Abstract
BACKGROUND
Amyotrophic lateral sclerosis (ALS) is an intractable neurodegenerative disease that primarily affects motor neurons in the cerebral cortex and the spinal cord. Recent evidence indicates that dysfunction of oligodendrocytes is implicated in the pathogenesis of ALS. The basic helix–loop–helix (bHLH) transcription factor Olig2 plays a pivotal role in the development of both motor neurons and oligodendrocytes in the progenitor of motor neuron (pMN) domain of the spinal cord, supporting evidence for the shared motor neuron/oligodendrocyte lineage. However, a comprehensive profile of Olig2 target genes in pMNs and oligodendrocyte progenitor cells (OPCs) with relevance to the pathogenesis of ALS remains to be characterized.
METHODS
By analyzing the ChIP-Seq datasets numbered SRP007566 and SRP015333 with the Strand NGS program, we identified genome-wide Olig2 target genes in pMNs and OPCs, followed by molecular network analysis using three distinct bioinformatics tools.
RESULTS
We identified 5966 Olig2 target genes in pMNs, including Nkx2.2, Pax6, Irx3, Ngn2, Zep2 (Cip1), Trp3, Mnx1 (Hb9), and Cdkn1a, and 1553 genes in OPCs. The genes closely related to the keyword “alternative splicing” were enriched in the set of 740 targets overlapping between pMNs and OPCs. Furthermore, approximately one-third of downregulated genes in purified motor neurons of presymptomatic mutant SOD1 transgenic mice and in lumbar spinal cord tissues of ALS patients corresponded to Olig2 target genes in pMNs. Molecular networks of Olig2 target genes indicate that Olig2 regulates a wide range of genes essential for diverse neuronal and glial functions.
CONCLUSIONS
These observations lead to a hypothesis that aberrant regulation of Olig2 function, by affecting biology of both motor neurons and oligodendrocytes, might be involved in the pathogenesis of ALS.
Keywords: amyotrophic lateral sclerosis, ChIP-Seq, GenomeJack, KeyMolnet, motor neurons, Olig2, oligodendrocytes, RNA-Seq, Strand NGS
Introduction
Amyotrophic lateral sclerosis (ALS) is an intractable neurodegenerative disease, characterized by rapid and progressive degeneration of motor neurons in the cerebral cortex and the spinal cord. Approximately 10% of ALS is of familial origin caused by genetic mutations of superoxide dismutase 1 (SOD1), TAR DNA binding protein (TARDBP, TDP-43), fused in sarcoma (FUS), optineurin (OPTN), ubiquilin 2 (UBQLN2), and chromosome 9 open reading frame 72 (C9orf72).1 At present, the precise molecular mechanism underlying motor neuron degeneration in sporadic ALS remains unknown. Increasing evidence indicates that dysfunction of oligodendrocytes plays a key role in pathological processes of ALS. The levels of expression of oligodendrocyte-specific monocarboxylate transporter 1, which transports lactate, an essential energy metabolite to motor neurons, are greatly reduced in the motor cortex of ALS patients and the spinal cord of SOD1 mutant mice.2 In the spinal cord of ALS patients, TDP-43-positive inclusions accumulate not only in motor neurons but also in oligodendrocytes.3 Extensive degeneration of gray matter oligodendrocytes is found in the motor cortex and the spinal cord of ALS patients.4 Furthermore, genetic deletion of mutant SOD1 selectively from oligodendrocytes markedly ameliorates the disease course in SOD1 mutant mice.4
The basic helix–loop–helix (bHLH) transcription factor Olig2, a downstream target of a ventralizing factor Sonic hedgehog (SHH) that is secreted from the notochord and floor plate, plays a pivotal role in the development and differentiation of both motor neurons and oligodendrocytes in the spinal cord.5 Olig2 is expressed in a restricted domain of the spinal cord ventricular zone named progenitor of motor neuron (pMN), which is composed of a pool of progenitor cells that sequentially generate motor neurons and oligodendrocytes, supporting evidence for the shared motor neuron/oligodendrocyte lineage.6,7 In contrast, V0, V1, V2, and V3 interneurons are separately generated from the p0, p1, p2, and p3 domains, respectively.8 The pMN domain is bounded ventrally by the p3 domain defined by the expression of NK2 homeobox 2 (Nkx2.2, Nkx2–2), a homeodomain transcription factor, and dorsally by the p2 domain, whose ventral boundary is defined by the expression of Iroquois related homeobox 3 (Irx3), another homeodomain protein.6 Both motor neurons and oligodendrocytes are almost completely lost in mice with a homozygous inactivation of Olig2.9 Olig2 specifies the pMN identity by direct repression of interneuron transcription programs.10 Notably, in Oligo1/2 double-mutant mice, pMN progenitors generate V2 interneurons and astrocytes.11 A microRNA mir-17–3p silences the expression of Olig2 in the p2 domain to generate V2 interneurons.12 Olig2 plays a decisive role in the specification of the pMN domain and initiates motor neuron differentiation by inducing expression of a proneural bHLH factor neurogenin 2 (Neurog2, Ngn2), which drives pMN progenitors to leave the cell cycle and express differentiation markers characteristic of post-mitotic neurons.13 Ngn2 expression is extinguished from the progeny destined to become oligodendrocytes, whereas the expression of Olig2 is downregulated in the progeny destined to become motor neurons.14 In contrast, the expression of Nkx2.2 is upregulated in pMN progenitors as they switch from producing motor neurons to generating oligodendrocytes.15
Structurally, Olig2 is characterized by the presence of the bHLH domain that mediates dimerization and binding to target DNA sequences, often containing the core hexanucleotide motif composed of CANNTG named the E-box.5 The bHLH domains of Olig1 and Olig2 exhibit more than 80% amino acid identities. However, Olig1 and Olig2 exert non-overlapping biological functions attributable to differential interactions with dimeric partners and co-regulator proteins.5 Olig2 favors forming a homodimer, while Ser147 phosphorylation of Olig2 by protein kinase A induces motor neuron differentiation by switching of dimerization partner from Olig2 to Ngn2.16 The levels of Olig2 expression in the developing spinal cord are much higher than those of Olig1, and Olig2 has a broader expression pattern in the embryonic forebrain.9 Furthermore, deletion of Olig1 has no visible impact on the generation of motor neurons and early oligodendrocyte progenitors.5,17 Olig2 generally acts as a transcriptional repressor of direct target genes.5,13 However, at present, the complete profile of Olig2 target genes in both pMNs and oligodendrocyte progenitor cells (OPCs) remains to be intensively characterized, particularly with relevance to the pathogenesis of ALS.
Recently, an advanced next-generation sequencing (NGS) technology termed chromatin immunoprecipitation, followed by deep sequencing (ChIP-Seq), encouraged us to characterize genome-wide profiles of DNA-binding proteins and histone modifications.18 ChIP-Seq, with the advantages of higher resolution, less noise, and greater coverage of the genome compared with microarray-based ChIP-Chip, serves as an innovative tool for studying gene regulatory networks on the whole-genome scale. To investigate a role of Olig2 in the pathogenesis of ALS, we attempted to characterize the comprehensive set of ChIP-Seq-based Olig2 direct target genes in pMNs and OPCs and their molecular networks by analyzing datasets retrieved from the public database.
Methods
ChIP-Seq dataset
We retrieved an Olig2 ChIP-Seq dataset of embryonic stem cells (ESCs) during motor neuron differentiation from the DDBJ Sequence Read Archive (DRA) (trace.ddbj.nig.ac.jp/DRASearch) under the accession number of SRP007566. The original experiment was carried out by the researchers in Dr. Hynek Wichterle’s Laboratory, Columbia University Medical Center.10 The relevant dataset numbered GSE30882 is open to public since November 12, 2011. They generated a mouse ESC line incorporated with a transgene composed of doxycycline (DOX)-inducible Olig2 tagged with the V5 epitope in the carboxyl terminal, named iOlig2-V5. Then, motor neuron differentiation was induced by exposure of embryoid bodies derived from ESCs to 1 μM all-trans retinoic acid (RA, Sigma) and 0.5 μM Smoothened Agonist (SAG, Merck/Calbiochem), an agonist of hedgehog signaling. This was followed by a 24-hour treatment with 1 μg/mL DOX starting on day 3, resulting in differentiation of motor neurons progenitors (pMNs) expressing an Olig2-V5 fusion protein, along with endogenous Olig2 (Olig2-V5+pMNs). In some experiments, pMN cells untreated with DOX (Olig2-V5− pMNs) were utilized to detect binding sites for an endogenous Olig2 protein. The cells were fixed with formaldehyde for ChIP. Sonicated nuclear extracts were immunoprecipitated with a rabbit polyclonal anti-Olig2 antibody (AB15328, Mil-lipore) for ChIP of Olig2-V5− pMNs (SRR315585), a rabbit polyclonal anti-V5 antibody (ab15828, Abcam) for ChIP of Olig2-V5+ pMNs (SRR315586), or with ab15828 for ChIP of Olig2-V5− pMNs serving as a negative control (SRR315590). NGS libraries constructed from adapter-ligated ChIP DNA fragments were processed for deep sequencing on Genome Analyzer IIx (Illumina).
To compare Olig2 target genes in pMNs with those in OPCs, we retrieved another Olig2 ChIP-Seq dataset from DRA under the accession number of SRP015333. The original experiment was carried out by the researchers in Dr. Qing Richard Lu’s Laboratory, University of Texas Southwestern Medical Center.19 The relevant dataset numbered GSE40506 is open to public since August 31, 2012. In their experiment, purified OPCs isolated from newborn rat cerebral cortices at P2 were grown in the OPC growth medium containing 10 ng/mL platelet-derived growth factor (PDGF)-AA and 20 ng/mL basic fibroblast growth factor (bFGF). Then, they were incubated for 3 days in the oligodendrocyte differentiation medium containing 15 nM triiodothyronine (T3) and 10 ng/mL ciliary neurotrophic factor (CNTF). The cells were fixed with formaldehyde for ChIP. Sonicated nuclear extracts were immunoprecipitated with a rabbit polyclonal anti-Olig2 antibody (ab136253, Abcam) for ChIP of the endogenous Olig2 protein expressed in differentiating oligodendrocytes (SRR548313), or with rabbit anti-human IgG (ab2410, Abcam) serving as a negative control (SRR548314). NGS libraries constructed from adapter-ligated ChIP DNA fragments were processed for deep sequencing on Genome Analyzer IIx.
RNA-Seq datasets
To study the involvement of Olig2 target genes in motor neuron degeneration in vivo in an animal model of ALS, we first retrieved a RNA-Seq dataset from DRA under the accession number of SRP013849. The original experiment was carried out by the researchers in Dr. Arthur L. Horwich’s Laboratory, Yale University School of Medicine.20 The relevant dataset numbered GSE38820 is open to public since December 4, 2012. In their experiment, motor neuron cell bodies were isolated by laser-captured microdissection (LCM) from spinal cords of transgenic mouse strains at 3 months of age in the presymptomatic stage, including wild-type SOD1-YFP (strain 592; SRR515121 and SRR515122) and mutant G85R SOD1-YFP (strain 737; SRR515123 and SRR515124). Total RNA extracted from pooled motor neuron cell bodies was subjected to polyA selection, fragmentation, cDNA synthesis, adaptor ligation, and library amplification. NGS libraries were processed for single-end sequencing on Genome Analyzer IIx.
To verify a pivotal role of Olig2 target genes in vivo in ALS patients, next we retrieved another RNA-Seq dataset numbered SRP033464 from DRA. The original experiment was carried out by the researchers in Dr. Oleg Butovsky’s Laboratory, Brigham and Women’s Hospital, Harvard Medical School.21 The relevant dataset numbered GSE52946 is open to public since November 24, 2014. In their experiment, whole lumbar spinal cord tissues were isolated from eight sporadic and two familial ALS (SOD1 A4V) patients and 10 healthy control subjects according to the guideline approved by the Institutional Review Boards at Massachusetts General Hospital and Brigham and Women’s Hospital. Total RNA was extracted by using mirVana™ miRNA isolation kit (Ambion) according to the manufacturer’s protocol. Paired-end RNA-Seq analysis was performed on HiSeq 2000 (Illumina) in the Harvard genetic facility.
NGS data analysis
First, we evaluated the quality of NGS short reads by searching on the FastQC program (www.bioinformatics.babraham.ac.uk/projects/fastqc). Then, we removed the reads of insufficient quality by filtering them out by the FASTX toolkit (hannonlab.cshl.edu/fastx_toolkit). For ChIP-Seq analysis, we mapped the cleaned data on the mouse genome reference sequence version mm9 or the rat genome reference sequence version rn4 by a mapping tool named COBWeb of the Strand NGS2.0 program, formerly named Avadis NGS (Strand Genomics) or by Bowtie version 1.0.0 or 2.1.0 (bowtie-bio.sourceforge.net). Then, we identified the peaks of the binding sites by using the Model-based Analysis of ChIP-Seq (MACS) program, as described previously.22 We determined the genes corresponding to the peaks by a neighboring gene analysis tool of Strand NGS in the setting within a distance of 5000 bp from peaks to genes. We identified the consensus motif sequences surrounding the peaks by using the GADEM program23 and the genomic location of binding peaks by a peak-finding tool of Strand NGS. We imported the processed data into a genome viewer named GenomeJack v1.4 (Mitsubishi Space Software).22 For RNA-Seq analysis, after removing poly-A tails and low-quality reads from NGS short-read data, we mapped them on mm9 or hg19 by TopHat version 2.0.9 (ccb.jhu.edu/software/tophat/index.shtml), and identified differentially expressed genes that satisfied the significance expressed as Q-value (FDR-adjusted P-value) ≤0.01 by Cufflinks version 2.1.1 (cufflinks.cbcb.umd.edu).
Molecular network analysis
To identify molecular networks biologically relevant to ChIP-Seq-based Olig2 target genes, we imported the corresponding Entrez Gene IDs into the Functional Annotation tool of Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (david.abcc.ncifcrf.gov).24 DAVID identifies relevant pathways constructed by the Kyoto Encyclopedia of Genes and Genomes (KEGG), composed of the genes enriched in the given set, followed by statistical evaluation by a modified Fisher’s exact test after correction by Bonferroni multiple comparison test. KEGG (www.kegg.jp) is a publicly accessible knowledge base that covers a wide range of pathway maps on metabolic, genetic, environmental, and cellular processes, as well as human diseases, and is currently composed of 381,274 pathways generated from 473 reference pathways.25
We also imported Entrez Gene IDs into Ingenuity Pathways Analysis (IPA) (Ingenuity Systems; www.ingenuity.com). IPA is a commercial knowledge base that contains approximately 3,000,000 biological and chemical interactions and functional annotations with definite scientific evidence. By uploading the list of Gene IDs, the network-generation algorithm identifies focused genes integrated in global molecular pathways and networks in the setting of 70 molecules per network. IPA calculates the score P-value that reflects the statistical significance of association between the genes and the pathways or networks by the Fisher’s exact test.
KeyMolnet (KM Data; www.km-data.jp), another commercial knowledge base, contains manually curated contents on 164,000 relationships among human genes and proteins, small molecules, diseases, pathways, and drugs.26 They include the core contents collected from selected review articles with the highest reliability. By importing the list of Gene ID, KeyMolnet automatically provides the corresponding molecules as nodes on the network. The neighboring network-search algorithm selected one or more molecules as starting points to generate the network of all kinds of molecular interactions around starting molecules, including direct activation/inactivation, transcriptional activation/repression, and the complex formation within one path from the starting points. The generated network was compared side by side with 501 human canonical pathways of the KeyMolnet library. The algorithm, which counts the number of overlapping molecular relations between the extracted network and the canonical pathway, makes it possible to identify the canonical pathway showing the most significant contribution to the extracted network.
Results
Identification of 5966 ChIP-Seq-based Olig2 target genes in motor neuron progenitor cells
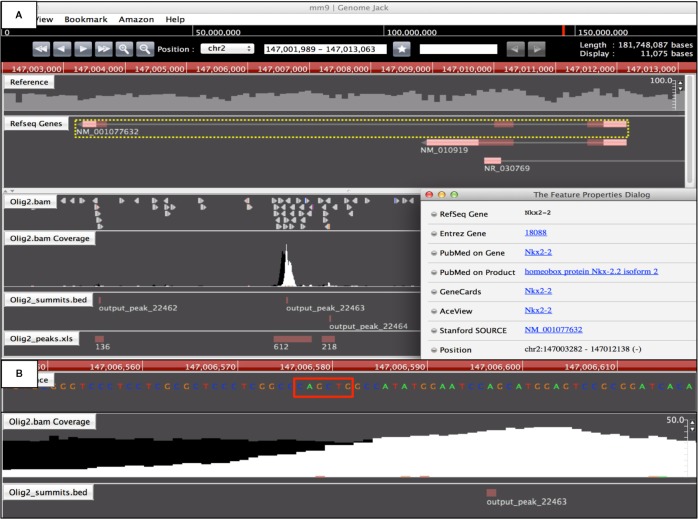
First, we evaluated the quality of ChIP-Seq data reanalyzed in the present study. After cleaning short-read data, the quality scores exceeded 20 across the bases on FastQC, indicating an acceptable quality of the data for downstream analysis (Supplementary Fig. 1). After mapping cleaned data on mm9 by COBWeb, we identified 43,419 ChIP-Seq peaks for the binding sites of the endogenous Olig2 protein and 24,112 ChIP-Seq peaks for binding sites of the Olig2-V5 fusion protein in mouse ESC-derived pMNs progenitors. We extracted the set of 20,043 peaks shared between both as the most reliable set of Olig2 ChIP-Seq peaks. From these, we collected the peaks located within a distance of 5000 bp from protein coding genes. Finally, we identified a set of 5966 Olig2 target genes that satisfied fold enrichment (FE) ≥10 and false discovery rate (FDR) ≤0.1 (Supplementary Table 1). Importantly, the list contained previously reported Olig2 direct targets,10,19 such as Nkx2.2 (FE = 353.7) (Fig. 1), Zeb2 (Sip1, FE = 88.5), Pax6 (FE = 87.4), and Irx3 (FE = 24.8). Furthermore, we identified Trp53 (FE = 108.4) and Ngn2 (EF = 28.6) as a category of unreported Olig2 targets (Supplementary Table 1). The peaks were distributed in the upstream (12.9%), 5′UTR (11.8%), 3′UTR (2.0%), coding exonic (13.1%), and intronic (54.3%) regions. The motif analysis by GADEM revealed a relative enrichment of the E-box consensus sequence defined as 5′-CAGCTG-3′ located within genomic regions surrounding ChIP-Seq peaks (Fig. 1 and Supplementary Fig. 2).
Figure 1.
Genomic location of Olig2 ChIP-Seq peak on the Nkx2-2 gene. The genomic location of Olig2 ChIP-Seq peak was determined by importing the processed data into GenomeJack. An example of homeobox protein Nkx2–2 (Nkx2.2; Entrez Gene ID 18088) is shown, where a MACS peak numbered 22463 in the Olig2.bam Coverage track is located in the intronic region of the Nkx2-2 gene, isoform 2 (panel A) with an E-box consensus sequence motif highlighted by orange square (panel B).
Identification of 1553 ChIP-Seq-based Olig2 target genes in OPCs
Next, we studied Olig2 target genes in rat OPCs by mapping ChIP-Seq data on rn4 by COBWeb. We identified 10,756 ChIP-Seq peaks in OPCs upon differentiation into oligodendrocytes. From these, we extracted the peaks located within a distance of 5000 bp from protein coding genes. Finally, we identified a set of 1553 Olig2 target genes that satisfied FE ≥10 and FDR ≤0.1 (Supplementary Table 2). The peaks were distributed in the upstream (9.2%), 5′UTR (11.8%), 3′UTR (1.3%), coding exonic (10.4%), and intronic (59.5%) regions. Importantly, the set of 740 genes among them were overlapped between pMNs and OPCs (Supplementary Table 3, underlined in Supplementary Tables 1 and 2). Importantly, functional annotation analysis by DAVID showed that 320 genes (43.2%) out of the set of 740 Olig2 target genes overlapping between pMNs and OPCs were significantly related to “alternative splicing” in the Swiss-Prot (SP)/Protein Information Resource (PIR) keyword (P = 3.73E–31 corrected by Bonferonni; underlined in Supplementary Table 3).
Molecular networks of ChIP-Seq-based Olig2 target genes in motor neuron progenitor cells and OPCs
Next, we studied molecular networks of Olig2 target genes by using three distinct pathway analysis tools of bioinformatics. By using DAVID, we identified functionally associated gene ontology (GO) terms. For 5966 Olig2 targets in pMNs, the most significant GO terms included “cell morphogenesis” (GO:0000902; P = 3.17E–16 corrected by Bonferroni) for biological process, “plasma membrane” (GO:0005886; P = 5.54E–18) for cellular component, and “metal ion binding” (GO:0046872; P = 6.46E–20) for molecular function. For 1533 Olig2 targets in OPCs, the most significant GO terms included “intracellular signaling cascade” (GO:0007242; P = 2.53E–05) for biological process, “plasma membrane” (GO:0005886; P = 1.13E–05) for cellular component, and “ion binding” (GO:0043167; P = 3.07E–04) for molecular function.
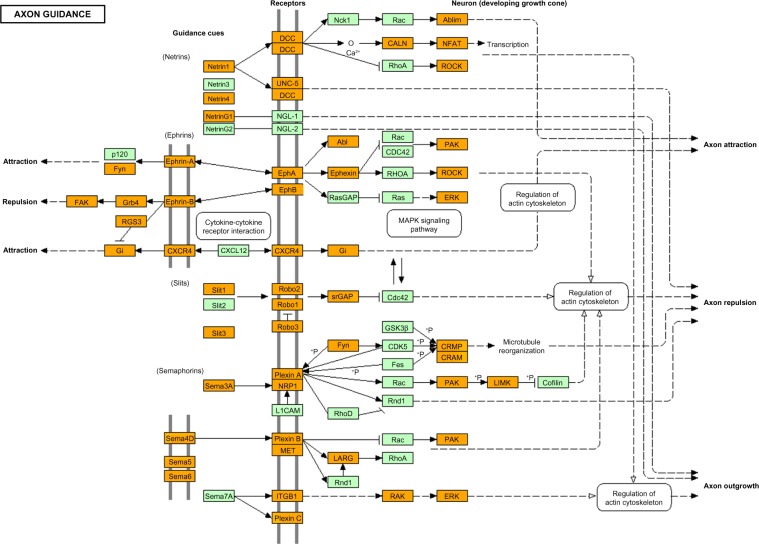
By KEGG pathway analysis, the set of 5966 Olig2 targets in pMNs showed a significant relationship with top three pathways defined as “Pathways in cancer” (mmu05200; P = 5.08E–08 corrected by Bonferonni), “Axon guidance” (mmu04360; P = 1.76E–09) (Fig. 2), and “Focal adhesion” (mmu04510; P = 2.90E–07) (Table 1). Importantly, they also exhibited significant association with the pathways defined as “Glioma” (mmu05214; rank 17, P = 0.024) (Supplementary Fig. 3) and “Hedgehog signaling pathway” (mmu04340; rank 18, P = 0.033), where patched homolog 1 (Ptch1), the receptor for SHH, corresponded to one of Olig2 targets (Table 1). From the 5966 genes described above, we extracted the set of 4717 Olig2 target genes that satisfied 10 times more stringent FDR (FDR ≤0.01) (Supplementary Table 4). We imported them into the Functional Annotation tool of DAVID to identify relevant KEGG pathways. Again, we found that the set of 4717 Olig2 target genes in pMN showed the most significant relationship with “Axon guidance” (mmu04360; P = 1.76E–09 corrected by Bonferroni), “Pathways in cancer” (mmu05200; P = 4.82E–09), “MAPK signaling pathway” (mmu04010; P = 7.11E–06), and “Focal adhesion” (mmu04510; P = 2.73–04), validating the reliability of initial results. For the 1533 Olig2 targets in OPCs, the top three KEGG pathways included “MAPK signaling pathway” (rno04010; P = 1.49E–05 corrected by Bonferroni), “Long-term depression” (rno04730; P = 3.34E–05), and “Vascular smooth muscle contraction” (rno04270; P = 3.57E–03).
Figure 2.
KEGG “Axon guidance” pathway relevant to Olig2 target genes in motor neuron progenitor cells. Entrez Gene IDs of 5966 Olig2 target genes in pMNs were imported into the Functional Annotation tool of DAVID. It extracted the KEGG “Axon guidance” pathway (mmu04360) as the second rank pathway (Table 1). Olig2 target genes are colored orange.
Table 1.
KEGG pathways relevant to ChIP-Seq-based Olig2 target genes in motor neuron progenitor cells.
| RANK | PATHWAYS | FOCUSED GENES | P-VALUE (BONFERRONI CORRECTED) | FDR |
|---|---|---|---|---|
| 1 | mmu05200: Pathways in cancer | Abl1, Acvr1c, Akt1, Akt3, Apc, Appl1, Arnt2, Axin2, Bax, Bcl2, Bcl2l1, Bcr, Casp3, Casp9, Cblc, Ccdc6, Ccnd1, Cdh1, Cdk2, Cdk4, Cdk6, Cdkn1a, Cdkn2a, Cdkn2b, Cebpa, Chuk, Cks1b, Col4a1, Col4a2, Col4a4, Col4a6, Crkl, Ctbp2, Ctnna1, Ctnna2, Ctnna3, Ctnnb1, Cycs, Dapk1, Dapk2, Dcc, E2f1, E2f3, Egfr, Egln2, Egln3, Erbb2, Fadd, Fas, Fgf1, Fgf10, Fgf11, Fgf12, Fgf14, Fgf18, Fgf2, Fgf21, Fgf7, Fgfr1, Fgfr2, Fgfr3, Flt3, Flt3l, Fn1, Fzd2, Fzd5, Fzd7, Fzd8, Fzd9, Gli1, Gli2, Gli3, Hhip, Hsp90ab1, Hsp90b1, Igf1, Igf1r, Itga2, Itga6, Itgav, Itgb1, Jak1, Jun, Jup, Kit, Kitl, Lama1, Lama3, Lama5, Lamb3, Lamc1, Lef1, Mapk1, Mapk10, Mapk3, Mecom, Met, Mitf, Mlh1, Msh3, Mtor, Nfkbia, Ntrk1, Pdgfrb, Pias2, Pias4, Pik3cb, Pik3r1, Pik3r2, Plcg1, Plcg2, Ppard, Pparg, Prkca, Prkcb, Ptch1, Ptch2, Ptk2, Ralgds, RarA, Rarb, Rb1, Ret, Runx1t1, Rxra, Rxrb, Rxrg, Sfpi1, Slc2a1, Smad2, Smad3, Sos1, Stat1, Stat3, Sufu, Tcf7l1, Tcf7l2, Tgfa, Tgfb2, Tpm3, Traf4, Traf5, Traf6, Trp53, Vegfc, Wnt1, Wnt10a, Wnt5a, Wnt5b, Wnt7a, Wnt8b, Wnt9a, Zbtb16 | 4.29E-14 | 2.78E-13 |
| 2 | mmu04360: Axon guidance | Abl1, Ablim1, Ablim2, Ablim3, Arhgef12, Cxcr4, Dcc, Dpysl2, Dpysl5, Efna5, Efnb1, Efnb2, Efnb3, Epha1, Epha2, Epha3, Epha4, Epha8, Ephb1, Ephb2, Ephb3, Fyn, Gnai2, Itgb1, Limk1, Mapk1, Mapk3, Met, Nck2, Nfatc1, Nfatc4, Ngef, Nrp1, Ntn1, Ntn4, Ntng1, Pak2, Pak3, Pak4, Pak6, Pak7, Plxna1, Plxna2, Plxna4, Plxnb2, Plxnc1, Ppp3ca, Ptk2, Rgs3, Robo1, Robo2, Robo3, Rock2, Sema3a, Sema3c, Sema3e, Sema3g, Sema4b, Sema5a, Sema5b, Sema6a, Sema6b, Sema6d, Slit1, Slit3, Srgap1, Srgap2, Srgap3, Unc5a, Unc5b, Unc5c, Unc5d | 1.76E-09 | 1.14E-08 |
| 3 | mmu04510: Focal adhesion | Actb, Actn1, Actn3, Actn4, Akt1, Akt3, Bcar1, Bcl2, Capn2, Cav1, Ccnd1, Ccnd2, Ccnd3, Col1a1, Col2a1, Col4a1, Col4a2, Col4a4, Col4a6, Col5a2, Col6a6, Crkl, Ctnnb1, Diap1, Dock1, Egfr, Elk1, Erbb2, Flna, Flnb, Flnc, Flt1, Fn1, Fyn, Igf1, Igf1r, Itga1, Itga2, Itga6, Itga8, Itga9, Itgav, Itgb1, Itgb5, Itgb6, Jun, Kdr, Lama1, Lama3, Lama5, Lamb3, Lamc1, Mapk1, Mapk10, Mapk3, Met, Myl7, Myl9, Mylk, Mylk3, Pak2, Pak3, Pak4, Pak6, Pak7, Parva, Parvb, Pdgfd, Pdgfrb, Pik3cb, Pik3r1, Pik3r2, Ppp1cc, Prkca, Prkcb, Ptk2, Rap1a, Rap1b, Reln, Rock2, Shc2, Sos1, Src, Thbs1, Thbs2, Tln2, Tnc, Tnr, Vav2, Vav3, Vegfc, Vwf | 2.90E-07 | 1.87E-06 |
| 4 | mmu04010:MAPK signaling pathway | Acvr1c, Akt1, Akt3, Arrb1, B230120H23Rik, Bdnf, Cacna1a, Cacna1b, Cacna1c, Cacna1d, Cacna1e, Cacna1g, Cacna1h, Cacna1i, Cacna1s, Cacna2d1, Cacna2d2, Cacna2d3, Cacna2d4, Cacnb2, Cacnb3, Cacng2, Cacng4, Cacng5, Casp3, Cdc25b, Chuk, Crkl, Daxx, Dusp5, Dusp6, Dusp7, Dusp8, Ecsit, Egfr, Elk1, Fas, Fgf1, Fgf10, Fgf11, Fgf12, Fgf14, Fgf18, Fgf2, Fgf21, Fgf7, Fgfr1, Fgfr2, Fgfr3, Fgfr4, Flna, Flnb, Flnc, Gadd45g, Gna12, Gng12, Hspa1a, Hspa1b, Hspa2, Hspa8, Jun, Map2k5, Map2k6, Map3k4, Map3k5, Map3k8, Map4k3, Map4k4, Mapk1, Mapk10, Mapk11, Mapk14, Mapk3, Mapk8ip1, Mapk8ip3, Mapkapk2, Mapkapk3, Mapt, Mecom, Mef2c, Mknk1, Nfatc4, Ntf3, Ntrk1, Ntrk2, Pak2, Pdgfrb, Pla2g1b, Pla2g2c, Pla2g2d, Pla2g5, Ppp3ca, Ppp5c, Prkacb, Prkca, Prkcb, Prkx, Rap1a, Rap1b, Rasgrf2, Rasgrp2, Rasgrp3, Rps6ka2, Rps6ka5, Rras2, Sos1, Srf, Stk3, Stmn1, Tab1, Tab2, Taok3, Tgfb2, Traf6, Trp53 | 7.28E-07 | 4.70E-06 |
| 5 | mmu05412: Arrhythmogenic right ventricular cardiomyopathy (ARVC) | Actb, Actn1, Actn3, Actn4, Atp2a2, Cacna1c, Cacna1d, Cacna1s, Cacna2d1, Cacna2d2, Cacna2d3, Cacna2d4, Cacnb2, Cacnb3, Cacng2, Cacng4, Cacng5, Cdh2, Ctnna1, Ctnna2, Ctnna3, Ctnnb1, Dag1, Dmd, Dsp, Emd, Itga1, Itga2, Itga6, Itga8, Itga9, Itgav, Itgb1, Itgb5, Itgb6, Jup, Lef1, Pkp2, Ryr2, Sgcb, Slc8a1, Tcf7l1, Tcf7l2 | 9.49E-06 | 6.13E-05 |
| 6 | mmu05222:Small cell lung cancer | Akt1, Akt3, Bcl2, Bcl2l1, Casp9, Ccnd1, Cdk2, Cdk4, Cdk6, Cdkn2b, Chuk, Cks1b, Col4a1, Col4a2, Col4a4, Col4a6, Cycs, E2f1, E2f3, Fhit, Fn1, Itga2, Itga6, Itgav, Itgb1, Lama1, Lama3, Lama5, Lamb3, Lamc1, Nfkbia, Pias2, Pias4, Pik3cb, Pik3r1, Pik3r2, Ptk2, Rarb, Rb1, Rxra, Rxrb, Rxrg, Traf4, Traf5, Traf6, Trp53 | 3.03E-05 | 1.95E-04 |
| 7 | mmu04520: Adherens junction | Actb, Actn1, Actn3, Actn4, Acvr1c, Baiap2, Cdh1, Ctnna1, Ctnna2, Ctnna3, Ctnnb1, Ctnnd1, Egfr, Erbb2, Fgfr1, Fyn, Igf1r, Iqgap1, Lef1, Mapk1, Mapk3, Met, Mllt4, Pard3, Ptprb, Ptprf, Ptprj, Ptprm, Pvrl1, Pvrl2, Pvrl3, Smad2, Smad3, Sorbs1, Src, Tcf7l1, Tcf7l2, Wasf2, Wasf3 | 0.00168756 | 0.010904142 |
| 8 | mmu05223: Non-small cell lung cancer | Akt1, Akt3, Casp9, Ccnd1, Cdk4, Cdk6, Cdkn2a, E2f1, E2f3, Egfr, Erbb2, Fhit, Foxo3, Mapk1, Mapk3, Pik3cb, Pik3r1, Pik3r2, Plcg1, Plcg2, Prkca, Prkcb, Rarb, Rb1, Rxra, Rxrb, Rxrg, Sos1, Tgfa, Trp53 | 0.003433104 | 0.0222011 |
| 9 | mmu04020: Calcium signaling pathway | Adcy1, Adcy3, Adcy9, Adora2a, Adra1a, Adra1b, Adra1d, Adrb1, Agtr1a, Atp2a2, Atp2b1, Atp2b2, Atp2b3, Atp2b4, Avpr1a, Cacna1a, Cacna1b, Cacna1c, Cacna1d, Cacna1e, Cacna1g, Cacna1h, Cacna1i, Cacna1s, Calm2, Calm3, Camk2a, Camk2b, Camk2d, Cckar, Chrna7, Ednrb, Egfr, Erbb2, Erbb4, F2r, Gna11, Gna14, Gna15, Gnaq, Gnas, Grm5, Htr2c, Itpr2, Itpr3, Lhcgr, Mylk, Mylk3, Nos1, Nos3, Ntsr1, P2rx1, P2rx4, P2rx7, Pde1a, Pde1b, Pde1c, Pdgfrb, Phka2, Plcb1, Plcb4, Plcd1, Plce1, Plcg1, Plcg2, Ppp3ca, Prkacb, Prkca, Prkcb, Prkx, Ptafr, Ptger3, Ryr1, Ryr2, Ryr3, Slc8a1, Slc8a3, Trhr | 0.005823665 | 0.037702607 |
| 10 | mmu04916: Melanogenesis | a, Adcy1, Adcy3, Adcy5, Adcy9, Calm2, Calm3, Camk2a, Camk2b, Camk2d, Creb1, Creb3l1, Ctnnb1, Dct, Edn1, Ednrb, Fzd2, Fzd5, Fzd7, Fzd8, Fzd9, Gnai2, Gnao1, Gnaq, Gnas, Kit, Kitl, Lef1, Mapk1, Mapk3, Mc1r, Mitf, Plcb1, Plcb4, Prkacb, Prkca, Prkcb, Prkx, Tcf7l1, Tcf7l2, Wnt1, Wnt10a, Wnt5a, Wnt5b, Wnt7a, Wnt8b, Wnt9a | 0.00819137 | 0.053090309 |
| 11 | mmu04144: Endocytosis | Acvr1c, Adrb1, Agap1, Agap3, Ap2a1, Ap2b1, Ap2s1, Arap2, Arfgap2, Arrb1, Asap1, Asap2, Asap3, Cblc, Chmp4c, Cxcr2, Cxcr4, Dab2, Dnajc6, Dnm1, Dnm3, Egfr, Ehd1, Ehd2, Ehd3, Epn1, Epn2, Epn3, Eps15, Erbb4, F2r, Fam125b, Fgfr2, Fgfr3, Fgfr4, Flt1, Git2, H2-K1, Hspa1a, Hspa1b, Hspa2, Hspa8, Igf1r, Iqsec1, Iqsec3, Kdr, Kit, Met, Nedd4, Nedd4l, Ntrk1, Pard3, Pard6g, Pip4k2b, Pip5k1b, Pld1, Prkcz, Psd, Psd3, Psd4, Rab11b, Rab11fip1, Rab11fip4, Rab22a, Rab31, Rab5b, Ret, Sh3gl1, Sh3gl3, Sh3glb1, Sh3kbp1, Smurf1, Src, Stam, Stambp, Traf6, Vps24, Vps25, Vps36, Vps37c, Wwp1 | 0.008343602 | 0.054080841 |
| 12 | mmu05210: Colorectal cancer | Acvr1c, Akt1, Akt3, Apc, Appl1, Axin2, Bax, Bcl2, Casp3, Casp9, Ccnd1, Ctnnb1, Cycs, Dcc, Egfr, Fzd2, Fzd5, Fzd7, Fzd8, Fzd9, Igf1r, Jun, Lef1, Mapk1, Mapk10, Mapk3, Met, Mlh1, Msh3, Pdgfrb, Pik3cb, Pik3r1, Pik3r2, Ralgds, Smad2, Smad3, Sos1, Tcf7l1, Tcf7l2, Tgfb2, Trp53 | 0.008624979 | 0.055912058 |
| 13 | mmu04540: Gap junction | Adcy1, Adcy3, Adcy5, Adcy9, Adrb1, Csnk1d, Drd2, Egfr, Gna11, Gnai2, Gnaq, Gnas, Grm5, Gucy1b2, Htr2c, Itpr2, Itpr3, lpar1, Map2k5, Mapk1, Mapk3, Pdgfd, Pdgfrb, Plcb1, Plcb4, Prkacb, Prkca, Prkcb, Prkg1, Prkx, Sos1, Src, Tuba1a, Tuba1b, Tubb2a, Tubb2b, Tubb2c, Tubb3, Tubb4, Tubb5, Tubb6 | 0.008624979 | 0.055912058 |
| 14 | mmu05414: Dilated cardiomyopathy | Actb, Adcy1, Adcy3, Adcy5, Adcy9, Adrb1, Atp2a2, Cacna1c, Cacna1d, Cacna1s, Cacna2d1, Cacna2d2, Cacna2d3, Cacna2d4, Cacnb2, Cacnb3, Cacng2, Cacng4, Cacng5, Dag1, Dmd, Emd, Gnas, Igf1, Itga1, Itga2, Itga6, Itga8, Itga9, Itgav, Itgb1, Itgb5, Itgb6, Myl3, Prkacb, Prkx, Ryr2, Sgcb, Slc8a1, Tgfb2, Tpm2, Tpm3, Tpm4 | 0.009683379 | 0.06280453 |
| 15 | mmu04012: ErbB signaling pathway | Abl1, Abl2, Akt1, Akt3, Camk2a, Camk2b, Camk2d, Cblc, Cdkn1a, Crkl, Egfr, Eif4ebp1, Elk1, Erbb2, Erbb4, Jun, Mapk1, Mapk10, Mapk3, Mtor, Nck2, Nrg1, Nrg2, Pak2, Pak3, Pak4, Pak6, Pak7, Pik3cb, Pik3r1, Pik3r2, Plcg1, Plcg2, Prkca, Prkcb, Ptk2, Rps6kb1, Shc2, Sos1, Src, Tgfa | 0.01193122 | 0.077465723 |
| 16 | mmu04270: Vascular smooth muscle contraction | Actg2, Adcy1, Adcy3, Adcy5, Adcy9, Adora2a, Adra1a, Adra1b, Adra1d, Agtr1a, Arhgef12, Avpr1a, Cacna1c, Cacna1d, Cacna1s, Calcrl, Cald1, Calm2, Calm3, Gna11, Gna12, Gnaq, Gnas, Gucy1b2, Itpr2, Itpr3, Kcnma1, Kcnmb1, Kcnmb2, Mapk1, Mapk3, Mrvi1, Myl9, Mylk, Mylk3, Pla2g1b, Pla2g2c, Pla2g2d, Pla2g5, Plcb1, Plcb4, Ppp1cc, Ppp1r14a, Prkacb, Prkca, Prkcb, Prkcd, Prkce, Prkch, Prkg1, Prkx, Ramp3, Rock2 | 0.016626811 | 0.108192921 |
| 17 | mmu05214: Glioma | Akt1, Akt3, Calm2, Calm3, Camk2a, Camk2b, Camk2d, Ccnd1, Cdk4, Cdk6, Cdkn1a, Cdkn2a, E2f1, E2f3, Egfr, Igf1, Igf1r, Mapk1, Mapk3, Mtor, Pdgfrb, Pik3cb, Pik3r1, Pik3r2, Plcg1, Plcg2, Prkca, Prkcb, Rb1, Shc2, Sos1, Tgfa, Trp53 | 0.023941745 | 0.15633521 |
| 18 | mmu04340: Hedgehog signaling pathway | Bmp6, Bmp7, Bmp8a, Btrc, Csnk1d, Csnk1e, Csnk1g1, Csnk1g2, Csnk1g3, Gas1, Gli1, Gli2, Gli3, Hhip, Lrp2, Prkacb, Prkx, Ptch1, Ptch2, Rab23, Sufu, Wnt1, Wnt10a, Wnt5a, Wnt5b, Wnt7a, Wnt8b, Wnt9a | 0.033019266 | 0.216549231 |
| 19 | mmu05218: Melanoma | Akt1, Akt3, Ccnd1, Cdh1, Cdk4, Cdk6, Cdkn1a, Cdkn2a, E2f1, E2f3, Egfr, Fgf1, Fgf10, Fgf11, Fgf12, Fgf14, Fgf18, Fgf2, Fgf21, Fgf7, Fgfr1, Igf1, Igf1r, Mapk1, Mapk3, Met, Mitf, Pdgfd, Pdgfrb, Pik3cb, Pik3r1, Pik3r2, Rb1, Trp53 | 0.039434735 | 0.259424773 |
| 20 | mmu04810: Regulation of actin cytoskeleton | Abi2, Actb, Actn1, Actn3, Actn4, Apc, Arhgef12, Arhgef7, Baiap2, Bcar1, Crkl, Cyfip2, Diap1, Diap3, Dock1, Egfr, Enah, Ezr, F2r, Fgf1, Fgf10, Fgf11, Fgf12, Fgf14, Fgf18, Fgf2, Fgf21, Fgf7, Fgfr1, Fgfr2, Fgfr3, Fgfr4, Fn1, Gna12, Gng12, Iqgap1, Itga1, Itga2, Itga6, Itga8, Itga9, Itgae, Itgam, Itgav, Itgb1, Itgb5, Itgb6, Limk1, Mapk1, Mapk3, Msn, Myh9, Myl7, Myl9, MylK, Mylk3, Nckap1l, Pak2, Pak3, Pak4, Pak6, Pak7, Pdgfd, Pdgfrb, Pik3cb, Pik3r1, Pik3r2, Pip4k2a, Pip4k2b, Pip5k1b, Ppp1cc, Ptk2, Rock2, Rras2, Slc9a1, Sos1, Ssh1, Tiam1, Tiam2, Vav2, Vav3, Wasf2 | 0.042152262 | 0.277667235 |
| 21 | mmu04512: ECM-receptor interaction | Agrn, CD44, Col1a1, Col2a1, Col4a1, Col4a2, Col4a4, Col4a6, Col5a2, Col6a6, Dag1, Fn1, Itga1, Itga2, Itga6, Itga8, Itga9, Itgav, Itgb1, Itgb5, Itgb6, Lama1, Lama3, Lama5, Lamb3, Lamc1, Reln, Sdc2, Sdc3, Sdc4, Sv2a, Sv2b, Sv2c, Thbs1, Thbs2, Tnc, Tnr, Vwf | 0.047806047 | 0.315776127 |
Notes: By importing Entrez Gene IDs of 5,966 ChIP-Seq-based Olig2 target genes in pMNs into the Functional Annotation tool of DAVID, KEGG pathways showing significant relevance to the set of imported genes were identified. They are listed with rank, pathways, focused genes, P-value corrected by Bonferroni multiple comparison test, and false discovery rate (FDR).
Next, we studied molecular networks of 5966 Olig2 targets in pMNs by using the Core Analysis tool of IPA. They showed a significant relationship with canonical pathways defined as “Axonal guidance signaling” (P = 3.70E–17) and “Molecular mechanisms of cancer” (P = 3.64E–15), well consistent with the results of KEGG. For the 1533 Olig2 targets in OPCs, they exhibited a significant relationship with canonical pathways defined as “Synaptic long term depression” (P = 2.98E–07) and “Wnt/β-catenin signaling” (P = 3.65E–06).
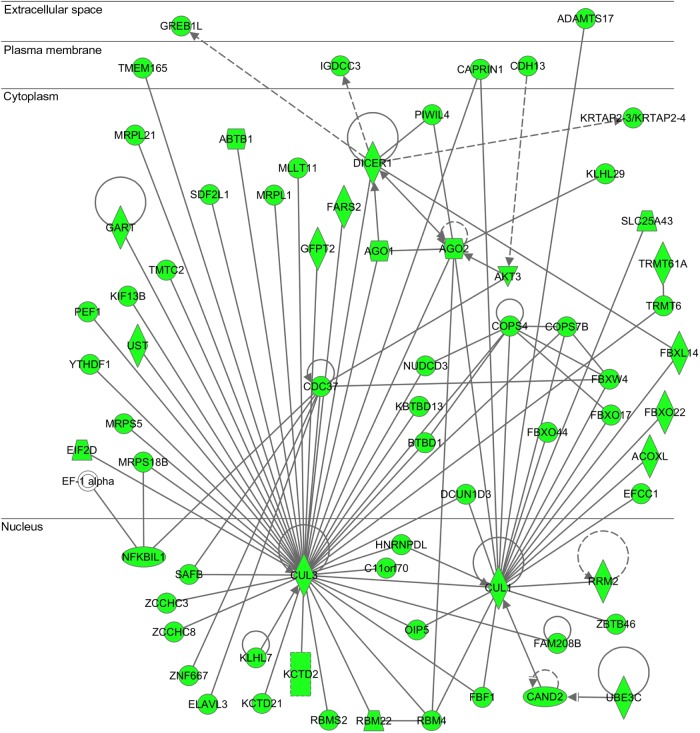
IPA also identified functional networks highly relevant to the 5966 Olig2 targets in pMNs, defined as “Cancer, cell-to-cell signaling and interaction, cellular assembly and organization” (P = 1.00E–54), “Auditory disease, connective tissue disorders, dermatological diseases and conditions” (P = 1.00E–54), “Cancer, gastrointestinal disease, organismal injury and abnormalities” (P = 1.00E–52), and “RNA post-transcriptional modification, embryonic development, hair and skin development and function” (P = 1.00E–52), where both Cul1 (cullin 1) and Cul3 (cullin 3), which constitute the CULLIN-RING ubiquitin ligase (CRL) complex, play a central role in the Olig2 target gene network (Fig. 3). For the 1533 Olig2 targets in OPCs, IPA identified a functional network defined as “Cell death and survival, cellular compromise, neurological disease” (P = 1.00E–83) as the top rank network.
Figure 3.
IPA “RNA post-transcriptional modification, embryonic development, hair and skin development and function” network relevant to Olig2 target genes in motor neuron progenitor cells. Entrez Gene IDs of 5966 Olig2 target genes in pMNs were imported into the Core Analysis tool of IPA. It extracted the “RNA post-transcriptional modification, embryonic development, hair and skin development and function” network as the third rank functional network. Olig2 target genes are colored green. Both CUL1 and CUL3 serve as a hub in the network.
Next, we studied molecular networks of the 5966 Olig2 targets in pMNs by using KeyMolnet. The neighboring network-search algorithm extracted the extremely complex network composed of 5839 molecules and 13,818 molecular relations (Supplementary Fig. 4). The network showed the most significant relationship with “Transcriptional regulation by p53” (P = 1.68E–237). For the 1533 Olig2 targets in OPCs, KeyMolnet identified the highly complex network showing the most significant relationship with “Transcriptional regulation by NF-κB” (P = 1.28E–181) (data not shown).
Downregulation of numerous Olig2 target genes in degenerating motor neurons of mutant SOD1 transgenic mice and spinal cord tissues of ALS patients
Finally, we studied a possible role of Olig2 target genes in the process of motor neuron degeneration in a mouse model of ALS and lumbar spinal cord tissues of ALS patients. We identified the set of 277 genes downregulated in LCM-purified spinal cord motor neurons derived from presymptomatic SOD1 G85R transgenic mice, which satisfied a Q-value ≤0.01 and log2 fold change ≤–1.0 (Supplementary Table 5). Among them, the set of 77 genes (27.8%) corresponded to Olig2 target genes in pMNs (Table 2, underlined in Supplementary Table 5). We also identified the set of 1583 genes downregulated in lumbar spinal cord tissues of ALS patients, which satisfied a Q-value ≤0.01 and log2 fold change ≤–1.0 (Supplementary Table 6). Among them, the set of 473 genes (29.9%) corresponded to Olig2 target genes in pMNs (underlined in Supplementary Table 6). Since Olig2 generally acts as a transcriptional repressor of direct target genes,5,13 these results suggest that downregulation of Olig2 target genes possibly due to overactivation of Olig2 plays a key role in progression of motor neuron degeneration both in mutant SOD mice and in ALS patients.
Table 2.
The set of 77 Olig2 target genes downregulated in motor neurons of mutant SOD1 transgenic mice.
| ENTREZ GENE ID | GENE SYMBOL | GENE NAME | LOCUS | LOG2(FOLD CHANGE) | Q-VALUE |
|---|---|---|---|---|---|
| 319239 | Npsr1 | neuropeptide S receptor 1 | chr9:23902461–24122130 | −3.78453 | 0.000161548 |
| 214642 | A430107O13Rik | RIKEN cDNA A430107O13 gene | chr6:21935909–22205606 | −3.64386 | 0.00288767 |
| 216974 | Proca1 | protein interacting with cyclin A1 | chr11:78006893–78019265 | −3.18442 | 0.00113794 |
| 94216 | Col4a6 | collagen, type IV, alpha 6 | chrX:137599945–137908619 | −3.09954 | 0.000161548 |
| 245038 | Dclk3 | doublecortin-like kinase 3 | chr9:111341584–111392115 | −2.66985 | 0.000161548 |
| 213980 | Fbxw10 | F-box and WD-40 domain protein 10 | chr11:62660624–62690964 | −2.62803 | 0.000161548 |
| 14120 | Fbp2 | fructose bisphosphatase 2 | chr13:62938244–62959730 | −2.49476 | 0.00587005 |
| 227327 | B3gnt7 | UDP-GlcNAc:betaGal beta-1,3-N- acetylglucosaminyltransferase 7 | chr1:88199795–88203880 | −2.365 | 0.000161548 |
| 331524 | Xkrx | X Kell blood group precursor related X linked | chrX:130683583–130696467 | −2.28876 | 0.000161548 |
| 22095 | Tshr | thyroid stimulating hormone receptor | chr12:92236931–92786285 | −2.0461 | 0.000161548 |
| 237759 | Col23a1 | collagen, type XXIII, alpha 1 | chr11:51103421–51397427 | −2.0398 | 0.000161548 |
| 20394 | Scg5 | secretogranin V | chr2:113616469–113669248 | −1.97998 | 0.000161548 |
| 140709 | Emid2 | EMI domain containing 2 | chr5:137217633–137358977 | −1.92523 | 0.000161548 |
| 228846 | D630003M21Rik | RIKEN cDNA D630003M21 gene | chr2:158008268–158054958 | −1.81897 | 0.000161548 |
| 71907 | Serpina9 | serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 | chr12:105234829–105251862 | −1.78003 | 0.00139798 |
| 67313 | 5730559C18Rik | RIKEN cDNA 5730559C18 gene | chr1:138110098–138130857 | −1.74702 | 0.000310928 |
| 22409 | Wnt10a | wingless related MMTV integration site 10a | chr1:74838592–74850749 | −1.74444 | 0.000161548 |
| 69047 | Atp2c2 | ATPase, Ca++ transporting, type 2C, member 2 | chr8:122223908–122281618 | −1.68066 | 0.000161548 |
| 102626 | Mapkapk3 | mitogen-activated protein kinase-activated protein kinase 3 | chr9:107157257–107192208 | −1.62955 | 0.000161548 |
| 18227 | Nr4a2 | nuclear receptor subfamily 4, group A, member 2 | chr2:56959636–56976414 | −1.60877 | 0.000161548 |
| 107371 | Exoc6 | exocyst complex component 6 | chr19:37611363–37767797 | −1.59946 | 0.00276769 |
| 380787 | A230065H16Rik | RIKEN cDNA A230065H16 gene | chr12:112644994–112650288 | −1.54378 | 0.000161548 |
| 432611 | Dnaic2 | dynein, axonemal, intermediate chain 2 | chr11:114588725–114619200 | −1.52146 | 0.000310928 |
| 67168 | Lpar6 | purinergic receptor P2Y, G-protein coupled, 5 | chr14:73595308–73725598 | −1.49476 | 0.000161548 |
| 13836 | Epha2 | Eph receptor A2 | chr4:140857135–140885299 | −1.45943 | 0.000161548 |
| 27428 | Shroom3 | shroom family member 3 | chr5:93112460–93394785 | −1.42323 | 0.000161548 |
| 243911 | Kirrel2 | kin of IRRE like 2 (Drosophila) | chr7:31232784–31242534 | −1.42158 | 0.000161548 |
| 320415 | Gchfr | GTP cyclohydrolase I feedback regulator | chr2:118993523–118998125 | −1.37907 | 0.000161548 |
| 224008 | 2310008H04Rik | RIKEN cDNA 2310008H04 gene | chr16:15887378–16146926 | −1.37707 | 0.000161548 |
| 66922 | Rras2 | related RAS viral (r-ras) oncogene homolog 2 | chr7:121190295–121261295 | −1.35527 | 0.000161548 |
| 23962 | Oasl2 | 2′-5′ oligoadenylate synthetase-like 2 | chr5:115346942–115362254 | −1.3523 | 0.000161548 |
| 24001 | Tiam2 | T-cell lymphoma invasion and metastasis 2 | chr17:3326572–3557713 | −1.35202 | 0.000161548 |
| 14403 | Gabrd | gamma-aminobutyric acid (GABA) A receptor, subunit delta | chr4:154759087–154772178 | −1.33198 | 0.000161548 |
| 72823 | Pard3b | par-3 partitioning defective 3 homolog B (C. elegans) | chr1:61685397–62688858 | −1.31316 | 0.00126983 |
| 12258 | Serping1 | serine (or cysteine) peptidase inhibitor, clade G, member 1 | chr2:84605516–84615586 | −1.31077 | 0.000161548 |
| 170788 | Crb1 | crumbs homolog 1 (Drosophila) | chr1:141094830–141273653 | −1.28699 | 0.00100681 |
| 27390 | Mmel1 | membrane metallo-endopeptidase- like 1 | chr4:154230308–154273345 | −1.2868 | 0.000161548 |
| 59010 | Sqrdl | sulfide quinone reductase-like (yeast) | chr2:122591094–122635287 | −1.28591 | 0.000161548 |
| 259300 | Ehd2 | EH-domain containing 2 | chr7:16534335–16552884 | −1.28353 | 0.000161548 |
| 231991 | Creb5 | cAMP responsive element binding protein 5 | chr6:53397542–53645826 | −1.27894 | 0.000161548 |
| 13824 | Epb4.1l4a | erythrocyte protein band 4.1-like 4a | chr18:33955980–34166860 | −1.26689 | 0.000161548 |
| 67578 | Patl2 | protein associated with topoisomerase II homolog 2 (yeast) | chr2:121945843–122011925 | −1.26105 | 0.000161548 |
| 212706 | N4bp3 | NEDD4 binding protein 3 | chr11:51456476–51471183 | −1.25585 | 0.000161548 |
| 320782 | Tmem154 | transmembrane protein 154 | chr3:84470113–84508497 | −1.24413 | 0.000161548 |
| 238725 | Gpr150 | G protein-coupled receptor 150 | chr13:76192298–76194444 | −1.23333 | 0.000161548 |
| 14675 | Gna14 | guanine nucleotide binding protein, alpha 14 | chr19:16510156–16685308 | −1.23149 | 0.000161548 |
| 216343 | Tph2 | tryptophan hydroxylase 2 | chr10:114515696–114622078 | −1.23133 | 0.000161548 |
| 226413 | Lct | lactase | chr1:130181340–130224895 | −1.21798 | 0.000161548 |
| 20148 | Dhrs3 | dehydrogenase/reductase (SDR family) member 3 | chr4:144482729–144517548 | −1.20253 | 0.000161548 |
| 52570 | Ccdc69 | coiled-coil domain containing 69 | chr11:54863239–54891633 | −1.18442 | 0.000161548 |
| 57915 | Tbc1d1 | TBC1 domain family, member 1 | chr5:64551449–64742725 | −1.17336 | 0.000161548 |
| 20975 | Synj2 | synaptojanin 2 | chr17:5941279–6079739 | −1.17159 | 0.000161548 |
| 380918 | Siah3 | seven in absentia homolog 3 (Drosophila) | chr14:75855781–75927095 | −1.16995 | 0.000161548 |
| 15208 | Hes5 | hairy and enhancer of split 5 (Drosophila) | chr4:154335031–154336480 | −1.15668 | 0.000161548 |
| 209225 | Zfp710 | zinc finger protein 710 | chr7:87169699–87237716 | −1.15627 | 0.000161548 |
| 12398 | Cbfa2t3 | core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) | chr8:125149035–125223009 | −1.1289 | 0.000161548 |
| 232680 | Cpa2 | carboxypeptidase A2, pancreatic | chr6:30491641–30514473 | −1.12326 | 0.000161548 |
| 18548 | Pcsk1 | proprotein convertase subtilisin/kexin type 1 | chr13:75227434–75269946 | −1.11356 | 0.000161548 |
| 70484 | Slc35d2 | solute carrier family 35, member D2 | chr13:64197617–64230638 | −1.113 | 0.000310928 |
| 56485 | Slc2a5 | solute carrier family 2 (facilitated glucose transporter), member 5 | chr4:149493432–149518277 | −1.10736 | 0.000161548 |
| 212377 | Mms22l | MMS22-like, DNA repair protein | chr4:24423608–24530095 | −1.10128 | 0.000161548 |
| 74762 | Mdga1 | MAM domain containing glycosylphosphatidylinositol anchor 1 | chr17:29964902–30024827 | −1.10036 | 0.000161548 |
| 24115 | Best1 | bestrophin 1 | chr19:10059661–10076123 | −1.09578 | 0.000161548 |
| 208213 | Tmem132c | transmembrane protein 132C | chr5:127722195–128046160 | −1.08022 | 0.000161548 |
| 320226 | 4930473A06Rik | RIKEN cDNA 4930473A06 gene | chr4:83171448–83511168 | −1.06949 | 0.000161548 |
| 68180 | Hyi | hydroxypyruvate isomerase homolog (E. coli) | chr4:118032603–118081868 | −1.06726 | 0.000161548 |
| 223864 | Rapgef3 | Rap guanine nucleotide exchange factor (GEF) 3 | chr15:97575200–97598097 | −1.06195 | 0.000161548 |
| 74376 | Myo18b | myosin XVIIIb | chr5:113117895–113325382 | −1.06154 | 0.0030034 |
| 18125 | Nos1 | nitric oxide synthase 1, neuronal | chr5:118317127–118406984 | −1.03899 | 0.000161548 |
| 239845 | Gpr156 | G protein-coupled receptor 156 | chr16:37916581–38007615 | −1.03242 | 0.000161548 |
| 56419 | Diap3 | diaphanous homolog 3 (Drosophila) | chr14:87055269–87540987 | −1.03241 | 0.000161548 |
| 18768 | Pkib | protein kinase inhibitor beta, cAMP dependent, testis specific | chr10:57351786–57460918 | −1.03089 | 0.000161548 |
| 320405 | Cadps2 | Ca2+-dependent activator protein for secretion 2 | chr6:23212773–23789421 | −1.0231 | 0.000161548 |
| 11554 | Adrb1 | adrenergic receptor, beta 1 | chr19:56796861–56799352 | −1.02296 | 0.00640302 |
| 26903 | Dysf | dysferlin | chr6:83958252–84161039 | −1.02138 | 0.000161548 |
| 11610 | Agtrap | angiotensin II, type I receptor- associated protein | chr4:147451169–147462173 | −1.01818 | 0.000161548 |
Notes: By analyzing RNA-Seq data numbered SRP013849, we identified the set of 277 genes significantly downregulated in LCM-purified spinal cord motor neurons of SOD1 G85R transgenic mice, which satisfy Q-value (FDR-adjusted P-value) ≤0.01 and log2 fold change (FC) ≤ −1.0 (Supplementary Table 5). Among them, the set of 77 genes corresponding to Olig2 target genes in pMNs (Supplementary Table 1) are listed with Entrez Gene ID, Gene Symbol, Gene Name, Locus, LOG2(FC), and Q-Value.
Discussion
ALS is a fatal neurodegenerative disease that chiefly affects motor neurons in the brain and spinal cord. All motor neurons in the spinal cord arise from a common set of progenitor cells located within the pMN domain.6,7 Recent studies have convincingly indicated an active involvement of oligodendrocyte dysfunction in the pathogenesis of ALS.2–4 Olig2, expressed selectively by pMN progenitor cells, plays an indispensable role in the development and differentiation of both motor neurons and oligodendrocytes in the spinal cord.5–9 Here, we characterized the comprehensive set of ChIP-Seq-based Olig2 direct target genes and their molecular networks in pMNs and OPCs with relevance to the pathogenesis of ALS.
First, we identified a reproducible set of 5966 Olig2 target genes in pMNs from the ChIP-Seq dataset numbered SRP007566. They included previously reported Olig2 direct targets in pMNs, such as Nkx2.2, Pax6, and Irx3,10 supporting the validity of our results. Next, we identified the cardinal set of 1553 Olig2 target genes in OPCs from the ChIP-Seq dataset numbered SRP015333. Then, we extracted the core set of 740 Olig2 target genes overlapping between pMNs and OPCs. Importantly, the genes closely related to “alternative splicing” in the category of SP/PIR keywords are highly enriched in Olig2 targets shared between pMNs and OPCs. Increasing evidence indicates that RNA metabolism, including regulation of transcription and alternative splicing, is profoundly disturbed in ALS.27,28 Notably, TDP-43 (TARDBP) and FUS, both of which are causative genes of familial ALS and play a key role in RNA processing and regulation of exon splicing, are mislocated from the nucleus to the cytoplasm in degenerating motor neurons of ALS.29 Most importantly, we found that approximately one-third of the downregulated genes in LCM-purified spinal cord motor neurons of presymptomatic SOD1 G85R transgenic mice, as well as in lumbar spinal cord tissues of ALS patients, correspond to Olig2 targets in pMNs, suggesting an involvement of overactivation of Olig2 in the pathogenesis of ALS, beginning from the very early stage of the disease.
We intensively studied molecular networks of Olig2 target genes in pMNs and OPCs. By analyzing with KEGG and IPA, the set of 5966 Olig2 targets in pMNs showed a significant relationship with “Molecular mechanisms and pathways in cancer” and “Axon guidance signaling”, while 1533 Olig2 target genes in OPCs exhibited relevance to “Synaptic long term depression”, “MAPK signaling pathway”, and “Wnt/β-catenin signaling”. Interestingly, synaptic plasticity regulated by a balance between long-term depression and potentiation is altered in the brain of SOD G93A transgenic mice.30 Conditional ablation of Erk1/2 in oligodendrocytes in the adult CNS reduces the expression of a key transcription factor named the myelin gene regulatory factor (Myrf), which is indispensable for myelin gene expression.31 Protein levels of Wnt4 and Wnt inhibitory factor-1 (Wif1) are elevated in the spinal cord of SOD1 G93A mice.32 Smad-interacting protein-1 (Sip1; Zeb2) activates Smad7 that antagonizes the bone morphogenetic protein (BMP) and Wnt/β-catenin signaling pathways pivotal for the development of astrocytes.19 Olig2 binds to the motor neuron and pancreas homeobox 1 (Mnx1, Hb9) promoter and represses its transcription to keep the replication-competent state of pMNs during neural tube development.14 Olig2 binds to the promoter of cyclin-dependent kinase inhibitor 1A (Cdkn1a, p21) and directly suppresses its expression.33 We identified Zeb2 (Sip1), Mnx1, and Cdkn1a as Olig2 targets in pMNs. These results suggest that Olig2 downregulates a wide range of target genes involved in diverse neuronal and glial functions under physiological conditions during development to avoid premature differentiation of Olig2-expressing cells and under pathological conditions to suppress oncogenesis of these cells. However, Olig2 is expressed in not only normal mature oligodendrocytes but also oligodendroglioma and diffuse glioma irrespective of their grades.34 Furthermore, we found a close relationship between Olig2 targets in pMNs and KEGG “Glioma” pathway. The apparent discrepancies might be derived from the proliferative functions of Olig2 affected by post-transcriptional modifications, which are largely controlled in neural progenitor cells by developmentally regulated phosphorylation of a triplet serine motif of Ser10, Ser13, and Ser14 located on the N-terminus of Olig2.35 Olig2, phosphorylated at the triplet serine motif and expressed in p53-positive human gliomas, inhibits biological responses to p53.36 Interestingly, KeyMolnet indicated that the molecular network of Olig2 targets in pMNs is the most relevant to the network of “Transcriptional regulation by p53”.
The ubiquitin-proteasome system, pivotal for protein turnover to prevent accumulation of abnormal proteins in the cell, is severely compromised in ALS.37 IPA showed that Olig2 target genes in pMNs have relevance to the functional network defined as “RNA post-transcriptional modification, embryonic development, hair and skin development and function”, where Cul1 and Cul3, both of which are central components of the CRL complex involved in regulation of protein quality control, serve as a hub in the Olig2 target gene network. These observations suggest the possibility that even subtle overactivation of Olig2 could induce a serious defect in physiological function of the CRL complex essential for cellular protein homeostasis, leading to neurodegeneration due to accumulation of disease-associated proteins.38
Conclusions
By reanalyzing ChIP-Seq datasets, we identified the set of 5966 Olig2 target genes in pMNs and the set of 1553 Olig2 target genes in OPCs. The genes related to the SP/PIR keyword “alternative splicing” are clustered in the core set of 740 targets overlapping between pMNs and OPCs. Approximately one-third of downregulated genes in purified motor neurons of presymptomatic mutant SOD1 transgenic mice and in lumbar spinal cord tissues of ALS patients correspond to Olig2 target genes in pMNs. Molecular network analysis suggests that Olig2 downregulates a wide range of target genes involved in diverse neuronal and glial functions. We identified a novel Olig2 target gene network composed of the CRL complex, which is possibly involved in clearance of disease-associated proteins. These observations lead to a novel hypothesis that aberrant regulation of Olig2 function, by affecting biology of both motor neurons and oligodendrocytes, might be involved in the pathogenesis of ALS.
Supplementary Data
Supplementary Figure 1. FastQC profile of five ChIP-Seq data analyzed in the present study. FASTQ format files of cleaned short read NGS data derived from SRR315585 (panel a), SRR315586 (panel b), SRR315590 (panel c), SRR548313 (panel d), and SRR548314 (panel e) were imported into the FastQC program. The per-base sequence quality score is shown with the median (red line), the mean (blue line), and the interquartile range (yellow box).
Supplementary Figure 2. Olig2-binding consensus sequence motif. The consensus motif sequence surrounding Olig2 ChIP-Seq peaks was identified by GADEM. The canonical E-box consensus sequence defined as 5′-CANNTG-3′ were found on 2376 peaks among a total of 20,043 peaks in pMNs detected by MACS.
Supplementary Figure 3. KEGG “Glioma” pathway relevant to Olig2 target genes in motor neuron progenitor cells. Entrez Gene IDs of 5966 Olig2 target genes in pMNs were imported into the Functional Annotation tool of DAVID. It extracted the KEGG “Glioma” pathway (mmu05214) as the 17th rank pathway (Table 1). Olig2 target genes are colored orange.
Supplementary Figure 4. KeyMolnet molecular network relevant to Olig2 target genes in motor neuron progenitor cells. Entrez Gene IDs of 5966 Olig2 target genes in pMNs were imported into KeyMolnet. The neighboring network-search algorithm extracted the extremely complex network composed of 5839 molecules and 13,818 molecular relations, showing the most significant relationship with “Transcriptional regulation by p53”. Red nodes indicate those closely related to imported genes. White nodes exhibit additional nodes extracted automatically from the core contents of KeyMolnet to establish molecular connections. The molecular relation is indicated by solid line with arrow (direct binding or activation), solid line with arrow and stop (direct inactivation), solid line without arrow (complex formation), dash line with arrow (transcriptional activation), and dash line with arrow and stop (transcriptional repression).
Supplementary Table 1. The set of 5,966 ChIP-Seq-based Olig2 target genes in motor neuron progenitor cells.
Supplementary Table 2. The set of 1,553 ChIP-Seq-based Olig2 target genes in oligodendrocyte progenitor cells.
Supplementary Table 3. The set of 740 Olig2 target genes overlapping bewteen motor neuron progenitor cells and oligodendrocyte progenitor cells.
Supplementary Table 4. The highly stringent set of 4,717 ChIP-Seq-based Olig2 target genes in motor neuron progenitor cells.
Supplementary Table 5. The set of 277 genes downregu-lated in motor neurons of mutant SOD1 transgenic mice
Supplementary Table 6. The set of 1,583 genes down-regulated in lumbar spinal cord tissues of ALS.
Acknowledgments
The authors thank Ms. Mutsumi Motouri for her invaluable help.
Footnotes
ACADEMIC EDITOR: Alexander Rotenberg, Editor in Chief
FUNDING: This work was supported by the JSPS KAKENHI (C22500322 and C25430054), the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan and a grant from the National Center for Geriatrics and Gerontology (NCGG26–20). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Designed the methods, analyzed the data, and drafted the manuscript: JS. Helped in data analysis: NA, SK, YK. All authors have read and approved the final manuscript.
REFERENCES
- 1.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Morrison BM, Li Y, et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature. 2012;487(7408):443–8. doi: 10.1038/nature11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Philips T, Bento-Abreu A, Nonneman A, et al. Oligodendrocyte dysfunction in the pathogenesis of amyotrophic lateral sclerosis. Brain. 2013;136(pt 2):471–82. doi: 10.1093/brain/aws339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang SH, Li Y, Fukaya M, et al. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat Neurosci. 2013;16(5):571–9. doi: 10.1038/nn.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijer DH, Kane MF, Mehta S, et al. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. 2012;13(12):819–31. doi: 10.1038/nrn3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31(5):791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- 7.Lu QR, Sun T, Zhu Z, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109(1):75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 8.Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7(1):11–8. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor Olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12(13):1157–63. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 10.Mazzoni EO, Mahony S, Iacovino M, et al. Embryonic stem cell-based mapping of developmental transcriptional programs. Nat Methods. 2011;8(12):1056–8. doi: 10.1038/nmeth.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 12.Chen JA, Huang YP, Mazzoni EO, Tan GC, Zavadil J, Wichterle H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69(4):721–35. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31(5):773–89. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 14.Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19(2):282–94. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci. 2003;23(29):9547–56. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, de Faria JP, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69(5):918–29. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paes de Faria J, Kessaris N, Andrew P, Richardson WD, Li H. New Olig1 null mice confirm a non-essential role for Olig1 in oligodendrocyte development. BMC Neurosci. 2014;15:12. doi: 10.1186/1471-2202-15-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10(10):669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng Q, Chen Y, Wang H, et al. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron. 2012;73(4):713–28. doi: 10.1016/j.neuron.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandyopadhyay U, Cotney J, Nagy M, et al. RNA-Seq profiling of spinal cord motor neurons from a presymptomatic SOD1 ALS mouse. PLoS One. 2013;8(1):e53575. doi: 10.1371/journal.pone.0053575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butovsky O, Jedrychowski MP, Cialic R, et al. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann Neurol. 2015;77(1):75–99. doi: 10.1002/ana.24304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoh J, Kawana N, Yamamoto Y. Pathway analysis of ChIP-Seq-based NRF1 target genes suggests a logical hypothesis of their involvement in the pathogenesis of neurodegenerative diseases. Gene Regul Syst Bio. 2013;7:139–52. doi: 10.4137/GRSB.S13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L. GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol. 2009;16(2):317–29. doi: 10.1089/cmb.2008.16TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh J, Tabunoki H. Comprehensive analysis of human microRNA target networks. BioData Min. 2011;4:17. doi: 10.1186/1756-0381-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabin SJ, Kim JM, Baughn M, et al. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet. 2010;19(2):313–28. doi: 10.1093/hmg/ddp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma A, Tandan R. RNA quality control and protein aggregates in amyotrophic lateral sclerosis: a review. Muscle Nerve. 2013;47(3):330–8. doi: 10.1002/mus.23673. [DOI] [PubMed] [Google Scholar]
- 29.Ito D, Suzuki N. Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNA-binding proteins TDP-43 and FUS. Neurology. 2011;77(17):1636–43. doi: 10.1212/WNL.0b013e3182343365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geracitano R, Paolucci E, Prisco S, et al. Altered long-term corticostriatal synaptic plasticity in transgenic mice overexpressing human CU/ZN superoxide dismutase (GLY93->ALA) mutation. Neuroscience. 2003;118(2):399–408. doi: 10.1016/s0306-4522(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 31.Ishii A, Furusho M, Dupree JL, Bansal R. Role of ERK1/2 MAPK signaling in the maintenance of myelin and axonal integrity in the adult CNS. J Neurosci. 2014;34(48):16031–45. doi: 10.1523/JNEUROSCI.3360-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L, Guan Y, Wu X, et al. Wnt Signaling is altered by spinal cord neuronal dysfunction in amyotrophic lateral sclerosis transgenic mice. Neurochem Res. 2013;38(9):1904–13. doi: 10.1007/s11064-013-1096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–17. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ligon KL, Alberta JA, Kho AT, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63(5):499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y, Meijer DH, Alberta JA, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69(5):906–17. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehta S, Huillard E, Kesari S, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19(3):359–71. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendotti C, Marino M, Cheroni C, et al. Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: implication for protein aggregation and immune response. Prog Neurobiol. 2012;97(2):101–26. doi: 10.1016/j.pneurobio.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3 s): cellular functions and disease implications. EMBO J. 2013;32(17):2307–20. doi: 10.1038/emboj.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. FastQC profile of five ChIP-Seq data analyzed in the present study. FASTQ format files of cleaned short read NGS data derived from SRR315585 (panel a), SRR315586 (panel b), SRR315590 (panel c), SRR548313 (panel d), and SRR548314 (panel e) were imported into the FastQC program. The per-base sequence quality score is shown with the median (red line), the mean (blue line), and the interquartile range (yellow box).
Supplementary Figure 2. Olig2-binding consensus sequence motif. The consensus motif sequence surrounding Olig2 ChIP-Seq peaks was identified by GADEM. The canonical E-box consensus sequence defined as 5′-CANNTG-3′ were found on 2376 peaks among a total of 20,043 peaks in pMNs detected by MACS.
Supplementary Figure 3. KEGG “Glioma” pathway relevant to Olig2 target genes in motor neuron progenitor cells. Entrez Gene IDs of 5966 Olig2 target genes in pMNs were imported into the Functional Annotation tool of DAVID. It extracted the KEGG “Glioma” pathway (mmu05214) as the 17th rank pathway (Table 1). Olig2 target genes are colored orange.
Supplementary Figure 4. KeyMolnet molecular network relevant to Olig2 target genes in motor neuron progenitor cells. Entrez Gene IDs of 5966 Olig2 target genes in pMNs were imported into KeyMolnet. The neighboring network-search algorithm extracted the extremely complex network composed of 5839 molecules and 13,818 molecular relations, showing the most significant relationship with “Transcriptional regulation by p53”. Red nodes indicate those closely related to imported genes. White nodes exhibit additional nodes extracted automatically from the core contents of KeyMolnet to establish molecular connections. The molecular relation is indicated by solid line with arrow (direct binding or activation), solid line with arrow and stop (direct inactivation), solid line without arrow (complex formation), dash line with arrow (transcriptional activation), and dash line with arrow and stop (transcriptional repression).
Supplementary Table 1. The set of 5,966 ChIP-Seq-based Olig2 target genes in motor neuron progenitor cells.
Supplementary Table 2. The set of 1,553 ChIP-Seq-based Olig2 target genes in oligodendrocyte progenitor cells.
Supplementary Table 3. The set of 740 Olig2 target genes overlapping bewteen motor neuron progenitor cells and oligodendrocyte progenitor cells.
Supplementary Table 4. The highly stringent set of 4,717 ChIP-Seq-based Olig2 target genes in motor neuron progenitor cells.
Supplementary Table 5. The set of 277 genes downregu-lated in motor neurons of mutant SOD1 transgenic mice
Supplementary Table 6. The set of 1,583 genes down-regulated in lumbar spinal cord tissues of ALS.