures S2B–S2D). The presence of mixed units (Figures 2G, 2K, S2D, and S2E) and the shift from basal-only units to mixed units during each involution-regeneration cycle suggest that most of the Lgr5+ basal cells were bipotent and giving rise to luminal cells. Moreover, these data suggest that indeed a subset of the Lgr5+-derived luminal progeny is castration resistant.
Single Lgr5+ Cells Can Generate Prostatic Cell Lineages
To further assess the capacity of Lgr5+ cells to repopulate the prostate, we used Lgr5DTR.EGFP mice, which express a knockin GFP fusion protein, to isolate single, viable Lin−/EpCAM+/GFP+ cells from castrated, regressed mouse prostates (Figure S3A). Single fluorescence-activated cell sorting (FACS)-sorted cells were recombined with rat urogenital mesenchyme (UGM) cells and implanted into renal capsules in immunodeficient male mice (Figure 4A). Seventy-five percent of Lgr5+ single-cell implantations generated large and translucent grafts after 3 months (Figures 4B and 4C). Histological analysis of the grafts showed that they had a branching morphology and expressed luminal cell, basal cell, and neuroendocrine markers (Figures 4D, 4E, and S3B), indicating that single Lgr5+ cells can generate prostatic structures containing all three epithelial cell lineages. We conclude that Lgr5 labels a subset of stem cells in the regressed prostate with the capacity to generate multilineage prostatic structures from a single cell in renal capsule transplantation experiments.
Figure 4.
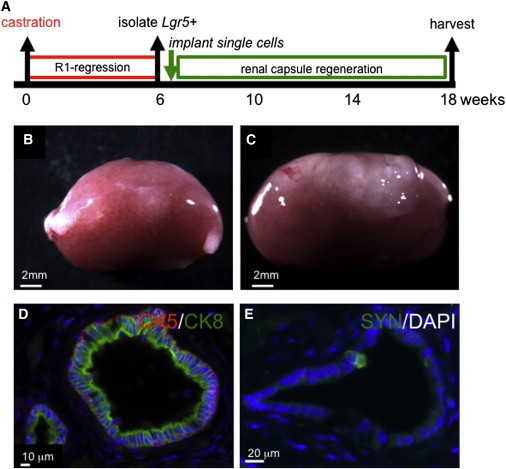
Single Lgr5+ Cells from Regressed Prostates Harbor Stem Cell Potential
(A) Experimental design.
(B and C) Six weeks after surgical castration, single Lin−/PI−/EpCAM+/GFP+ cells were isolated and implanted into the renal capsule of athymic nu/nu male mice together with rat UGM cells for 3 months, and the prostate regeneration capacity of UGM-only control (B) and the single Lin−/PI−/EpCAM+/GFP+ cell implant (C) was assessed. Successful grafts demonstrating both CK5 and CK8 immunofluorescence positivity were 0/18 transplanting UGM-only and 3/4 transplanting Lin−/PI−/EpCAM+/GFP+ cells; ∗∗∗∗p < 0.0001.
(D and E) Successful engraftment and expansion was confirmed by immunofluorescence staining for CK5 (red), CK8 (green) (D), and synaptophysin (Syn, in green) (E).
See also Figure S3.
Lgr5+ Cells Are Necessary for Prostate Regeneration
We next wanted to ascertain whether the castration-resistant Lgr5+ cells were required for prostate regeneration. To this end, we took advantage of the Lgr5-targeted knockin animal, which expresses human diphtheria toxin receptor (DTR) from the endogenous Lgr5 locus (Lgr5DTR.EGFP). Through systemic administration of diphtheria toxin (DT), we selectively culled Lgr5+ cells. Animals with fully regressed prostates were then challenged to regenerate in the absence of Lgr5+ cells by concurrent administration of DT and testosterone for 9 days (Figures 5A and S4A). Compared with wild-type littermates, the DT-treated Lgr5DTR.EGFP animals had significantly smaller prostates (Figure 5B). To confirm that the heterozygous state of Lgr5 in Lgr5DTR.EGFP mice did not influence prostate regeneration, we treated Lgr5DTR.EGFP mice with vehicle (PBS) concurrently with testosterone, and in contrast to the DT-treated Lgr5DTR.EGFP mice, the PBS-treated Lgr5DTR.EGFP mice showed normal prostate regeneration (Figures 5B and S4B). A closer examination of the epithelial composition showed that the ratio of CK18+ luminal cells to CK5+ basal cells in the anterior lobe was significantly lower in the DT-treated cohort (DT-treated: CK18/CK5, 0.49; PBS-treated: CK18/CK5, 1.82; Figures 5C and S4C). This suggests that Lgr5+ cell depletion leads to impaired luminal cell expansion and the prostate remains in an early stage of regeneration in which basal cells are enriched. Interestingly, BrdU incorporation revealed that the percentage of BrdU+ cells was ∼5-fold higher in the DT-treated cohort, again reminiscent of an early stage of regeneration (DT, 8.6%; PBS, 1.5%; Figures 6A–6C). These data suggest that depletion of Lgr5+ cells leads to altered luminal cell differentiation and expansion, and impaired prostate regeneration. Gene expression analysis further revealed that cluster of differentiation 31 (CD31), vascular endothelial growth factor receptor 1 (VEGFR1), and Wnt pathway target genes (e.g., β-catenin and Axin 2) were significantly elevated in Lgr5-depleted prostates (Figure 6D). This expression profile is reminiscent of an earlier stage (days 1–3) of prostate regeneration (Wang et al., 2007, 2008), pointing to a delayed regeneration response upon Lgr5+ cell depletion in the prostate. Taken together, these findings demonstrate that Lgr5+ cells are required for prostate regeneration, and depletion of Lgr5+ cells leads to impaired luminal cell expansion and prostate regeneration.
Figure 5.
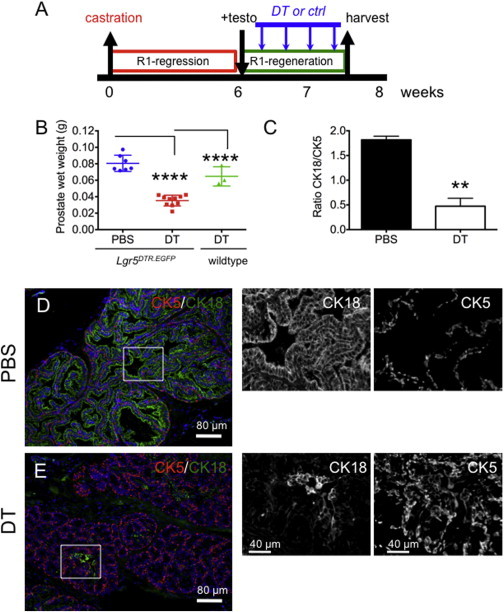
Depletion of Lgr5+ Cells Impairs Prostate Regeneration
(A) Experimental design summary. Six weeks after surgical castration, testosterone pellet implantation was performed concurrently with diphtheria toxin (DT) treatment (DT treatment every other day).
(B) Quantification of prostate wet weights from Lgr5DTR.EGFP mice fully regressed and administered PBS + testo or DT + testo.
(C) Quantification of the ratio of CK18/CK5 in anterior lobes of prostates (n = 3 mice per group). Two coronal prostatic sections per mouse were analyzed by whole-slide scan, using the imaging protocol described in Experimental Procedures.
(D and E) Immunofluorescence of CK5 (red) and CK18 (green) in anterior lobes from prostates treated with PBS (D) or DT (E). Right: enlargements of designated areas showing only single channels. ∗∗p < 0.01, ∗∗∗∗p ≤ 0.0001.
See also Figure S4.
Figure 6.
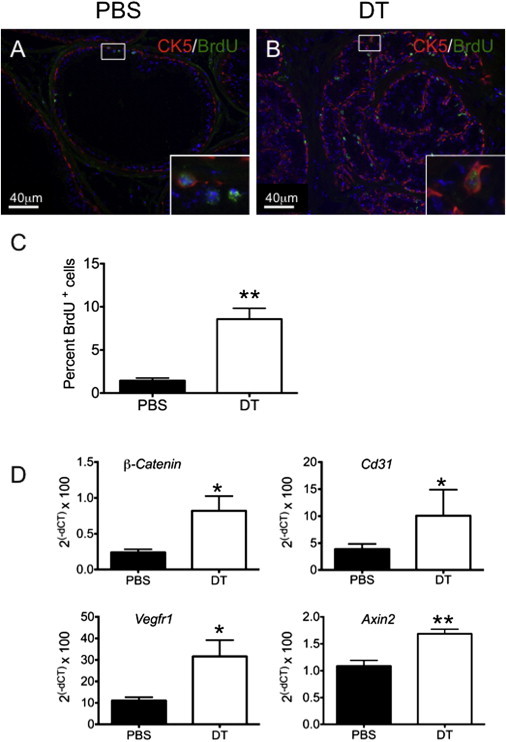
Delayed Regenerative Profiles following Depletion of Lgr5+ Cells
(A and B) Immunofluorescence of BrdU in the anterior lobes of prostate sections from Lgr5DTR.EGFP mice administered either PBS (A) or DT (B). The insets are enlarged images of the boxed areas.
(C) Quantification of BrdU+ cells as a percentage of total epithelial cells in the anterior lobes of prostates: DT, 8.6%, cn = 11,242, n = 3 mice; PBS, 1.5%, cn = 9145, n = 3 mice; p = 0.005.
(D) Transcriptional analysis of prostate tissue following Lgr5+ cell depletion; n = 4 animals per group.
All values are represented as mean and SEM. ∗p < 0.05, ∗∗p < 0.01.
Discussion
Experimental evidence has shown that Lgr5 is a marker of stem cells in the skin, small intestine, ovary, and mammary gland. Herein, we report the finding of a subpopulation of Lgr5-expressing stem cells in the mouse prostate that are long-lived and can generate both luminal and basal cells both in situ during prostate regeneration and within renal capsule implantation studies using single Lgr5+ cells. Lgr5+ cells, particularly the basal Lgr5+ cells, meet the two criteria of stem cells (longevity and multipotency) and thus are identified as stem cells in the prostate. Our in situ hybridization and pulse-label studies indicate that Lgr5+ cells are a rare cell population in the adult prostate. In contrast to the basal expression pattern of Lgr5+ cells in adult mammary glands (Rios et al., 2014), prostatic Lgr5+ cells are mostly located in the luminal compartment. Although most luminal cells are androgen dependent and undergo apoptosis following androgen depletion by castration, castration-resistant luminal Lgr5+ cells were observed. The Lgr5+ cells and their progeny survived two rounds of regression and regeneration, and maintained a similar regeneration capacity over time. Our in situ regeneration data demonstrated that basal Lgr5+ cells were bipotent, but the vast majority of the luminal Lgr5+ cells were unipotent. However, we cannot exclude the possibility that a small portion of luminal Lgr5+ cells can also be bipotent in situ. Previous work has established that stem cells within the basal compartment divide asymmetrically, generating one stem cell and one committed progenitor cell that differentiates to ultimately yield a luminal or neuroendocrine cell during prostate postnatal development (Ousset et al., 2012; Pignon et al., 2013). However, there is some controversy about the cellular hierarchy that governs adult prostate regeneration following castration-induced involution. Some studies have described multipotency for both luminal (Wang et al., 2009) and basal (Wang et al., 2014a) cells, and others have supported committed lineage-specified progenitors for both luminal and basal cells during adult prostatic regeneration (Choi et al., 2012; Liu et al., 2011). All of these studies used similar genetic lineage-tracing methodologies. Our data suggest that by tracing from the rare Lgr5+ cells obtained from castrated, regressed prostates during regeneration, we obtained evidence of basal cell bipotency in situ, and observed unipotent division of luminal cells only. Therefore, our work seems to agree with the most recent work by Wang et al. (2014a), in which basal cells demonstrated both symmetric and asymmetric divisions leading to distinct cell fates, and luminal cells only exhibited symmetric divisions during adult prostate regeneration.
Renal capsule implantation of Lgr5+ cells further supported the high repopulation capacity of single Lgr5+ cells within the regressed prostate. A limiting-dilution analysis indicated that every prostatic Lgr5+ cell could be a stem cell in regressed prostates, meaning that both luminal and basal Lgr5+ cells in regressed prostates could obtain a bipotent capacity in the transplantation setting. Importantly, cell types have been shown to acquire a facultative plasticity in single-cell ex vivo and renal-capsule assays that is not readily observable in vivo (Choi et al., 2012; Wang et al., 2013, 2014b). This is consistent with recent work that identified multipotent basal and luminal cells in a human organoid culture system (Karthaus et al., 2014). It seems that prostatic Lgr5+ cells are capable of bipotentiality depending on the environmental cues or assay conditions, i.e., these cells can generate luminal progeny in response to a demand for luminal epithelial cells in androgen manipulation studies in situ, or give rise to both luminal and basal progeny to repopulate the prostate structure from single cells in transplantation studies.
Moreover, in this study we demonstrate that Lgr5+ cells are required for prostate regeneration after castration and subsequent androgen substitution, as depletion of Lgr5+ cells led to the generation of significantly smaller prostates. Considering the low frequency of Lgr5+ cells in the prostate and the profound impact of Lgr5+ depletion on prostate regeneration, we speculated that Lgr5+ cells could give rise to other progenitor cells to mediate the regeneration process. Indeed, we observed an increase in labeled cells during two rounds of regression and regeneration. The average unit size in the second-round regressed prostates was two cells, compared with one in the first-round regressed prostate. The additional labeled cell is most likely derived from the single Lgr5+ cell from the first round of regression. This cell is also castration resistant and its expansion rate is similar to that of the original Lgr5+ cell, suggesting that Lgr5+ stem cells in the regressed prostate are self-renewing and the newly generated stem cell also takes part in the regeneration process. More studies will be required to elucidate the networking and the symmetry of divisions of stem cells and progenitor cells in the prostate.
With regard to human prostate cancer, tumor cells are largely characterized by luminal phenotypes, and loss of basal cells is the hallmark of prostate adenocarcinoma. However, there is evidence for both basal and luminal cell lineages initiating the disease. Initially, prostate tumors respond to androgen deprivation therapy well, but eventually tumors progress in the absence of androgen, suggesting the possible presence of resistant cells that are responsible for repopulating the tumor. Lgr5+ cells have been shown to be a cell of origin for intestinal tumors (Barker et al., 2009). Although LGR5 is expressed in both human prostate tissues and prostate tumors (Figure S5), it is unclear what role LGR5+ cells play in prostate cancer initiation or maintenance. Interestingly, the majority of castration-resistant Lgr5+ cells are of luminal origin, and it was recently demonstrated that luminal cells are the preferred cell of origin for preclinical prostate murine tumor models (Wang et al., 2014b). If castration-resistant Lgr5+ cells can serve as the cell of origin for castration-resistant prostate cancer (CRPC), then selectively targeting these Lgr5+ cells during androgen-deprivation treatment should be considered as a therapeutic strategy to prevent CRPC. It will be important to continue to explore the role of Lgr5+ cells in prostate cancer biology to assess their potential for tumor initiation, especially given that our studies demonstrate that Lgr5+ cells are castration-resistant, long-lived stem cells that are required for complete prostate regeneration following castration.
Experimental Procedures
Animals
E18 pregnant rats and athymic nu/nu male mice at 8 weeks of age were purchased from Harlan Sprague Dawley. Lgr5DTR.EGFP and Lgr5CreER mice were generated as previously described (Tian et al., 2011). Lgr5CreER mice were interbred with the Rosa26.LSL.tdTomato mouse line to generate Lgr5CreER;Rosa26.LSL.tdTomato (LT) mice. The animals were dosed and monitored according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) at Genentech.
Antibodies
The following antibodies were used: CK5 (1:1,000, PRB-160P; Covance), P63 (1:600, clone 4A4; Santa Cruz Biotechnology), CK8 (1:1,000, MMS-162P; Covance), CK18 (1:500, ab82254; Abcam), Ki67 (1:200, RM-9106; Thermo Scientific), anti-BrdU (1:1,000, #347580; Becton Dickinson), and Alexa Fluor 488 donkey anti-rabbit IgG (H+L, 1:500, A-21206; Life Technologies).
Castration, Tamoxifen Labeling, and Prostate Regeneration
Surgery was conducted using aseptic procedures. Following preparation of the surgical site, an incision was made in the scrotum. Then an incision was made in the tunica of the first testicle. The testis, vas deferens, and attached testicular fat pad were pulled out of the incision. The blood vessels supplying the testis were cauterized. The testis, vas deferens, and fatty tissue were severed just below the site of cauterization. The tunica on the contralateral side was similarly penetrated and the procedure was repeated. The scrotum incision was closed with a 4-0 absorbable Vicryl suture. Four weeks after castration, LT mice were administered tamoxifen at 5 mg/20 g, or corn oil by oral gavage daily for four consecutive days. A 12.5 mg/pellet/mouse testosterone pellet was implanted 2 weeks after the last dose of tamoxifen.
Renal Capsule Grafting and Testosterone Pellet Implantation
Animals were anesthetized using ketamine/xylazine. The back of the skin (dorsal midline over the lumbar) was prepped with povidone iodine and 70% ethanol. The back skin was lifted with a pair of blunt forceps and a lumbar incision was made with a pair of scissors, creating an incision approximately 1 inch long. The animal was placed on its side and a small incision was made in the body wall just along the long axis of the kidney. A pair of fine forceps (#5) was used to gently pinch and lift the capsule from the parenchyma of the kidney so that a 2–4 mm incision was made in the capsule with fine spring-loaded scissors. Grafts were transferred to the surface of the kidney using forceps. The body wall was closed with a 4-0 Vicryl suture. A testosterone pellet (12.5 mg/pellet, one pellet/mouse) was placed between the body wall and skin. After grafting, the edges of the back skin were aligned and closed with a 4-0 absorbable Vicryl suture.
UGM Preparation
UGM cells were dissociated from embryonic day 18 (E18) embryos from pregnant Sprague Dawley rats as described previously (Leong et al., 2008; Marker et al., 2003; Tsujimura et al., 2002). Briefly, UGM cells from E18 embryos were dissociated with 1 mg/ml of collagenase/dispase (Roche) in DMEM plus 10% fetal bovine serum (FBS), 2 mM glutamine, and 100 U/ml of penicillin and streptomycin for 1 hr at 37°C. UGM cells were cultured in DMEM with 10% FBS, 2 mM glutamine, 10 mg/ml insulin, 5.5 mg/ml transferrin, 6.7 ng/ml selenium, 1 nM testosterone (Innovative Research of America), 100 U/ml penicillin, and 100 mg/ml streptomycin. UGM cells were passaged with trypsin twice and used within 7 days after tissue dissociation.
Flow Cytometry
Five to eight prostates were harvested and pooled together for one FACS sorting or analysis experiment. Prostate cells were dissociated as described previously (Leong et al., 2008; Taylor et al., 2010; Wang et al., 2009). Cells were non-lineage depleted using a mouse lineage cell depletion kit (130-090-858; Miltenyi Biotec), stained with propidium iodide (PI), and analyzed on an LSR-II flow cytometer (Becton Dickinson). Single cells were sorted into individual wells of U-bottom 96-well plates containing 20 μl of collagen type I by an LSR-II flow cytometer (Becton Dickinson), and confirmed with an inverted Olympus Ix81 microscope.
DT Treatment and BrdU Labeling In Vivo
Adult male mice were castrated and 4 weeks later were implanted with a testosterone pellet (as described above) administered concurrently with DT (catalog #322326; EMD) at 50 ng/g, intraperitoneally (i.p.). The DT treatment was administered every other day, for a total of four times. BrdU (550891; BD PharMingen) was administered at 200 μl/mouse, 10 mg/ml, i.p., 16 hr before the mice were euthanized. The mice were euthanized and prostates were harvested on day 9 after the first DT treatment.
Immunohistochemistry and Image Acquisitions
Prostates were fixed with 4% paraformaldehyde for 2–3 hr on ice, followed by 30% sucrose overnight at 4°C, embedded in OCT, and sectioned at 5–6 μm. Sections were blocked with 10% normal goat or donkey serum for 1 hr at room temperature, incubated with primary antibodies over night at 4°C, followed by secondary antibody incubation for 30 min at room temperature. Serial sections of each mouse prostate were observed and imaged on either a Zeiss Axioplan 2 imaging microscope with an ORCA-ER digital camera (Hamamatsu), or on Olympus BX61 upright microscope. Or, whole slide images were acquired by the Ariol SL-50 automated slide scanning platform (Leica Microsystems) at 100× final magnification using standard fluorescence filters. Scanned slides were analyzed in the MATLAB software package (version R2012b; The MathWorks). Individual cell nuclei were segmented using a radial symmetry based method applied to the DAPI channel (Veta et al., 2013) and then scored for the presence of CK5, CK18 signal above a global intensity threshold in the area immediately surrounding each nucleus.
RNA Extraction
Tumor samples were placed into RNALater (Ambion) at 4°C for ∼24 hr. Then, 30 mg of tissue was excised from the total tissue collected and total RNA was extracted with the QIAGEN AllPrep Kit (QIAGEN) following the manufacturer’s protocol. RNA quantity was determined using Nanodrop (Thermo Scientific) and the quality was assessed by Agilent 2100 Bioanalyzer (Agilent Technologies) according to the manufacturers’ protocol.
RNA Reverse Transcription
The 20 μl reverse transcription reaction contained 10 μl of 10 ng/μ of total RNA, 2 μl of 10× RT buffer, 0.8 μl of 100 mM dNTPs, 2 μl of 10× RT Random Hexamer Primers; 1 μl of RNase inhibitor (20 U/μl), 1 μl of Multiscribe Reverse Transcriptase (50 U/μl), and 3.2 μl of H2O. All reagents were purchased from Applied Biosystems (part number 4374966). The reaction mixture was mixed with the RNA and incubated as follows; 25°C for 10 min, 37°C for 120 min and then 85°C for 5 min.
Preamplification of cDNA
A total of 96 TaqMan assays (Life Technologies) were pooled at a final concentration of 0.2× for each assay. Pre-PCR amplification reaction was done at 5 μl containing 2.5 μl TaqMan PreAmp Master Mix (2×, part number 4391128), 1.25 μl of 96-pooled TaqMan assay mix (0.2×) and 1.25 μl of cDNA from the reverse transcription reaction described above. The pre-amplification PCR was incubated as follows: one cycle at 95°C for 10 min, and 14 cycles at 95°C for 15 s and then at 60°C for 4 min. After preamplification PCR, the product was diluted 1:5 with TE buffer (Ambion, part number 9849) and stored at −80°C until needed.
RNA Expression Analysis Using a 96.96 Dynamic Array
Reverse transcription and preamplification were performed as described above. Quantitative PCR was carried out using the 96.96 dynamic array platform (Fluidigm) following the manufacturer’s protocol. Specifically, a 6 μl sample mixture was prepared for each sample containing 3 μl of TaqMan Universal Master Mix (part number 4304437), 0.3 μl of Sample Loading Reagent (part number 85000746) and 2.7 μl of diluted, preamplified cDNA. A 6 μl assay mixture was prepared with 3 μl of each TaqMan assay and 3 μl of Assay Loading Reagent (part number 85000736). An Integrated Fluidic Circuit (IFC) controller was used to prime the 96.96 Dynamic Array IFC chip with control line fluid, and 5 μl of both the sample and assay mixes were loaded into the appropriate inlets. After loading, the chip was placed in the BioMark Instrument for PCR at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The data were analyzed with Real-Time PCR Analysis Software (Fluidigm).
Quantitative Real-Time RT-PCR
Quantitative real-time RT-PCR was performed on a 7900HT thermal cycler (Applied Biosystems) with the following primers and probes: Lgr5 (cat# Mm00438890_m1, Applied Biosystems); Ctnnb1 (cat# Mm00483039_m1, Applied Biosystems); CD31 (cat# Mm01242584_m1, Applied Biosystems); VEGFR1 (cat# Mm00438980_m1, Applied Biosystems); Axin2: forward primer, TGGCTTTGACTACGCCCAC; reverse primer, GGGAGCTGAAGCGCTGG, probe, CCCAACGCGCCCTCTTTGATCTG; RPL19: forward 5′-AGA AGG TGA CCT GGA TGA GAA-3′, reverse 5′- TGA TAC ATA TGG CGG TCA ATC T-3′, probe 5′- CTT CTC AGG AGA TAC CGG GAA TCC AAG-3′.
Quantification and Statistics
Three to 11 mice were used for each time point of analysis. tdTomato+ cells and the ratio of CK18+/CK5+ cells were analyzed using the MATLAB software package (version R2012b; The MathWorks) following whole-slide imaging as described above. The numbers of luminal and basal cells per tdTomato+ unit were counted based on CK5-Alexa 488+, tdTomato fluorescence, and DAPI+ on Adobe Photoshop CS6. BrdU+ cells were counted manually using 20× images. The t test, F test, and chi-square test were used for statistical analysis. Three or more animals were analyzed for each experiment.
Author Contributions
B.W. and M.R.J. conceived and designed studies. B.W. performed in vitro experiments. B.W. and X.W. performed in vivo experiments. R.F. provided histopathology support. J.E.-A. and B.W. performed image quantitation and analysis. B.W. and M.R.J. wrote and edited the manuscript.
Acknowledgments
We thank the staff of the Genentech Animal Care Facility for excellent technical support. We also thank Kevin Leong, Felipe de Sousa e Melo, and Joerg Hoeck for critical input regarding the manuscript, and Fred de Sauvage and Joerg Hoeck for insightful discussions. All authors are full-time employees of Genentech and own shares in Roche.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Choi N., Zhang B., Zhang L., Ittmann M., Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.S., Lawson D.A., Cheng D., Sun W., Garraway I.P., Witte O.N. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl. Acad. Sci. USA. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs J.T., Coffey D.S. Etiology and disease process of benign prostatic hyperplasia. Prostate Suppl. 1989;2:33–50. doi: 10.1002/pros.2990150506. [DOI] [PubMed] [Google Scholar]
- Jaks V., Barker N., Kasper M., van Es J.H., Snippert H.J., Clevers H., Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K.G., Wang B.-E., Johnson L., Gao W.-Q. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- Liu J., Pascal L.E., Isharwal S., Metzger D., Ramos Garcia R., Pilch J., Kasper S., Williams K., Basse P.H., Nelson J.B. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol. Endocrinol. 2011;25:1849–1857. doi: 10.1210/me.2011-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker P.C., Donjacour A.A., Dahiya R., Cunha G.R. Hormonal, cellular, and molecular control of prostatic development. Dev. Biol. 2003;253:165–174. doi: 10.1016/s0012-1606(02)00031-3. [DOI] [PubMed] [Google Scholar]
- Ng A., Tan S., Singh G., Rizk P., Swathi Y., Tan T.Z., Huang R.Y.-J., Leushacke M., Barker N. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat. Cell Biol. 2014;16:745–757. doi: 10.1038/ncb3000. [DOI] [PubMed] [Google Scholar]
- Ousset M., Van Keymeulen A., Bouvencourt G., Sharma N., Achouri Y., Simons B.D., Blanpain C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- Pignon J.-C., Grisanzio C., Geng Y., Song J., Shivdasani R.A., Signoretti S. p63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc. Natl. Acad. Sci. USA. 2013;110:8105–8110. doi: 10.1073/pnas.1221216110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V., Brenot A., Lawson D.A., Linnemann J.R., Van Kappel E.C., Wong K.C., de Sauvage F., Klein O.D., Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Taylor R.A., Toivanen R., Risbridger G.P. Stem cells in prostate cancer: treating the root of the problem. Endocr. Relat. Cancer. 2010;17:R273–R285. doi: 10.1677/ERC-10-0145. [DOI] [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura A., Koikawa Y., Salm S., Takao T., Coetzee S., Moscatelli D., Shapiro E., Lepor H., Sun T.-T., Wilson E.L. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J. Cell Biol. 2002;157:1257–1265. doi: 10.1083/jcb.200202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veta M., van Diest P.J., Kornegoor R., Huisman A., Viergever M.A., Pluim J.P.W. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS ONE. 2013;8:e70221. doi: 10.1371/journal.pone.0070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-M., Kovalenko B., Huang Y., Moscatelli D. Vascular endothelial growth factor and angiopoietin are required for prostate regeneration. Prostate. 2007;67:485–499. doi: 10.1002/pros.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.-E., Wang X.-D., Ernst J.A., Polakis P., Gao W.-Q. Regulation of epithelial branching morphogenesis and cancer cell growth of the prostate by Wnt signaling. PLoS ONE. 2008;3:e2186. doi: 10.1371/journal.pone.0002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kruithof-de Julio M., Economides K.D., Walker D., Yu H., Halili M.V., Hu Y.-P., Price S.M., Abate-Shen C., Shen M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Mitrofanova A., Bergren S.K., Abate-Shen C., Cardiff R.D., Califano A., Shen M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhu H.H., Chu M., Liu Y., Zhang C., Liu G., Yang X., Yang R., Gao W.-Q. Symmetrical and asymmetrical division analysis provides evidence for a hierarchy of prostate epithelial cell lineages. Nat Commun. 2014;5:4758. doi: 10.1038/ncomms5758. [DOI] [PubMed] [Google Scholar]
- Wang Z.A., Toivanen R., Bergren S.K., Chambon P., Shen M.M. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 2014;8:1339–1346. doi: 10.1016/j.celrep.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Smedts F., Debruyne F.M., de la Rosette J.J., Schalken J.A. Identification of intermediate cell types by keratin expression in the developing human prostate. Prostate. 1998;34:292–301. doi: 10.1002/(sici)1097-0045(19980301)34:4<292::aid-pros7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


