Abstract
The crystal structure of the protein product of the gene locus At1g05000, a hypothetical protein from A. thaliana, was determined by the multiple-wavelength anomalous diffraction method and was refined to an R factor of 20.4% (Rfree = 24.9%) at 3.3 Å. The protein adopts the α/β fold found in cysteine phosphatases, a superfamily of phosphatases that possess a catalytic cysteine and form a covalent thiol-phosphate intermediate during the catalytic cycle. In At1g05000, the analogous cysteine (Cys150) is located at the bottom of a positively-charged pocket formed by residues that include the conserved arginine (Arg156) of the signature active site motif, HCxxGxxRT. Of 74 model phosphatase substrates tested, purified recombinant At1g05000 showed highest activity toward polyphosphate (poly-P12–13) and deoxyribo- and ribonucleoside triphosphates, and less activity toward phosphoenolpyruvate, phosphotyrosine, phosphotyrosine-containing peptides, and phosphatidyl inositols. Divalent metal cations were not required for activity and had little effect on the reaction.
Keywords: phosphatase, Arabidopsis, polyphosphate, At1g05000, structure, crystallography
INTRODUCTION
Although it is generally agreed that protein phosphorylation is regulated by the equal and balanced action of protein kinases and protein phosphatases, much more research has been focused on kinases.1 However, recent findings have led to the realization that protein phosphatases play active and perhaps dominant roles in the regulation of many physiological processes.2–4 “Cysteine phosphatases” are a large superfamily of phosphatases that possess an active site cysteine that acts as a nucleophile to form a covalent thiolphosphate intermediate.5, 6 The catalytic cysteine is located in a highly conserved active site motif, HCxxGxxR, that is present with only slight variations in several hundred individual proteins in the NCBI database (http://www.ncbi.nlm.nih.gov). A pocket that binds and positions the phosphate(s) is formed by the arginine of the active site motif and other nearby positively charged amino acids.7
Known cysteine phosphatase substrates are limited to proteins/peptides containing phosphotyrosine/serine/threonine,8, 9 ribo/deoxyribonucleotide 5′-triphosphates,10–12 phosphoinositides,13 pyrophosphate/triphosphate,14 and phosphorylated complex carbohydrates.15, 16
Phosphatases that dephosphorylate proteins play important roles in signal transduction pathways related to metabolism, cell cycle progression, ion transport, developmental control, and stress responses.1, 4 This diverse spectrum of functions is reflected by the large number of intracellular proteins regulated by phosphorylation/dephosphorylation and the large number of protein kinases and phosphatases that catalyze the reactions.
Proteins are commonly phosphorylated at tyrosine, serine, or threonine residues. Known cysteine phosphatases are specific either for tyrosine alone (protein tyrosine phosphatases or PTPases, EC3.1.3.48) or for tyrosine and serine/threonine (dual-specificity phosphatases or DSPases, EC3.3.1.16). Phosphatases that target serine and threonine but not tyrosine do not belong to the cysteine phosphatase superfamily and bear no significant resemblance to it by sequence or structure.4 PTPases are characterized by their sensitivity to vanadate, ability to hydrolyze p-nitrophenyl phosphate, and lack of metal ion requirement for catalysis.4 A number of PTPases and DSPases have been well-characterized; for example, the human cyclin-dependent kinase-associated phosphatase is a DSPase that appears to dephosphorylate a kinase essential in cell cycle control.8
Nucleotide triphosphatases/RNA 5′-triphosphatases (NTPases/RTPases, EC3.1.3.31 and EC2.7.7.50) of the cysteine phosphatase superfamily are thought to act in mRNA capping, removing the γ-phosphate of 5′-triphosphorylated mRNA as the first step in mRNA 5′cap formation; the GMP cap is then added by a mRNA guanylyltransferase present either as a second domain of the same enzyme or as an independent protein. A much-studied NTPase/RTPase is the “BVP” baculovirus enzyme, which has also been shown to act on pyrophosphate and triphosphate, albeit at a very low rate.11
Cysteine phosphoinositol phosphatases (PIPases, EC3.1.3.36) act on phospholipid second-messenger signaling molecules such as phosphatidylinositol (2,4,5)-triphosphate which are involved in many cellular processes including membrane trafficking, and release of phosphate from storage molecules such as inositol hexakisphosphate (phytate) (EC3.1.3.72). A well-characterized PIPase is the human PTEN tumor suppressor.13
Carbohydrate phosphatases function in the formation of soluble storage carbohydrates such as glycogen.15, 16 They are taxonomically widespread and consist of two domains, a carbohydrate-binding domain and a DSP-like phosphatase domain. The first described member of this class of enzymes was the human “Laforin” carbohydrate phosphatase.
In this work, we present the crystal structure of the A. thaliana phosphatase encoded by the gene locus At1g05000 and an investigation of the enzymatic activity of this protein. At1g05000 is a 215 amino acid protein of native molecular weight 24,537 Da. We show that At1g05000 has both sequence and structural similarity to cysteine phosphatases, however it was largely unnaffected by several SH-group inhibitors. Enzymatic activity was confirmed with a general phosphatase substrate and a number of model substrates. We show that the favored substrate of At1g05000 among those tested was polyphosphate (poly-P12–13). The structure was determined under the National Institutes of Health NIGMS Protein Structure Initiative.
MATERIALS AND METHODS
Cloning, protein expression, and purification
The At1g05000 gene was cloned and the selenomethione-labeled protein was expressed and purified according to the standard CESG pipeline procedures for cloning,17 protein expression,18 protein purification,19 and information management.20 Briefly, the open reading frame was amplified by PCR from a cDNA pool created by reverse transcription from extracts of the T87 Arabidopsis thaliana ecotype Columbia callus cell line. It was cloned into the pDONR221 Gateway vector (Invitrogen, Carlsbad, CA). The gene sequence was verified by DNA sequencing and then transferred to expression vector pVP13, a vector created by CESG through modification of a pQE80 (Qiagen, Valencia, CA) backbone to contain Gateway recombination cloning regions and to add an N-terminal tag consisting of His6/maltose-binding protein tag cleavable from the target with tobacco etch virus (TEV) protease. The protein was over-expressed in E. coli strain B834(DE3) containing the pRARE plasmid (Invitrogen) in seleno-methionine containing self-inducing medium.18 Purification was performed by subtractive Ni2+ metal affinity chromatography. The tag was removed using TEV protease, leaving native sequence except that the N-terminal methionine was replaced with serine. The protein was concentrated to 10 mg mL−1 in a final storage buffer consisting of 5 mM MES, 250 mM NaCl, and 0.3 mM TCEP, pH 6.0.
Structure determination
Crystals of At1g05000 were grown by the hanging-drop method using protein solution in the storage buffer mixed with an equal amount of well solution containing 0.6 M (NH4)2SO4 and 100 mM PIPES, pH 6.5, at 293 K. The selenomethionyl crystals of At1g05000 belong to spacegroup P213, with unit cell dimensions a = b = c = 124.5 Å, α = β = γ = 90°.
Diffraction data was collected at BioCARS 14-ID and SBC 19-BM beamlines at Argonne National Laboratory Advance Photon Source for phasing and refinement datasets, respectively. The datasets were integrated and scaled using the HKL2000 suite.21 The selenium substructure was determined using HySS22; four anomalous sites could be reliably identified suggesting the presence of one molecule in the asymmetric unit. The protein structure was phased in SOLVE23 to 3.5 Å by multiple anomalous diffraction using two wavelength datasets from crystal one and density modified in RESOLVE.24 Inspection of the electron density map at this stage revealed the existence of the two molecules in the asymmetric unit. To further improve phase information, density modification was performed in RESOLVE against a higher resolution dataset (from crystal 2, 3.3 Å) with two-fold averaging based on positions of six sites obtained from analysis of residual anomalous maps. The resulting electron density map was of sufficient quality to allow straightforward manual model building in XFIT.25 The structure was completed in several cycles of model building and refinement using CNS.26 Tight stereochemical restraints were used during the refinement. All refinement steps were monitored using an Rfree value based on 9.2% of the independent reflections. The stereochemical quality of the final model was assessed using ProCheck27 and MOLPROBITY.28
Enzymatic screening and assays
Enzymatic screens for various activities (phosphatase, phosphodiesterase, esterase, protease, dehydrogenase, and oxidase) were performed in 96-well microplates at 37°C as previously described.29 Secondary screens for phosphatase activity against model phosphatase substrates were performed using a panel of 74 phosphorylated compounds (Supplementary materials Table I, all from Sigma) in 96-well microplates using the Malachite Green reagent.29, 30 Phosphatase activity against polyphosphate was measured using the mild ascorbate assay.31 The pH dependence of phosphatase activity of At1g05000 was determined using polyphosphate as substrate and the mixed buffer system (pH range 4.5–9.0).32 Phosphatase activity toward selected substrates or phosphorylated peptides (substrate profiles) was assayed in 96-well microplates using 160 µL reaction mixtures containing 50 mM buffer (mixed buffer, pH 5.0; or HEPES-K buffer, pH 7.0), 0.14 mM poly-P, 0.1 mM phosphatidyl inositols, or 1 mM other substrates (for Malachite Green assay), and 0.3–0.6 µg of enzyme. After 20 min incubation at 37°C, the reaction was terminated by the addition of 40 µL of Malachite Green reagent, and after 5 min the production of Pi was measured at 630 nm. Phosphorylated peptides were purchased from EZBiolab (www.ezbiolab.com) with following composition, where (pX) indicates the phosphorylated residue: TSTEPQ(pY)QPGENL, RRLIEDAE(pY)AARG, KR(pT)IRR, RRA(pS)VA, RRA(pT)VA, DADE(pY)LIPQQG, and END(pY)INASL.
For determination of Km and kcat, the phosphatase assays contained substrates at concentrations 0.005–4 mM. Kinetic parameters were determined by non-linear curve fitting using GraphPad Prism software (version 4.00 for Windows, GraphPad Software, San Diego, CA, www.graphpad.com).
RESULTS
Structure of At1g05000
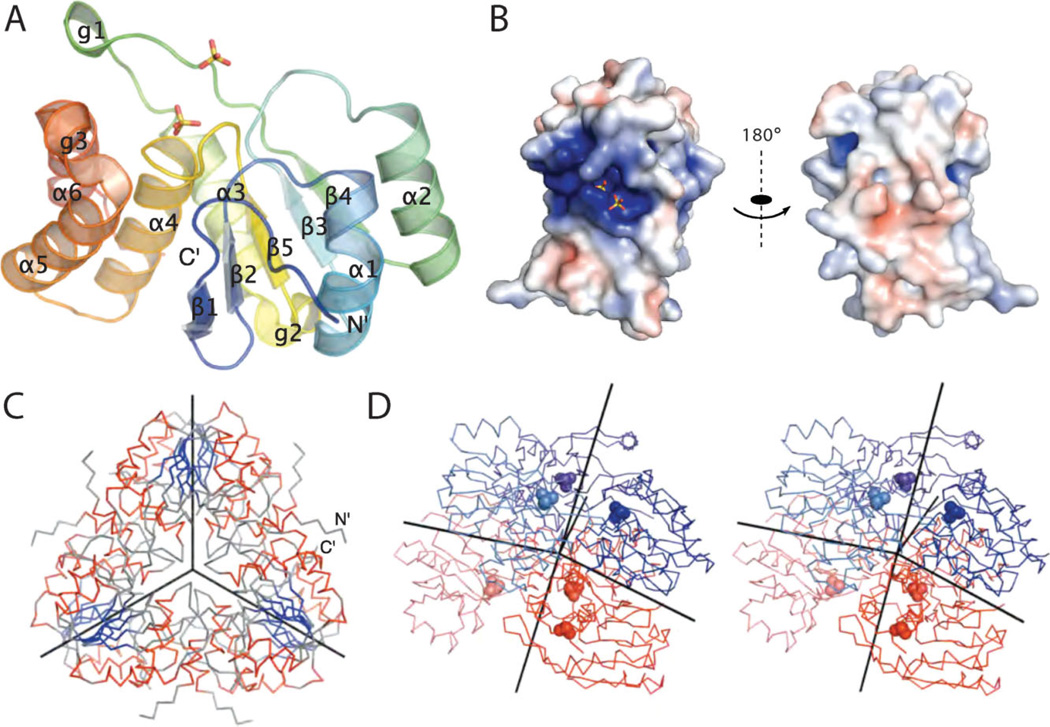
The structure of At1g05000 was solved by multiple wavelength anomalous diffraction using datasets collected at two wavelengths and refined to a resolution of 3.3 Å. Data collection, refinement, and model statistics are summarized in Table I. Residues 52–202 of this 215 residue protein form a well-defined globular domain [Fig. 1(A)]; however, interpretable electron density was not observed for either the 51 amino-terminal residues or for the 13 carboxyl-terminal residues. Two sulfates per molecule of At1g05000 were located in the positively charged active site cavity [Fig. 1(B)]. At1g05000 belongs to the α/β class of proteins with a complex α/β architecture and protein-tyrosine phosphatase topology.33 Specifically, the observed core of At1g05000 comprises eight extended residues followed by antiparallel β-strands β1 and β2, three alternating α/β pairs (α1/β3, α2/β4, and α3/β5), and three successive α-helices (α4-α6) [(Fig. 1(A)]. The five β-strands form a mixed central β-sheet with strand β1 antiparallel to β-strands β2-β5. The central β-sheet is sandwiched between two α-helices on one side and four on the other. Based on interpretation of the crystal packing, At1g05000 forms a hexamer with perfect C3 point-group symmetry [Fig. 1(C)]. The three-fold symmetry axis of the hexamer coincides with the crystallographic three-fold symmetry axes present in the cubic space-group P213 of the At1g05000 crystals. The asymmetric unit of crystals contains two molecules of At1g05000 related by a 2-fold noncrystallographic axis, which is approximately perpendicular to the 3-fold axis of the hexamer (the axes intersect with an angle of 85.55°). The hexamer thus consists of two layers of three At1g05000 molecules related by a three-fold axis [Fig. 1(D)]. The idealized point-group symmetry that the At1g05000 hexamer could achieve is D3; however, this is not the case and the observed deviations from D3 symmetry might have resulted from crystallographic packing effects. The interface involved in formation of the asymmetric unit dimer buries 825 Å2 per molecule, whereas the total buried surface (per monomer) due to formation of the hexamer is 1840 Å2. These values are consistent with the energetically favorable formation of multimeric species in solution. Importantly, the active sites of all molecules are easily accessible because they are located on the top and the bottom of the hexamer [Fig. 1(D)]. Also, the non-observed 51 amino-terminal and several carboxy-terminal residues can extend from the sides of the hexamer into the surrounding solution (or solvent channels of At1g05000 crystals) [Fig. 1(C)]. The hexameric form determined by structural analysis contrasts with the results of gel filtration experiments, where ~95% of the protein eluted at in a peak with apparent molecular weight corresponding to a dimer (47.6 ± 4.0 kDa) whereas the remainder eluted in a peak with apparent molecular weight close to that predicted for a hexamer (164 ± 3.0 kDa) (data not shown).
Table I.
Crystal Parameters, X-ray Data Collection, and Refinement Statistics
| Crystal 2 PEAK | Crystal 1 HREM | Crystal 1 PEAK | |
|---|---|---|---|
| Space group | P213 | P213 | P213 |
| Unit-cell parameters (Å, deg) | a = b = c 124.5 | a = b = c 125.0 | a = b = c 124.5; α = β = γ = 90 |
| Data collection statistics | |||
| Wavelength (Å) | 0.97932 | 0.96374 | 0.97896 |
| Energy (keV) | 12.660 | 12.865 | 12.665 |
| Resolution range (Å)a | 44.01–3.30 (3.38–3.30) | 33.42–3.50 (3.58–3.50) | 33.40–3.50 (3.58–3.50) |
| No. of reflections (measured/unique) | 215783/9989 | 126372/8476 | 126588/8477 |
| Completeness (%) | 100.0 (100.0) | 99.8 (100.0) | 99.8 (100.0) |
| Rmergeb | 0.115 (0.291) | 0.144 (0.359) | 0.138 (0.319) |
| Redundancy | 21.6 (22.2) | 14.9 (14.9) | 14.9 (15.2) |
| Mean I/sigma(I) | 20.8 (10.3) | 14.6 (7.2) | 14.2 (7.7) |
| Refinement and model statistics | |||
| No. of reflections (work/test) | 18486/1736 | ||
| Rcrystc | 0.204 (0.220) | ||
| Rfreed | 0.249 (0.285) | ||
| R.m.s.d. bonds (°) | 0.007 | ||
| R.m.s.d. angles (°) | 1.20 | ||
| ESD from cross-validated sigma A (Å) | 0.43 | ||
| B factor, protein/solvent/ligands (Å2) | 29.3/19.3/36.9 | ||
| No. of protein molecules/atoms | 2/2448 | ||
| No. of waters | 60 | ||
| No. of sulfates | 4 | ||
| Validation by MOLPROBITY | |||
| Ramachandrand plot | |||
| Favored | 91.61% | ||
| Allowed | 8.39% | ||
| Outliers | 0% | ||
| Rotamer outliers | 2.2% | ||
| PBD code | 1xri |
Values in parentheses are for the highest resolution shell.
Rmerge = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and 〈I(h)〉 is the mean intensity of the reflection.
Rcryst = Σh||Fobs|Fcalc||/Σh|Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using ~9.2% of the randomly selected unique reflections that were omitted from structure refinement.
Figure 1.
Structure of At1g05000. (A) Ribbon diagram of At1g05000 is shown in a rainbow color scheme from the amino terminus (blue) to the carboxy terminus (red). Secondary structure elements are annotated as α1-α6 for a helices; β1-β5 for the β-strands of the central 5-stranded β sheet; g1-g3 for auxiliary 310 helices. (B) Electrostatic surface potential of At1g05000 contoured from 210 kT (red) to 10 kT (blue). Sulfates (sticks) are clearly buried in the deep active site cavity. The active site/substrate binding portion of the surface forms the positively charged patch on the surface of the protein. (C) a Cα-trace of the At1g05000 hexamer viewed along its 3-fold rotational axis. (D) A stereo figure of a Cα-trace of a At1g05000 hexamer with two layers of trimers shown in different color families (blue and red shades). The 3-fold rotation axis and three 2-fold axes, which relate two molecules in the asymmetric units, intersect with 85.55°. The active sites of individual molecules with bound sulfates (spheres) are accessible from the top and the bottom of the hexamer.
Structural homology search
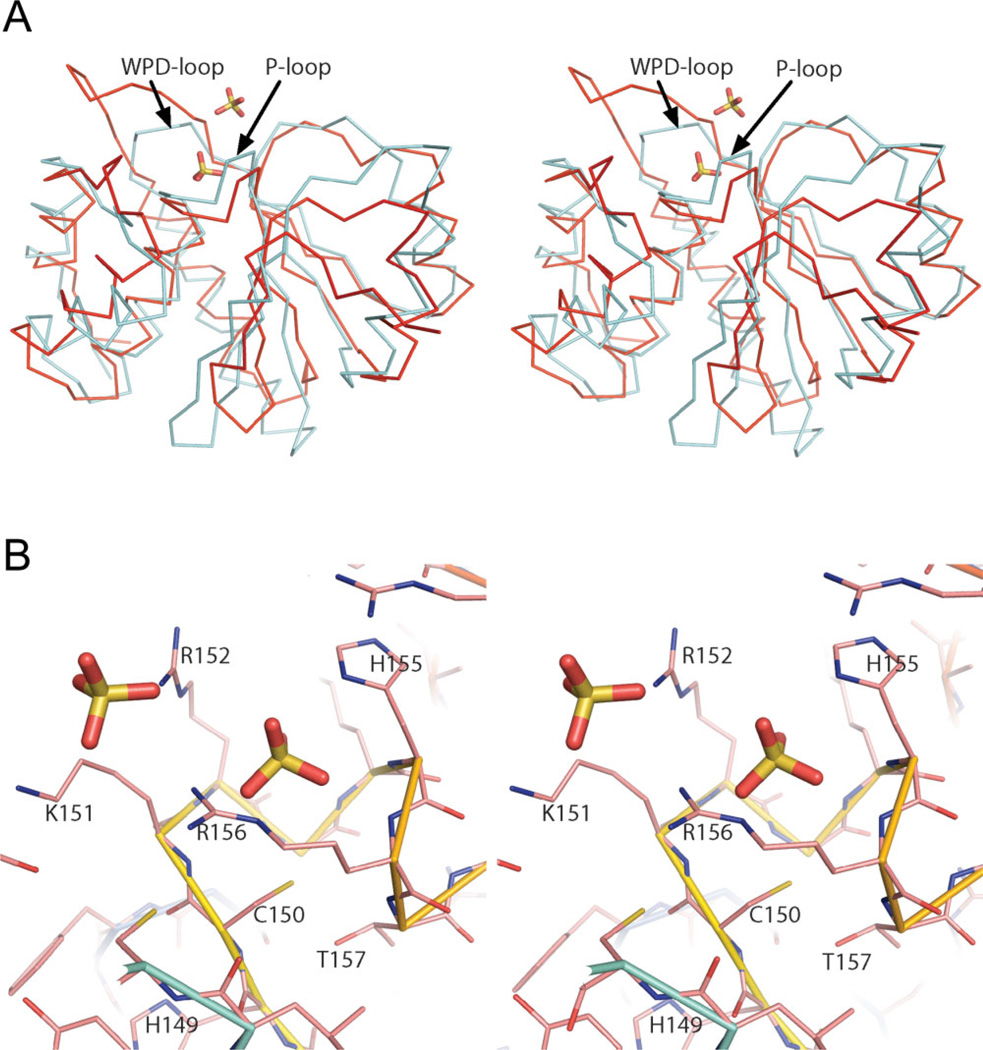
Comparison of the At1g05000 structure to those in the Protein Data Bank (PDB)34 using the VAST structural alignment tool35 yielded 36 structural neighbors. The best match by VAST score was the rat DSPase PRL-1 in complex with sulfate (PDB code 1ZCL, VAST score 15.7, 11% sequence identity with the 149 residue At1g05000 phosphatase domain).7 The second best match was to the human MTMR2 myotubularin family PIPase crystallized in complex with phosphatidylinositol 3,5-bisphosphate (PDB 1ZVR, VAST score 15.6, 11% sequence identity with the At1g05000 phosphatase domain).36 The best match by sequence identity was the PIPase/DSPase PTEN Tumor Suppressor (1D5R, 24% sequence identity over 126 aligned residues). Alignment with PRL-1 is shown in [Fig. 2(A)]. The most obvious structural differences are the shorter loop of At1g05000 between β1 and β2 [see also Fig. 1(A)], the shorter loop between β3 and α2, which includes auxiliary 310 helix g2, and the extended loop between β4 and α3, which includes auxiliary 310 helix g1.
Figure 2.
(A) Structural superposition of At1g05000 (red) and rat dual-specificity protein phosphatase PRL-1 (PDB code 1ZCL) (cyan), which was identified as the closest structural homolog of At1g05000 by VAST.35 (B) Stereoview model of the active site of At1g05000. Selected residues of the active site motif sequence 149HCKRGKHRT157 (conserved residues are underlined) The view is rotated approximately 180 degrees left relative to the model in Figure 1(A). Two bound sulfates are represented by red and yellow sticks.
Sequence homology search
A search of the NCBI Conserved Domain Database37 established that the catalytic core of At1g05000 (residues 53–205) shows homology to the Pfam domain family Y_ phosphatase2 (59% identity against the domain consensus sequence, E = 6 × 10−59), annotated as a tyrosine phosphatase domain. This domain contains the characteristic cysteine phosphatase motif, which is present in At1g05000 as 149HCKRGKHR156. In At1g05000, this motif is located in the loop connecting β5 and α4 of [Fig. 2(A)] which is analogous to the “P loop” of cysteine phosphatases, while the loop between and β4 and α3 is analogous “WPD loop” of cysteine phosphatases.12, 13, 38, 39
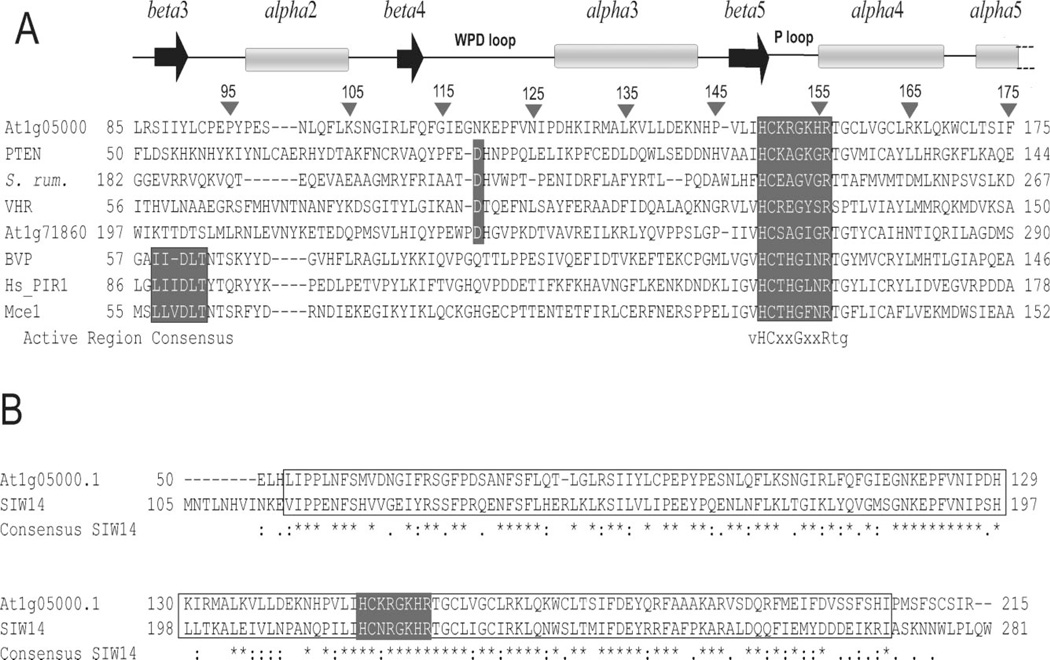
Few of the hundreds of proteins that contain the conserved cysteine phosphatase motif have been tested for catalytic activity and substrate specificity. Selected proteins with demonstrated activity are aligned with At1g05000 in [Fig. 3(A)]. These were chosen to include four of the five known classes of cysteine phosphatases (phosphotyrosine, phosphotyrosine/serine/threonine, nucleotide triphosphates/RNA 5′-triphosphates, and phosphoinositide phosphatases). Each enzyme contains the consensus active site motif (I/V)HCxxGxxR. The protein most similar to At1g05000 is the human PTEN PIPase (consensus IHCKxGKxRTG). All represented phosphatases have little sequence homology to At1g05000 apart from the active site consensus sequence. The NTPases/ RTPases form a cohesive group with an active site region consensus sequence GVHCTHG(I/F)NRTG and an upstream motif consensus sequence (I/L)(I/L)xDLTxT.11
Figure 3.
Sequence alignment of At1g05000 and representative cysteine phosphatases. (A) At1g05000 sequence and secondary structure aligned, using MultAlign, with representative enzymatically characterized cysteine phosphatases40; PTEN is human PTEN Tumor Suppressor phosphoinositol phosphatase/dual-specificity protein phosphatase (PDP code 1DR5); S. rum. is a Selonomonas ruminantium phosphoinositol phosphatase (PDP 1U24); VHR is human Vaccinia VH-related dual-specificity protein phosphatase (PDP 1VHR); At1g71860 is an Arabidopsis thaliana phosphotyrosine phosphatase (Genbank protein ID NP_ 177331.1); BVP is baculovirus nucleotide phosphatase (PDP 1YN9); Hs_ PIR1 is a human nucleotide triphosphatase/5′-RNA triphosphatase (Genbank protein ID AAC39925)41; Mce1 is a mouse nucleotide triphosphatase/5′-RNA triphosphatase (PDP 1I9S). Highlighted are a conserved Asp residue, a sequence conserved among nucleotide triphosphatases/5′-RNA triphosphatases, and the characteristic cysteine phosphatase active site motif. (B) The At1g05000 sequence aligned with that of the S. cerevisiae phosphatase SIW14 (shared cysteine phosphatase domains are boxed, the active site motif highlighted, and the consensus shown).
A sequence search of the NCBI database revealed ~100 proteins with significant (34–85%) identity to At1g05000. Those that were most similar were from the plants Arabidopsis thaliana, Capsicum annum, Oryza sativa, and Medicago truncatula, closely followed by proteins of the amoeba Dictyostelium discoideum and the fungi Ustilago maydis and Saccharomyces cerevisiae. Of these, functional information is available only for the S. cerevisiae protein SIW14, which is 58% identical to the At1g05000 phosphatase domain region [Fig. 3(B)]. There is little homology outside of the conserved domain and the N-terminus of At1g05000 is significantly shorter (not shown). A predicted tyrosine phosphatase, SIW14 has been shown by gene disruption to contribute to the ability of the cell to halt cell division under conditions of nutrient limitation and other stresses, apparently through control of cytoskeleton actin filament organization.42 The At1g05000 domain is the closest non-fungal homolog of the SIW14 domain.
Active site architecture
Residues Lys151, Arg152, His155, and Arg156 of the active site motif (149HCKRGKHR156) make up the positively-charged pocket [Fig. 2(B)] that binds the sulfates in the crystal structure. The residue analogous to the catalytic cysteine of cysteine phosphatases is Cys150, which is located near the bottom of the pocket. Residues analogous to At1g05000 His149 are highly conserved among cysteine phosphatases as are a serine or threonine positioned immediately after the active site motif. These residues are positioned close to the active site [Fig. 2(B)] and may be important to catalysis.43, 44
Enzymatic studies
First, recombinant At1g05000 was tested for phosphatase, phosphodiesterase, esterase, protease, dehydrogenase, and oxidase activities29 (Table II). Significant activity (>1 µmole min−1 mg−1 protein) was detected with p-nitrophenyl phosphate, a model phosphatase substrate. Notably, this activity was little affected by three different SH-group inhibitors that would be expected to inhibit a cysteine phosphatase; iodoacetate (0.06–10.0 mM) and 5, 5′-dithiobis(2-nitrobenzoic acid) (DTNB) (0.04–3.0 mM) produced no effect, whereas N-ethylmaleimide (0.06–10.0 mM) produced 30% inhibition at 10 mM.
Table II.
Results of General Enzyme Assays
| Activity assay | Substrate | Absorbancea |
|---|---|---|
| Phosphodiesterase | bis-pNPP | 0.078 |
| Dehydrogenase | Amino acids | 0 |
| Dehydrogenase | Acids | 0.022 |
| Dehydrogenase | Alcohols | 0.043 |
| Dehydrogenase | Aldehyde | 20.021 |
| Dehydrogenase | Sugars | 0.007 |
| Thioesterase | palmitoyl-CoA | 0.12 |
| Oxidase | NAD(P)H Ox | 0.035 |
| Protease | Protease Mix | 0.046 |
| Phosphatase | pNPP | 4.701 |
Absorbance values at the wavelengths at which the catalytic products are measured. 0.15 AU is usually used as a cutoff for a positive signal.
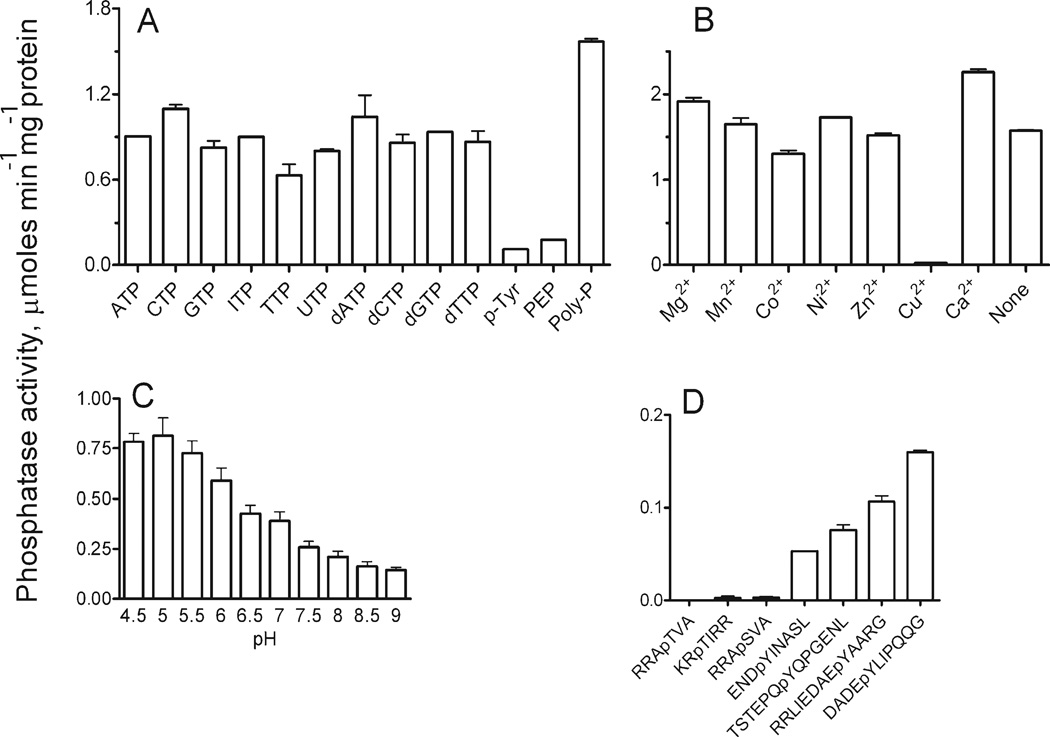
Activity of At1g05000 was next evaluated against a panel of 74 common phosphatase substrates (Supplementary materials, Table I). At1g05000 was found to have the highest activity against polyphosphate (poly-P12–13) followed by the array of ribo- and deoxyribonucleoside triphosphates [Fig. 4(A)]. Low but detectable activity was found against P-Tyr, phosphoenolpyruvate. In addition, low levels of activity (0.006–0.009 µmoles min−1 mg−1) were found with the phosphoinositols l-α-phosphotidyl-d-myo-inositol 5-monophosphate and l-α-phosphotidyl-d-myo-inositol 3-monophosphate (data not shown). The reaction conditions were optimized using poly-P as a substrate. Phosphatase activity of At1g05000 was highest at pH 5.0 [Fig. 4(C)] and was not dependent on the presence of divalent cation [Fig. 4(B)]; similar levels of activity were observed in the presence or absence of Mg2+, Mn2+, Co2+, Ni2+, Zn2+, or Ca2+, indicating that this enzyme is a metal independent phosphatase. Only Cu2+ was found to inhibit the phosphatase activity of At1g05000.
Figure 4.
Phosphatase activity of At1g05000 with various substrates, metals, and pH conditions. (A) Phosphatase activity with various model substrates. p-Tyr is phosphotyrosine, PEP is phosphoenolpyruvate, and Poly-P is polyphosphate (poly-P12–13). (B) Metal dependence was measured with poly-P as substrate (0.14 mM) and metal concentrations were 5 mM Mg2+, 1.0 mM Mn2+, and 0.5 mM for other metals. (C) Phosphatase activity against poly-P with variation in pH (D) Phosphatase activity with phosphorylated peptides. The reaction contained 50 mM HEPES-K (pH 7.0), 5 mM Mg2+, 0.5 mM peptide, and 0.6 µg of At1g05000. Other experimental conditions were as described under “Materials and Methods.”
At1g05000 was tested for phosphatase activity against seven phosphorylated oligopeptides [Fig. 4(D)] (39); ENDpYINASL is derived from a highly conserved region of the T-cell phosphatase TC.PTP45; TSTEPQpYQPGENL is the C-terminal phosphorylation site of c-Src46; RRLIEDAEpYAARG is based on the sequence surrounding the human p60src tyrosine kinase autophosphorylation site47; DADEpYLIPQQG is based on human epidermal growth factor receptor (EGFR), a substrate of the SHP2 protein tyrosine phosphatase.48 At1g05000 showed detectable activity only toward P-Tyr-containing substrates. Highest specific activity was toward (a) the human SHP2 substrate (0.16 µmoles min−1 mg−1) and (b) the p60src kinase site (0.11 µmoles min−1 mg−1).
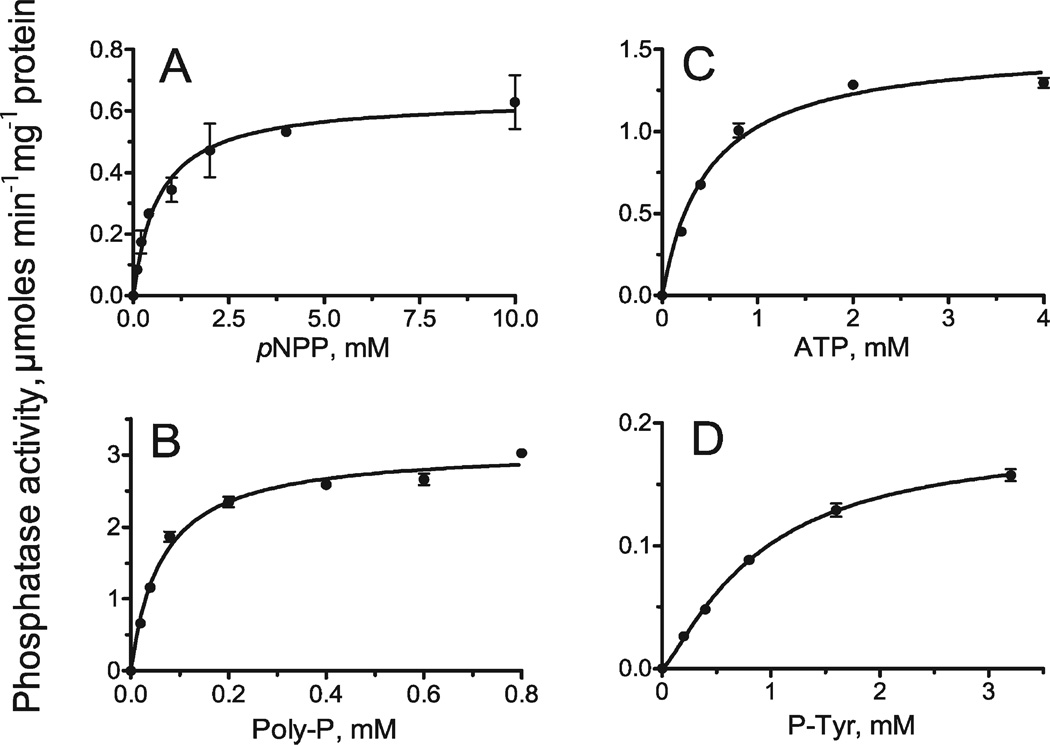
Using the optimized reaction conditions (pH 5.0, no metal), we determined the specific activity of this protein against the range of substrates (see Fig. 5). With pNPP, poly-P and ATP, At1g05000 showed classical hyperbolic saturations that were not affected by the addition of metals. Analysis of the kinetic parameters revealed that At1g05000 had the highest affinity toward poly-P (Table III) with a Km of 0.063 mM. The Km and catalytic efficiency (kcat/Km) were ~10 times lower with nucleotide substrates than with poly-P (Table III). With P-Tyr, the protein showed saturation at substrate concentrations 2.0–3.0 mM [Fig. 5(D)], where it had a specific activity of ~0.20 µmol min−1 mg−1 protein (kcat = 0.077 s−1). Phosphatase activity of At1g05000 was inhibited by P-Tyr concentrations >3.0 mM (data not shown).
Figure 5.
Phosphatase activity of At1g05000 toward four different substrates as a function of substrate concentration: (A) pNPP; (B) poly-P(10–12); (C) ATP; (D) P-Tyr. The reaction conditions were as described under “Materials and Methods.”
Table III.
Kinetic Parameters of At1g05000 with Various Substrates
| Variable substrate | Metal | Km (mM) | Vmax (U/mg)a | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|
| Poly-P(12–13) | None | 0.063 ± 0.006 | 3.09 ± 0.07 | 1.26 ± 0.03 | 2.00 × 104 |
| Poly-P(12–13) | Mg2+ | 0.052 ± 0.006 | 2.63 ± 0.07 | 1.07 ± 0.03 | 2.06 × 104 |
| Poly-P(12–13) | Ca2+ | 0.063 ± 0.006 | 3.47 ± 0.08 | 1.41 ± 0.03 | 2.24 × 104 |
| ATP | None | 0.48 ± 0.05 | 1.52 ± 0.04 | 0.62 ± 0.02 | 1.29 × 103 |
| ATP | Ca2+ | 0.38 ± 0.04 | 1.50 ± 0.04 | 0.61 ± 0.02 | 1.61 × 103 |
| P-Tyr | None | 0.88 ± 0.10 | 0.19 ± 0.01 | 0.077 ± 0.004 | 8.75 × 10 |
| pNPPb | None | 0.67 ± 0.14 | 0.64 ± 0.04 | 0.26 ± 0.02 | 3.92 × 102 |
U/mg, µmoles min−1 mg−1 protein.
p-nitrophenyl phosphate.
Table IV.
Comparison of Kinetic Parameters
| Phosphatase | Substrate | kcat (s−1) |
kcat/Km (10−5 × M−1 s−1) |
|---|---|---|---|
| At1g05000 | Polyphosphate | 1.26 | 0.20 |
| VHR | pNPP | 7.14 | 1.25 |
| Yersinia | Peptide | 75.7 | 223 |
| Rat | Peptide | 1314 | 228 |
DISCUSSION
A survey of the genome of the plant Arabidopsis thaliana by Kerk et al.49 identified 112 genes predicted to encode phosphatases, with protein Ser/Thr phosphatases dominating. Of 18 predicted DSPases and two PTPases, 12 are cysteine phosphatases.
The structure of At1g05000 shows a protein fold that is typical for cysteine phosphatases. Cysteine phosphatases are frequently found in multimeric states,7, 8, 38 although monomers have been described as well.10, 50 At1g05000 was crystallized as an apparent hexamer and is, to the best of our knowledge, the first cysteine phosphatase-like enzyme to demonstrate a hexameric form. However, gel filtration experiments indicated a mixture dominated by a dimeric form but containing a small fraction of hexamer, and it was presumably this mixture that demonstrated phosphatase activity. Interchange between dimer and hexamer may well be a concentration-dependent phenomenon.
Cysteine phosphatase structures often contain phosphate(s)12 or sulfate(s)38 within a positively-charged pocket that includes the HCxxGxxR motif; sulfate presumably substitutes for the physiological phosphate. Two sulfate groups were observed in the catalytic site cavity of At1g05000, suggesting the possibility that a multiply phosphorylated physiological substrate may be recognized by the active site. Sulfates/phosphates in At1g05000 and in the baculovirus BVP12 and human VHR38 enzymes are rather distant (>3 Å) from the cysteine of the cysteine phosphatase active site motif, bringing their physiological relevance into doubt.
The closest structural homolog of At1g05000 (as identified by VAST) was the rat DSPase PRL-1 (Phosphatase of Regenerating Liver 1). This protein belongs to a small class of proposed PTPases that are involved in the regulation of cell growth and proliferation and are overexpressed in several types of metastatic tumors.7 A potentially important difference between these structures is the loop between β4 and α3 (Figs. 1 and 2); in many cysteine phosphatases, this region corresponds to the functionally important “WPD loop” that moves toward the active site during catalysis, positioning a conserved aspartate residue in the loop [Fig. 2(A)] to act as a general acid.51 The analogous loop in At1g05000 is longer and located far away from the active site in At1g05000. In addition, it lacks the aspartate residue, suggesting that this residue is not involved in catalysis in At1g05000. Instead, His155 is well-positioned to carry out the role of general acid [Fig. 2(B)]; analogous histidines are present in the yeast SIW14 protein [Fig. 3(B)] and many other cysteine phosphatases related to At1g05000 including four uncharacterized Arabidopsis proteins (At4g03960, At3g02800, At5g16480, and At2g32960), which share significant sequence similarity with At1g05000 (65–75% identity).
Another active site motif residue that likely takes part in catalysis is Arg156. In the baculovirus NTPase/RTPase, the arginine residue analogous to Arg156 forms bidentate contact with two phosphate oxygens, and in vivo activity is abolished upon mutation of this residue to alanine.10, 12 The position of Arg156 in At1g05000 suggests that it could bind the second phosphate of a multiply phosphorylated substrate [Fig. 3(A)]. The importance of residues analogous to this arginine as well as the cysteine have been demonstrated in the closely related SIW14 protein through mutations of SIW14 residues C214S and R220K, both of which resulted in proteins that were non-functional in vivo.42
Thr157 immediately C-terminal to the established active site motif (Figs. 2 and 3), matches the highly conserved Ser/Thr found at this position in many cysteine phosphatases; the hydroxyl is thought to be necessary for the rapid hydrolysis of the thiol phosphate intermediate44 Mutagenesis of the residue analogous to At1g05000 residue Gly153 was found to eliminate in vivo activity of SIW1442; although the function of this glycine is unknown, it is universally conserved in cysteine phosphatase active sites. The highly conserved histidine residue of the cysteine phosphatase active site motif (HCxxGxxR), represented by His149 in At1g05000, has been shown in the mouse PIPase10 and Yersinia PTPase52 to form a hydrogen bond with the backbone carboxyl of the active site cysteine and is proposed to stabilize the thiolate anion.43 Mutation of this histidine to alanine in the Yersinia PTPase and the baculovirus NTPase/RTPase reduced kcat by two to three orders of magnitude.43, 53 In At1g05000, the distance between the side chains of His149 and Cys150 are compatible with the possible hydrogen bonding.
Known NTPases/RTPases feature a conserved active site asparagine (HXxxGxNR; see also [Fig. 3(A)] that has been shown to be essential in the mouse enzyme.10 NTPases/RTPases also share the sequence (I/L)IxDLT(N/Y)T [Fig. 3(A)],10, 53 which has been shown to form a surface loop near the entrance of the active site pocket in the baculovirus enzyme, and several residues of this loop have been shown to be important for in vivo activity. At1g05000 lacks both the asparagine and this conserved sequence. Three hypothetical proteins in A. thaliana are better NTPase/RTPase candidates than is At1g05000; At5g01290.1, At5g28210.1, and At3g09100.1 share the Asn-containing active site sequence VHCTGxNRTG as well as the upstream consensus sequence VIDLTNT. The differences between At1g05000 and NTPases/RTPases suggest a different role or at least a different mechanism for At1g05000.
At1g05000 was able to act on phosphotyrosine in free form or within peptides (Table III), although affinity (0.8 mM) and efficiency (4.0 × 102 M−1 s−1) were less than with polyphosphate or nucleotides. In contrast to the PTPases from Yersinia and rats,43 phosphatase activity of At1g05000 was inhibited by P-Tyr concentrations greater than 3.0 mM. At1g05000 appears not to be a DSPase, as phosphoserine, phosphothreonine, or peptides containing them, were not substrates.
Surprisingly, several SH-group inhibitors (iodoacetate, DTNB, and N-ethylmaleimide) were largely ineffective in inhibiting activity of At1g05000 with pNPP as substrate. This suggests either that the cysteine of the cysteine-phosphatase active site-like pocket is inaccessible to these reagents, or that the enzyme acts acts via a mechanism different from that of described cysteine phosphatases.
PIPases were among the proteins with the greatest structural similarity to At1g05000, and the active site region of the PIPase PTEN shows significant sequence homology to that of At1g05000 [Fig. 3(A)], therefore it was of interest that several phosphatidyl inositols (l-α-phosphotidyl-d-myo-inositol 5-monophosphate and l-α-phosphotidyl-d-myo-inositol 3-monophosphate) were substrates for At1g05000, albeit with low levels of activity. This result is suggestive that a more extensive survey of phosphatidyl inositols might find some that support greater activity.
At1g05000 showed neither sequence nor structural similarity to any polyphosphate phosphatase (PPPase) except for the baculovirus NPTase/mRTPase, which catalyzes this reaction at a very low rate14; hence, it was surprising that polyphosphate (poly-P12–13) was a substrate and, in fact, the preferred substrate among those tested (Table III). Turnover rate (kcat) with poly-P was relatively slow and catalytic efficiency (kcat/Km) was weak when compared to some cysteine phosphatases with their preferred substrates (Table IV; data for the VHR DSPase are from Denu and Dixon44 and that for the Yersinia and rat PTPases are from Zhang et al.47), suggesting that poly-P or this particular chain length is not the physiological substrate.
Nevertheless, this is the first detection of significant PPPase activity in a cysteine phosphatase-like enzyme. Polyphosphate functions in a number of important cellular processes, including phosphate and energy storage, regulation of osmotic pressure, substitution for ATP in certain enzymes, and pH buffering.54 PPPases hydrolyze phosphate bonds of pyrophosphate, oligophosphates, long-chain phosphates, or combinations of these, and are characterized as exo-phosphatases (EC3.6.1.11) or endophosphatases (EC3.6.1.10). We are aware of only two enzymes demonstrated to function as PPPases in vivo that have also been characterized for enzymatic activity, the Saccharomyces cerevisiae endopolyphosphatase PPN155 and the E. coli exopolyphosphatase PPX.56 PPN1 was shown to function as a PPPase in vivo through mutagenesis of the ppn1 gene, which resulted in the accumulation of polyphosphate in the cell and loss of the ability to grow on minimal medium. PPN1, in contrast to At1g05000 and cysteine phosphatases, is a metallophosphatase and requires Mn2+ or Mg2+ for activity.57 PPN1 shows 340-fold greater affinity for poly-P750 (Km = 185 nM) than did At1g05000 for poly-P12–13 (63 µM) and its catalytic efficiency was approximately four orders of magnitude greater (kcat/Km for chain lengths polyP100–750 = 3 × 108 M−1 s−1 to 8 × 108 M−1 s−1 for PPN1 compared to 2 × 104 M−1 s−1 for At1g05000). The E. coli exopolyphosphatase PPX, which likely acts as a PPPase in vivo based on the location of the ppx gene adjacent to a gene for polyphosphate kinase56 and by cellular degradation of polyphosphate upon artificial induction of PPX,58 also requires a divalent cation for activity. Furthermore, it differs from At1g05000 in that it does not use ATP as a substrate. The Km of PPX for polyP(500) was 9 nM,56 nearly 4 orders of magnitude greater than that of At1g05000 for poly-P12–13. These comparisons again suggest that polyphosphate either is not the physiological substrate of At1g05000 or that its preferred polyP chain length has not been found.
CONCLUSIONS
Up to this point, the only solved structure of a plant enzyme with tyrosine phosphatase activity is the Arabidopsis cysteine phosphatase CDC25, a DSPase.59 Therefore, the structure of At1g05000 presented here is the first of a plant protein with tyrosine-specific phosphatase activity. At1g05000 bears a strong structural and sequence similarity to cysteine phosphatases; however, its activity was highly resistant to inhibition by SH-group inhibitors, suggesting that it may act via a different mechanism from the cysteine phosphatases. Whether the polyphosphatase and other activities detected are relevant to its in vivo function will need to be established by additional investigation.
Supplementary Material
ACKNOWLEDGMENTS
The Center for Eukaryotic Structural Genomics (CESG) is an NIH-funded structural genomics project with the goal of filling in protein structure space by solving protein structures by both X-ray Crystallography and Nuclear Magnetic Resonance (NMR). Special thanks goes to members of the Center for Eukaryotic Structural Genomics team including Todd Kimball, John Kunert, Nicholas Dillon, Rachel Schiesher, Juhyung Chin, Megan Riters, Andrew C. Olson, Jason M. Ellefson, Janet E. McCombs, Brendan T. Burns, Blake W. Buchan, Holalkere V. Geetha, Zhaohui Sun, Ip Kei Sam, Eldon L. Ulrich, Bryan Ramirez, Zsolt Zolnai, Peter T. Lee, Jianhua Zhang, Won Bae Jeon, John Primm, Michael R. Sussman, and Brian F. Volkman.
Grant sponsor: National Institutes of Health; Grant number: P50 GM64598; Grant sponsor: National Institute for General Medical Sciences; Grant number: U54 GM074901; Grant sponsor: Ontario Genomics Institute.
Abbreviations
- DSPase
dual-specificity phosphatase
- NTPase/RTPase
nucleotide triphosphatase/RNA 5′-triphosphatase
- ORF
open reading frame
- PIPase
phosphoinositol phosphatase
- pNPP
p-nitrophenyl phosphate
- PPPase
polyphosphate phosphatase
- PTPase
phosphotyrosine phosphatase
- RMSD
root mean square deviation
- SeMet
selenomethionine
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Mustelin T, Abraham RT, Rudd CE, Alonso A, Merlo JJ. Protein tyrosine phosphorylation in T cell signaling. Front Biosci. 2002;7:d918–d969. doi: 10.2741/A821. [DOI] [PubMed] [Google Scholar]
- 3.Mustelin T, Tasken K. Positive and negative regulation of T-cell activation through kinases and phosphatases. Biochem J. 2003;371(Part 1):15–27. doi: 10.1042/BJ20021637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan S. Protein phosphatases in plants. Annu Rev Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 5.Zhou G, Denu JM, Wu L, Dixon JE. The catalytic role of Cys124 in the dual specificity phosphatase VHR. J Biol Chem. 1994;269:28084–28090. [PubMed] [Google Scholar]
- 6.Guan KL, Dixon JE. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J Biol Chem. 1991;266:17026–17030. [PubMed] [Google Scholar]
- 7.Sun JP, Wang WQ, Yang H, Liu S, Liang F, Fedorov AA, Almo SC, Zhang ZY. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochemistry. 2005;44:12009–12021. doi: 10.1021/bi0509191. [DOI] [PubMed] [Google Scholar]
- 8.Song H, Hanlon N, Brown NR, Noble ME, Johnson LN, Barford D. Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Mol Cell. 2001;7:615–626. doi: 10.1016/s1097-2765(01)00208-8. [DOI] [PubMed] [Google Scholar]
- 9.Diamond RH, Cressman DE, Laz TM, Abrams CS, Taub R. PRL-1, a unique nuclear protein tyrosine phosphatase, affects cell growth. Mol Cell Biol. 1994;14:3752–3762. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Changela A, Ho CK, Martins A, Shuman S, Mondragon A. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. Embo J. 2001;20:2575–2586. doi: 10.1093/emboj/20.10.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martins A, Shuman S. Mutational analysis of baculovirus phosphatase identifies structural residues important for triphosphatase activity in vitro and in vivo. Biochemistry. 2002;41:13403–13409. doi: 10.1021/bi0265426. [DOI] [PubMed] [Google Scholar]
- 12.Changela A, Martins A, Shuman S, Mondragon A. Crystal structure of baculovirus RNA triphosphatase complexed with phosphate. J Biol Chem. 2005;280:17848–17856. doi: 10.1074/jbc.M500885200. [DOI] [PubMed] [Google Scholar]
- 13.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 14.Martins A, Shuman S. The domain order of mammalian capping enzyme can be inverted and baculovirus phosphatase can function in cap formation in vivo. Virology. 2002;304:167–175. doi: 10.1006/viro.2002.1606. [DOI] [PubMed] [Google Scholar]
- 15.Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thao S, Zhao Q, Kimball T, Steffen E, Blommel PG, Riters M, Newman CS, Fox BG, Wrobel RL. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J Struct Funct Genomics. 2004;5:267–276. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- 18.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr, Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40:256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Jeon WB, Aceti DJ, Bingman CA, Vojtik FC, Olson AC, Ellefson JM, McCombs JE, Sreenath HK, Blommel PG, Seder KD, Burns BT, Geetha HV, Harms AC, Sabat G, Sussman MR, Fox BG, Phillips GN., Jr High-throughput purification and quality assurance of Arabidopsis thaliana proteins for eukaryotic structural genomics. J Struct Funct Genomics. 2005;6:143–147. doi: 10.1007/s10969-005-1908-7. [DOI] [PubMed] [Google Scholar]
- 20.Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: sesame. J Struct Funct Genomics. 2003;4:11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59(Part 11):1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 23.Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr D Biol Crystallogr. 1998;54(Part 4):487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56(Part 4):965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McRee DE. XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Part 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Lovell SC, Davis IW, Arendall WB, III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 29.Kuznetsova E, Proudfoot M, Sanders SA, Reinking J, Savchenko A, Arrowsmith CH, Edwards AM, Yakunin AF. Enzyme genomics: application of general enzymatic screens to discover new enzymes. FEMS Microbiol Rev. 2005;29:263–279. doi: 10.1016/j.femsre.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 31.Saheki S, Takeda A, Shimazu T. Assay of inorganic phosphate in the mild pH range, suitable for measurement of glycogen phosphorylase activity. Anal Biochem. 1985;148:277–281. doi: 10.1016/0003-2697(85)90229-5. [DOI] [PubMed] [Google Scholar]
- 32.Heering HA, Weiner JH, Armstrong FA. J Am Chem Soc. 1997;119:11628–11638. [Google Scholar]
- 33.Pearl FM, Bennett CF, Bray JE, Harrison AP, Martin N, Shepherd A, Sillitoe I, Thornton J, Orengo CA. The CATH database: an extended protein family resource for structural and functional genomics. Nucleic Acids Res. 2003;31:452–455. doi: 10.1093/nar/gkg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madej T, Gibrat JF, Bryant SH. Threading a database of protein cores. Proteins. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- 36.Begley MJ, Taylor GS, Brock MA, Ghosh P, Woods VL, Dixon JE. Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc Natl Acad Sci U S A. 2006;103:927–932. doi: 10.1073/pnas.0510006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 39.Chu HM, Guo RT, Lin TW, Chou CC, Shr HL, Lai HL, Tang TY, Cheng KJ, Selinger BL, Wang AH. Structures of Selenomonas ruminantium phytase in complex with persulfated phytate: DSP phytase fold and mechanism for sequential substrate hydrolysis. Structure. 2004;12:2015–2024. doi: 10.1016/j.str.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deshpande T, Takagi T, Hao L, Buratowski S, Charbonneau H. Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5′-triphosphatase and diphosphatase activities. J Biol Chem. 1999;274:16590–16594. doi: 10.1074/jbc.274.23.16590. [DOI] [PubMed] [Google Scholar]
- 42.Care A, Vousden KA, Binley KM, Radcliffe P, Trevethick J, Mannazzu I, Sudbery PE. A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics. 2004;166:707–719. doi: 10.1093/genetics/166.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang ZY, Dixon JE. Active site labeling of the Yersinia protein tyrosine phosphatase: the determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry. 1993;32:9340–9345. doi: 10.1021/bi00087a012. [DOI] [PubMed] [Google Scholar]
- 44.Denu JM, Dixon JE. A catalytic mechanism for the dual-specific phosphatases. Proc Natl Acad Sci U S A. 1995;92:5910–5914. doi: 10.1073/pnas.92.13.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daum G, Solca F, Diltz CD, Zhao Z, Cool DE, Fischer EH. A general peptide substrate for protein tyrosine phosphatases. Anal Biochem. 1993;211:50–54. doi: 10.1006/abio.1993.1231. [DOI] [PubMed] [Google Scholar]
- 46.Harder KW, Owen P, Wong LK, Aebersold R, Clark-Lewis I, Jirik FR. Characterization and kinetic analysis of the intracellular domain of human protein tyrosine phosphatase beta (HPTP beta) using synthetic phosphopeptides. Biochem J. 1994;298(Part 2):395–401. doi: 10.1042/bj2980395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang ZY, Maclean D, Thieme-Sefler AM, Roeske RW, Dixon JE. A continuous spectrophotometric and fluorimetric assay for protein tyrosine phosphatase using phosphotyrosine-containing peptides. Anal Biochem. 1993;211:7–15. doi: 10.1006/abio.1993.1224. [DOI] [PubMed] [Google Scholar]
- 48.Agazie YM, Hayman MJ. Development of an efficient “substrate-trapping” mutant of Src homology phosphotyrosine phosphatase 2 and identification of the epidermal growth factor receptor. Gab1, and three other proteins as target substrates. J Biol Chem. 2003;278:13952–13958. doi: 10.1074/jbc.M210670200. [DOI] [PubMed] [Google Scholar]
- 49.Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002;129:908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray CH, Good VM, Tonks NK, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. Embo J. 2003;22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denu JM, Zhou G, Guo Y, Dixon JE. The catalytic role of aspartic acid-92 in a human dual-specific protein-tyrosine-phosphatase. Biochemistry. 1995;34:3396–3403. doi: 10.1021/bi00010a031. [DOI] [PubMed] [Google Scholar]
- 52.Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, Saper MA. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate. Nature. 1994;370:571–575. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 53.Martins A, Shuman S. Mechanism of phosphoanhydride cleavage by baculovirus phosphatase. J Biol Chem. 2000;275:35070–35076. doi: 10.1074/jbc.M005748200. [DOI] [PubMed] [Google Scholar]
- 54.Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 55.Sethuraman A, Rao NN, Kornberg A. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:8542–8547. doi: 10.1073/pnas.151269398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 57.Kumble KD, Kornberg A. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J Biol Chem. 1996;271:27146–27151. doi: 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- 58.Van Dien SJ, Keasling JD. Effect of polyphosphate metabolism on the Escherichia coli phosphate-starvation response. Biotechnol Prog. 1999;15:587–593. doi: 10.1021/bp990067u. [DOI] [PubMed] [Google Scholar]
- 59.Landrieu I, Hassan S, Sauty M, Dewitte F, Wieruszeski JM, Inze D, De Veylder L, Lippens G. Characterization of the Arabidopsis thaliana Arath;CDC25 dual-specificity tyrosine phosphatase. Biochem Biophys Res Commun. 2004;322:734–739. doi: 10.1016/j.bbrc.2004.07.182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.