Abstract
Members of the myocyte enhancer factor–2 (MEF2) family of MADS (MCM1, agamous, deficiens, serum response factor)–box transcription factors bind an A-T–rich DNA sequence associated with muscle-specific genes. The murine MEF2C gene is expressed in heart precursor cells before formation of the linear heart tube. In mice homozygous for a null mutation of MEF2C, the heart tube did not undergo looping morphogenesis, the future right ventricle did not form, and a subset of cardiac muscle genes was not expressed. The absence of the right ventricular region of the mutant heart correlated with down-regulation of the dHAND gene, which encodes a basic helix-loop-helix transcription factor required for cardiac morphogenesis. Thus, MEF2C is an essential regulator of cardiac myogenesis and right ventricular development.
The mechanisms that regulate heart formation during embryogenesis are only beginning to be elucidated (1). Members of the MEF2 family of transcription factors bind a conserved A-T–rich DNA sequence associated with most cardiac muscle structural genes (2) and are expressed in cardiogenic precursor cells and differentiated cardiomyocytes during embryogenesis (3). MEF2 factors are also expressed in skeletal and smooth muscle cell lineages (3, 4), and MEF2 binding sites are essential for expression of muscle genes in all three muscle cell types (5).
There are four MEF2 genes in vertebrate species, designated MEF2A, -B, -C, and -D, share homology in an NH2-terminal MADS-box and an adjacent motif known as the MEF2 domain (5). These protein domains mediate DNA binding, homo- and heterodimerization, and interaction with basic helix-loop-helix (bHLH) transcription factors (6–8).
In the mouse, MEF2B and MEF2C are co-expressed in the precardiogenic mesoderm beginning at embryonic day 7.75 (E7.75), and MEF2A and MEF2D are expressed about 12 hours later (3, 8). MEF2 gene expression is detected in skeletal muscle precursors in the somites and in smooth muscle cells beginning at about E9.0. Loss-of-function mutations in a single MEF2 gene in Drosophila, D-mef2, prevent differentiation of cardiac, skeletal, and visceral muscle cells (9), but the functions of the vertebrate MEF2 genes in the embryo have not been determined.
To investigate MEF2C function during mouse embryogenesis, we inactivated this gene with a targeting vector (10) that deleted the second protein-coding exon, which encodes amino acids 18 to 86 (Fig. 1). The MADS and MEF2 domains are contained in residues 1 to 56 and 57 to 86, respectively, and the residues deleted by the mutation are essential for DNA binding and dimerization (7). The vector was introduced into embryonic stem (ES) cells by electroporation, clones were isolated after positive-negative selection (11), and genomic DNA was analyzed by Southern blot analysis for gene replacement at the MEF2C locus (12). The frequency of ES cell clones bearing a targeted MEF2C allele was 1: 7. Three targeted ES cell clones were injected into blastocysts isolated from C57BL/6J mice to generate chimeras, two of which transmitted the mutation through the germ line. Mice heterozygous for the targeted allele showed no discernible phenotype and were intercrossed to obtain MEF2C homozygous null offspring. The genotypes of offspring from heterozygous intercrosses were determined within 1 to 3 weeks after birth by Southern blot analysis of DNA obtained from tail biopsies. No neonates homozygous for the MEF2C mutation were found among 189 offspring and no neonatal lethality was observed, indicating that the homozygous mutation resulted in embryonic lethality.
Fig. 1.
Targeting strategy for the MEF2C locus. Diagram of MEF2C, which contains 465 amino acids, is shown at the top, above a diagram of the mouse MEF2C locus around coding exon 2, which encodes residues 18 to 86. The first intron in the coding region is >10 kb. In the targeting vector (10), the neomycin-resistance gene (neo) was transcribed in the same direction as MEF2C. Homologous recombination resulted in deletion of coding exon 2. The structure of the targeted allele is shown at the bottom. Insertion of neo introduced an Eco RI site that could be used to distinguish the wild-type and mutant alleles. Position of the 3′ probe used for Southern blot analysis of yolk sac DNA is indicated. Hybridization of Eco RI– digested DNA with the 3′ probe yielded fragments of 11.5 and 3.2 kb from the wild-type and mutant loci, respectively.
We determined the genotypes of embryos between E6.5 and E12.5 by Southern blot or polymerase chain reaction (PCR) analysis (12) of DNA isolated from yolk sacs. Homozygous mutants were detected at roughly the predicted Mendelian frequency between E6.5 and E9.5. Wild-type and mutant embryos were similar in size up to the 14-somite stage (E9.0), and the first branchial arch appeared to form normally in the mutant. However, the mutants developed no more than 20 somites and showed retarded growth after E9.0. No viable mutants were observed by E10.5, indicating that the lethality of the MEF2C mutation was fully penetrant by that stage.
The most obvious morphologic defects in mutant embryos were within the heart. The heart develops from bilaterally symmetric cardiogenic primordia that coalesce at the midline to form a primitive heart tube (13). Mutant and wild-type embryos were microscopically indistinguishable at the linear heart tube stage (E8.0–8.25), and contractions were initiated in the heart tubes of mutants, although they were slower and less rhythmic than normal. At E9.0, the heart rate of wild-type and MEF2C heterozygous embryos was 58 ± 3 beats per minute, compared with 26 ± 4 beats per minute for mutants (14). In wild-type embryos, the heart tube initially exhibits a peristaltic beat that becomes sequential after looping, with strong atrial contraction preceding ventricular contraction. In mutants, the atrial chamber exhibited weak contractions, and the hypoplastic ventricular chamber appeared to vibrate only in response to atrial contractions; there was no evidence of independent ventricular contractions. Mutant embryos also exhibited severe pericardial effusion (Fig. 2, A and B), indicative of hemodynamic insufficiency.
Fig. 2.
Cardiac defects in MEF2C null embryos. (A) Wild-type and (B) mutant embryos at E9.0. The lower portion of the wild-type embryo was removed for a clear view of the heart. Note severe pericardial effusion in the mutant. Hearts of the wild-type (C) and mutant (D) embryos are shown. Scanning electron micrographs of wild-type (E) and mutant (F) embryos at E9.0. Abbreviations: a, Atrium; bc, bulbus cordis; lv, future left ventricle; ps, pericardial sac; and v, ventricular chamber. Bars = 2 µm.
At E8.5, the heart tube normally initiates rightward looping, a process that is necessary to orient the atrial and ventricular chambers and to align the outflow tract with the vasculature. Looping morphogenesis is accompanied by formation of the conotruncus in the anteriormost region and later by formation of the common ventricular and atrial chambers. Further maturation of the looped cardiac tube yields the future right and left ventricles and the common atrial chamber. In mutant embryos, the heart tube did not undergo rightward looping (Fig. 2, C to F), and there was no morphologic evidence of the future right ventricle. Rather, a single hypoplastic ventricular chamber was fused directly to an enlarged atrial chamber. Because of the apparent absence of the future right ventricle, the remaining portion of the ventricular chamber was displaced slightly to the left. The atrioventricular (AV) demarcation was apparent in the mutant, but the AV canal did not elongate. Also, the sinus venosus, which normally forms from the posterior portion of the atrial chamber, did not form in the mutant.
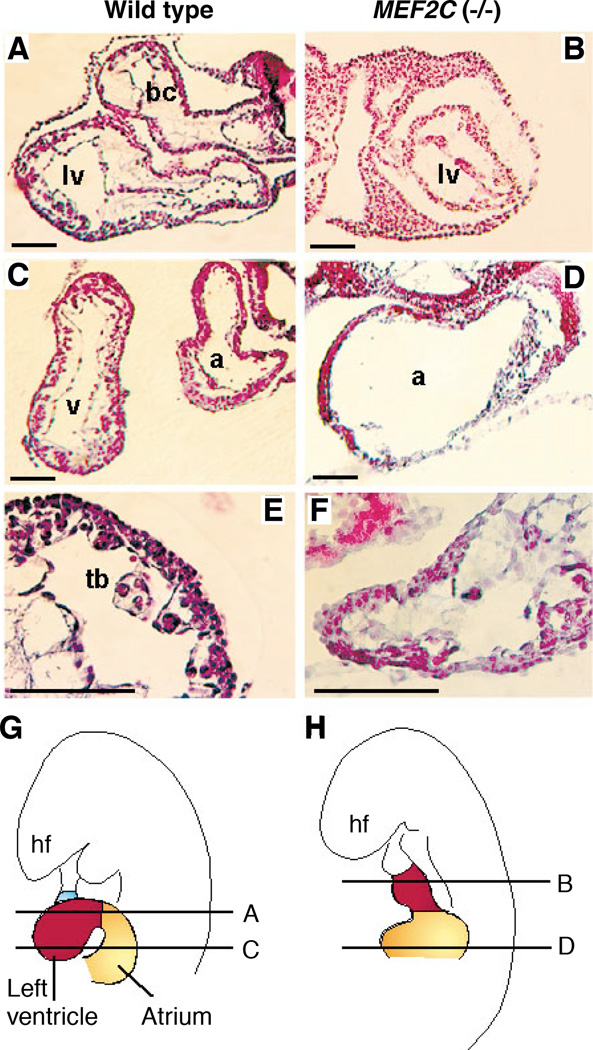
As the ventricular chambers develop, the trabeculae form as fingerlike projections along the inner myocardial wall, which becomes clearly separated from the endocardial layer by a loose mesenchyme called the cardiac jelly. Histological sections of wild-type hearts at E9.0 revealed well-defined atrial and ventricular chambers, with trabeculae projecting from the inner ventricular walls (Fig. 3, A, C, and E). The endocardium was also evident in wild-type embryos at this stage. In mutant hearts, the trabeculae were poorly developed and the lumen of the ventricular chamber was extremely narrow (Fig. 3, B and F). The cardiomyocytes within the ventricular wall and the endocardial cells also appeared to be disorganized. In the atrial chamber of the mutant, the myocardial wall was thin and the cardiac jelly was absent (Fig. 3D). The AV canal was present in the mutant; however, the endocardial cushions did not form. Red blood cells were seen in the mutant embryos, but very few were present in the heart, suggesting that circulation was impaired. Schematic diagrams showing the morphologies of wild-type and mutant hearts at E9.0 are shown in Fig. 3, G and H, respectively.
Fig. 3.
Hematoxylin and eosin staining of thin sections of hearts of wild-type and mutant embryos. Wild-type (A, C, and E) and mutant (B, D, and F) embryos at E9.0 were fixed, sectioned, and stained with hematoxylin and eosin (27). (G and H) Diagrams of planes of section. Abbreviations: a, Atrium; bc, bulbus cordis; hf, headfold; lv, left ventricle; tb, trabeculae; and v, ventricular chamber. Bars = 1 µm.
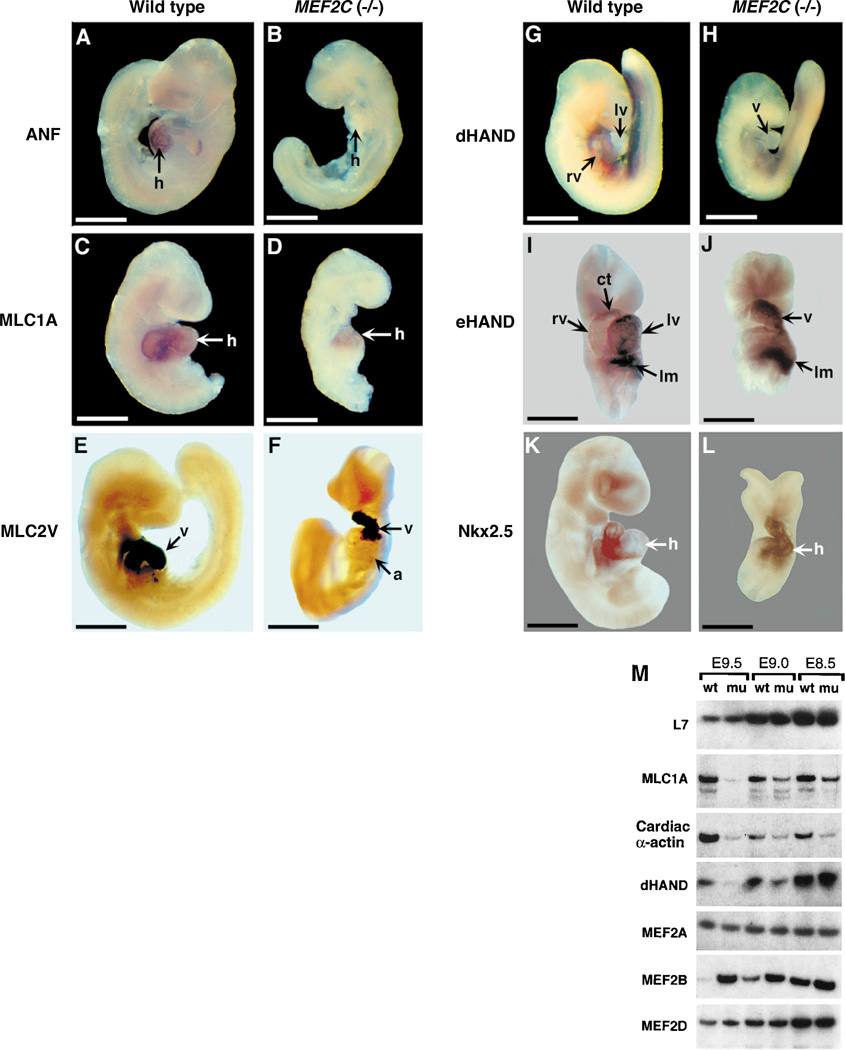
Whole mount in situ hybridization of embryos at E9.0, before the mutants began to show retarded growth, demonstrated a reduction in expression of several markers of cardiac differentiation. Transcripts for atrial natriuretic factor (ANF), cardiac α-actin, and α-myosin heavy chain were down-regulated to background levels, and myosin light chain (MLC)–1A expression was decreased significantly but was still detectable at a low level in the hearts of the mutants (Fig. 4, A to D) (15). In contrast, other cardiac muscle genes, including MLC2V (Fig. 4, E and F) and MLC2A (15), were expressed at normal levels in the mutants. Expression of MLC2V is normally localized to the ventricular region of the developing heart by E9.0 (16). In the mutants, MLC2V expression was localized to the hypoplastic ventricular chamber, indicating that the AV chambers were specified correctly.
Fig. 4.
Analysis of cardiac gene expression by whole mount in situ hybridization. Wild-type (A, C, E, G, I, and K) and mutant (B, D, F, H, J, and L) embryos were stained by in situ hybridization for the indicated transcripts (3, 28). For a clearer view of the heart, heads and tails were dissected away from the embryos in (I), (J), and (L). Note that dHAND expression is restricted to the right ventricular region of the heart in the wild-type embryo and is not expressed in the heart of the mutant. eHAND is expressed in the conotruncus and left ventricular region in the wild-type heart; in the mutant, there is no gap in expression that would represent the right ventricle. Tails were dissected away from embryos in (C), (D), and (K). (F), (I), (J), and (L) are frontal views; other embryos are viewed from the side. Abbreviations: a, Atrium; ct, conotruncus; h, heart; lm, lateral mesoderm; lv, left ventricle; rv, right ventricle; and v, ventricular chamber. Bars = 5 µm. (M) Quantitative RT-PCR analysis of gene expression in wild-type (wt) and MEF2C mutant (mu) embryos. PCR amplifications were performed with cDNA synthesized from RNA isolated from E9.5 embryonic heart or whole embryos at E8.5 and E9.0 and gene-specific primers for the indicated transcripts. L7 was used as a loading control. Transcripts for dHAND were down-regulated in the heart by E9.0, as determined by in situ hybridization. The expression of dHAND transcripts in the E8.5 and E9.0 RNA samples used for RT-PCR reflects its expression in the lateral mesoderm and not in the heart. RNA concentrations were quantitated by PhosphorImager analysis.
Quantitative reverse transcriptase–PCR (RT-PCR) (17) confirmed the results of whole mount in situ hybridization and showed that several muscle-specific transcripts were down-regulated four- to fivefold in the mutants. Down-regulation of the cardiac muscle genes in MEF2C mutant embryos was a specific response to the absence of MEF2C rather than a secondary result of embryonic demise, because it preceded growth retardation of the embryo and was specific to a certain subset of genes.
The bHLH genes dHAND and eHAND are expressed in complementary patterns that demarcate distinct regions of the developing mouse heart (18–20). dHAND is normally expressed throughout the heart tube before expression becomes localized predominantly to the right side of the looping heart tube (19, 20). In contrast, eHAND is expressed in the conotruncus and future left ventricle, but expression is excluded from the region of the heart tube that gives rise to the right ventricle (18–20). In the MEF2C mutant, dHAND was expressed at normal levels in the heart tube before looping (Fig. 4M) (15). However, dHAND transcripts were down-regulated at the time of looping, concomitant with failure of the right ventricular region to form (Fig. 4, G, H, and M). eHAND was expressed in the mutant, but expression was contiguous throughout the heart tube without the gap in expression that normally characterizes the future right ventricle (Fig. 4, I and J). This distortion in eHAND expression and the lack of dHAND expression in the heart tube of the mutant are consistent with absence of the future right ventricle. The cardiac homeobox gene Nkx-2.5 (21) was expressed throughout the developing heart in wild-type and mutant embryos (Fig. 4, K and L). MEF2B, which is normally coexpressed with MEF2C during the earliest stages of cardiogenesis (3, 8), was up-regulated more than sevenfold in the hearts of mutants at E9.5 (Fig. 4M), whereas MEF2A and MEF2D transcripts were expressed at comparable levels in wild-type and mutant hearts.
The phenotype of MEF2C mutant embryos demonstrates that MEF2C is required for looping of the cardiac tube, development of the right ventricle, and expression of a subset of cardiac muscle genes. The cardiogenic defects seen in MEF2C mutant embryos are distinct from those seen in other mouse mutants (22) and reveal a cardiogenic regulatory program for right ventricular development.
MEF2C is expressed uniformly throughout the heart tube. Therefore, the selective ablation in the MEF2C mutant of the segment of the heart tube that gives rise to the right ventricle suggests that MEF2C regulates expression or activity of a regionally restricted regulatory factor required for development of this region of the heart. A candidate for such a factor is the cardiogenic transcription factor dHAND, which is expressed throughout the linear heart tube and becomes restricted to the future right ventricle as looping is initiated. Mice homozygous for a null mutation in dHAND exhibit a cardiac phenotype similar to that of the MEF2C mutant (19), suggesting that MEF2C regulates dHAND expression in the future right ventricular region of the heart tube or that these two transcription factors cooperate to control right ventricular development, as has been described for MEF2 and myogenic bHLH proteins in the skeletal muscle lineage (23). MEF2C and dHAND may control right ventricular development by specifying a subpopulation of cardiogenic cells within the cardiac tube or by regulating the expansion of such a population.
In the absence of MEF2C, a subset of structural genes for cardiac muscle was not up-regulated, whereas others were expressed normally. Some of the cardiac genes that were independent of MEF2C, such as MLC2V, contain essential MEF2-binding sites in their control regions (24), suggesting that downstream genes in the pathway for cardiomyocyte differentiation can discriminate between different members of the MEF2 family and that another member of the MEF2 family supports activation of those cardiac genes that are expressed in the MEF2C mutant. MEF2B is coexpressed with MEF2C throughout the early stages of cardiogenesis (3, 8), and it was up-regulated in the MEF2C mutant, consistent with the possibility that it may partially substitute for MEF2C. That cardiac development occurs normally in MEF2B-null mice (25) also suggests that MEF2C shares functions with MEF2B.
In addition to their expression in the developing heart, members of the MEF2 family are expressed later in development in skeletal and smooth muscle cells (3, 4, 8), in the nervous system (26), and eventually in a wide range of cell types. MEF2C may have additional functions in these other cell types.
Acknowledgments
We thank J. Martin for isolating the MEF2C genomic clone, A. Tizenor for assistance with graphics, and D. Srivastava and members of the Olson laboratory for helpful discussions. Supported by grants from NIH, the Muscular Dystrophy Association, the American Heart Association, and the Human Frontiers Sciences Program to E.N.O.
Contributor Information
Qing Lin, Department of Molecular Biology and Oncology, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75235–9148, USA.
John Schwarz, Division of Cardiology, Department of Internal Medicine, University of Texas Medical School, Houston, TX 77030, USA.
Corazon Bucana, Department of Cell Biology, University of Texas M. D. Anderson Cancer Center, Houston, TX 77030, USA.
Eric N. Olson, Department of Molecular Biology and Oncology, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75235–9148, USA
REFERENCES AND NOTES
- 1.Olson EN, Srivastava D. Science. 1996;272:671. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]; Fischman MC, Stanier DYR. Circ. Res. 1994;74:757. doi: 10.1161/01.res.74.5.757. [DOI] [PubMed] [Google Scholar]
- 2.Gossett LA, Kelvin DJ, Sternberg EA, Olson EN. Mol. Cell. Biol. 1989;9:5022. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edmondson DG, Lyons GE, Martin JF, Olson EN. Development. 1994;120:1251. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 4.Chambers AE, et al. Genes Dev. 1994;8:1324. doi: 10.1101/gad.8.11.1324. [DOI] [PubMed] [Google Scholar]; Wong MW, Pisegna M, Lu MF, Leibham D, Perry M. Dev. Biol. 1994;166:683. doi: 10.1006/dbio.1994.1347. [DOI] [PubMed] [Google Scholar]; Nadal-Ginard B, Subramanian SV. Mech. Dev. 1996;57:103. doi: 10.1016/0925-4773(96)00542-4. [DOI] [PubMed] [Google Scholar]; Firulli AB, et al. Circ. Res. 1996;78:196. doi: 10.1161/01.res.78.2.196. [DOI] [PubMed] [Google Scholar]
- 5.Olson EN, Perry M, Schulz RA. Dev. Biol. 1995;172:280. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 6.Pollock R, Treisman R. Genes Dev. 1991;5:2327. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]; Martin JF, et al. Mol. Cell. Biol. 1994;14:1647. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]; Martin JF, Schwarz JJ, Olson EN. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5282. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu YT, et al. Genes Dev. 1995;9:1388. [Google Scholar]; Breitbart RE, et al. Development. 1993;118:1095. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]; Leifer D, et al. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1546. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mc-Dermott JC, et al. Mol. Cell. Biol. 1993;13:2564. [Google Scholar]
- 7.Molkentin JD, et al. Mol. Cell. Biol. 1996;16:2627. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu Y-T. J. Biol. Chem. 1996;271:24675. [PubMed] [Google Scholar]
- 8.Molkentin J, et al. Mol. Cell. Biol. 1996;16:3814. doi: 10.1128/mcb.16.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lilly B, et al. Science. 1995;267:688. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]; Bour BA, et al. Genes Dev. 1995;9:730. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]; Ranganayakulu G, et al. Dev. Biol. 1995;171:169. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 10.The MEF2C targeting vector was constructed as shown in Fig. 1, with the neomycin resistance gene under control of the phosphoglycerol kinase promoter in the same transcriptional orientation as MEF2C. A thymidine kinase gene under control of the herpes simplex virus promoter pMC1-HSVtk [S. L. Mansour, K. R. Thomas, M. R. Capecchi, Nature 336, 348 (1988)] was cloned into an Xho I site immediately 5′ of the 5′ arm of genomic homology. All the cloning junctions in the targeting vector were confirmed by DNA sequencing. The targeting vector was linearized by digestion with Not I before electroporation.
- 11.The MEF2C targeting vector was introduced by electroporation into 129 ES cells [A. P. McMahon and A. Bradley, Cell 62, 1073 (1990)]. After positive-negative selection, surviving clones were isolated and replica-plated onto SN76/7 fibroblasts in 96-well microtiter plates, and genomic DNA was analyzed on Southern blots.
- 12.Homologous recombination at the MEF2C locus was detected by Southern blot or PCR analysis of genomic DNA isolated from ES cells, yolk sacs, or tail biopsies. For Southern blots, genomic DNA was digested with Eco RI and hybridized to a probe from a region immediately 3′ of the region of homology used for targeting. For PCR, the following oligonucleotide primers were used: neo primer, 5′-GGCAT-GCTGGGGATGCGGTGGGCTC-3′; MEF2C MADS box primer, 5′-AGTACAACGAGCCGCACGAGAG-CCG-3′; and MEF2C short-arm primer, 5′-GTCAC-CTTAAGACATAAAGCACCCTCC-3′.
- 13.DeHaan RL. In: Organogenesis. DeHaan RL, Ursprung H, editors. New York: Holt Rinehart Winston; 1964. pp. 377–419. [Google Scholar]
- 14.Heart rates were determined in E9.0 embryos obtained from timed matings of MEF2C heterozygotes. Pregnant females were killed by cervical dislocation; embryos were removed and placed in phosphate-buffered saline at 37°C. Fourteen wild-type and six mutant embryos were analyzed. The average values were statistically significant, as assessed by a standard t test.
- 15.Lin Q, Olson EN. unpublished data. [Google Scholar]
- 16.O’Brien TX, Lee KJ, Chien KR. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5157. doi: 10.1073/pnas.90.11.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.For RT-PCR, total RNA was prepared from isolated hearts at E9.5 and from whole embryos at E8.5 and E9.0, treated with ribonuclease-free deoxyribonuclease I, and resuspended in 20 µl of water. First strand cDNA synthesis was performed with 10 µl of RNA with Moloney murine leukemia virus reverse transcriptase (BRL) and random primers. PCR amplification was performed with 0.5 µl of the cDNA, 0.1 µCi of [32P]deoxycytidine triphosphate (dCTP), and gene-specific primers under conditions of linearity for each individual primer set. Most oligonucleotide primers spanned introns. To confirm that samples were not contaminated with genomic DNA, we also performed duplicate PCRs without RT. All PCR products corresponded to the size predicted for the corresponding transcript. PCR cycles were as follows: 99°C for 2 min then 18 to 25 cycles of 96°C for 30 s, 58°C for 30 s, and 72°C for 45 s. PCR products were separated by 6% polyacrylamide gel electrophoresis and quantitated by analysis on a phosphor-Imager (Molecular Dynamics).
- 18.Srivastava D, Cserjesi P, Olson EN. Science. 1995;270:1995. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]; Cserjesi P, Brown D, Lyons GE, Olson EN. Dev. Biol. 1995;170:664. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava D, Thomas T, Lin Q, Brown D, Olson EN. Nature Genetics. doi: 10.1038/ng0697-154. in press. [DOI] [PubMed] [Google Scholar]
- 20.Biben C, Harvey RP. personal communication [Google Scholar]
- 21.Lints T, et al. Development. 1993;119:419. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 22.Lyons I, et al. Genes Dev. 1995;9:1654. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]; Rossant J. Circ. Res. 1996;78:349. doi: 10.1161/01.res.78.3.349. [DOI] [PubMed] [Google Scholar]
- 23.Molkentin JD, Black BL, Martin JF, Olson EN. Cell. 1995;83:1125. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]; Kaushal S, Schneider JW, Nadal-Ginard B, Mahdavi V. Science. 1994;266:1236. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin JD, Markham BE. J. Biol. Chem. 1993;268:19512. [PubMed] [Google Scholar]; Navankasattusas S, Zhu H, Garcia AV, Evans SM, Chien KR. Mol. Cell. Biol. 12:1469. doi: 10.1128/mcb.12.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molkentin J, Lin Q, Olson EN. unpublished data. [Google Scholar]
- 26.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. J. Neurosci. 1995;15:5727. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin JF, Bradley A, Olson EN. Genes Dev. 1995;9:1237. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- 28.Sources of probes were as follows: ANF Zeller R, et al. Genes Dev. 1987;1:693. doi: 10.1101/gad.1.7.693. MLC1A Barton P, et al. J. Biol. Chem. 1988;263:12669. MLC2V (16); eHAND and dHAND (18); and Nkx-2.5 (21).