Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating neurological disease with no effective treatment. Here we report the results of a moderate-scale sequencing study aimed at identifying new genes contributing to predisposition for ALS. We performed whole exome sequencing of 2,874 ALS patients and compared them to 6,405 controls. Several known ALS genes were found to be associated, and the non-canonical IκB kinase family TANK-Binding Kinase 1 (TBK1) was identified as an ALS gene. TBK1 is known to bind to and phosphorylate a number of proteins involved in innate immunity and autophagy, including optineurin (OPTN) and p62 (SQSTM1/sequestosome), both of which have also been implicated in ALS. These observations reveal a key role of the autophagic pathway in ALS and suggest specific targets for therapeutic intervention.
Amyotrophic lateral sclerosis (ALS) is a fatal, progressive neurodegenerative disease characterized by loss of motor neuron function for which there is no effective treatment or definitive diagnostic test (most cases are diagnosed clinically) (1). Approximately 10% of ALS cases are familial and inherited in an autosomal dominant, autosomal recessive, or X-linked mode, while the remaining cases are apparently sporadic (2, 3). Approximately 20 genes collectively explain a majority of familial cases, but these genes can explain only a minority (about 10%) of sporadic cases (2, 3) (Table 1).
Table 1.
Variants in previously described and presently reported ALS genes
| Gene | Reported inheritance modelǂ | Reported FALS explainedǂ | Reported SALS explainedǂ | Best model with case enrichment in present study (p-value)† | Cases with variant in best model† | Controls with variant in best model† | Potential ALS cases explained†† |
|---|---|---|---|---|---|---|---|
| TBK1 | N/A | N/A | N/A | Dom not benign (D=1.13×10−5; R=5.78×10−7; C=3.63×10−11) | D=23 (0.8%); R=23 (1.745%); C=46 (1.097%) | D=12 (0.187%); R=5 (0.211%); C=17 (0.194%) | 0.904% |
| NEK1 | N/A | N/A | N/A | Dom LoF (D=1.08×10−6; R=0.001; C=3.20×10−9) | D=25 (0.870%); R=10 (0.759%); C=35 (0.835%) | D=6 (0.094%); R=2 (0.084%); C=8 (0.091%) | 0.744% |
| SOD1 | AR/AD | 12% | 1.50% | Dom coding (7.23×10−8) | 25 (0.870%) | 5 (0.078%) | 0.792% |
| TARDBP | AD | 4% | 1% | Dom coding (2.97×10−6) | 19 (0.661%) | 6 (0.094%) | 0.567% |
| OPTN | AR/AD | <1% | <1% | Dom not benign (D=0.023; R=0.002; C=0.002) | D=18 (0.626%); R=8 (0.607%); C=26 (0.620%) | D=16 (0.25%); R=4 (0.169%); C=20 (0.228%) | 0.392% |
| SPG11 | AR | <1% | <1% | Dom LoF (D=0.015; R=0.183; C=0.017) | D=21 (0.731%); R=5 (0.379%); C=26 (0.620%) | D=20 (0.312%); R=7 (0.295%); C=27 (0.308%) | 0.313% |
| VCP | AD | 1% | 1% | Dom coding (0.022) | 8 (0.278%) | 4 (0.062%) | 0.216% |
| HNRNPA1 | AD | <1% | <1% | Dom coding (0.103) | 6 (0.209%) | 5 (0.078%) | 0.131% |
| ATXN2** | AD | <1% | <1% | Rec coding (0.206) | 4 (0.139%) | 2 (0.031%) | 0.108% |
| ANG | AD | <1% | <1% | Dom LoF (0.217) | 2 (0.070%) | 1 (0.016%) | 0.054% |
| CHCHD10 | AD | <1% | <1% | Dom coding (0.226) | 2 (0.070%) | 0 (0%) | 0.070% |
| SIGMAR1 | AR | <1% | <1% | Dom LoF (0.226) | 1 (0.035%) | 0 (0%) | 0.035% |
| FIG 4 | AR/AD | <1% | <1% | Dom LoF (0.233) | 9 (0.313%) | 12 (0.187%) | 0.126% |
| SS18L1 | AD | <1% | <1% | Dom LoF (0.241) | 1 (0.035%) | 0 (0%) | 0.035% |
| GRN | AD | <1% | <1% | Dom not benign (0.357) | 14 (0.487%) | 24 (0.375%) | 0.112% |
| SETX | AD | <1% | <1% | Rec not benign (0.380) | 3 (0.104%) | 4 (0.062%) | 0.042% |
| HNRNPA2B1 | AD | <1% | <1% | Dom not benign (0.423) | 3 (0.104%) | 4 (0.062%) | 0.042% |
| SQSTM1 | AD | 1% | <1% | Dom LoF (0.546) | 1 (0.035%) | 2 (0.031%) | 0.004% |
| TAF15 | AR/AD | <1% | <1% | Rec not benign (0.555) | 2 (0.070%) | 1 (0.016%) | 0.054% |
| FUS | AR/AD | 4% | 1% | Dom LoF (0.612) | 2 (0.070%) | 3 (0.047%) | 0.023% |
| ALS2 | AR | <1% | <1% | Rec coding (0.655) | 2 (0.070%) | 4 (0.062%) | 0.007% |
| VAPB | AD | <1% | <1% | Dom not benign (0.688) | 3 (0.104%) | 5 (0.078%) | 0.026% |
| NEFH | AD | <1% | <1% | Dom coding (0.777) | 22 (0.765%) | 37 (0.578%) | 0.188% |
| C9orf72** | AD | 40% | 7% | Dom not benign (1.000) | 4 (0.139%) | 7 (0.109%) | 0.030% |
| CHMP2B | AD | <1% | <1% | Rec coding (1.000) | 1 (0.035%) | 1 (0.016%) | 0.019% |
| MATR3 | AD | <1% | <1% | Dom coding (1.000) | 19 (0.661%) | 35 (0.546%) | 0.115% |
| PFN1 | AD | <1% | <1% | Rec coding (1.000) | 9 (0.313%) | 15 (0.234%) | 0.079% |
| PRPH | AD | <1% | <1% | Dom LoF (1.000) | 1 (0.035%) | 2 (0.031%) | 0.004% |
| SPAST | AD | <1% | <1% | Dom coding (1.000) | 6 (0.209%) | 12 (0.187%) | 0.021% |
| TUBA4A* | AD | 1% | <1% | Dom coding (0.743) | 3 (0.104%) | 7 (0.109%) | 0% |
| ELP3* | Allelic | <1% | <1% | Rec coding (1.000) | 0 (0%) | 0 (0%) | 0% |
| DAO* | AD | <1% | <1% | Rec coding (1.000) | 0 (0%) | 0 (0%) | 0% |
| DCTN1* | AD | <1% | <1% | Dom coding (0.668) | 32 (1.113%) | 76 (1.187%) | 0% |
| EWSR1* | AD | <1% | <1% | Dom coding (0.375) | 10 (0.348%) | 28 (0.437%) | 0% |
| GLE1* | AD | <1% | <1% | Rec LoF (1.000) | 0 (0%) | 0 (0%) | 0% |
| UBQLN2* | XD | <1% | <1% | Dom LoF (1.000) | 0 (0%) | 0 (0%) | 0% |
No model showed case enrichment.
Because the known causal variants are repeat expansions that are not generally captured by next generation sequencing, no case enrichment is expected.
Based on discovery dataset for genes not included in the replication dataset, and otherwise D=discovery, R=replication, and C=combined.
Calculated as Cases with variant in best model - Controls with variant in best model; as case variants are risk factors for disease and may not be causal, this represents the potential percentage of cases for which this gene plays a role in disease.
Protein and protein/RNA aggregates are a common feature of ALS pathology. These aggregates often include proteins encoded by genes that cause ALS when mutated, including those encoding SOD1, TARDBP (TDP-43), and FUS(4). Multiple genes (e.g. C9orf72, GRN, VCP, UBQLN2, OPTN, NIPA1, SQSTM1) in addition to TARDBP harbor variants pathogenic for TARDBP proteinopathy manifesting as ALS. This pathological TARDBP is part of a common pathway linked to neurodegeneration caused by diverse genetic abnormalities (5). While murine models of ALS are limited, silencing certain ALS genes can cause regression of the disease phenotypes and clearance of the protein aggregates (6).
Identifying ALS genes
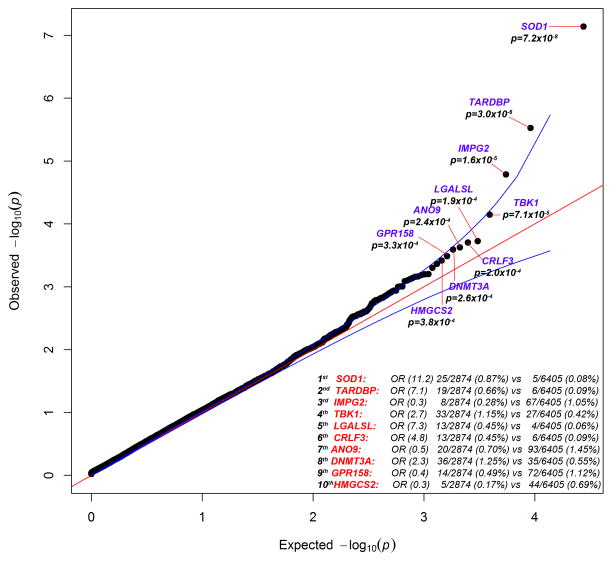
To identify genetic variants associated with ALS, we sequenced the exomes of 2,874 patients with ALS and 6,405 controls. We ran a standard collapsing analysis where the gene was the unit of analysis, and we coded individuals based on the presence or absence of “qualifying” variants in each sequenced gene, where qualifying was defined based on one of six different genetic models (7). A total of 17,248 genes had more than one case or control sample with a genetic variant meeting the inclusion criteria for at least one of the genetic models tested (Figs. 1, S1, S2). After correcting for multiple tests, the known ALS gene SOD1 (p=7.23×10−8; dominant coding model) was found to have a study-wide significant enrichment of rare variants in ALS cases as compared to controls, with qualifying variants in 0.870% of cases and 0.078% of controls. The genes HLA-B, ZNF729, SIRPA, and TP53 were found to have a significant enrichment of variants in controls; however, these associations appear to be due to sequencing differences and to subsets of the controls having been ascertained on the basis of relevant phenotypes.
Fig. 1. QQ plot of discovery results for dominant coding model.
The results for the analysis of 2,874 case and 6,405 control exomes are shown. 16,491 covered genes passed QC with more than one case or control carrier for this test. The genes with the top 10 associations are labeled. The genomic inflation factor, lambda (λ), is 1.061. The association with SOD1 passed correction for multiple tests.
Based on their associations with ALS in a preliminary discovery phase analysis utilizing 2,843 cases and 4,310 controls, we chose 51 genes (Table S4) for analysis in an additional 1,318 cases and 2,371 controls (sequenced using either whole exome or custom capture)(7). This analysis definitively identified TANK-Binding Kinase 1 (TBK1) as an ALS gene with a discovery association p=1.13×10−5, a replication p=5.78×10−7, and a combined p=3.63×10−11 (dominant not benign model). In the combined dataset, dominant not benign variants in this gene were found in 1.097% of cases and 0.194% of controls, with loss-of-function (LoF) variants occurring in 0.382% of cases and 0.034% of controls.
Analysis of clinical features
We also performed gene-based collapsing analyses to identify genes associated with patients’ age of onset, site of onset, and survival time. No genes showed genome-wide significant association with any of these features. When applying multiple-test correction to only known ALS predisposition genes and TBK1, we found that D-amino acid oxidase (DAO) significantly correlated with survival times, with variant carriers showing shorter survival times (p=5.5×10−7, dominant coding model). In mice, DAO is required for the clearance of D-serine. Indeed, D-serine levels are increased in SOD1 mutant mice and spinal cords from humans with familial (FALS) or sporadic ALS (SALS)(8, 9). Known FALS mutations seem to reduce DAO activity, leading to neurotoxicity (10).
ALS patients with mutations in more than one known ALS gene are reported to have a younger age of onset (11). While we did not replicate this finding in our dataset, our lack of sequence data for known C9orf72 carriers, by far the most common ALS variant, as well as our lack of information about ATXN2 expansions inhibits our ability to adequately assess such an association.
Associations with other ALS genes
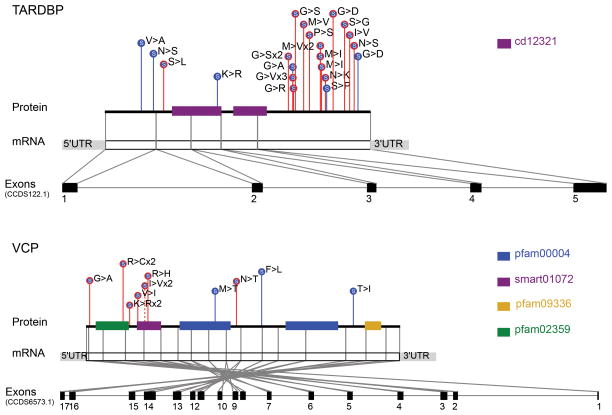
Although SOD1 was the only already known ALS gene to attain a genome-wide significant association in our data, many other known ALS genes showed strong associations. For example, rare coding variants in TARDBP occurred in 0.661% of the ALS cases and 0.094% of controls in our study, ranking this gene second to SOD1 genome-wide under the dominant coding model (discovery data, p=2.97×10−6; Fig. 1). Consistent with previous reports and the ALS pathogenic TARDBP “DM” variants in the Human Genome Mutation Database (HGMD) database (3, 12), we observed that the implicated non-synonymous variants were generally predicted to be benign by PolyPhen-2(13) and were clearly concentrated in the 3′ protein-coding portion of the gene in the cases as compared with the controls (Fig. 2).
Fig. 2. Variants in TARDBP and VCP.
Dominant coding variants are shown in TARDBP and VCP (discovery dataset). Case variants are enriched at the 3′ end of the gene in TARDBP and near the cell division protein 48 domain 2 region in VCP. LoF variants are filled in red, non-synonymous variants are filled in blue, and splice variants are filled in purple. Case variants are shown with red lines, control variants are shown with blue lines, and variants found in both cases and controls are shown with dashed lines.
In the case of OPTN, we observed rare damaging variants in 0.620% of ALS cases and 0.228% of controls (combined dominant not benign model; p=0.002). The greatest enrichment was for LoF variants, which occur in 0.334% of cases and 0.114% of controls (combined dominant LoF model, p=0.013). Whereas the initial studies of OPTN in ALS found a role in only a few families with a recessive genetic model, subsequent studies identified dominant mutations (14, 15). Here, dominant acting variants appeared to make a substantial contribution to sporadic disease.
Finally, we also observed a modest excess of qualifying variants in VCP (discovery dominant coding model; p=0.022) and of LoF variants in SPG11 (combined dominant LoF model; p=0.017). The former was driven by variants near the cell division protein 48 domain 2 region, where variants were found in 71% of case variants as compared to 25% of control variants (Fig. 2). Similar to OPTN, SPG11 has previously been reported as a cause of recessive juvenile ALS, but based on our data, it could play a broader role because these cases did not have early onset (16).
We did not identify even a nominal association with other previously reported ALS genes in our dataset, including the recently reported TUBA4A, MATR3, GLE1, SS18L1, and CHCHD10 (Table 1)(17–21). A fraction of our samples were genetically screened for some of the known genes and had positive cases removed prior to sequencing, which may partially explain the lack of signal (7). Additionally, a comparison with genes implicated in a recent assessment of the role of 169 previously reported and candidate ALS genes in 242 sporadic ALS cases and 129 controls showed no overlap beyond signals for SOD1 and SPG11 (22). Some of these previously studied genes are mutated so rarely that even the sample size presented here is not sufficient to detect causal variant enrichment, while others simply show comparable proportions of rare variants among cases and controls. Finally, certain genes did not show associations owing to the nature of the causal variation: most known pathogenic variants in ATXN2 and C9orf72 are repeats that cannot be identified in our sequence data.
TBK1, autophagy and neuroinflammation
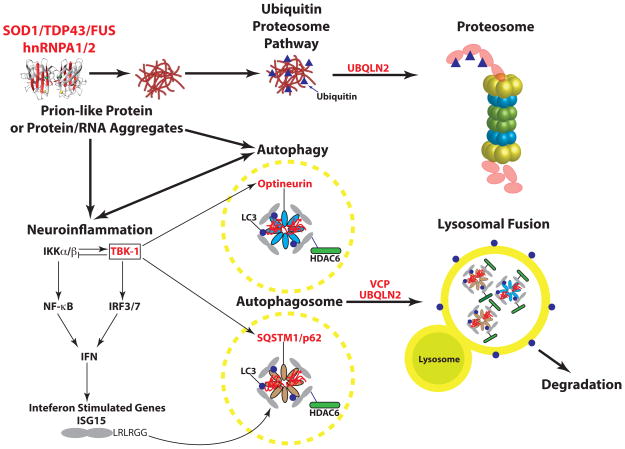
Previous studies have implicated both OPTN (optineurin)(23) and SQSTM1 (p62)(24) in ALS. The current study implicates TBK1 and suggests that OPTN is a more important disease gene than previously recognized. These genes play important and interconnected roles in both autophagy and inflammation, emerging areas of interest in ALS research (Fig. 3)(25–27). Mutations in SOD1, TARDBP and FUS result in the formation of protein aggregates that stain with anti-SQSTM1 and OPTN antibodies (28). These aggregates are thought to lead to a cargo-specific subtype of autophagy involved in the degradation of ubiquitinated proteins through the lysosome (29). The SQSTM1 and OPTN proteins function as cargo receptors, recruiting ubiquitinated proteins to the autophagosome via their LC3-interaction region (LIR) motifs. TBK1 phosphorylates both OPTN and SQSTM1 (30, 31) and enhances the binding of OPTN to the essential autophagosome protein LC3, thereby facilitating the autophagic turnover of infectious bacteria coated with ubiquitylated proteins, a specific cargo of the OPTN adaptor (32, 33). Considering that TBK1 co-localizes with OPTN and SQSTM1 in autophagosomes, it is possible that all three proteins associate with protein aggregates in ALS(32). Indeed, TBK1 appears to play a role in the degradation of protein aggregates by autophagy (34). Additionally, OPTN also functions in the autophagic turnover of damaged mitochondria via the PARKIN ubiquitin ligase pathway (35). Finally, VCP, encoded by another gene with mutations that cause ALS, also binds to ubiquitinated protein aggregates. VCP and autophagy are required for the removal of stress granules, dense cytoplasmic protein/RNA aggregates which are a common feature of ALS pathology (36). Thus, OPTN, SQSTM1, VCP and TBK1 may be critical components of the aggresome pathway required for the removal pathological ribonucleoprotein inclusions (37). It appears that defects in this pathway can be selective for motor neuron death, in some cases apparently sparing other neuronal cell types.
Fig. 3. A diagram showing the genes and pathways implicated in ALS disease progression.
Genes known to have sequence variants that cause or are associated with ALS are indicated in red. These mutations can lead to the formation of protein, or protein-RNA aggregates that appear as inclusion bodies in postmortem samples from both familial and sporadic ALS patients. Some of the mutant proteins adopt “prion-like” structures (see text for more detail). The misfolded proteins activate the ubiquitin/proteasome autophagy pathways to remove the misfolded proteins. Ubiquilin2 functions in both the Ub-proteosome and autophagy pathways. TBK-1 (boxed) lies at the interface between autophagy and inflammation and associates with and phosphorylates both optineurin and p62, which can, in turn, enhance inflammation. ISG15 is induced by type I interferons (α & β) and interacts with p62 and HDAC6 in the autophagosome.
In addition to their roles in autophagy, OPTN, SQSTM1 and TBK1 all function in the NF-κB pathway (Fig. 3)(27, 38). For example, IκB Kinases (IKKα and IKKβ) phosphorylate the IKK-related kinase TBK1, which in turn phosphorylates the IκB kinases, suppressing their activity in a negative autoregulatory feedback loop (39). TBK1 also phosphorylates and activates the transcription factor IRF3(40–42) and the critical innate immunity signaling components MAVS and STING(43). The coordinate activation of NF-κB and IRF3 turns on the transcription of many inflammatory genes, including interferon-β(44). The innate immune pathway and neuroinflammation in general are thought to be an important aspect of neurodegenerative disease progression (45). Thus, pathogenic variants in OPTN, SQSTM1, or TBK1 would be expected to lead to defects in autophagy and in key innate immunity signaling pathways. Mutations in these genes might therefore interfere with the normal function of these pathways in maintaining normal cellular riboproteostasis (37).
The simple observation of enrichment of qualifying variants in patients shows that some of the variants we have identified influence risk of disease. We cannot determine, however, the extent to which they may interact with any other variants or other risk factors in determining risk. We therefore focus on estimating the proportion of patients in which variants in the relevant genes either “cause or contribute” to disease by subtracting the proportion of controls with qualifying variants in a gene from the proportion of cases with such variants. While we saw no enrichment of case variants in SQSTM1, variants in OPTN and TBK1 were estimated to explain or contribute to 1.30% of cases in our dataset when taken together (combined data), suggesting an important subgroup of patients that may have a common biological etiology. No individual ALS cases had qualifying variants in more than one of these three genes.
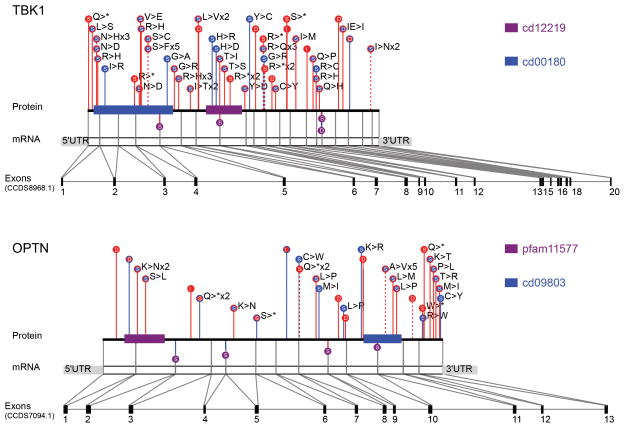
The case variants found in OPTN and TBK1 were largely heterozygous and LoF, suggesting that a reduction in trafficking of cargo through the autophagosomal pathway or disruption of autophagosomal maturation may promote disease. While the most obvious enrichment of case variants in TBK1 was seen for LoF, there was also a signal for non-synonymous variants, which were found in 1.026% of cases and 0.365% of controls (combined data). If any of these non-synonymous variants are selective LoF for specific TBK1 functions as opposed to complete LoF variants, they may help elucidate which TBK1 function is most relevant to disease. We did not observe any clear concentration of qualifying variants in any part of the TBK1 gene (Fig. 4).
Fig. 4. Variants in TBK1 and OPTN.
Dominant not benign variants are shown in TBK1 and OPTN (combined datasets). LoF variants are filled in red, non-synonymous variants are filled in blue, and splice variants are filled in purple. Case variants are shown with red lines, control variants are shown with blue lines, and variants found in both cases and controls are shown with dashed lines.
NEK1 associates with ALS2 and VAPB
Although no additional genes showed sufficiently strong evidence to be definitively declared disease genes at this point, some of the strongly associated genes identified here may be securely implicated as sample sizes increase. One gene of particular interest is NIMA-Related Kinase 1 (NEK1). This gene just reached experiment-wide significance in the combined discovery and replication data sets (discovery p=1.08×10−6; replication p=0.001, combined p=3.20×10−9; dominant LoF model). In the combined dataset, dominant LoF variants in this gene were found in 0.835% of cases and 0.091% of controls (Fig. S3). Additional studies are needed to confirm this suggestive association. Even if LoF variants in this gene do predispose to ALS, their relatively high prevalence in our controls and in public databases indicates that such variants have quite low penetrance given that the lifetime prevalence of ALS is approximately 0.2%.
NEK1 is a widely expressed multi-functional kinase linked to multiple cellular processes, but it has not been linked to ALS. In an unbiased proteomic search for NEK1-interacting proteins in HEK293T cells, we discovered an interaction between NEK1 and two widely expressed proteins previously found to be mutated in familial ALS – the RAB guanine nucleotide exchange factor ALS2 (also called Alsin) involved in endosomal trafficking and the endoplasmic reticulum protein VAPB involved in lipid trafficking to the plasma membrane (Fig. S4A,B, Table S5) (46). ALS2 reciprocally associated with NEK1 in HEK293T cells, and both ALS2 and VAPB associated with NEK1 reciprocally in a mouse neuronal cell line NSC-34 (Fig. S4C–E).
Other top genes showing interesting association patterns but not obtaining genome-wide significance included ENAH, with variants in 0.262% of cases and 0.011% of controls (combined dataset) (discovery p=1.83×10−5; replication p=0.133; combined p=9.59×10−6; recessive not benign model); CRLF3, with variants in 0.452% of cases and 0.094% of controls (discovery p= 0.0002; dominant coding model); DNMT3A, with variants in 1.002% of cases and 0.456% of controls (combined dataset) (discovery p=0.0002; replication p=0.261; combined p=0.0002; dominant not benign model); and LGALSL, with variants in 0.382% of cases and 0.068% of controls (combined data) (discovery p=0.0002; replication p=0.356; combined p=0.0002; dominant coding model).
Conclusions
Here, we have implicated TBK1 as an ALS gene, providing insight into disease biology and suggesting possible directions for drug screening programs. We have also provided evidence that OPTN plays a broader role in ALS than previously recognized. Both TBK1 and OPTN are involved in autophagy, with TBK1 possibly playing a crucial role in autophagosome maturation as well as the clearance of pathological aggregates (31, 34). These observations highlight a critical role of autophagy and/or inflammation in disease predisposition. It is also noteworthy that many drugs have been developed that act on TBK1-mediated pathways owing to their role in tumor cell survival (47) and can therefore be used to investigate the effects of drug-dependent loss of function of the kinase.
We also provide a large genetic dataset for ALS, which suggests other possible ALS genes and provides a substantial collection of pathogenic variants across ALS genes. After removing the expected number of variants to be seen based on the frequencies of rare variants in controls, we identify more than 70 distinct pathogenic mutations across SOD1, OPTN, TARDBP, VCP, SPG11, and TBK1 that can be used in future efforts to functionally characterize the role of these ALS genes. The identification of TBK1 and the expanded role for OPTN as ALS genes reinforce the growing recognition of the central role of autophagy and neuroinflammation in the pathophysiology of ALS (Fig. 3). These pathways appear to be activated in response to the formation of various types of cellular inclusions, the most prominent of which appear to be ribonucleoprotein complexes, which has led to the proposal that the control of protein (proteostasis) or ribonucleoprotein/RNA misfolding (“ribostasis”) plays a key role in neurodegenerative diseases (37). Cellular RNP inclusions can be caused by mutations in low complexity sequence domains or “prion” domains of RNA binding proteins (37, 48) and exacerbated by mutations that diminish the autophagy pathway. Remarkably, a hallmark of motor neuron pathology in >95% of sporadic and familial ALS patients is the formation of TARDBP inclusions, suggesting that defects in ribostasis is a common feature of the disease (5, 49). The prominence of this disease mechanism in ALS has been proposed to be the consequence of the normal function of low complexity domains in RNA binding proteins in the assembly of functional “RNA granules” such as P-bodies and stress granules (see (37) for detailed discussion).
Here, an exome sequencing study has successfully identified variants that definitively predispose humans to a sporadic, complex human disease. Suggestive evidence in genes that do not yet achieve significant associations strongly motivates performing even larger exome sequencing studies in ALS. There is reason for optimism that such studies will begin to fill in an increasingly complete picture of the key genes implicated in ALS, providing multiple entry points for therapeutic intervention (Fig. 3). It is also likely that whole genome sequencing (especially with longer reads) will prove of particular value in ALS, given that there are many causal variants refractory to identification using contemporary exome sequencing. Finally, we note that effective studies will depend critically on the control samples available. For example, here we used the recently released ExAC dataset of >60,000 samples to hone in on extremely rare variants in our samples (50). Well-characterized, publically available control sample sets will be of great importance for further discovery of variants associated with complex traits, in particular for whole genome sequencing studies.
Supplementary Material
Acknowledgments
J.W.H. is a consultant for Biogen-Idec and Millennium: the Takeda Oncology Company. R.B. is a consultant for Biogen, Idec and a cofounder of AviTx. F.B. is a founder of Regenesance. D.B.G. is a consultant for Biogen, Idec. R.M.M. is a consultant for Biogen, Idec.
Footnotes
FALS Sequencing Consortium: Peter C. Sapp1, Claire S. Leblond2, Diane McKenna-Yasek1, Kevin P. Kenna3, Bradley N. Smith4, Simon Topp4, Jack Miller4, Athina Gkazi4, Ammar Al-Chalabi4, Leonard H. van den Berg5, Jan Veldink5, Vincenzo Silani6, Nicola Ticozzi6, John Landers1, Frank Baas7, Christopher E. Shaw4, Jonathan D. Glass8, Guy A. Rouleau9, Robert Brown1; other consortium members can be found in the supplementary materials.
Department of Neurology, University of Massachusetts Medical School, Worcester, MA 01655, USA.
Montreal Neurological Institute, Department of Neurology and Neurosurgery, McGill University, Montreal Qc H3A 2B4, Canada.
Academic Unit of Neurology, Trinity Biomedical Sciences Institute, Trinity College Dublin, Dublin, Republic of Ireland.
Department of Basic and Clinical Neuroscience, King’s College London, Institute of Psychiatry, Psychology and Neuroscience, London, SE5 8AF
Department of Neurology, Brain Center Rudolf Magnus, University Medical Centre Utrecht, 3508 GA Utrecht, The Netherlands.
Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, Milan 20149, Italy; Department of Pathophysiology and Transplantation, ‘Dino Ferrari’ Center - Università degli Studi di Milano, Milan 20122 Italy.
Department of Genome Analysis, Academic Medical Center. Meibergdreef 9, 1105AZ Amsterdam, The Netherlands.
Department of Neurology, Emory University, Atlanta, GA 30322, USA.
Montreal Neurological Institute, Department of Neurology and Neurosurgery, McGill University, Montreal Qc H3A 2B4, Canada.
The results presented in this study can be found in Table S6.
Please see the Supplementary Materials for additional acknowledgements.
Materials and Methods
References and Notes
- 1.Poppe L, Rue L, Robberecht W, Van Den Bosch L. Translating biological findings into new treatment strategies for amyotrophic lateral sclerosis (ALS) Experimental neurology. 2014 doi: 10.1016/j.expneurol.2014.07.001. published online EpubJul 11. [DOI] [PubMed] [Google Scholar]
- 2.Chen S, Sayana P, Zhang X, Le W. Genetics of amyotrophic lateral sclerosis: an update. Molecular neurodegeneration. 2013;8(28) doi: 10.1186/1750-1326-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. published online EpubJan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Chalabi A, et al. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta neuropathologica. 2012;124:339–352. doi: 10.1007/s00401-012-1022-4. published online EpubSep. [DOI] [PubMed] [Google Scholar]
- 5.Neumann M. Frontotemporal lobar degeneration and amyotrophic lateral sclerosis: molecular similarities and differences. Revue neurologique. 2013;169:793–798. doi: 10.1016/j.neurol.2013.07.019. published online EpubOct. [DOI] [PubMed] [Google Scholar]
- 6.Smith RA, et al. Antisense oligonucleotide therapy for neurodegenerative disease. The Journal of clinical investigation. 2006;116:2290–2296. doi: 10.1172/JCI25424. published online EpubAug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Information on materials and methods is available at the Science Web site.
- 8.Sasabe J, et al. D-serine is a key determinant of glutamate toxicity in amyotrophic lateral sclerosis. The EMBO journal. 2007;26:4149–4159. doi: 10.1038/sj.emboj.7601840. published online EpubSep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson M, et al. Paradoxical roles of serine racemase and D-serine in the G93A mSOD1 mouse model of amyotrophic lateral sclerosis. Journal of neurochemistry. 2012;120:598–610. doi: 10.1111/j.1471-4159.2011.07601.x. published online EpubFeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul P, de Belleroche J. The role of D-amino acids in amyotrophic lateral sclerosis pathogenesis: a review. Amino acids. 2012;43:1823–1831. doi: 10.1007/s00726-012-1385-9. published online EpubNov. [DOI] [PubMed] [Google Scholar]
- 11.Cady J, et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Annals of neurology. 2014 doi: 10.1002/ana.24306. published online EpubNov 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenson PD, et al. Human Gene Mutation Database (HGMD): 2003 update. Human mutation. 2003;21:577–581. doi: 10.1002/humu.10212. published online EpubJun. [DOI] [PubMed] [Google Scholar]
- 13.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iida A, et al. Novel deletion mutations of OPTN in amyotrophic lateral sclerosis in Japanese. Neurobiol Aging. 2012;33:1843 e1819–1824. doi: 10.1016/j.neurobiolaging.2011.12.037. published online EpubAug. [DOI] [PubMed] [Google Scholar]
- 15.van Blitterswijk M, et al. Novel optineurin mutations in sporadic amyotrophic lateral sclerosis patients. Neurobiol Aging. 2012;33:1016 e1011–1017. doi: 10.1016/j.neurobiolaging.2011.05.019. published online EpubMay. [DOI] [PubMed] [Google Scholar]
- 16.Daoud H, et al. Exome sequencing reveals SPG11 mutations causing juvenile ALS. Neurobiology of aging. 2012;33:839 e835–839. doi: 10.1016/j.neurobiolaging.2011.11.012. published online EpubApr. [DOI] [PubMed] [Google Scholar]
- 17.Smith BN, et al. Exome-wide Rare Variant Analysis Identifies TUBA4A Mutations Associated with Familial ALS. Neuron. 2014;84:324–331. doi: 10.1016/j.neuron.2014.09.027. published online EpubOct 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson JO, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. published online EpubMay. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneb HM, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu545. published online EpubOct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaussenot A, et al. Screening of CHCHD10 in a French cohort confirms the involvement of this gene in frontotemporal dementia with amyotrophic lateral sclerosis patients. Neurobiol Aging. 2014 doi: 10.1016/j.neurobiolaging.2014.07.022. published online EpubJul 24. [DOI] [PubMed] [Google Scholar]
- 21.Bannwarth S, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–2345. doi: 10.1093/brain/awu138. published online EpubAug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couthouis J, Raphael AR, Daneshjou R, Gitler AD. Targeted exon capture and sequencing in sporadic amyotrophic lateral sclerosis. PLoS genetics. 2014;10:e1004704. doi: 10.1371/journal.pgen.1004704. published online EpubOct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. published online EpubMay 13. [DOI] [PubMed] [Google Scholar]
- 24.Rea SL, Majcher V, Searle MS, Layfield R. SQSTM1 mutations--bridging Paget disease of bone and ALS/FTLD. Experimental cell research. 2014;325:27–37. doi: 10.1016/j.yexcr.2014.01.020. published online EpubJul 1. [DOI] [PubMed] [Google Scholar]
- 25.Maruyama H, Kawakami H. Optineurin and amyotrophic lateral sclerosis. Geriatrics & gerontology international. 2013;13:528–532. doi: 10.1111/ggi.12022. published online EpubJul. [DOI] [PubMed] [Google Scholar]
- 26.Thomas M, Alegre-Abarrategui J, Wade-Martins R. RNA dysfunction and aggrephagy at the centre of an amyotrophic lateral sclerosis/frontotemporal dementia disease continuum. Brain. 2013;136:1345–1360. doi: 10.1093/brain/awt030. published online EpubMay. [DOI] [PubMed] [Google Scholar]
- 27.Kachaner D, Genin P, Laplantine E, Weil R. Toward an integrative view of Optineurin functions. Cell cycle. 2012;11:2808–2818. doi: 10.4161/cc.20946. published online EpubAug 1. [DOI] [PubMed] [Google Scholar]
- 28.Keller BA, et al. Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta neuropathologica. 2012;124:733–747. doi: 10.1007/s00401-012-1035-z. published online EpubNov. [DOI] [PubMed] [Google Scholar]
- 29.Scotter EL, et al. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. Journal of cell science. 2014;127:1263–1278. doi: 10.1242/jcs.140087. published online EpubMar 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS letters. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. published online EpubMar 19. [DOI] [PubMed] [Google Scholar]
- 31.Pilli M, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. published online EpubAug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. published online EpubJul 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gleason CE, Ordureau A, Gourlay R, Arthur JS, Cohen P. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon beta. The Journal of biological chemistry. 2011;286:35663–35674. doi: 10.1074/jbc.M111.267567. published online EpubOct 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korac J, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. Journal of cell science. 2013;126:580–592. doi: 10.1242/jcs.114926. published online EpubJan 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–4448. doi: 10.1073/pnas.1405752111. published online EpubOct 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. published online EpubJun 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. published online EpubAug 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu M, Kageyama S, Ichimura Y. p62/SQSTM1/A170: physiology and pathology. Pharmacol Res. 2012;66:457–462. doi: 10.1016/j.phrs.2012.07.004. published online EpubDec. [DOI] [PubMed] [Google Scholar]
- 39.Clark K, et al. Novel cross-talk within the IKK family controls innate immunity. The Biochemical journal. 2011;434:93–104. doi: 10.1042/BJ20101701. published online EpubFeb 15. [DOI] [PubMed] [Google Scholar]
- 40.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. published online EpubMay 16. [DOI] [PubMed] [Google Scholar]
- 41.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nature immunology. 2003;4:491–496. doi: 10.1038/ni921. published online EpubMay. [DOI] [PubMed] [Google Scholar]
- 42.Hemmi H, et al. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. The Journal of experimental medicine. 2004;199:1641–1650. doi: 10.1084/jem.20040520. published online EpubJun 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015 doi: 10.1126/science.aaa2630. published online EpubJan 29. [DOI] [PubMed] [Google Scholar]
- 44.Wathelet MG, et al. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. published online EpubMar. [DOI] [PubMed] [Google Scholar]
- 45.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. published online EpubMar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nature genetics. 2001;29:160–165. doi: 10.1038/ng1001-160. published online EpubOct. [DOI] [PubMed] [Google Scholar]
- 47.Li J, et al. Selective TBK1/IKKi dual inhibitors with anticancer potency. International journal of cancer. Journal international du cancer. 2014;134:1972–1980. doi: 10.1002/ijc.28507. published online EpubApr 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. published online EpubMay 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee EB, Lee VM, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nature reviews Neuroscience. 2012;13:38–50. doi: 10.1038/nrn3121. published online EpubJan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ExomeAggregationConsortium(ExAC) http://exac.broadinstitute.org.
- 51.Van Damme P, Robberecht W. Clinical implications of recent breakthroughs in amyotrophic lateral sclerosis. Current opinion in neurology. 2013;26:466–472. doi: 10.1097/WCO.0b013e328364c063. published online EpubOct. [DOI] [PubMed] [Google Scholar]
- 52.Munch C, Rolfs A, Meyer T. Heterozygous S44L missense change of the spastin gene in amyotrophic lateral sclerosis. Amyotrophic lateral sclerosis : official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases. 2008;9:251–253. doi: 10.1080/17482960801900172. published online EpubAug. [DOI] [PubMed] [Google Scholar]
- 53.Meyer T, et al. Early-onset ALS with long-term survival associated with spastin gene mutation. Neurology. 2005;65:141–143. doi: 10.1212/01.wnl.0000167130.31618.0a. published online EpubJul 12. [DOI] [PubMed] [Google Scholar]
- 54.Chesi A, et al. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. published online EpubJul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. published online EpubNov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. American journal of human genetics. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. published online EpubNov 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. published online EpubJul 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. published online EpubSep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.NHLBI GO Exome Sequencing Project (ESP) ExomeVariantServer; Seattle, WA: http://evs.gs.washington.edu/EVS/ [Google Scholar]
- 60.C Genomes Project et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. published online EpubNov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS genetics. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R. CoreTeam. A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. http://www.R-project.org/ [Google Scholar]
- 63.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. published online EpubJul 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. published online EpubJul 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. published online EpubDec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.