Abstract
Mammalian hybrids often show abnormal growth, indicating that developmental inviability may play an important role in mammalian speciation. Yet it is unclear if this recurrent phenotype reflects a common genetic basis. Here we describe extreme parent-of-origin dependent growth in hybrids from crosses between two species of dwarf hamsters, Phodopus campbelli and P. sungorus. One cross type resulted in massive placental and embryonic overgrowth, severe developmental defects, and maternal death. Embryos from the reciprocal cross were viable and normal sized but adult hybrid males were relatively small. These effects are strikingly similar to patterns from several other mammalian hybrids. Using comparative sequence data from dwarf hamsters and several other hybridizing mammals, we argue that extreme hybrid growth can contribute to reproductive isolation during the early stages of species divergence. Next we tested if abnormal growth in hybrid hamsters was associated with disrupted genomic imprinting. We found no association between imprinting status at several candidate genes and hybrid growth, though two interacting genes involved in embryonic growth did show reduced expression in overgrown hybrids. Collectively, our study indicates that growth-related hybrid inviability may play an important role in mammalian speciation but that the genetic underpinnings of these phenotypes remain unresolved.
Keywords: reproductive isolation, genetic imprinting, Haldane's rule, speciation, Phodopus
Introduction
Considerable progress has been made on understanding the evolution of genetic interactions that lead to reduced fertility or viability of hybrids (i.e., intrinsic postzygotic reproductive isolation). These efforts have yielded several fundamental insights into the evolution of intrinsic reproductive isolation (reviewed in Coyne and Orr 2004), including that hybrid incompatibilities often result from deleterious interactions between divergent genes (Dobzhansky 1937; Muller 1942; Orr 1996; Brideau et al. 2006; Tang and Presgraves 2009) and that epistatic interactions involving the sex chromosomes evolve very rapidly (Coyne and Orr 1989b; Masly and Presgraves 2007). Some progress has also been made in linking genetic patterns underlying hybrid male sterility to the disruption of specific developmental processes during spermatogenesis (Good et al. 2010; Meiklejohn et al. 2011; Campbell et al. 2013; Bhattacharyya et al. 2013). However, much less headway has been made on the evolution of inviability and we remain relatively ignorant to the simple question of whether specific developmental pathways are predisposed to disruption in animal hybrids (Coyne and Orr 2004).
It is unclear whether we should even expect hybrid inviability to manifest during early development as embryogenesis tends to be widely conserved between species (Coyne and Orr 2004). Consistent with this, hybrid lethality tends to evolve more slowly than hybrid sterility (Coyne and Orr 1989a; 1997), leading some to question the relevance of inviability to the early stages of speciation (Sobel et al. 2010). Nonetheless, regulatory changes influencing diverse aspects of morphological development can evolve very rapidly (Abzhanov et al. 2004; Shapiro et al. 2004; Mallarino et al. 2012), and thus could also play an important role in the evolution of reproductive isolation between closely related species. In particular, the mammalian radiation is a compelling system in which to study the evolution of development in the context of speciation. Mammals show great morphological diversity between species, hybrid inviability arises at a comparatively rapid rate (Prager and Wilson 1975; Fitzpatrick 2004), and several mammalian hybrids show abnormal growth (Table 1; Gray 1972). Interestingly, many of these hybrids show parent-of-origin dependent growth where reciprocal hybrids differ in size and are either larger or smaller than the parent species (Dawson 1965; Allen 1969; Rogers and Dawson 1970; Allen et al. 1993; Zechner et al. 1996). The recurrent nature of these phenotypes suggests a common developmental and/or genetic basis, yet their relevance to mammalian speciation remains unclear because they have not been considered in an evolutionary context. Indeed, some of the best-known examples of hybrid growth effects come from hybridizing species pairs separated by millions of years of divergence (e.g., lions × tigers; ~4 MY to the most recent common ancestor; Davis et al. 2010). To resolve these issues, we need to determine if the evolution of growth-related hybrid inviability in mammals reflects the disruption of a common developmental process and if the underlying intrinsic incompatibilities evolve rapidly enough to play a role in mammalian speciation.
Table 1.
Mammal hybrids with observed growth effects. Phylogenetically independent crosses are in parentheses. Data are from Allen et al. (1993), Dawson (1965), Gray (1972), Sokolov (1993), and Zechner (1996). Details of the exact crosses can be found in Supplemental Table 1.
| Order | Reciprocal crosses |
Single crosses |
|||
|---|---|---|---|---|---|
| Both hybrids larger than parent species | Parent of origin growth | Both hybrids smaller than parent species | Larger than parent species | Smaller than parent species | |
| Cetartiodactyla | 5 (3) | 1 (1) | - | 4 (4) | 1 (1) |
| Carnivora | 1 (1) | 2 (2) | 1 (1) | 3 (3) | 1 (1) |
| Perissodactyla | 1 (1) | 1 (1) | - | 3 (3) | 1 (1) |
| Primates | - | - | - | 1 (1) | 2 (2) |
| Rodentia | 1 (1) | 6 (4) | - | 6 (6) | - |
| Total | 8 (5) | 10 (8) | 1 (1) | 17 (17) | 5 (5) |
1P<0.001, Fisher's exact test versus all other crosses, Bonferroni corrected α =0.008.
2F3,43= 12.811, P<0.001. Also significant in all pairwise Wilcoxon rank-sum tests, P<0.0073, Bonferroni corrected α =0.013.
Most of the evidence for abnormal growth in mammals derives from casual descriptions of captive hybrids that showed extreme adult sizes relative to their parent species (Gray 1972). However, a few in depth studies have shown that hybrid growth effects are associated with abnormal placentation during mid-gestation (Dawson 1965; Rogers and Dawson 1970; Zechner et al. 1996; Vrana et al. 1998). The placenta is derived largely from embryonic tissue and acts as a conduit for the transfer of maternal nutrients to the embryo. Both growth factors and their antagonists (i.e., growth repressors) are expressed in the placenta. The dosage-ependent interaction between these two classes of genes influences nutrient allocation and ultimately regulates growth in developing embryos (Haig 1996; Reik et al. 2003; Saukkonen 2004). Moreover, many placental expressed genes are controlled by an unusual mode of regulation called genomic imprinting, which results in the epigenetic silencing of one allele depending on its parent of origin (Surani et al. 1990). Imprinted genes are commonly involved in placental formation (Piedrahita 2011) and show a functional bias where paternally expressed (i.e., maternally imprinted) genes tend to promote embryonic growth while maternally expressed (i.e., paternally imprinted) genes repress growth (Morison et al. 2005). Thus, abnormal growth in hybrid mammals may generally reflect disrupted placental function caused by failed interactions between imprinted genes (Vrana 2007).
There are three key reasons why imprinted placental genes are intriguing in the context of speciation. First, the placenta is likely subject to intense evolutionary conflict over resource allocation (Burt and Trivers 1998) because it mediates interactions between two different genomes (maternal and paternal/offspring). Conflict is particularly relevant in the case of multiple paternity (Haig 1999) where offspring strategies should evolve to garner more resources at the expense of their hal^siblings and maternal countermeasures are expected to assure even allocation to all offspring (Zeh and Zeh 2000; Haig 2002; Crespi and Semeniuk 2004). Consistent with these predictions, the placenta shows the highest rate of structural evolution of all mammali3an tissues (Leiser and Kaufmann 1994). Second, loss of imprinting at a single gene can skew the dosage balance between growth factors and repressors, causing abnormal development and pronounced growth (Li et al. 1999). In turn, divergence of imprinting patterns between species is predicted to cause dosage imbalance in hybrids (Varmuza 1993). Third, imprinted genes are expressed from a single chromosome and are thus functionally haploid. Similar to the well-known differential exposure of recessive X-linked hybrid incompatibilities in males (Muller 1942; Turelli and Orr 1995; 2000), haploid expression could expose recessive incompatibilities that would otherwise be masked in hybrids. Furthermore, epistatic interactions between recessive hybrid incompatibilities would also be differentially exposed in F1 hybrids because many paternally and maternally imprinted genes directly interact (Czech 1989; Haig and Graham 1991).
Patterns of placental gene expression have been evaluated in three systems that show abnormal hybrid growth. Disrupted gene expression is associated with parent-of-origin effects on morphology and size of the placenta in both deer mice (Vrana et al. 1998; Duselis and Vrana 2007; 2010) and house mice (Zechner et al. 1996; 1997; Shi et al. 2004; Brown et al. 2012). These two systems show similar placental phenotypes that appear to have different genetic bases (Zechner et al. 2004). Placental dysgenesis in deer mice is caused by an epistatic interaction between loss of imprinting in at least one paternally expressed gene and the maternally expressed X chromosome (Vrana et al. 2000; Loschiavo et al. 2007), though widespread loss of maternal imprinting is also apparent in overgrown offspring (Vrana et al. 2000). Reciprocal growth effects in the placenta of hybrid house mice also are caused by an X-autosome interaction (Zechner et al. 1996; Hemberger et al. 1999) and there is some evidence for disrupted imprinting (Shi et al. 2004; 2005). However, these regulatory effects appear to be less pronounced and genetically distinct from those described in deer mice (Zechner et al. 2004). In a third system, a hybrid cross between horses and donkeys results in parent-of-origin effects for abnormal placental morphology (Allen 1969; Allen et al. 1993), but with no evidence for disrupted genomic imprinting (Wang et al. 2013). As these three systems show similar phenotypes caused by different genetic mechanisms, it remains to be seen how often disrupted imprinting underlies the evolution of mammalian reproductive isolation in general and parent-of-origin growth effects in particular.
Here we describe patterns of reproductive isolation between two species of dwarf hamsters, Phodopus sungorus and P. campbelli. Dwarf hamsters are native to the xeric habitats of central Asia with P. sungorus inhabiting the Kazakh steppe (Ross 1998) and P. campbelli inhabiting the semi-deserts of Mongolia, northern China, and southern Russia (Ross 1995). They are sister species (Neumann et al. 2006) that have only recently been elevated from subspecies status based primarily on evidence of sterility in hybrid males (Sokolov and Vasil'eva 1993). The two species have very similar karyotype morphologies (2n=28), though hybrid male sterility is associated with extensive autosomal and sex chromosome asynapsis during meiosis I (Safronova and Vasil'eva 1996; Safronova et al. 1999). In addition, one direction of the cross (female P. sungorus × male P. campbelli) has been reported to result in “heterotic” hybrids with exaggerated growth and an increased incidence of birth defects (Safronova and Vasil'eva 1996). These observations suggest that dwarf hamsters may provide a novel system with which to evaluate the developmental basis of abnormal hybrid growth between mammal species still in the early stages of divergence. We have three primary objectives. First, we use reciprocal crosses to test for hybrid inviability phenotypes in dwarf hamsters, focusing on parent-of-origin growth effects throughout the lifecycle of F1 hybrids. Second, we examine levels and allele-specific patterns of expression at eight candidate genes to test for disrupted expression and/or genomic imprinting in hybrid placenta. Third, we present DNA divergence estimates between dwarf hamsters and other species pairs exhibiting disrupted hybrid growth to assess the evolutionary tempo of growth-related hybrid inviability in mammals.
Methods
Animals
Outbred dwarf hamster colonies were established at the University of Montana in the fall of 2011 using six mating pairs of P. campbelli provided by Robert Johnston and six mating pairs of Phodopus sungorus provided by Ned Place, both from Cornell University. These stocks were derived from natural populations sampled by Catherine Wynne-Edwards in 1981 and most recently supplemented with additional wild hamsters in 1990 (Scribner and Wynne-Edwards 1994). We have maintained our breeding colonies using a crossing scheme designed to minimize inbreeding (Wright 1921). All animals were housed in 14L:10D light/dark regimen and in accordance with IACUC regulations.
Experimental crosses and phenotypic analyses
We conducted a total of 331 experimental crosses within and between the two species: 1) 110 P. campbelli × P. campbelli, 2) 88 P. campbelli × P. sungorus, 3) 32 P. sungorus × P. campbelli, and 4) 101 P. sungorus × P. sungorus. Here and elsewhere the female is given first following standard mouse genetics crossing notation. These crosses were used to collect a suite of developmental phenotypes described below.
First, we collected late-term embryos and placentas from euthanized pregnant females to determine the frequency and extent of developmental defects. Dwarf hamsters have discoid, labyrinthine hemochorial placentas (Elliot and Crespi 2009) with an 18-day gestation period and a facultative delay of up to four days due to developmental diapause and/or delayed implantation (Newkirk et al. 1997). To control for this variation, dissected embryos were developmentally staged according to a suite of established characters in golden hamsters (Boyer 1953) and mice (Butler and Juurlink 1987). For our analysis, we only used late-term embryos corresponding roughly to Theiler's Stages 24-27 of mouse development (Theiler 1972). Litter size was recorded and dissected embryos and placentas were photographed, weighed and given a presence/absence score for several developmental defects including the occurrence of molar conceptuses (hydatiform moles), embryo reabsorption, and embryo swelling (edema). All embryos and placenta were then snap-frozen on dry ice to preserve RNA for gene expression analyses.
Second, we allowed several crosses to proceed to term to determine if growth phenotypes identified in utero persisted throughout the animal's life cycle and to test for the emergence of new phenotypes in adults. To quantify mating isolation, we tested for differences in the number of successful crosses and latency to birth for adult females paired with a hetero- or conspecific male for up to 40 days. To quantify postnatal growth, we generated a standard growth curve for each cross type by weighing each offspring every ten days after birth until day 100. We modeled growth with an asymptotic curve and tested for differences in the asymptote (final adult size) between each of the cross types. Female P. sungorus × male P. campbelli hybrids could not be brought to term and were excluded from these experiments.
Phenotypic data has been deposited in Dryad and all statistical analyses were performed using R version 3.0.2 (R Core Team 2008). We calculated both one-way analyses of variance (ANOVA) and non-parametric pairwise Wilcoxon rank-um tests for all comparisons between cross types. Results of the Wilcoxon test are reported for phenotypes with large differences in variance between the groups (e.g., embryo and placental weights). Multiple comparisons were accounted for by using a Bonferroni correction when appropriate.
Genetic sex-typing of embryos
The sex of hybrid embryos was determined by Polymerase Chain Reaction (PCR) amplification and Sanger sequencing of a 764 bp fragment of the X-linked gene Zfx. Degenerate primers were designed for dwarf hamsters by modifying the generic LGL331 and LGL335 primers of Shaw and colleagues (2003) based on an alignment of Zfx and Zfy sequences from rat, house mouse, guinea pig, Golden hamster, and Chinese hamster (see Supplemental Table 2 for accession numbers). The Zfx/Zfy sex-typing system usually relies upon a diagnostic intron length polymorphism between homologous genes on the X (Zfx) and Y (Zfy)(Shaw et al. 2003), but we were unable to amplify Zfy in Phodopus. Therefore, we sequenced Zfx and identified five fixed nucleotide differences between the species that we then assayed in hybrids. Heterozygous hybrids were classified as female and homozygous hybrids possessing the expected maternal genotype were classified as male. We verified the accuracy of our assay by typing several adult hybrids of known sex; however, the sex of non-hybrid individuals could not be determined using this approach. All primer sequences and PCR reaction conditions used in this study can be found in Supplemental Table 2. Sequence alignments were performed using the program Geneious (Drummond et al. 2005).
Survey of candidate gene expression
We targeted eight genes that show imprinted expression in the placenta of house mice (Morison et al. 2005), including four paternally expressed genes (maternally imprinted Igf2, Mest, Peg3, Snrpn) and four maternally expressed genes (paternally imprinted H19, Igf2r, Grb10, and Mash2). These candidates were selected because several of them show disrupted placental imprinting in hybrid deer mice (Vrana et al. 1998). Primers were designed using Primer3 (Rozen and Skaletsky 2000) based on exon sequences aligned between mouse, human, rat, and guinea pig (Supplemental Table 2). Amplicons were designed to span at least one intron in five of the genes to minimize the risk of genomic DNA contamination. PCR products for Peg3, H19 and Mash2 did not span introns because either no conserved priming sites could be found or no diagnostic site was present near the intron-exon boundary.
We assayed allele-specific expression of these genes by sequencing complementary DNA (cDNA) from 18 late-gestation placentas, including three from each species and six (three male, three female) from each reciprocal hybrid. Whole placentas were homogenized in liquid nitrogen with a mortar and pestle and total RNA was extracted using an E.Z.N.A. Total RNA Kit (Omega), treated with DNase, and converted to cDNA with the cDNA Supermix Kit (Quantas). Exonic regions were then PCR amplified from cDNA, Sanger sequenced, and examined for fixed differences between the species. All eight loci are autosomal in house mice; therefore hybrid individuals should be heterozygous at all diagnostic positions in the absence of imprinting. Using this rationale, we classified gene expression in hybrids as imprinted (homozygous for the maternal or paternal allele) or biallelic (heterozygous). As with the sex-typing assay, this assay is only effective in the F1 hybrids. Imprinted expression was called only when a single peak from the expected allele was visible on the chromatogram (Supplemental Figure 1). This is a conservative metric given that imprinting sometimes results in skewed biallelic expression (Babak et al. 2008). To evaluate the sensitivity of our assay to detect skewed expression, we tested each primer pair by sequencing a series of “mock hybrid” cDNA pools constructed by combining P. campbelli and P. sungorus cDNA in the following ratios: 1:99, 25:75, 50:50, 75:25, and 99:1. All PCR products were Sanger sequenced at the University of Montana Murdock Lab DNA Sequencing Facility or the University of Arizona Genetics Core and have been deposited in GenBank under the accession numbers JX217832-JX217849, JX436485-JX436486, and KF673394.1-KF673395.1.
We assayed relative expression level using a DyNAmo Flash SYBR Green qPCR kit (Thermo Scientific) on an Agilent Mx3000 in six of each hybrid type and six of each parent species. ROX was used at a concentration of 0.3X as a passive reference dye. All primer pairs had efficiencies between 90% and 105%, which were determined with a four point dilution series. All reactions were carried out in triplicate in 20μl reaction volumes with final primer concentrations of 1mM. Quantitative PCR reaction conditions are reported in Supplemental Table 2. Melt curves were collected after 40 cycles and single peaks were found for all genes. The gene Ywhaz was used as a normalizing control (Kumar et al. 2012). Crossing thresholds (CT) were generated with Agilent's MxPro software and analyzed with custom R scripts using the ΔCT method (Schmittgen and Livak 2008).
Genetic divergence between hybridizing mammal species
We compiled mitochondrial cytochrome b (cyt b) sequence data for 36 species pairs that have been reported to show abnormal hybrid growth (Supplemental Table 1). For dwarf hamsters, we designed primers to amplify and sequence 910 base pairs (bp) of cyt b in both species (Supplemental Table 2). For the other 35 species pairs we used previously published cyt b sequences from GenBank (see Supplemental Table 1 for accession numbers); five species pairs did not have available data for cyt b. After trimming positions with missing data, 34 species pairs shared a common 718 bp alignment that we used to calculate Kimura two-parameter-corrected pairwise divergences (Dxy) with the program Phylip (version 3.6a3, Felsenstein 2002).
We also calculated pairwise synonymous divergence (Ks) between P. campbelii and P. sungorus using partial coding sequences from the eight nuclear genes sequenced in our expression assays. For comparison, orthologous sequences were retrieved from available genomic data from Mus musculus and M. spretus (Keane et al. 2011). Each nuclear sequence was aligned, trimmed to the same length and codons were assigned. Then the genes were concatenated into a 5,669-base fragment to calculate Ks with the program DNAsp (version 5.10.1, Librado and Rozas 2009).
Results
Reduced pregnancy rate in hybrid crosses
Pregnancy rates were similar and relatively high within each species. Females became pregnant in 83% (67 of 81) of P. campbelli crosses and 84% (68 of 81) of P. sungorus crosses. In contrast, P. campbelli females were successfully impregnated by a P. sungorus male only 68% of the time (43 of 63 crosses; P=0.017, Fisher's Exact Test (FET) versus pooled species, Bonferroni corrected α =0.025). The pregnancy rate was also marginally reduced in the reciprocal cross (P. sungorus × P. campbelli; 66% or 19 of 29 crosses; P=0.038 FET versus pooled species). When considering the subset of pairs that reached parturition, we found no reduction in the average latency from pairing to birth for successful heterospecific pregnancies relative to conspecific matings. Phodopus campbelli and P. sungorus averaged 24.2 days (62 crosses) and 23.3 (63 crosses) days respectively from pairing to birth, while the hybrid cross P. campbelli × P. sungorus averaged 22.2 days (37 crosses). The reciprocal hybrid cross, P. sungorus × P. campbelli, did not yield any successful births.
Parent-of-origin effects with extreme asymmetric hybrid overgrowth
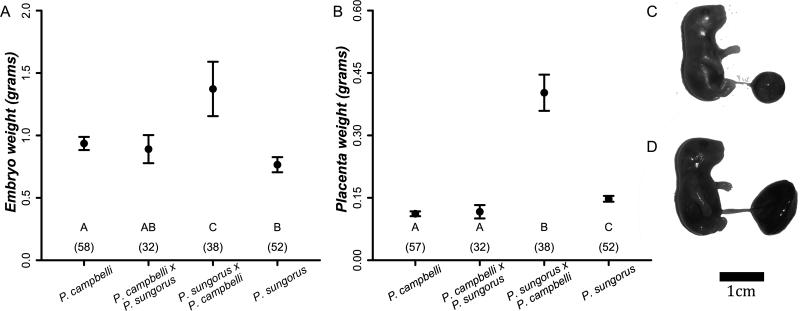
We found that the mean weight of hybrid embryos from a P. sungorus mother and a P. campbelli father was ~38% larger than any other cross type (Figure 1A, P<0.001, Wilcoxon rank-sum test, Bonferroni corrected α=0.008). Parent-of-origin dependent growth was even more striking in the placenta (Figure 1B, Supplemental Figure 2); placentas derived from a female P. sungorus and a male P. campbelli father were ~300% heavier than placentas from any other cross type (Figure 1B, P<0.001, Wilcoxon rank-sum test, Bonferroni corrected α =0.008). We found no sex-specific differences in the placenta or embryo weights of either reciprocal hybrid type (P. sungorus × P. campbelli: placenta P=0.175, embryo P=0.109; P. campbelli × P. sungorus: placenta P=0.880, embryo P=0.880; Wilcoxon rank-sum tests, Bonferroni corrected α =0.013). Following Vrana and colleagues (1998) we will hereafter refer to the large P. sungorus × P. campbelli hybrids as “S×C” and the reciprocal P. campbelli × P. sungorus hybrids as “c×s”, where the species are designated by the first letter of their specific epithet, the maternal species is listed first, and the capitalization reflects the relative size of the hybrid offspring.
Figure 1.
Average weights in grams (±2 SE) of late-term embryos (A) and placentas (B) were much higher in female P. sungorus × male P. campbelli (S×C) hybrids when compared to other cross types. Letters designate significant differences between groups based on pairwise Wilcoxon rank-sum tests and group sample sizes are in parentheses. Numbers in parentheses are sample sizes. Insets show (C) a normal P. campbelli offspring with average sized placenta and (D) an overgrown P. sungorus × P. campbelli (S×C) offspring with an enlarged placenta.
The overgrown S×C offspring were more often affected by severe developmental defects. We found an elevated proportion of molar conceptuses and reabsorbing embryos in S×C crosses relative to all other cross types (Table 2). Also known as hydatiform moles, molar conceptuses are a form of placental pathology characterized by excessive extra-embryonic (placenta) tissue and no embryonic tissue (Supplemental Figure 2D: Lindor et al. 1992). Twenty-five percent of S×C embryos (18 of 73) showed edema, characterized by mild to extreme swelling (Supplemental Figure 2C), whereas embryonic edema was comparably rare in all other crosses (Table 2). Thus, abnormal in utero development was largely restricted to S×C hybrids, with ~70% (53 out of 73) of S×C embryos afflicted by severe developmental defects (even when excluding embryos with edema, S×C offspring were significantly overgrown). In contrast, cxs hybrids were not significantly different than P. campbelli or P. sungorus for any of the in utero developmental phenotypes that we considered. However, late-gestation hybrid litters were smaller than intraspecific litters (Table 2).
Table 2.
Late gestation litter size, sex ratio, and developmental defects of dwarf hamsters and their hybrids. Viable embryos are defined as those that were not molar conceptuses (Molar) or reabsorbing (Reab.), but may have edema. Only the viable embryos were used to calculate litter sizes and sex ratios. Significant values are in bold.
| Viable Embryos |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cross | Total | Litters | Females | Males | Unk. Sex | Molar | Reab. | % Male | Litter Size ±se | Females / Litter ±se | Males / Litter ±se |
| P. campbelli | 60 | 10 | NA | NA | 58 | 0 | 2 | n.a. | 5.8.±0.7 | n.a. | n.a. |
| P. sungorus | 52 | 9 | NA | NA | 52 | 0 | 0 | n.a. | 5.8±0.5 | n.a. | n.a. |
| P. campbelli × P. sungorus | 36 | 12 | 13 | 19 | 1 | 0 | 3 | 59.3 | 2.8±0.5 2 | 1.1±0.3 | 1.6±0.3 |
| P. sungorus × P. campbelli | 73 | 16 | 16 | 18 | 4 | 18 1 | 17 1 | 52.9 | 2.4±0.4 2 | 1.0±0.2 | 1.1±0.2 |
P<0.001, Fisher's exact test versus all other cross types, Bonferroni-corrected α =0.008.
F3,43= 12.811, P<0.001. Also significant in all pairwise Wilcoxon rank-sum tests, P<0.0073, Bonferroni-corrected α =0.013.
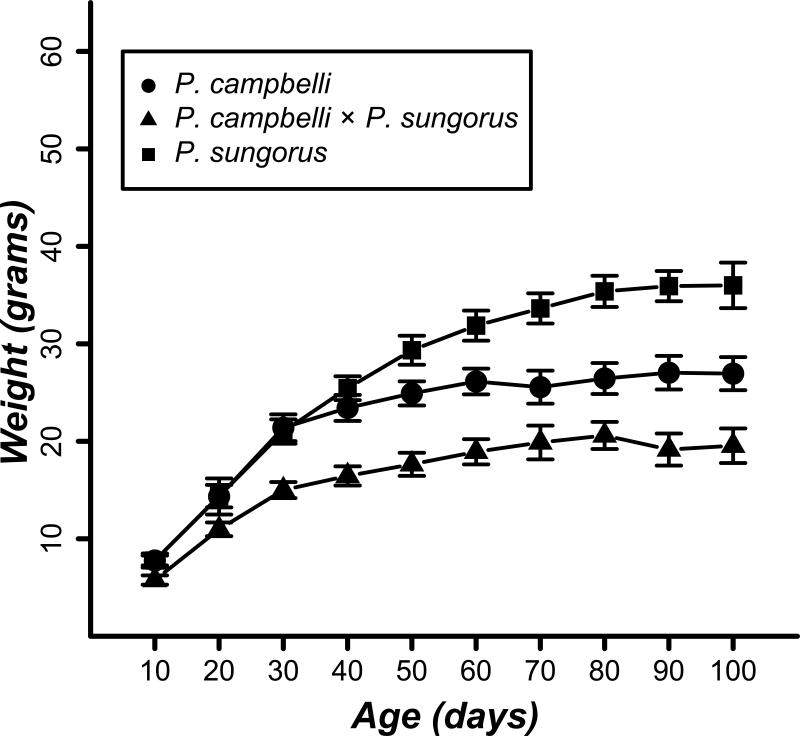
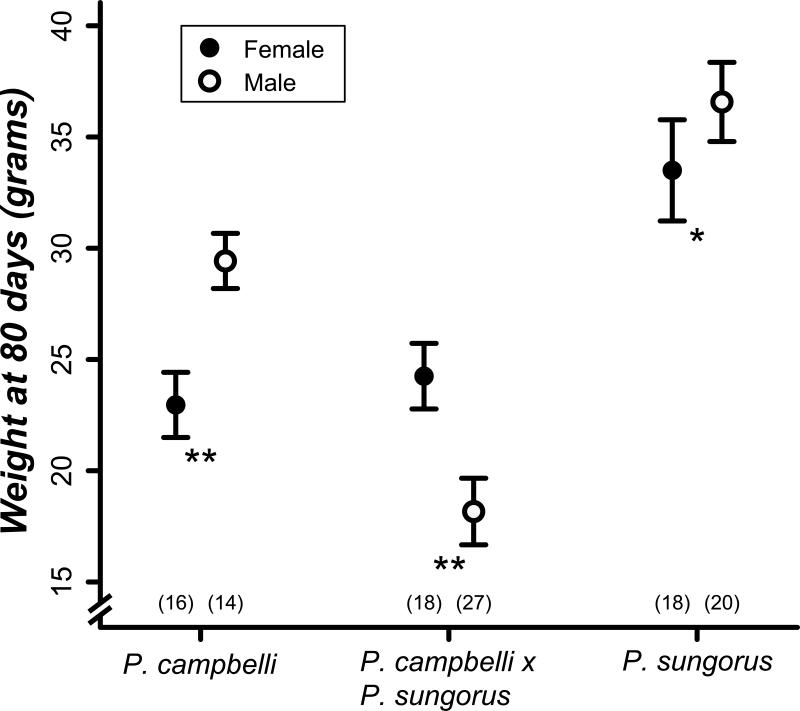
The lack of significant undergrowth phenotypes in late-term cxs hybrids (Figure 1) contrasts with described parent-of-origin effects in hybrid deer mice (Vrana et al. 1998) and house mice (Zechner et al. 1996), where significant but opposite growth effects are manifest in utero in reciprocal hybrid embryos and/or placentas. Our study lacks power to detect subtle weight differences at late gestation (e.g., we have only ~12% power to detect an effect size of 0.2). Because parent-of-origin effects often persist or even become exaggerated into adulthood, we next compared the postnatal growth curves of both species to the cxs hybrids (Figure 2). This experiment was initiated with 26 individuals from 6 crosses for P. campbelli, 38 individuals from 6 crosses for P. sungorus, and 45 individuals from 15 crosses for P. campbelli × P. sungorus hybrids. Though approximately the same size at late gestation (Figure 1A), adult cxs hybrids were much smaller than either parent species (Figure 2; F2,14=77.116, P<0.001). Specifically, we found a significant reduction in adult weights of cxs males versus males of either species (Figure 3; P<0.001, t-test, Bonferroni corrected α =0.017). As is typical for mammals, dwarf hamsters are sexually dimorphic as adults (80 days) with males ~10% larger than the females (Figure 3). In contrast, cxs hybrid females were larger than hybrid males (Figure 3); 80-day old cxs females were approximately the same size as their P. campbelli mothers whereas the males were only half as large as their fathers (Figure 3). Thus, reduced adult growth of cxs hybrids (Figure 2) appears to be driven mostly by a male-specific reduction in body weight. We did not measure postnatal growth of S×C hybrids because three attempts to birth the overgrown hybrids failed and resulted in maternal death.
Figure 2.
Growth curves for P. campbelli, P. sungorus, and female P. campbelli × male P. sungorus (cxs) offspring. Average weights (±2 SE) are shown every 10 days. Hybrids show a significant reduction in adult size compared to the parents.
Figure 3.
Average weights in grams (±2 SE) of hamsters at 80 days. Filled circles represent females, empty circles represent males, and sample sizes are given in parentheses. Hybrid cxs hamsters show reversed sexual dimorphism where males are smaller than females. **P<0.001, *P<0.05, pairwise t-test, Bonferroni corrected α =0.017.
Reduced postnatal growth only in cxs hybrid males is consistent with Haldane's rule, which states that inviability should affect males more often than females in male-heterogametic taxa (Haldane 1922). To further test for sex-specific inviability we analyzed the sex ratios of late-term and adult hybrids. We found no significant bias in the sex ratio of either hybrid type in utero (Table 2; cxs χ2=1.125, df=1, P=0.288; S×C χ2=0.118, df=1, P=0.537, chi-squared test), but we did find significantly male-biased adult sex ratios in cxs hybrids (Table 3; 61.8% male, χ2=8.463, df=1, P<0.001, chi-squared test). Further inspection of the average counts for each sex suggests that this male-biased skew primarily reflects a reduction in the number of females per litter (Table 3). Finally, we tested whether one sex was more susceptible to molar conceptuses, reabsorption, and/or edema in the S×C hybrids. We found approximately equal numbers of males and females affected by each of these phenotypes (molar conceptuses: P=0.715; reabsorbing embryos: P=1.0; edema: P=0.169, n=25 females and 30 males, FET). Thus, we find a surprising pattern in adult cxs hybrids where males are more common but significantly smaller than females.
Table 3.
Adult sex ratios in dwarf hamsters and their hybrids partitioned by litter. Significant values are in bold. Sex ratio was tested with a chi-square test using the pooled sex ratio of the species (50.6% male) as the null expectation.
| Cross | Females | Males | Litters | % Male | Litter Size ±se | Females/Litter ±se | Males/Litter ±se |
|---|---|---|---|---|---|---|---|
| P. campbelli | 199 | 205 | 85 | 50.7 | 4.8±0.2 | 2.3±0.2 | 2.4±0.2 |
| P. sungorus | 229 | 226 | 77 | 49.7 | 5.9±0.2 | 3.0±0.2 | 2.9±0.2 |
| P. campbelli × P. sungorus | 60 | 97 | 43 | 61.8 1 | 3.7±0.2 2 | 1.4±0.2 | 2.3±0.2 |
Chi-square test, X2=8.463, df=1, P<0.001.
F2,202=23.665, P<0.001. Also significant in all pairwise Wilcoxon rank-sum tests, P<0.004, Bonferroni corrected α =0.025.
No widespread disruption of imprinting associated with parent-of-origin growth effects
The existence of strongly asymmetric placental growth phenotypes between reciprocal hybrids (Figure 1) provides a powerful framework with which to differentiate between gene expression changes associated with abnormal growth versus differences that manifest between all hybrid genotypes and the parental species. Species-specific disruption of placental imprinting has been put forth as a general explanation for such parent-of-origin dependent growth effects in reciprocal hybrids (Vrana et al. 1998; Vrana 2007). Specifically, this dosage model predicts that (i) hybrid overgrowth results from maternal expression of one or more growth factors that are normally silenced through imprinting and (ii) that undergrowth results in the reciprocal cross when growth repressors are expressed from the normally silenced paternal genes. For a given gene, this simple model predicts that disrupted imprinting will result in biallelic expression in one hybrid while expression in the reciprocal hybrid remains properly imprinted. Extreme overgrowth occurs in our dwarf hamster crosses when P. sungorus is the mother and undergrowth is manifest when P. sungorus is the father, albeit undergrowth is not apparent until after birth (Figures 1 and 2). Thus, if disrupted imprinting is the cause of these parent-of-origin effects then we should see the gain of expression of P. sungorus (maternal) growth factor alleles in S×C placentas, P. sungorus (paternal) growth repressors in cxs placentas, and proper imprinting of P. campbelli alleles in both hybrid cross types. Gene expression patterns that are the same between the reciprocal hybrids are by definition not associated with parent-of-origin dependent phenotypes.
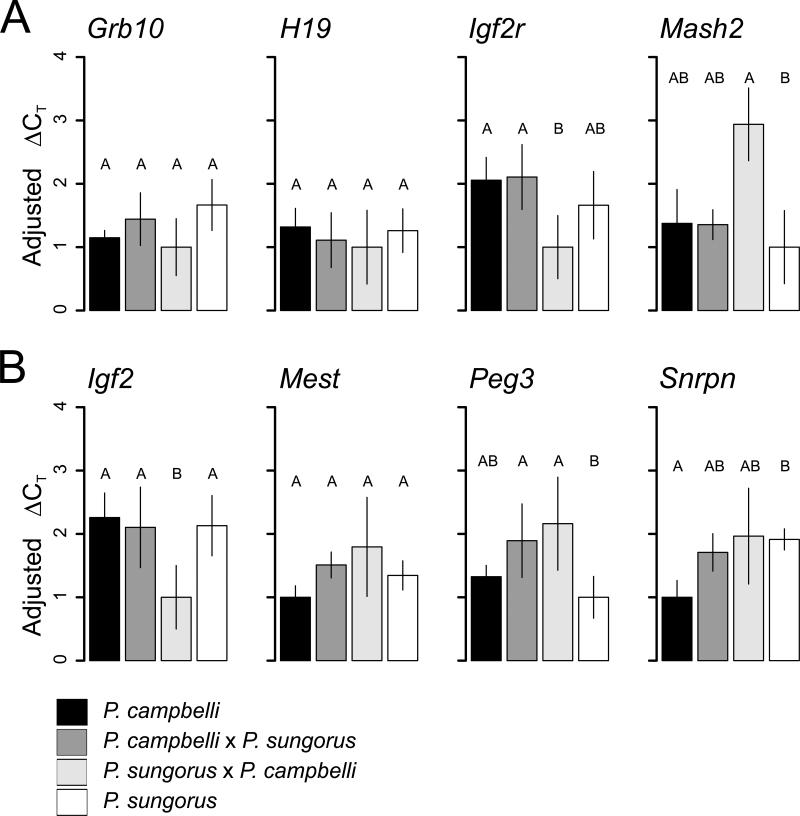
To test this general model, we assayed the expression of eight genes that are imprinted in mice and have been shown to influence embryonic growth. For each gene, we tested six placentas (three per sex) from each of the reciprocal hybrids for imprinted (monoallelic) expression of the maternal or paternal allele (Supplemental Figure 1). Seven of the eight candidate genes were found to contain one or more fixed differences between the species. The paternally expressed gene Mest showed no fixed differences and therefore could not be assayed for allele-specific expression in hybrids. The maternally imprinted candidate Igf2 showed expression of only the paternal allele, consistent with imprinted expression patterns in Mus (Table 4). Two maternally imprinted genes, Peg3 and Snrpn, showed proper imprinting in cxs hybrids, but exhibited some variation in imprinting status among SxC hybrids. In Peg3, one male and one female SxC hybrid showed biallelic expression while the other four offspring only expressed the paternal allele. In Snrpn, a single Sx C hybrid female showed biallelic expression while the other five individuals showed proper imprinting. Three of the four maternally expressed genes (Grb10, Igf2r, and Mash2) showed biallelic expression in both reciprocal hybrids, while H19 showed only maternal expression (Table 4, Supplemental Figure 1; Morison et al. 2005).
Table 4.
Hybrid expression of eight candidate imprinted genes in hamsters and deer mice. Predicted expression is based on the known M.musculus expression of these genes described in the Parent-of-Origin Effect Database (www.otago.ac.nz/IGC). At Peg3, one S×C male and one female show biallelic expression while all other hybrids show paternal expression. Similarly, Snrpn is biallelic in a single female, while the other five individuals showed imprinted expression. Deer mouse data are from Vrana (1998)
| Hamster hybrid expression |
Deer mouse hybrid expression |
||||
|---|---|---|---|---|---|
| Gene name | Predicted expression | P. campbelli × P. sungorus (normal-sized) | P. sungorus × P. campbelli (large) | P. maniculatus × P. polionotus (small) | P. polionotus × P. maniculatus (large) |
| Grb10 | Maternal | Biallelic | Biallelic | Biallelic | Biallelic |
| H19 | Maternal | Maternal | Maternal | Maternal | Biallelic |
| Igf2r | Maternal | Biallelic | Biallelic | Biallelic | Maternal |
| Mash2 | Maternal | Biallelic | Biallelic | Biallelic | Biallelic |
| Igf2 | Paternal | Paternal | Paternal | Paternal | Paternal |
| Mest | Paternal | Not Diagnostic | Not Diagnostic | Paternal | Biallelic |
| Peg3 | Paternal | Paternal | Paternal1 | Paternal | Biallelic |
| Snrpn | Paternal | Paternal | Paternal2 | Paternal | Biallelic |
One male and one female showed biallelic expression, four other hybrids showed unambiguous paternal expression.
One female showed biallelic expression, five other individuals showed unambiguous paternal expression.
Patterns of allelic expression were identical between males and females for all genes except at Snrpn as noted above. Sequencing results from our constructed mock hybrid pools confirmed that the species-specific alleles amplified with similar efficiency for all genes. We found approximately equal allele representation at diagnostic positions in 50:50 mock hybrid pools. Skewed biallelic expression was detected for reciprocal 75:25 pools while 99:1 pools were scored as monoallelic, indicating that our assays were sensitive to detecting biallelic expression at genes with moderately (but not highly) skewed allele-specific expression.
We used qPCR to test for changes in expression level in the same eight candidate genes. These assays test for changes in expression level irrespective of changes in imprinting status and so provide an additional means by which to test for regulatory disruption in the hybrids. Three genes (Grb10, H19, Mest) showed similar expression between the species and the reciprocal hybrids (Figure 4; Grb10, F3,20=2.158, P=0.125; H19, F3,20=0.754, P=0.533; Mest, F3,20=1.733, P=0.193). Snrpn showed significantly different expression between the species with the hybrids showing intermediate expression (F3,20=3.205, P=0.045). In all cases, cxs hybrids showed similar expression to at least one of the parental species. Peg3 tended towards higher expression in both hybrids, but this difference was only significant relative to P. sungorus (F3,20 =4.7 5 0, P=0.012). Igf2 and Igf2r were the only genes that showed significantly different expression between the reciprocal hybrids, and in both cases the cxs hybrid was similar to the parent species (Igf2, F3,20=5.749, P=0.005; F3,20=3.913, P=0.024 for Igf2r).
Figure 4.
Placental expression levels of candidate maternally (A) and paternally (B) expressed genes. Expression was normalized using the gene Ywhaz with the ΔCT method and subsequently adjusted to set the expression level of the lowest expressed cross type to one (±2 SE). Bars represent the mean of each cross (n=6), significant differences were tested with an ANOVA (P < 0.05), and the lettered groupings were assigned based on a Tukey test.
Divergence estimates for taxa showing abnormal growth effects
We estimated Dxy (Kimura-two-parameter-corrected) sequence divergence for 34 species pairs that show abnormal hybrid growth and had cyt b sequence data available from GenBank. Pairwise divergence estimates ranged between 19.3% and 0.0% (Supplemental Table 1). Mitochondrial divergence for dwarf hamsters at cyt b was 3.7%. We also estimated pairwise silent site divergence (Ks) for dwarf hamsters and house mice (M. musculus and M. spretus) in a concatenated alignment of eight candidate nuclear genes to be 0.9% and 1.4%, respectively.
Discussion
Parent-of-origin growth effects in dwarf hamsters and other mammals
Dwarf hamster hybrids display strong parent-of-origin growth effects that manifest a wide range of inviability phenotypes. When a P. sungorus female was crossed with a P. campbelli male, embryo and placenta overgrowth was so extreme that it was ultimately lethal to the mother and offspring during birth. However, despite the potentially high rates of maternal mortality suggested by our study, viable S×C hybrids have been reported (Sokolov and Vasil'eva 1993). In this previous study, the average adult weight of 13 S×C hybrids was 55.2 grams — nearly 200% the weight of either species. It is unclear what the probability of a successful S×C birth is based on these limited data, but they do demonstrate that the striking placental and embryonic overgrowth that we observed during late gestation (Figure 1) persist through to adulthood. Alternatively, adult cxs hybrid males were approximately 40% smaller than their male parents though they appeared normal at birth with respect embryo and placenta sizes. Thus the overgrown S×C cross yields more severe hybrid inviability phenotypes but both cross types show evidence for growth effects and reproductive isolation as indicated by significantly reduced litter sizes (Table 2).
Parent-of-origin growth effects in hybrid dwarf hamsters are strikingly similar to several previously described examples in other hybrid mammals (Vrana 2007). Extreme and often lethal hybrid overgrowth also occurs in crosses between female Peromyscus polionotus and male P. maniculatus (Dawson 1965; Rogers and Dawson 1970; Dawson et al. 1993; Vrana 2007). Likewise, the well-known example of reciprocal crosses between lions (Panthera leo) and tigers (P. tigris) results in strong parent-of-origin growth phenotypes that persist into adulthood. So-called ligers (hybrids from a female tiger × male lion) reach adult weights approaching 150% the size of a tiger (the larger of the parents). Gray (1972) quotes a report of one liger that “weighed as much as both parents together”, which is striking if not strongly quantitative. The reciprocal cross (i.e., tigons) is smaller than either species (Vrana 2007). No data are available for liger or tigon placentas but we predict that their growth would be similarly affected.
Interestingly, not all hybrid crosses presenting abnormal placental growth also manifest embryonic or adult growth phenotypes. For example, parent-of-origin developmental effects in several mouse crosses (genus Mus) are restricted to the placenta, and do not strongly influence embryonic or adult growth (Zechner et al. 1996; Kurz et al. 1999; Zechner et al. 2004). Reciprocal crosses between horses and donkeys also yield parent-of-origin effects on placental size and morphology but not embryo size (Allen 1969; Allen et al. 1993). In both of these systems abnormal placentation impacts embryonic viability in hybrids (West et al. 1977; Zechner et al. 1996; Kurz et al. 1999; Allen 2001). Artificial insemination has recently been used to achieve a more divergent Mus cross that results in extreme placental and embryonic growth (M. musculus and M. caroli: Brown et al. 2012). Thus abnormal placentation appears to represent an important but not sufficient first step in the evolution of parent-of-origin growth effects in adult hybrid mammals. Most of the phenotypic data from mammalian hybrids derive from qualitative differences in postnatal body size (Gray 1972) and placental phenotypes are rarely collected. Therefore, it is possible that the disruption of hybrid placentation is much more rapidly evolving and widespread than is commonly appreciated. Consistent with this prediction, the rate at which reproductive isolation evolves across different mammal groups has been shown to correlate with physiological aspects of placental morphology (Elliot and Crespi 2006).
The hybrid growth effects noted by Gray (1972) derive from crosses between species pairs spanning a broad range of taxonomic (intraspecific to intergeneric) and genetic divergence (0 to ~20% pairwise divergence at cyt b; Supplemental Table 1). Some of these qualitative growth effects are anecdotal and require further validation. Other examples likely reflect heterosis generated through the masking of deleterious recessive alleles and thus do not reflect true intrinsic incompatibilities. In this context, parent-of-origin growth effects likely provide the strongest and most relevant examples of hybrid inviability. The P. sungorus × P. campbelli cross is especially intriguing because they are among the most closely related species that show asymmetric hybrid growth (3.7% pairwise divergence at cyt b). This is on the low end of divergence typically found between sister mammalian species (Bradley and Baker 2001). By comparison, deer mice (P. polionotus and P. maniculatus; 4.0%), horses and donkeys (7.7%), lions and tigers (11.6%), and house mice (M. musculus and M. spretus; 9.6%) are all more divergent. Admittedly, mitochondrial DNA often does not accurately reflect genomic divergence between species (Ballard and Whitlock 2004) but information on nuclear divergence is not yet available for most of the species pairs considered here. Pairwise synonymous divergence (Ks) between P. sungorus × P. campbelli at the eight candidate imprinted genes was 0.9%, indicating that they also show very low divergence across the nuclear genome. For comparison, the same eight genes show considerably more synonymous divergence (Ks=1.4%) between two house mouse species (M. musculus and M. spretus) estimated to share a most recent common ancestor ~1.5 MYA (Suzuki et al. 2004). These data indicate that parent-of-origin growth effects can manifest between closely related species and may contribute to the early stages of mammalian speciation.
Our data also support the observation that the evolution of abnormal hybrid growth in mammals tends to follow Haldane's rule. Hybrid mice, hamsters, and deer mice all show male-specific growth phenotypes at some point during development. Deer mouse hybrids show a strongly female-biased sex ratio (Dawson et al. 1993) due to more extreme overgrowth in male placentas and embryos (Vrana et al. 2000; Vrana 2007). Male placentas also tend to be much larger in mouse hybrids between M. musculus and M. spretus or M. macedonicus, (Zechner et al. 1996). Hamster hybrids do not show any sex-specific placental or embryonic differences, but adult cxs hybrid males are severely growth restricted when compared to the females (Figure 3). However, we did observe a male bias in cxs litters (~60%) that could reflect differential female inviability and thus an exception to Haldane's rule. However, we believe that a maternal effect in this cross provides the simplest explanation for this pattern. Maternal effects that give rise to skewed sex ratios are common in mammals (Clutton-Brock and Iason 1986) and there were no embryonic phenotypes that indicated females were less viable in utero.
Resolving the genetic and epigenetic bases of parent-of-origin growth
If we assume that abnormal hybrid growth in mammals generally follows the Dobzhansky-Muller model for intrinsic incompatibilities (Dobzhansky 1937; Muller 1942), then it is likely caused by the evolution of incompatible interactions between growth-related genes that have diverged between the hybridizing species. Such failed interactions could disrupt the epigenetic regulation of imprinting and change the expression of genes that control offspring growth. Alternatively, hybrid incompatibilities may cause abnormal growth independent of disrupted imprinting. Differentiating between these models remains a fundamental problem in mammalian speciation. Epigenetic disruption of imprinting has emerged as the predominant model to explain parent-of-origin dependent growth in mammal hybrids (Vrana 2007; Crespi and Nosil 2013). This model is compelling because errors in imprinting have the ability to explain growth effects (many imprinted genes regulate growth), parent-of-origin effects (imprinting is a parent-of-origin dependent process), and why these phenotypes are common in mammals (mammals are the only vertebrates where imprinting has been found). Indeed, an analogous regulatory process has been described in the endosperm of angiosperms (Lin 1982; Haig and Westoby 1989). Hybrid endosperm development sometimes shows parent-of-origin growth effects (Ishikawa et al. 2011), and disrupted imprinting has been associated with abnormal endosperm development (Erilova et al. 2009). Nonetheless, despite the broad appeal of this model, no consensus has been reached on the role of disrupted imprinting in the evolution of parent-of-origin dependent growth effects in mammals (Zechner et al. 2004; Brown et al. 2012; Wang et al. 2013).
Our candidate gene data do not support the hypothesis that changes in imprinting status contribute to abnormal hybrid growth in hamsters. Imprinting is maintained at some genes in reciprocal dwarf hamster hybrids while other genes are biallelically expressed in both hybrids (Table 4). Gene expression patterns that are the same between the reciprocal hybrids cannot by definition explain phenotypic differences between those hybrids, even if they are a pleiotropic consequence of underlying hybrid incompatibilities. None of the candidate genes showed differences in imprinting status between the reciprocal hybrids. This strongly contrasts with the extensive changes in imprinting status associated with similarly extreme parent-of-origin growth effects between reciprocal deer mouse hybrids (Table 4; Vrana et al. 1998). In our study, biallelic expression of Igf2r, Grb10, and Mash2 may simply reflect that these genes are not imprinted in hamsters or that imprinting is disrupted at these genes in hybrids independent of the observed parent-of-origin growth effects. Our qPCR results support the former hypothesis; biallelic expression of Igf2r, Grb10, and Mash2 was not accompanied by concomitant increases in expression level in both hybrids relative to the parental species as would be predicted for loss of imprinting (Figure 4). Likewise, apparent variation in the imprinting at Peg3 (two out of six S×C offspring have biallelic expression) and Snrpn (biallelic in a single S×C female) could reflect polymorphism for imprinting at these genes in dwarf hamsters. In agreement with these patters, Mash2 does not appear to be imprinted in deer mice (Vrana et al. 1998), imprinting of Igf2r is polymorphic in humans (Xu et al. 1993), and Grb10 is imprinted in opposite parental directions in a tissue-specific manner in mice (Garfield et al. 2011). Collectively, these data argue that imprinting status is labile within and between species and underscore that much more work is needed to understand the evolution of genomic imprinting.
The limitations of our allele-specific expression assay aside, monoallelic expression at Igf2, H19, Peg3, and Snrpn clearly demonstrates that imprinting is not widely disrupted in placenta of Phodopus hybrids. Moreover, several genes with disrupted imprinting in deer mouse hybrids show normal imprinted expression in hamster hybrids (Table 4; Vrana et al. 1998). When considered in light of other studies, this result indicates that both the identity and genomic extent of disrupted imprinting is highly variable across mammalian hybrids (Vrana et al. 1998; O'Neill et al. 1998; Roemer et al. 1999; Schutt et al. 2003; Wang et al. 2013). However, our candidate gene approach does not exclude disrupted imprinting as the ultimate cause of parent-of-origin dependent growth in hamsters. Our experiment has only considered ~10% of the approximately 80 imprinted genes expressed in the mouse placenta (Morison et al. 2005). Imprinted genes also tend to occur in clusters in mammalian genomes (Verona et al. 2003) and cluster-specific imprinting breakdown has been described in deer mice (Wiley et al. 2008). The diagnostic seven genes that we surveyed represent only five of the eighteen clusters in house mice (Morison et al. 2005). We are currently collecting genome-wide expression data to determine if cluster-specific breakdown of imprinting also occurs in hybrid dwarf hamsters.
While allele-specific expression differences in hamster hybrids did not correlate with parent-of-origin growth effects, we did observe significant differences in expression levels at Igf2 and Igf2r between reciprocal hybrids (Figure 4). These genes are antagonists to each other in the fetal growth pathway. Igf2 encodes a protein hormone (IGF2) that promotes growth while Igf2r encodes a transmembrane receptor that sequesters IGF2 out of circulation. The observation that two interacting genes involved in regulating fetal growth are differentially expressed in hybrids with severe growth defects is intriguing. We naively predicted that Igf2 (and other paternally expressed growth factors) would show increased expression in overgrown hybrids both Igf2 and Igf2r show lower expression in S×C. This result suggests that our simple models based on imprinted gene dosage and functions are unlikely to effectively explain the complex phenotypes of parent-of-origin growth. Recent work in house mice has shown that complex epistatic interactions between imprinted and non-imprinted genes can lead to pervasive disruption of gene regulatory networks and manifest in strong parent-of-origin effects (Mott et al. 2014). Such effects are likely to be even more pervasive when considering the extensive epistatic interactions that are exposed when two divergent genomes are combined in a hybrid.
Finally, our data also establish that sex-specific effects are recurrent in the evolution of parent-of-origin dependent hybrid growth. At face value this is not surprising given Haldane's rule (Haldane 1922) and the general predictions of dominance theory (Turelli and Orr 2000). However, sex-specific effects are not expected in the placenta because the paternal X chromosome is silenced in extra-embryonic tissues in rodents (imprinted X chromosome inactivation or XCI; Lyon 1961; 1962). Given imprinted XCI, X-linked genes expressed in the placenta are expected to be effectively hemizygous in both sexes and thus a recessive incompatibility on X chromosome should affect both sexes similarly. Several hypotheses have been proposed to account for unexpected sex-specific effects in hybrid placenta (Hemberger et al. 2001). Recessive X-linked incompatibilities may be partially masked in females due to incomplete silencing of the paternal X chromosome (i.e., leaky imprinted XCI) or through some contribution of X-linked gene products expressed in female embryos where XCI is random (Payer and Lee 2008). Likewise, disruption of imprinted XCI in the placenta could also mask deleterious recessive interactions in females; though it seems unlikely that breakdown of a major epigenetic process would generally result in increased viability. Finally, the male-specific effects could reflect the action of the Y chromosome (Hemberger et al. 2001), though Y-linked effects usually are restricted to male reproductive phenotypes. Differentiating among these potential models will be crucial for resolving the ultimate causes of male-biased developmental abnormalities in mammals.
Supplementary Material
Acknowledgments
We would like to thank Catherine Wynne-Edwards for bringing to our attention the presence of parent-of-origin growth effects in hamsters. We would also like to thank Robert Johnston, Mary Timonin, and Ned Place for supplying the animals, and Kelly Carrick and the staff of the UM laboratory animal research facilities. Colin Prather, Sara Keeble, and Lindy Henry assisted with dissections and animal care. Ted Cosart retrieved orthologous DNA sequences from M. spretus. Ryan Bracewell, Doug Emlen, Lila Fishman, Erica Larson, John McCutcheon, Dan Vanderpool, Paul Vrana, and Kris Crandell provided helpful discussions during the development of this project. Finally we would like to thank Robb Brumfield and three anonymous reviewers for their helpful comments. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number R01HD73439 (JMG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no conflict of interest. Additional funding was provided by the University of Montana (JMG) and the Rosemary Grant Student Research Award from the Society for the Study of Evolution (TDB).
Literature Cited
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Allen WR. Factors influencing pregnant mare serum gonadotrophin production. Nature. 1969;223:64–66. doi: 10.1038/223064a0. [DOI] [PubMed] [Google Scholar]
- Allen WR. Fetomaternal interactions and influences during equine pregnancy. Reproduction. 2001;121:513–527. [PubMed] [Google Scholar]
- Allen WR, Skidmore JA, Stewart F, Antczak DF. Effects of fetal genotype and uterine environment on placental development in equids. J. Reprod. Fertil. 1993;98:5560. doi: 10.1530/jrf.0.0980055. [DOI] [PubMed] [Google Scholar]
- Babak T, DeVeale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 2008;18:1735–1741. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, Denny P, Simecek P, Forejt J. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc. Natl. Acad. Sci. 2013;110:E468–E477. doi: 10.1073/pnas.1219126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer CC. Chronology of development for the golden hamster. J. Morphol. 1953;92:1–37. [Google Scholar]
- Bradley RD, Baker RJ. A test of the genetic species concept: cytochrome-b sequences and mammals. J. Mammal. 2001;82:960–973. doi: 10.1644/06-MAMM-F-038R2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Brown JD, Piccuillo V, O'Neill RJ. Retroelement demethylation associated with abnormal placentation in Mus musculus x Mus caroli hybrids. Biol. Reprod. 2012;86:88–88. doi: 10.1095/biolreprod.111.095273. [DOI] [PubMed] [Google Scholar]
- Burt A, Trivers R. Genetic conflicts in genomic imprinting. Proc. Roy. Soc. B. 1998;265:2393–2397. doi: 10.1098/rspb.1998.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler H, Juurlink BHJ. An atlas for staging mammalian and chick embryos. CRC Press; Boca Raton, FL: 1987. [Google Scholar]
- Campbell P, Good JM, Nachman MW. Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics. 2013;193:819–828. doi: 10.1534/genetics.112.148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock TH, Iason GR. Sex ratio variation in mammals. Q. Rev. Biol. 1986;61:339–374. doi: 10.1086/415033. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989a;43:362381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates Inc; Sunderland, MA: 2004. [Google Scholar]
- Coyne JA, Orr HA. In: Speciation and its consequences. Otte D, Endler JA, editors. Sinauer Associates Inc; Sunderland, MA.: 1989b. pp. 180–207. [Google Scholar]
- Coyne JA, Orr HA. “Patterns of speciation in Drosophila” revisited. Evolution. 1997;51:295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- Crespi BJ, Semeniuk C. Parent-offspring conflict in the evolution of vertebrate reproductive mode. Am. Nat. 2004;163:635–653. doi: 10.1086/382734. [DOI] [PubMed] [Google Scholar]
- Crespi BJ, Nosil P. Conflictual speciation: species formation via genomic conflict. Trends Ecol. Evol. 2013;1:48–57. doi: 10.1016/j.tree.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Czech MP. Signal transmission by the insulin-like growth factors. Cell. 1989;59:235–238. doi: 10.1016/0092-8674(89)90281-x. [DOI] [PubMed] [Google Scholar]
- Davis BW, Li G, Murphy WJ. Supermatrix and species tree methods resolve phylogenetic relationships within the big cats, Panthera (Carnivora: Felidae). Mol. Phylogenet. and Evol. 2010;56:64–76. doi: 10.1016/j.ympev.2010.01.036. [DOI] [PubMed] [Google Scholar]
- Dawson WD. Fertility and size inheritance in a Peromyscus species cross. Evolution. 1965;19:44–55. [Google Scholar]
- Dawson WD, Sagedy MN, En-yu L, Kass DH, Crossland JP. Growth regulation in Peromyscus species hybrids: a test for mitochondrial-nuclear genomic interaction. Growth Dev. Aging. 1993;57:121–133. [PubMed] [Google Scholar]
- Dobzhansky TG. Genetics and the origin of species. Columbia Univ. Press; New York, NY.: 1937. [Google Scholar]
- Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious Version 6.1.5 . 2005 Available from http://www.geneious.com/ [Google Scholar]
- Duselis AR, Vrana PB. Aberrant growth and pattern formation in Peromyscus hybrid placental development. Biol. Reprod. 2010;83:988–996. doi: 10.1095/biolreprod.110.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duselis AR, Vrana PB. Assessment and disease comparisons of hybrid developmental defects. Hum. Mol. Gen. 2007;16:808–819. doi: 10.1093/hmg/ddm025. [DOI] [PubMed] [Google Scholar]
- Elliot MG, Crespi BJ. Phylogenetic evidence for early hemochorial placentation in eutheria. Placenta. 2009;30:949–967. doi: 10.1016/j.placenta.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Elliot MG, Crespi BJ. Placental invasiveness mediates the evolution of hybrid inviability in mammals. Am. Nat. 2006;168:114–120. doi: 10.1086/505162. [DOI] [PubMed] [Google Scholar]
- Erilova A, Brownfield L, Exner V, Rosa M, Twell D, Scheid OM, Hennig L, Kohler C. Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLOS Genet. 2009;5:e1000663. doi: 10.1371/journal.pgen.1000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny inference package) version 3.6a3. 2002 Available from http://evolution.genetics.washington.edu/phylip.html.
- Fitzpatrick BM. Rates of evolution of hybrid inviability in birds and mammals. Evolution. 2004;58:1865. doi: 10.1111/j.0014-3820.2004.tb00471.x. [DOI] [PubMed] [Google Scholar]
- Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, Gilroy K, Baker S, Xia J, Dalley JW, Hurst LD, Wilkinson LS, Isles AR, Ward A. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469:534–538. doi: 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Giger T, Dean MD, Nachman MW. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLOS Genet. 2010;6:e1001148. doi: 10.1371/journal.pgen.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray AP. Mammalian hybrids. 2nd ed. Commonwealth Agricultural Bureau. Slough; UK.: 1972. [Google Scholar]
- Haig D. Genomic imprinting and kinship. Rutgers Univ. Press; Piscataway, NJ.: 2002. [Google Scholar]
- Haig D. Multiple paternity and genomic imprinting. Genetics. 1999;151:1229–1231. doi: 10.1093/genetics/151.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Placental hormones, genomic imprinting, and maternal–fetal communication. J. Evol. Biol. 1996;9:357–380. [Google Scholar]
- Haig D, Graham C. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell. 1991;64:1045. doi: 10.1016/0092-8674(91)90256-x. [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M. Parent-specific gene expression and the triploid endosperm. Am. Nat. 1989;134:147–155. [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Hemberger MC, Pearsall RS, Zechner U, Orth A, Otto S, Rüschendorf F, Fundele RH, Elliott RW. Genetic dissection of X-linked interspecific hybrid placental dysplasia in congenic mouse strains. Genetics. 1999;153:383–390. doi: 10.1093/genetics/153.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Kurz H, Orth A, Otto S, Lüttges A, Elliott RW, Nagy A, Tan S-S, Tam P, Zechner U, Fundele RH. Genetic and developmental analysis of X-inactivation in interspecific hybrid mice suggests a role for the Y chromosome in placental dysplasia. Genetics. 2001;157:341–348. doi: 10.1093/genetics/157.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T. Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J. 2011;65:798806. doi: 10.1111/j.1365-313X.2010.04466.x. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Leverence J, Bick D, Sampath V. Ontogeny of growth-regulating genes in the placenta. Placenta. 2012;33:94–99. doi: 10.1016/j.placenta.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Kurz H, Zechner U, Orth A, Fundele RH. Lack of correlation between placenta and offspring size in mouse interspecific crosses. Anat. Embryol. 1999;200:335–343. doi: 10.1007/s004290050284. [DOI] [PubMed] [Google Scholar]
- Leiser R, Kaufmann P. Placental structure: in a comparative aspect. Exp. Clin. Endocrinol. 1994;102:122–134. doi: 10.1055/s-0029-1211275. [DOI] [PubMed] [Google Scholar]
- Li LL, Keverne EB, Aparicio SA, Ishino F, Barton SC. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lin BY. Association of endosperm reduction with parental imprinting in maize. Genetics. 1982;100:475–486. doi: 10.1093/genetics/100.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindor NM, Ney JA, Gaffey TA, Jenkins I, Thibodeau SN, DeWald GW. A genetic review of complete and partial hydatidiform moles and nonmolar triploidy. Mayo Clin. Proc. 1992;67:791–799. doi: 10.1016/s0025-6196(12)60805-2. [DOI] [PubMed] [Google Scholar]
- Loschiavo M, Nguyen QK, Duselis AR, Vrana PB. Mapping and identification of candidate loci responsible for Peromyscus hybrid overgrowth. Mamm. Genome. 2007;18:75–85. doi: 10.1007/s00335-006-0083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 1962;14:135–148. [PMC free article] [PubMed] [Google Scholar]
- Mallarino R, Campás O, Fritz JA, Burns KJ, Weeks OG, Brenner MP, Abzhanov A. Closely related bird species demonstrate flexibility between beak morphology and underlying developmental programs. Proc. Natl. Acad. Sci. 2012;109:16222–16227. doi: 10.1073/pnas.1206205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLOS Biol. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Landeen EL, Cook JM, Kingan SB, Presgraves DC. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLOS Biol. 2011;9:e1001126. doi: 10.1371/journal.pbio.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Mott R, Yuan W, Kaisaki P, Gan X, Cleak J, Edwards A, Baud A, Flint J. The architecture of parent-of-origin effects in mice. Cell. 2014;156:332–342. doi: 10.1016/j.cell.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution, and temperature. Biol. Symp. 1942;6:71–125. [Google Scholar]
- Neumann K, Michaux J, Lebedev V, Yigit N, Colak E, Ivanova N, Poltoraus A, Surov A, Markov G, Maak S, Neumann S, Gattermann R. Molecular phylogeny of the Cricetinae subfamily based on the mitochondrial cytochrome b and 12S rRNA genes and the nuclear vWF gene. Mol. Phylogenet. and Evol. 2006;39:135–148. doi: 10.1016/j.ympev.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Newkirk KD, McMillan HJ, Wynne-Edwards KE. Length of delay to birth of a second litter in dwarf hamsters (Phodopus): Evidence for post-implantation embryonic diapause. J. Exp. Zool. 1997;278:106–114. [PubMed] [Google Scholar]
- O'Neill RJW, O'Neill MJ, Graves JAM. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- Orr HA. Dobzhansky, Bateson, and the genetics of speciation. Genetics. 1996;144:13311335. doi: 10.1093/genetics/144.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the 36 balance. Annu. Rev. Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Piedrahita JA. The role of imprinted genes in fetal growth abnormalities. Birth Defects Res. A. Clin. Mol. Teratol. 2011;91:682–692. doi: 10.1002/bdra.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager EM, Wilson AC. Slow evolutionary loss of the potential for interspecific hybridization in birds: a manifestation of slow regulatory evolution. Proc. Natl. Acad. Sci. 1975;72:200–204. doi: 10.1073/pnas.72.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. URL: http://www.R-project.org/ [Google Scholar]
- Reik W, Constancia M, Fowden A, Anderson N, Dean W, Ferguson-Smith A, Tycko B, Sibley C. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J. Physiol. 2003;547:35–44. doi: 10.1113/jphysiol.2002.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer I, Grutzner F, Winking H, Haaf T, Orth A. Genome evolution: global methylation in eutherian hybrids. Nature. 1999;401:131–132. doi: 10.1038/43607. [DOI] [PubMed] [Google Scholar]
- Rogers JF, Dawson WD. Foetal and placental size in a Peromyscus species cross. J. Reprod. Fertil. 1970;21:255–262. doi: 10.1530/jrf.0.0210255. [DOI] [PubMed] [Google Scholar]
- Ross PD. Phodopus campbelli. Mamm. Species. 1995;503:1–7. [Google Scholar]
- Ross PD. Phodopus sungorus. Mamm. Species. 1998;595:1–9. [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz SA, editors. Bioinformatics Methods and Protocols: Methods for Molecular Biology. Humana Press; Totowa, NJ.: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Safronova LD, Vasil'eva NY. Meiotic abnormalities in interspecific hybrids between Phodopus sungorus (Pallas, 1773) and Ph. campbelli (Thomas, 1905). Russ. J. Genet. 1996;32:486–494. [Google Scholar]
- Safronova LD, Cherepanova EV, Vasil'eva NY. Specific features of the first meiotic division in hamster hybrids obtained by backcrossing Phodopus sungorus and Phodopus campbelli. Russ. J. Genet. 1999;35:184–188. [PubMed] [Google Scholar]
- Saukkonen T. Dose-dependent effects of recombinant human insulin-like growth factor (IGF)-I/IGF binding protein-3 complex on overnight growth hormone secretion and insulin sensitivity in type 1 diabetes. J. Clin. Endocr. Metab. 2004;89:46344641. doi: 10.1210/jc.2004-0243. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schutt S, Florl AR, Shi W, Hemberger M, Orth A. DNA methylation in placentas of interspecies mouse hybrids. Genetics. 2003;165:223–228. doi: 10.1093/genetics/165.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scribner SJ, Wynne-Edwards KE. Disruption of body temperature and behavior rhythms during reproduction in dwarf hamsters ( Phodopus). Physiol. Behav. 1994;55:361–369. doi: 10.1016/0031-9384(94)90147-3. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Shaw CN, Wilson PJ, White BN. A reliable molecular method of gender determination for mammals. J. Mammal. 2003;84:123–128. [Google Scholar]
- Shi W, Krella A, Orth A, Yu Y, Fundele RH. Widespread disruption of genomic imprinting in adult interspecies mouse (Mus) hybrids. genesis. 2005;43:100–108. doi: 10.1002/gene.20161. [DOI] [PubMed] [Google Scholar]
- Shi W, Lefebvre L, Yu Y, Otto S, Krella A, Orth A, Fundele RH. Loss-of-imprinting of Pegl in mouse interspecies hybrids is correlated with altered growth. genesis. 2004;39:65–72. doi: 10.1002/gene.20027. [DOI] [PubMed] [Google Scholar]
- Sobel JM, Chen GF, Watt LR, Schemske DW. The biology of speciation. Evolution. 2010;64:295–315. doi: 10.1111/j.1558-5646.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- Sokolov VE, Vasil'eva NI. Hybridologic analysis confirms the species specificity of Phodopus sungorus (Pallus, 1773) and Phodopus campbelli (Thomas, 1905). Dokl. Akad. Nauk. 1993;332:120–123. [PubMed] [Google Scholar]
- Surani MA, Allen ND, Barton SC, Fundele RH, Howlett SK, Norris ML, Reik W. Developmental consequences of imprinting of parental chromosomes by DNA methylation. Philos. Trans. R. Soc. London. B. 1990;326:313–327. doi: 10.1098/rstb.1990.0014. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. and Evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Tang S, Presgraves DC. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 2009;323:779–782. doi: 10.1126/science.1169123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler K. The house mouse. Development and normal stages from fertilization to 4 weeks of age. Springer-Verlag; New York, NY.: 1972. [Google Scholar]
- Turelli M, Orr HA. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. The dominance theory of Haldane's rule. Genetics. 1995;140:389402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmuza S. Gametic imprinting as a speciation mechanism in mammals. J. Theor. Biol. 1993;164:1–13. doi: 10.1006/jtbi.1993.1137. [DOI] [PubMed] [Google Scholar]
- Verona RI, Mann MRW, Bartolomei MS. Genomic imprinting: intricacies of 39 epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- Vrana PB. Genomic imprinting as a mechanism of reproductive isolation in mammals. J. Mammal. 2007;88:5–23. [Google Scholar]
- Vrana PB, Fossella JA, Matteson P, del Rio T, O'Neill MJ, Tilghman SM. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat. Genet. 2000;25:120–124. doi: 10.1038/75518. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Guan XJ, Ingram RS, Tilghman SM. Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nat. Genet. 1998;20:362–365. doi: 10.1038/3833. [DOI] [PubMed] [Google Scholar]
- Wang X, Miller DC, Harman R. Paternally expressed genes predominate in the placenta. Proc. Natl. Acad. Sci. 2013;110:10705–10710. doi: 10.1073/pnas.1308998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JD, Frels WI, Papaioannou VE. Development of interspecific hybrids of Mus. J. Embryol. Exp. Morph. 1977;41:233–243. [PubMed] [Google Scholar]
- Wiley CD, Matundan HH, Duselis AR, Isaacs AT, Vrana PB. Patterns of hybrid loss of imprinting reveal tissue- and cluster-specific regulation. PLOS ONE. 2008;3:e3572. doi: 10.1371/journal.pone.0003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Systems of mating. II. The effects of inbreeding on the genetic composition of a population. Genetics. 1921;6:124–143. doi: 10.1093/genetics/6.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Goodyer CG, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem. Biophys. Res. Comm. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- Zechner U, Reule M, Orth A, Bonhomme F, Strack B, Guenet J-L, Hameister H, Fundele RH. An X-chromosome linked locus contributes to abnormal placental development in mouse interspecific hybrids. Nat. Genet. 1996;12:398–403. doi: 10.1038/ng0496-398. [DOI] [PubMed] [Google Scholar]
- Zechner U, Reule M, Burgoyne PS, Schubert A, Orth A, Hameister H, Fundele RH. Paternal transmission of X-linked placental dysplasia in mouse interspecific hybrids. Genetics. 1997;146:1399–1405. doi: 10.1093/genetics/146.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner U, Shi W, Hemberger M, Himmelbauer H, Otto S, Orth A, Kalscheuer V, Fischer U, Elango R, Reis A, Vogel W, Ropers H, R schendorf F, Fundele RH. Divergent genetic and epigenetic post-zygotic isolation mechanisms in Mus and Peromyscus. J. Evol. Biol. 2004;17:453–460. doi: 10.1046/j.1420-9101.2003.00656.x. [DOI] [PubMed] [Google Scholar]
- Zeh DW, Zeh JA. Reproductive mode and speciation: the viviparity-driven conflict hypothesis. Bioessays. 2000;22:938–946. doi: 10.1002/1521-1878(200010)22:10<938::AID-BIES9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.