Abstract
The reverse transcriptase from human immunodeficiency virus type 1 is a heterodimer consisting of one 66-kDa and one 51-kDa subunit. The p66 subunit contains both a polymerase and an RNase H domain; proteolytic cleavage of p66 removes the RNase H domain to yield the p51 subunit. Although the polymerase domain of p66 folds into an open, extended structure containing a large active-site cleft, that of p51 is closed and compact. The connection subdomain, which lies between the polymerase and RNase H active sites in p66, plays a central role in the formation of the reverse transcriptase heterodimer. Extensive and very different intra- and intersubunit contacts are made by the connection subdomains of each of the subunits. Together, contacts between the two connection domains constitute approximately one-third of the total contacts between subunits of the heterodimer. Conversion of an open p66 polymerase domain structure to a closed p51-like structure results in a reduction in solvent-accessible surface area by 1600 A2 and the burying of an extensive hydrophobic surface. Thus, the monomeric forms of both p66 and p51 are proposed to have the same closed structure as seen in the p51 subunit of the heterodimer. The free energy required to convert p66 from a closed p51-like structure to the observed open p66 polymerase domain structure is generated by the burying of a large, predominantly hydrophobic surface area upon formation of the heterodimer. It is likely that the only kind of dimer that can form is an asymmetric one like that seen in the heterodimer structure, since one dimer interaction surface exists only in p51 and the other only in p66. We suggest that both p51 and p66 form asymmetric homodimers that are assembled from one subunit that has assumed the open conformation and one that has the closed structure.
Full text
PDF


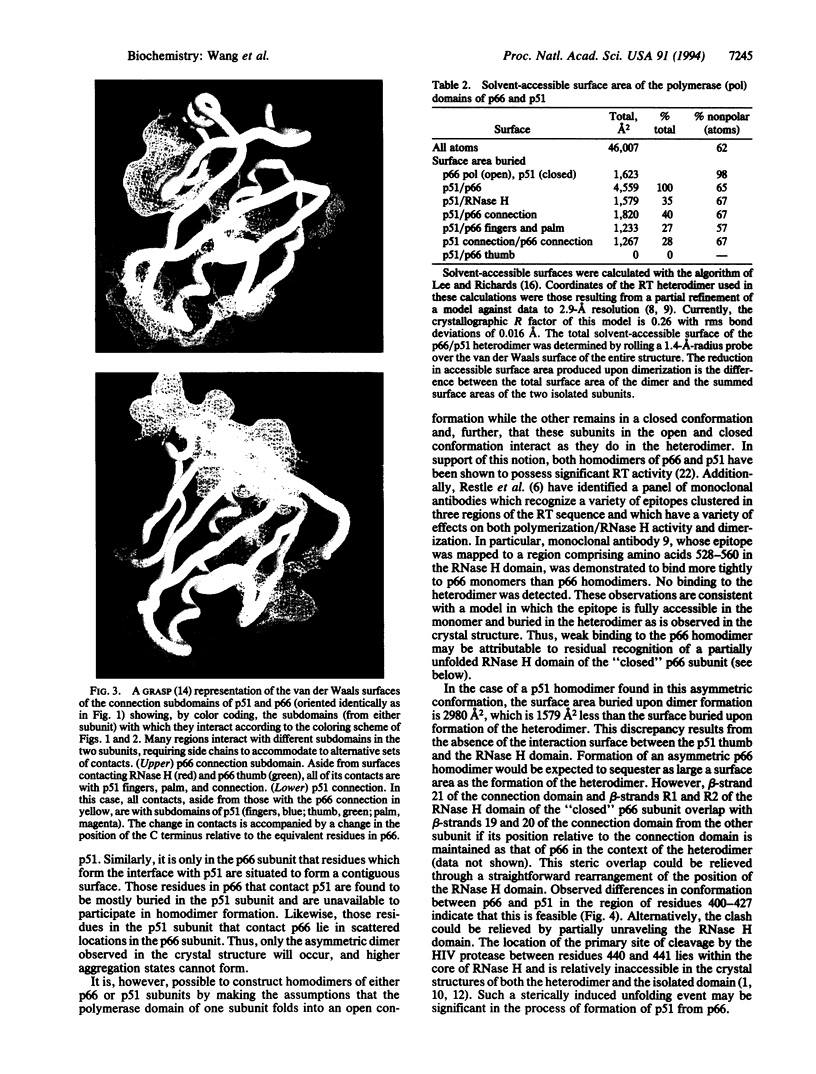

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavand M. R., Wagner R., Richmond T. J. HIV-1 reverse transcriptase: polymerization properties of the p51 homodimer compared to the p66/p51 heterodimer. Biochemistry. 1993 Oct 12;32(40):10543–10552. doi: 10.1021/bi00091a003. [DOI] [PubMed] [Google Scholar]
- Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974 Mar 22;248(446):338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- Chothia C., Janin J. Principles of protein-protein recognition. Nature. 1975 Aug 28;256(5520):705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Divita G., Restle T., Goody R. S. Characterization of the dimerization process of HIV-1 reverse transcriptase heterodimer using intrinsic protein fluorescence. FEBS Lett. 1993 Jun 14;324(2):153–158. doi: 10.1016/0014-5793(93)81383-b. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. V., Janin J. The price of lost freedom: entropy of bimolecular complex formation. Protein Eng. 1989 Oct;3(1):1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Fu T. B., Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J Virol. 1992 May;66(5):3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P. S., Wöhrl B. M., Howard K. J., Le Grice S. F. Modulation of HIV-1 reverse transcriptase function in "selectively deleted" p66/p51 heterodimers. J Biol Chem. 1994 Jan 14;269(2):1388–1393. [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991 Dec;10(12):3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Page M. I., Jencks W. P. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restle T., Müller B., Goody R. S. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J Biol Chem. 1990 Jun 5;265(16):8986–8988. [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Smerdon S. J., Jäger J., Wang J., Kohlstaedt L. A., Chirino A. J., Friedman J. M., Rice P. A., Steitz T. A. Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3911–3915. doi: 10.1073/pnas.91.9.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Smerdon S., Jäger J., Wang J., Kohlstaedt L. A., Friedman J. M., Beese L. S., Rice P. A. Two DNA polymerases: HIV reverse transcriptase and the Klenow fragment of Escherichia coli DNA polymerase I. Cold Spring Harb Symp Quant Biol. 1993;58:495–504. doi: 10.1101/sqb.1993.058.01.056. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Warren T. C., Adams J., Proudfoot J., Skiles J., Raghavan P., Perry C., Potocki I., Farina P. R., Grob P. M. A novel dipyridodiazepinone inhibitor of HIV-1 reverse transcriptase acts through a nonsubstrate binding site. Biochemistry. 1991 Feb 26;30(8):2022–2026. doi: 10.1021/bi00222a003. [DOI] [PubMed] [Google Scholar]