Abstract
Background
In view of the prominent role in cancer cell biology and alteration in substantial numbers of ESCC, defining EGFR molecular characteristics relevant to patient prognosis is of great importance. Therefore, we analyzed the protein expression and gene copy variation of the epithelial growth factor receptor (EGFR) in Chinese esophageal squamous cell carcinoma (ESCC) and explored the possible associations with various features of the tumors and survival of the patients.
Methods
Sections were made from tissue microarray composed of 96 ESCC, and examined for EGFR expression by means of immunohistochemistry (IHC) and for EGFR gene amplification by means of fluorescence in situ hybridization (FISH). The results of IHC were evaluated with six different reported scoring systems. Correlation with clinical features and survival was evaluated using chi-square test and Kaplan–Meier analysis.
Results
EGFR overexpression according to scoring system 1 significantly correlated with advanced lymph node involvement (P = 0.046), patient disease specific free survival (DFS) (P = 0.006) and overall survival (OS) (P = 0.007). No such association was observed using other 5 scoring systems (P > 0.05 ). EGFR amplification was associated with lymph node metastasis (P = 0.028), but not correlated with DFS and OS until 20 months.
Conclusions
EGFR IHC overexpression evaluated by scoring system 1 might be suitable to be used in predicting patients survival in ESCC. EGFR gene amplification showed delayed prognostic information after 20 months.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1393-8) contains supplementary material, which is available to authorized users.
Keywords: Esophageal squamous cell carcinoma, Epidermal growth factor receptor, Immunohistochemistry scoring system, Fluorescence in situ hybridization
Background
Esophageal carcinoma is one of the most common malignancies in China, and squamous cell carcinoma is the main histological type [1, 2]. It generally has a poor prognosis because it is usually in an advanced stage at the time of diagnosis. Despite the progress in chemotherapeutic, radiotherapeutic and surgical treatment, the five-year survival rate is still less than 20 % [3-6]. In recent years, molecular targeted therapy has become an important treatment [7-10]. With the aim of increasing the clinical benefit–risk ratio of anticancer treatments, consideration is increasingly given to the identification of predictive tumour biomarkers.
One potential group of useful protein biomarkers is the epidermal growth factor receptor (EGFR) family of receptors. This family contains four members, EGFR, ErbB2/human epidermal growth factor receptor-2 (HER2), ErbB3/HER3, and ErbB4/HER4, that act as receptor tyrosine kinases and have a well-defined function in cell signaling, controlling cell proliferation and differentiation. Esophageal cancers frequently show EGFR or HER2 gene amplification and overexpression [11, 12]. And esophageal squamous cell carcinomas (ESCCs) predominantly show alterations of EGFR, whereas esophageal (Barrett’s) adenocarcinomas (EACs) frequently show HER2 gene amplification and protein overexpression. In view of the prominent role in cancer cell biology and (over-)expression in substantial numbers of ESCC, EGFR represents valuable therapeutic target. Defining EGFR molecular characteristics relevant to patient prognosis is an important step toward deciding treatment.
At present, the literatures about EGFR expression in ESCC contain conflicting data on the relationship between overexpression and survival [13-15]. This variability may be due to heterogeneity of study populations or lack of a standardized assay for determining EGFR status. Here, we collected a cohort of Chinese patients with ESCC, and evaluated their protein expression using 6 representative scoring systems. To the best of our knowledge and available literature data, so far such comparisons of different EGFR-IHC scoring systems in ESCC patients are sparse.
EGFR gene copy number variation may be more reliable than protein expression in predicting prognosis. However, reports on the influence of EGFR gene variation in ESCC patients have been equivocal [13, 16-18]. In general, the relationships between tumor EGFR gene variation and protein expression have not been clearly defined, and the prognostic value of these tumor characteristics has not been well evaluated for ESCC.
Therefore, the aims of this study are to compare the six different scoring systems for EGFR expression, to explore the cut off value in assessing EGFR gene variation, and to investigate their prognostic significance in ESCC.
Methods
Patients and specimens
A total of 96 ESCC samples were treated in the Department of Thorax Surgery, Zhongshan Hospital during March to October 2010. All patients had not received chemotherapy or radiotherapy prior to surgical resection. Prior written informed consent was obtained from all patients. The present study has been carried out in accordance with the Declaration of Helsinki, and was approved by Human Research Ethics Committee of Zhongshan hospital, Fudan University.
Sections were stained with hematoxylin and eosin and reviewed by two pathologists to confirm the ESCC diagnosis. The following patient characteristics were collected: gender, age, tumor site (upper, middle, and lower region of esophagus), histological grade, coagulative necrosis, nerve and vascular infiltration, mitotic index (numbers recorded as ≤20 per 10 high power fields [HPF], 20-50/10HPF, or ≥ 50/10HPF), lymph node metastasis, and stage, as previously reported [19].
Tissue microarrays
The tissue microarray (TMA) was constructed as previously described [20]. Briefly, the region of interest (2 mm wide and 6 mm long) was extracted and then vertically planted into the recipient block one by one according to the corresponding location indicated by letters and numbers. The planting surface was aggregated on the aggregation instrument.
Immunohistochemistry
The TMA recipient block was sectioned on a routine microtome machine. The IHC assay using EGFR rabbit monoclonal antibody (EGFR.25, Leica Biosystems Newcastle Ltd, Newcastle, UK) was performed with the Ventana iView DAB Detection Kit on a BenchMark XT automated staining system (Ventana Medical Systems, Tucson, AZ). Normal IgG from the same species of primary antibody diluted to match the concentration of the primary antibody was used as the negative control. For EGFR negative cases, the experiment was repeated on the whole section in order to exclude heterogeneity.
EGFR expression was evaluated according to published scoring system, summarized as follows:1) The percentage of positive tumor cells (0 % to 100 %) was multiplied by the staining intensity (SI) (1, negative or trace; 2, weak; 3, moderate; 4, intense). Scores 0 to 200, 201 to 300, and 301 to 400 were respectively classified as having negative or low, intermediate, and high levels of expression [21]. 2) 0, negative, no discernible staining or background type staining; 1+, definite cytoplasmic staining and/or equivocal discontinuous membrane staining; 2+, unequivocal membrane staining with moderate intensity; 3+, strong and complete plasma membrane staining. Samples exhibiting 2+ or 3+ were classified as overexpression [13]. 3) a = 0 % (score 0); 1–20 % (score 1); 21–40 % (score 2); 41–60 % (score 3); 61– 80 % (score 4); or 81–100 % (score 5). i = absent (score 0); faint (score 1); moderate (score 2); or strong (score 3). A final score was calculated by multiplying i by a, using the score of 8 as the cutoff [22]. 4) 1 × (percentage of cells staining weakly [1 +]) + 2 × (percentage of cells staining moderately [2 +]) + 3 × (percentage of cells staining strongly [3 +]). Score of 200 is a cutoff [23]. 5) SI was classified as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). An area of SI was defined as 0 if <10 %, 1 if 10 %–25 %, 2 if 26 %–50 %, 3 if 51 %–75 %, and 4 if >75%. Immunostaining intensity was divided into 0 negative (−), 1–3 weakly positive (+), 4–5 moderately positive (2+), and 6–7 strong positive (3+); EGFR overexpression was defined as positive staining of tumor cells reaching 2+ or 3 + [24]. 6) loss of expression: SI = 0; weak expression: SI = 1 in < 70 % or SI = 2 in < 30 % of cells in a tumor spot, moderate expression: SI = 1 in > 70 % or SI =2 in > 30 % of cells in a tumor spot and strong expression: SI = 2 in > 70 % or SI = 3 in > 30 % of cells in a tumor spot [25].
Fluorescence in situ hybridization
TMA sections were dewaxed and dehydrated. Dual color EGFR FISH was performed with the Spectrum Orange locus-specific identifier EGFR probe (Vysis, Abbott Molecular Inc, Des plaines, USA) specific for the EGFR locus (7p12) and the Spectrum Green CEP7 chromosome 7 centromeric probe (7p11.1 to q11.1; Vysis). The specific steps were similar to HER2-FISH procedure, reported previously [26].
EGFR signals were counted from at least 100 cancer cell nuclei, and were divided into six types: 1) disomy was an EGFR to CEP7 ratio ≤2 copies in >90 % of cells; 2) low trisomy was ≤2 copies in ≥40 % of cells, 3 copies in 10 %–40 % of the cells, ≥4 copies in <10 % of cells; 3) high trisomy was ≤2 copies in ≥40 % of cells, 3 copies in >40 % of cells, ≥4 copies in <10 % of cells; 4) low polysomy was ≥4 copies in 10 %–40 % of cells; 5) high polysomy was ≥4 copies in >40 % of cells; 6) gene amplification was defined by the presence of tight EGFR gene clusters, or a ratio of EGFR gene to chromosome7 ≥ 2, or ≥15 copies of EGFR per cell in ≥10 % of tumor cells. EGFR FISH-positive was defined as EGFR high polysomy or gene amplification [27].
Follow-up information
Follow-up information for the 96 patients after surgery and treatment was provided by the referring clinicians, or else obtained directly from patients and their family members as standard procedure. The date of last follow up was May 16, 2014. Disease-free survival (DFS) and overall survival (OS) were measured from the time of surgery to the time of first recurrence (or most recent follow-up) or death.
Statistical analysis
A χ2 test was used for univariate analysis, the agreement of different scoring systems was measured by the index Kappa and the statistical differences were analyzed by the McNemar test. Kaplan-Meier analysis was used to calculate DFS and OS. Log-rank test of survival analysis was used to compare DFS and OS as functions of variables and to identify significant differences. P < 0.05 were recorded as significant.
Results
Characterization of ESCC patients
The clinicopathological features of the 96 ESCC patients are summarized previously [28]. The majority of the patients were males (83.3 %). The median age of patients was 62 years. By anatomic site, 1 was located in the upper esophagus, 33 in the middle and 62 in the lower area. Most of the tumor differentiation was grade II (63.5 %), 36.5 % was grade III and none was grade 1. Five tumors had invaded to the submucosa, 24 to the muscularis propria and 67 to the adventitia. Fifty-three tumors were associated with nerve or vascular infiltration and 44 with lymph node metastases.
EGFR IHC analysis
Among the 96 ESCC cases analyzed, the EGFR IHC staining results are evaluated using six scoring systems (Additional file 1: Table S1). The scoring system 1 and 6 has low, intermediate and high level of EGFR expression, while 2, 3, 4 and 5 only has low and high level (Table 1).
Table 1.
Comparison of EGFR-IHC results of 6 scoring systems
| Pairs | Levels | Number | P | Kappa |
|---|---|---|---|---|
| System 1 vs. System 6 | (L/M/H) vs. (L/M/H) | (49/39/8) vs. (14/33/49) | 0.001 | 0.037 |
| System 2 vs. System 3 | (L/H) vs. (L/H) | (42/54) vs. (48/48) | 0.031 | 0.875 |
| System 2 vs. System 4 | (L/H) vs. (L/H) | (42/54) vs. (88/8) | 0.001 | 0.132 |
| System 2 vs. System 5 | (L/H) vs. (L/H) | (42/54) vs. (12/84) | 0.001 | 0.31 |
| System 3 vs. System 4 | (L/H) vs. (L/H) | (48/48) vs. (88/8) | 0.001 | 0.167 |
| System 3 vs. System 5 | (L/H) vs. (L/H) | (48/48) vs. (12/84) | 0.001 | 0.25 |
| System 4 vs. System 5 | (L/H) vs. (L/H) | (88/8) vs. (12/84) | 0.001 | 0.026 |
The McNemar Test, P < 0.05 was considered statistically significant difference
Kappa > =0.75 was regarded as better concordance, Kappa < 0.4 indicated a poor concordance
System 1–6, EGFR-IHC scoring system 1 to 6
L, low level of EGFR expression; M, intermediate level; H, high level
According to scoring system 1 to 6, overexpression of EGFR were observed in 8 (8.3 %), 64 (66.7 %), 48 (50 %), 8 (8.3 %), 84 (87.5 %), and 49 (51.0 %) cases, respectively (Table 1). Within the 6 scoring systems, 17 cases had the same level of EGFR expression (7 in high level, 10 in low level, and none in intermediate level) (Additional file 1: Table S1). Fig. 1a showed the negative control, 1b, 1c, and 1d were low, intermediate and high level of EGFR expression according to system 1.
Fig. 1.
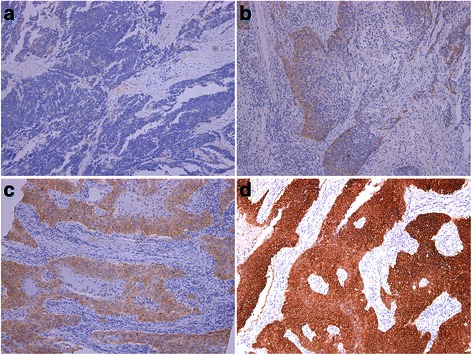
Examples of different immunohistochemical EGFR expression in ESCC according to system 1: a = negative control, b = low, c = intermediate, d = high level of EGFR expression
There were significant difference among the six IHC score results (P < 0.001) (Table 1). The scoring system 2 and 3 are highly in agreement with each other (k = 0.87). No comparison could be conducted between 1 and 4.
EGFR gene copy variation
The average gene copy number per cell and the EGFR-to-chromosome 7 ratio for the major FISH patterns are listed in Additional file 2: Table S2. EGFR FISH-positive was seen in 29 (30.2 %) cases. The EGFR genes were amplified in 7 (7.3 %) cases (6 were clustered-type signals and 1 were multiple scattered signals) (Figs. 2a and b). High polysomy (≥4 copies in >40 % of cells) was present in the other 22 (22.9 %) cases, with the averages of EGFR and chromosome 7 signals per cell ranging between 3.11 and 5.10 and the gene-to-chromosome ratio ranging from 0.82 to 1.84 (Fig. 2c). Low polysomy was present in 39 (40.6 %) cases with the averages of gene and chromosome copies per cell ranging from 2.43 to 3.27 and the chromosome-to-gene ratio ranging from 0.92 to 1.14 (Fig. 2d), disomy in 3 (3.1 %) cases, with the averages per cell for the gene and chromosome 7 copies ranging from 2.02 to 2.17 and the ratio of gene-to-chromosome from 0.97 to 1.04 (Fig. 2f), low trisomy in 19 (19.8 %) cases with the averages per cell for the gene and chromosome 7 copies ranging from 1.95 to 2.49 and the ratio of gene-to-chromosome from 0.74 to 1.12, and high trisomy in 6 (6.3 %) cases with the averages per cell for the gene and chromosome 7 copies ranging from 2.56 to 2.91 and the ratio of gene-to-chromosome from 0.95 to 1.02 (Fig. 2e). These cases were categorized as a FISH-negative group.
Fig. 2.
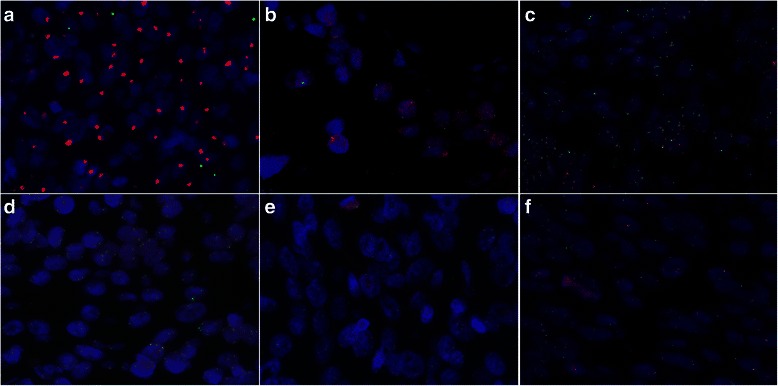
The representative EGFR (red) and chromosome 7 (green) FISH for tumors with EGFR gene amplification (a and b, n = 7/96), High polysomy (c, n = 22/96), Low polysomy ( d, n = 39/96), high trisomy (e, n = 6/96) and disomy (f, n = 3/96)
Correlation between EGFR protein expression and gene variation
EGFR expression and gene copy number are analyzed and showed in Table 2. EGFR gene amplification was associated with EGFR protein overexpression in scoring system 1, 2, 3, 4, and 6, EGFR-FISH positive only in scoring system 1, 2 and 4.
Table 2.
Correlation between EGFR protein expression and gene variation
| EGFR-FISH result | ||||||
|---|---|---|---|---|---|---|
| EGFR-IHC | FISH positive | Amplification | ||||
| No | Yes | P | No | Yes | P | |
| System 1 | 0.034 | 0.001 | ||||
| L | 39 | 10 | 9 | 0 | ||
| M | 25 | 14 | 37 | 2 | ||
| H | 3 | 5 | 3 | 5 | ||
| System 2 | 0.036 | 0.015 | ||||
| L | 34 | 8 | 42 | 0 | ||
| H | 31 | 21 | 47 | 7 | ||
| System 3 | 0.120 | 0.006 | ||||
| L | 37 | 11 | 48 | 0 | ||
| H | 30 | 18 | 41 | 7 | ||
| System 4 | 0.038 | 0.001 | ||||
| L | 64 | 24 | 86 | 2 | ||
| H | 3 | 5 | 3 | 5 | ||
| System 5 | 0.674 | 0.299 | ||||
| L | 9 | 3 | 12 | 0 | ||
| H | 58 | 26 | 77 | 7 | ||
| System 6 | 0.175 | 0.027 | ||||
| L | 11 | 3 | 14 | 0 | ||
| M | 26 | 7 | 33 | 0 | ||
| H | 30 | 19 | 42 | 7 | ||
| Total | 67 | 29 | 89 | 7 | ||
System 1–6, EGFR-IHC scoring system 1–6
L, low level of EGFR expression; M, intermediate level; H, high level
FISH positive, EGFR gene amplification or high polysomy
On the basis of scoring system 1, 3 and 4, patients with trisomy and polysomy showed low mean IHC scores (206 and 197, 6.6 and 6.5, 94.6 and 100 respectively), whereas the mean IHC score increased when FISH abnormalities became more severe. The mean score was 348.6, 13.7 and 254.3 for patients with gene amplification (Additional file 3: Table S3).
Prognostic implication of EGFR protein expression levels and gene variation
EGFR protein overexpression and gene amplification were statistically evaluated for correlation with established clinicopathological factors (Table 3). EGFR overexpression according to scoring system 1 and 4 was significantly correlated with the vascular invasion, lymph node metastasis (P < 0.05, Table 3). No such correction was observed using other scoring systems. EGFR amplification was associated with the lymph node metastasis (P = 0.028), while high polysomy wasn’t associated with this factor (P = 0.227).
Table 3.
Relationship of status of EGFR in ESCC with the clinicopathological parameters
| EGFR-IHC result | EGFR-FISH result | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| System 1 | System 4 | FISH Positive | Amplification | |||||||||||
| N | L | M | H | P | L | H | P | No | Yes | P | No | Yes | P | |
| Gender | ||||||||||||||
| Male | 80 | 42 | 32 | 6 | 0.724 | 74 | 6 | 0.509 | 52 | 28 | 0.022 | 74 | 6 | 0.861 |
| Female | 16 | 7 | 7 | 2 | 14 | 2 | 15 | 1 | 15 | 1 | ||||
| Age | ||||||||||||||
| <60 | 34 | 24 | 8 | 2 | 0.017 | 22 | 12 | 0.422 | 22 | 12 | 0.422 | 32 | 2 | 0.694 |
| >60 | 62 | 25 | 31 | 6 | 45 | 17 | 45 | 17 | 57 | 5 | ||||
| Tumor site | ||||||||||||||
| Upper | 1 | 0 | 1 | 0 | 0.181 | 1 | 0 | 0.607 | 1 | 0 | 0.313 | 1 | 0 | 0.413 |
| Middle | 33 | 12 | 17 | 4 | 29 | 4 | 20 | 13 | 29 | 4 | ||||
| Lower | 62 | 37 | 21 | 4 | 58 | 4 | 46 | 16 | 59 | 3 | ||||
| T-stage | ||||||||||||||
| T1 | 5 | 2 | 3 | 0 | 0.889 | 5 | 0 | 0.499 | 3 | 2 | 0.552 | 5 | 0 | 0.602 |
| T2 | 24 | 13 | 9 | 2 | 23 | 1 | 15 | 9 | 23 | 1 | ||||
| T3 | 67 | 34 | 27 | 6 | 60 | 7 | 49 | 18 | 61 | 6 | ||||
| Vaso invasion | ||||||||||||||
| No | 77 | 34 | 35 | 8 | 0.020 | 69 | 8 | 0.142 | 54 | 23 | 0.884 | 70 | 7 | 0.172 |
| Yes | 19 | 15 | 4 | 0 | 19 | 0 | 13 | 6 | 19 | 0 | ||||
| Nerve invasion | ||||||||||||||
| No | 62 | 34 | 24 | 4 | 0.497 | 59 | 3 | 0.094 | 44 | 18 | 0.735 | 59 | 3 | 0.212 |
| Yes | 34 | 15 | 15 | 4 | 29 | 5 | 23 | 11 | 30 | 4 | ||||
| LN metastases | ||||||||||||||
| No | 52 | 29 | 22 | 1 | 0.046 | 51 | 1 | 0.013 | 39 | 13 | 0.227 | 51 | 1 | 0.028 |
| Yes | 44 | 20 | 17 | 7 | 37 | 7 | 28 | 16 | 38 | 6 | ||||
| Necrosis | ||||||||||||||
| No | 38 | 21 | 16 | 1 | 0.258 | 36 | 2 | 0.378 | 27 | 11 | 0.828 | 37 | 1 | 0.155 |
| Yes | 58 | 28 | 23 | 7 | 52 | 6 | 40 | 18 | 52 | 6 | ||||
| Mitoses (/10HPF) | ||||||||||||||
| ≤20 | 29 | 12 | 15 | 2 | 0.645 | 27 | 2 | 0.307 | 18 | 11 | 0.06 | 27 | 2 | 0.768 |
| 20~50 | 37 | 20 | 13 | 4 | 32 | 5 | 31 | 6 | 35 | 2 | ||||
| ≥50 | 30 | 17 | 11 | 2 | 29 | 1 | 18 | 12 | 27 | 3 | ||||
| Tumour differentiation | ||||||||||||||
| G2 | 61 | 33 | 26 | 2 | 0.061 | 43 | 18 | 0.844 | 43 | 18 | 0.844 | 57 | 4 | 0.715 |
| G3 | 35 | 16 | 13 | 6 | 24 | 11 | 24 | 11 | 32 | 3 | ||||
System 1and System 4, EGFR-IHC scoring system 1 and 4
L, low level of EGFR expression; M, intermediate level; H, high level
FISH positive , EGFR gene amplification or high polysomy
EGFR overexpression and gene amplification were evaluated for their potential prognostic significance. The Kaplan-Meier survival curves for patients in the different scoring systems of EGFR expression or gene numbers are shown in Figs. 3 and 4. Protein overexpression, on the basis of scoring system 1, had poorer DFS (P = 0.006) (Fig. 3a) and OS (P = 0.004) (Fig. 3b). However, other systems had no prognostic value whether in DFS or in OS (Fig. 3c and d). And gene amplification did not represent a statistically significant adverse prognosis until 20 months (Fig. 4). No significant difference in survival rates with respect to high polysomy was observed.
Fig. 3.
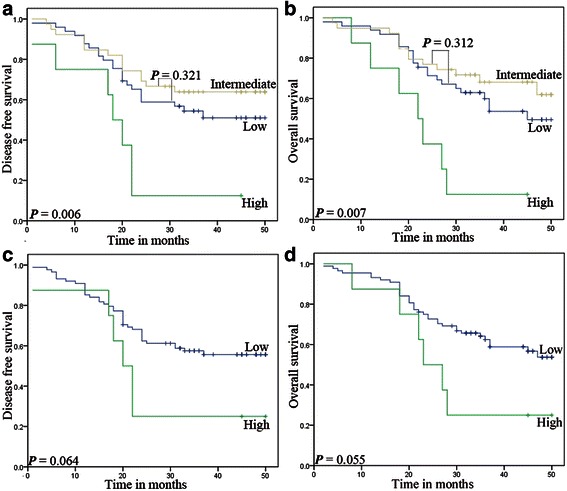
Association between EGFR overexpression and survival in ESCC. Protein overexpression, on the basis of scoring system 1, had poorer DFS (a, P = 0.006) and OS (b, P =0.007), with system 2 (c and d) no prognostic value
Fig. 4.
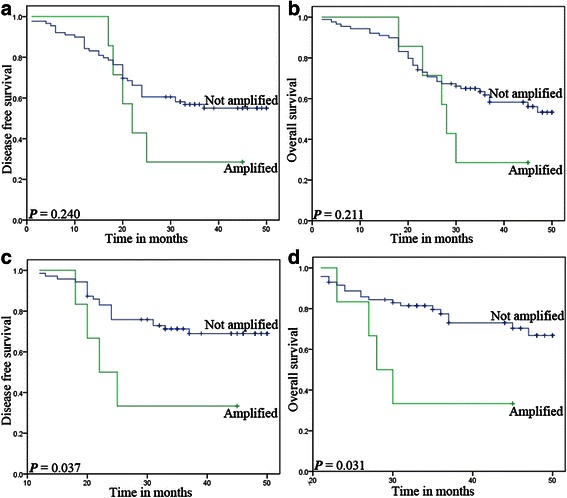
Association between EGFR gene variation and survival in ESCC. The gene amplification (a and b) was not significantly associated with DFS or OS (P = 0.240 or 0.211). However, gene amplification (c and d) did represent delayed prognostic information (P = 0.037 and 0.031)
Discussion
The epithelial growth factor receptor (EGFR) is a 170-kDa transmembrane glycoprotein and a tyrosine kinase receptor expressed in various human tissues, especially in cells of epithelial origin, which plays important roles in modulating cell proliferation, survival, migration, and differentiation [29]. EGFR alterations in cancer can be divided mostly in two categories: mutations in exons 18–21 mainly identified in Asia lung adenocarcinoma [30, 31], and gene and protein overexpression [32]. It’s known to us, ESCC predominantly show EGFR gene copy number alterations and protein overexpression [11, 13, 24, 33], with little EGFR mutation [16, 26, 34]. EGFR gene variation and protein overexpression might be the candidate for predictive biomarker in ESCC. There have been several IHC studies examining EGFR protein expression in ESCC, the expression rate ranged from 4 % to 86 % [13, 24, 33]. We found the most important discrepancies might be due to the selected threshold for positivity, which may induce conflicting results in different laboratories and authors. Therefore, we selected six different scoring systems presented in literature to focus upon the same ESCC samples with EGFR antibody.
Evaluation of EGFR expression by six scoring systems in ESCC
Firstly, the overexpression of EGFR were observed in 8 (8.3 %), 64 (66.7), 48 (50 %), 8 (8.3 %), 84 (87.5 %), and 49 (51.0 %) cases according to system 1 to 6, with ranging from 8.3 % to 87.5 %. These results were in agreement with our speculation that EGFR expression was obviously influenced by the selected threshold. Secondly, the correlation of EGFR expression with clinical features and prognosis were evaluated by scoring system 1 to 6, 1 and 4 related to lymph node metastasis, however, only 1 showed a statistically significant prognosis with DFS (0.006) and OS (0.007). Therefore, scoring system 1 for EGFR expression seems to be more valuable for predicting tumor aggressiveness and prognosis.
Evaluation of EGFR gene variation in ESCC
Firstly, EGFR gene status disclosed by our FISH included disomy, low trisomy, high trisomy, low polysomy, high polysomy and gene amplification, which was consistent with previous reports [24]. Secondly, EGFR gene amplification was associated with EGFR expression evaluated by scoring system 1, 2, 3, 4, 6 except 5, whereas EGFR-FISH positive was only associated with scoring system 1, 2 and 4. Thirdly, EGFR-FISH positive had no relationship with clinical features and prognosis; however, EGFR amplification was associated with lymph node metastasis, and patients with EGFR amplification had poorer prognosis whether in DFS or OS after 20 months survival. Therefore, EGFR amplification, not EGFR-FISH positive or high polysomy, seems to be a suitable cut off value in clinical practice.
Conclusion
This study firstly compared six scoring system evaluation for EGFR IHC overexpression used in ESCC, and found scoring system 1 might be suitable to be adopted in clinical practice since the value in predicting patients’ survival. EGFR gene amplification was associated with protein overexpression in ESCC, and indicated poorer prognosis after 20 months survival.
Acknowledgments
We gratefully acknowledge the support of the patients and their families who agreed to contribute to this research program.
Abbreviations
- ESCC
Esophageal squamous cell carcinoma
- EAC
Esophageal adenocarcinoma
- EGFR
Epithelial growth factor receptor
- HER2
Human epidermal growth factor receptor-2
- IHC
Immunohistochemistry
- FISH
Fluorescent in situ hybridization
- TMA
Tissue microarray
- SI
Staining intensity
- DFS
Disease-free survival
- OS
Overall survival
Additional files
EGFR immunohistochemical staining results evaluated using six scoring systems.
EGFR-FISH results of all cases.
EGFR IHC scores and gene variation.
Footnotes
Dongxian Jiang and Xiaojing Li contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DJ and XL carried out the molecular analysis and drafted the manuscript; HW and YS had significant roles in guiding the project direction and aided with the molecular aspects of the study; HZ and JS performed the IHC studies whereas JH and YX established FISH approach and made substantial contributions to the analysis; CX and SL were histopathologist responsible for all histopathological support and IHC second scoring; YH and LT formulated the research question, obtained the funding for the project, supervised the research program, and closely edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Dongxian Jiang, Email: jiangdongxian3@aliyun.com.
Xiaojing Li, Email: li.xiaojing@zs-hospital.sh.cn.
Haixing Wang, Email: wang.haixing@zs-hospital.sh.cn.
Yuan Shi, Email: shi.yuan@zs-hospital.sh.cn.
Chen Xu, Email: xu.chen@zs-hospital.sh.cn.
Shaohua Lu, Email: lu.shaohua@zs-hospital.sh.cn.
Jie Huang, Email: huang.jie@zs-hospital.sh.cn.
Yifan Xu, Email: xu.yifan@zs-hospital.sh.cn.
Haiying Zeng, Email: zeng.haiying@zs-hospital.sh.cn.
Jieakesu Su, Email: su.jieakesu@zs-hospital.sh.cn.
Yingyong Hou, Email: houyingyong@hotmail.com.
Lijie Tan, Email: tan.lijie@zs-hospital.sh.cn.
References
- 1.Montgomery EF, Boffetta P, Daigo Y, Shimizu M, Shimoda T. WHO classification of tumors of the oesophagus. 4. Lyon: IARC Press; 2010. [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–12. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 3.Dipetrillo T, Suntharalingam M, Ng T, Fontaine J, Horiba N, Oldenburg N, et al. Neoadjuvant paclitaxel poliglumex, cisplatin, and radiation for esophageal cancer: a phase 2 trial. Am J Clin Oncol. 2012;35:64–7. doi: 10.1097/COC.0b013e318201a126. [DOI] [PubMed] [Google Scholar]
- 4.Jatoi A, Soori G, Foster NR, Hiatt BK, Knost JA, Fitch TR, et al. Phase II study of preoperative pemetrexed, carboplatin, and radiation followed by surgery for locally advanced esophageal cancer and gastroesophageal junction tumors. J Thorac Oncol. 1994;2010:5. doi: 10.1097/JTO.0b013e3181fb5c3e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge HM, Wijnhoven BP, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;2074–2084:366. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 6.Ruhstaller T, Widmer L, Schuller JC, Roth A, Hess V, Mingrone W, et al. Popescu R:Multicenter phase II trial of preoperative induction chemotherapy followed by chemoradiation with docetaxel and cisplatin for locally advanced esophageal carcinoma (SAKK 75/02) Ann Oncol. 2009;20:1522–8. doi: 10.1093/annonc/mdp045. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed S, Sami A, Xiang J. HER2-directed therapy: current treatment options for HER2-positive breast cancer. Breast Cancer. 2015;22:101–16. doi: 10.1007/s12282-015-0587-x. [DOI] [PubMed] [Google Scholar]
- 8.Folprecht G, Gruenberger T, Bechstein WO, Raab HR, Lordick F, Hartmann JT, et al. Tumour response and secondary resectability of colorectal liver metastases following neoadjuvant chemotherapy with cetuximab: the CELIM randomised phase 2 trial. Lancet Oncol. 2010;11:38–47. doi: 10.1016/S1470-2045(09)70330-4. [DOI] [PubMed] [Google Scholar]
- 9.Sanford M, Scott LJ. Imatinib: as adjuvant therapy for gastrointestinal stromal tumour. Drugs. 2010;70:1963–72. doi: 10.2165/11205000-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Crosby T, Hurt CN, Falk S, Gollins S, Mukherjee S, Staffurth J, et al. Chemoradiotherapy with or without cetuximab in patients with oesophageal cancer (SCOPE1): a multicentre, phase 2/3 randomised trial. Lancet Oncol. 2013;14:627–37. doi: 10.1016/S1470-2045(13)70136-0. [DOI] [PubMed] [Google Scholar]
- 11.Fichter CD, Timme S, Braun JA, Gudernatsch V, Schopflin A, Bogatyreva L, et al. EGFR, HER2 and HER3 dimerization patterns guide targeted inhibition in two histotypes of esophageal cancer. Int J Cancer. 2014;135:1517–30. doi: 10.1002/ijc.28771. [DOI] [PubMed] [Google Scholar]
- 12.Wei QC, Chen LR, Sheng LM, Nordgren H, Wester K, Carlsson J. EGFR, HER2 and HER3 expression in esophageal primary tumours and corresponding metastases. Int J Oncol. 2007;31:493–9. [PubMed] [Google Scholar]
- 13.Hanawa M, Suzuki S, Dobashi Y, Yamane T, Kono K, Enomoto N, et al. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer. 2006;118:1173–80. doi: 10.1002/ijc.21454. [DOI] [PubMed] [Google Scholar]
- 14.Gibault L, Metges JP, Conan-Charlet V, Lozac'H P, Robaszkiewicz M, Bessaguet C, et al. Diffuse EGFR staining is associated with reduced overall survival in locally advanced oesophageal squamous cell cancer. Br J Cancer. 2005;93:107–15. doi: 10.1038/sj.bjc.6602625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itakura Y, Sasano H, Shiga C, Furukawa Y, Shiga K, Mori S, et al. Epidermal growth factor receptor overexpression in esophageal carcinoma. An immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer. 1994;74:795–804. doi: 10.1002/1097-0142(19940801)74:3<795::AID-CNCR2820740303>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Arao T, Matsumoto K, Fujita Y, Kimura H, Hayashi H, et al. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. Int J Oncol. 2013;42:1151–8. doi: 10.3892/ijo.2013.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunpaweravong P, Sunpaweravong S, Puttawibul P, Mitarnun W, Zeng C, Baron AE, et al. Epidermal growth factor receptor and cyclin D1 are independently amplified and overexpressed in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2005;131:111–9. doi: 10.1007/s00432-004-0610-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitagawa Y, Ueda M, Ando N, Ozawa S, Shimizu N, Kitajima M. Further evidence for prognostic significance of epidermal growth factor receptor gene amplification in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 1996;2:909–14. [PubMed] [Google Scholar]
- 19.Kaifi JT, Gusani NJ, Jiang Y, Mackley HB, Dye CE, Mathew A, et al. Multidisciplinary management of early and locally advanced esophageal cancer. J Clin Gastroenterol. 2011;45:391–9. doi: 10.1097/MCG.0b013e3182049949. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, He D, Hou Y, Hu Q, Xu C, Liu Y, et al. An alternative high output tissue microarray technique. Diagn Pathol. 2013;8:9. doi: 10.1186/1746-1596-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch FR, Varella-Garcia M, Bunn PJ, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 22.Cronin J, McAdam E, Danikas A, Tselepis C, Griffiths P, Baxter J, et al. Epidermal growth factor receptor (EGFR) is overexpressed in high-grade dysplasia and adenocarcinoma of the esophagus and may represent a biomarker of histological progression in Barrett's esophagus (BE) Am J Gastroenterology. 2011;106:46–56. doi: 10.1038/ajg.2010.433. [DOI] [PubMed] [Google Scholar]
- 23.Pirker R, Pereira JR, von Pawel J, Krzakowski M, Ramlau R, Park K, et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 2012;13:33–42. doi: 10.1016/S1470-2045(11)70318-7. [DOI] [PubMed] [Google Scholar]
- 24.Yang YL, Xu KL, Zhou Y, Gao X, Chen LR. Correlation of epidermal growth factor receptor overexpression with increased epidermal growth factor receptor gene copy number in esophageal squamous cell carcinomas. Chin Med J (Engl) 2012;125:450–4. [PubMed] [Google Scholar]
- 25.Grobe A, Eichhorn W, Fraederich M, Kluwe L, Vashist Y, Wikner J, et al. Immunohistochemical and FISH analysis of EGFR and its prognostic value in patients with oral squamous cell carcinoma. J Oral Pathol Med. 2014;43:205–10. doi: 10.1111/jop.12111. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Jiang D, Li X, Lv J, Xie L, Zheng L, et al. Establishment and characterization of esophageal squamous cell carcinoma patient-derived xenograft mouse models for preclinical drug discovery. Lab Investig. 2014;94:917–26. doi: 10.1038/labinvest.2014.77. [DOI] [PubMed] [Google Scholar]
- 27.Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–55. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 28.Hou J, Jiang DX, Zhang JC, Gavine PR, Xu ST, Liu YL, et al. Frequency, characterization, and prognostic analysis of PIK3CA gene mutations in Chinese esophageal squamous cell carcinoma. Hum Pathol. 2014;45:352–8. doi: 10.1016/j.humpath.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–62. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–19. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai K, Takaoka A. Comparing antibody and small-molecule therapies for cancer. Nat Rev Cancer. 2006;6:714–27. doi: 10.1038/nrc1913. [DOI] [PubMed] [Google Scholar]
- 33.Gonzaga IM, Soares-Lima SC, de Santos PT, Blanco TC, de Reis BS, Quintella DC, et al. Alterations in epidermal growth factor receptors 1 and 2 in esophageal squamous cell carcinomas. BMC Cancer. 2012;12:569. doi: 10.1186/1471-2407-12-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abedi-Ardekani B, Dar NA, Mir MM, Zargar SA, Lone MM, Martel-Planche G, et al. Epidermal growth factor receptor (EGFR) mutations and expression in squamous cell carcinoma of the esophagus in central Asia. BMC Cancer. 2012;12:602. doi: 10.1186/1471-2407-12-602. [DOI] [PMC free article] [PubMed] [Google Scholar]


