Summary
The changing environments strongly affect plants growth and development. Phytohormones, endogenous plant-made small molecules such as ethylene, regulate a wide range of processes throughout the lifetime of plants[1, 2]. The ability of plants to integrate external signals with endogenous regulatory pathways is vital for their survival [3, 4]. Ethylene was found to suppress hypocotyl elongation in darkness[5], while promote it in light[6, 7]. How ethylene regulates hypocotyl elongation in such an opposite way is largely unknown. In particular, how light modulates and even reverses the function of ethylene has yet to be characterized. Here we show that the bHLH transcription factor Phytochrome-Interacting Factor 3 (PIF3), is directly activated by Ethylene-Insensitive 3 (EIN3), and is indispensible for ethylene-induced hypocotyl elongation in light. Ethylene via EIN3 concomitantly activates two contrasting pathways: the PIF3-dependent growth-promoting pathway and an Ethylene-Response Factor 1 (ERF1)-mediated growth-inhibiting pathway. The PIF3 pathway is saturated in dark but progressively fortified with light de-stabilizing PIF proteins to reduce their redundancy. While the ERF1 pathway is mainly functional in dark but gradually saturated with light dramatically stabilizing ERF1 protein. Our findings provide a mechanistic insight into how light modulates internal hormone-regulated plant growth.
Results and Discussions
PIF3 acts downstream of EIN3/EIL1 in mediating ethylene-stimulated hypocotyl elongation in light
Ethylene signaling pathway has been uncovered that receptors and CTR1 are negative regulators while EIN2, EIN3 and EIN3-like 1(EIL1) are downstream activators[2, 8, 9]. Previous studies showed that the ethylene insensitive mutant ein2 exhibited shorter hypocotyls, whilst the constitutive ethylene response mutant ctr1 displayed longer hypocotyls than the wild type (WT) seedlings grown in light [6, 7]. We found that the light-grown ethylene-insensitive mutant ein3 eil1, which lacks both EIN3 and EIL1 transcription factors, also displayed shortened hypocotyls (Figure S1A and S1B), whereas the transgenic plants over-expressing EIN3 (EIN3OX) constitutively exhibited elongated hypocotyls compared with Columbia-0 WT (Col-0) (Figure S1A and S1B). Moreover, treatment with ACC, a biosynthetic precursor of ethylene, promoted hypocotyl elongation of light-grown WT seedlings but not ein3 eil1 mutant seedlings (Figure 1A and S1C). Therefore, EIN3/EIL1 are required for ethylene-induced hypocotyl elongation in light.
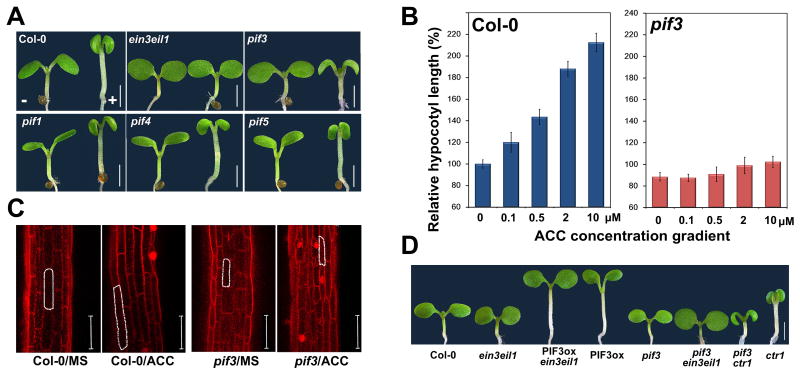
Figure 1. PIF3 acts downstream of EIN3/EIL1 in mediating ethylene-stimulated hypocotyl elongation in light.
(A) Phenotype of 5-day-old light-grown seedlings on MS medium (MS) supplemented with (+, right) or without (-, left) 10μM ACC.
(B) Hypocotyl lengths of 7-day-old light-grown seedlings on MS supplemented with increasing concentrations of ACC. Mean ±s.d., n>20.
(C) Propidium iodide (PI) staining of hypocotyl cells of 7-day-old light-grown seedlings.
(D) Hypocotyl phenotype of 5-day-old light-grown seedlings on MS.
See also Figure S1.
Recent studies revealed that Phytochrome-Interacting factors (PIFs) play central roles in repressing photomorphogenesis [10-12]. As ethylene-induced hypocotyl elongation antagonizes light-regulated plant development, we investigated whether PIFs are involved in ethylene-induced hypocotyl elongation. Strikingly, upon ACC treatment, although pif1, pif4 and pif5 mutants displayed no obvious difference from WT, the pif3 mutant was almost entirely impaired in ethylene-induced hypocotyl elongation, mimicking the ein3 eil1 mutant (Figure 1A and S1C). A dosage response experiment further confirmed that pif3 was virtually insensitive to ACC in the regulation of hypocotyl elongation (Figure 1B). Microscopic observation demonstrated that ethylene promoted hypocotyl length mainly by inducing cell elongation, which was abolished in the pif3 mutant (Figure 1C). These results indicate that PIF3 is an essential component required for ethylene-induced hypocotyl elongation in light.
We next assessed the genetic relationship between these two classes of transcription factors, EIN3/EIL1 and PIF3. Over-expression of PIF3 (PIF3OX) led to robust hypocotyl elongation even in the absence of ethylene, and it completely rescued the shortened hypocotyl of ein3 eil1 (Figure 1D and S1D). Conversely, pif3 fully suppressed the elongated hypocotyls of ctr1, whereas pif3 ein3 eil1 triple-mutant displayed the same short hypocotyl phenotype as pif3 or ein3 eil1 (Figure 1D and S1D). These genetic interactions suggest that PIF3 acts in the same pathway downstream of EIN3/EIL1 in regulating ethylene-promoted hypocotyl elongation in light.
EIN3 activates PIF3 gene expression by directly binding to the promoter elements of PIF3
To reveal how EIN3/EIL1 regulate PIF3, we first examined whether ethylene regulates PIF3 transcription via EIN3/EIL1 in light. Upon ACC treatment, the PIF3 transcription was notably elevated in WT, but not in the ein3 eil1 mutant (Figure 2A). Consistently, PIF3 transcript level was almost eliminated in ein2 and ein3 eil1 but drastically increased in ctr1 (Figure 2A). In addition, we also examined the PIF3 transcription in transgenic plants that expressing EIN3 protein under the estradiol-inducible promoter in the ein3 eil1 ebf1 ebf2 quadruple mutant background (iE/qm) [13]. PIF3 expression was up-regulated rapidly (in 10 min) and in a dosage-dependent manner following the induction of EIN3 protein in the iE/qm seedlings (Figure 2B). Notably, the patterns of EIN3-induced PIF3 expression were similar to that of EBF2 and ERF1, two known direct target genes of EIN3 (Figure S2A) [14, 15]. By contrast, the transcript levels of PIF1, PIF4 or PIF5, which plays no role in ethylene-induced hypocotyl elongation, were not significantly regulated by EIN3/EIL1 or ethylene (Figure S2B). GUS staining of the PIF3p: GUS transgenic plants further demonstrated that the PIF3 promoter was active in cotyledons and hypocotyls, where its activity was markedly enhanced by ACC treatment or in the ctr1 mutant, but clearly decreased in the ein3 eil1 mutant (Figure 2C). Therefore, ethylene stimulates PIF3 gene expression via the action of EIN3/EIL1.
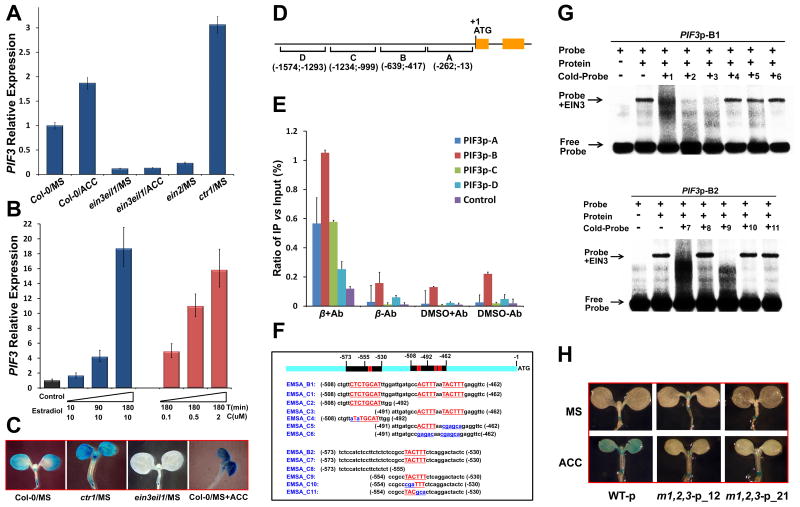
Figure 2. EIN3 activates PIF3 gene expression by directly binding to the promoter elements of PIF3.
(A) qRT-PCR analysis of PIF3 gene expression in 5-day-old continuous light-grown seedlings on MS (MS) or MS supplemented with 10μM ACC (ACC). Mean ±s.d., n=3.
(B) qRT-PCR analysis of PIF3 gene expression in 5-day-old continuous light-grown seedlings on MS medium with gradually induced EIN3 protein synthesis. Mean ±s.d., n=3. The seedlings expressing estradiol-inducible EIN3 protein in ein3 eil1 ebf1 ebf2 mutant background (iE/qm) were treated with estradiol at gradient concentrations (C) or treatment time (T).
(C) GUS staining of 5-day-old continuous light-grown PIF3-promoter:GUS seedlings.
(D) Diagram of the four PIF3 promoter regions, named A-D.
(E) ChIP results using anti-FLAG antibody (± Ab) to first precipitate the induced EIN3-FLAG protein (β means estradiol-induced, DMSO is un-induced control), followed by qPCR detection of the four PIF3 promoter regions in 5-day-old continuous light-grown iE/qm seedlings. Mean ±s.d., n=3.
(F) The sequences of the probes used in (G). Underlined red bases in capitals indicate the predicted EIN3 binding motifs, and the base changes in the mutant sequences are marked in blue and in lower case.
(G) EMSA assays of recombinant EIN3 protein (1-314 aa) and PIF3 promoter regions. Cold-probes (un-labeled probes) used for competition were in 200-fold excess over labeled probes.
(H) GUS staining of 5-day-old continuous light-grown PIF3-promoter:GUS transgenic seedlings. WT-p indicates intact PIF3 promoter fused with GUS reporter gene, whereas M-p#12 and M-p#21 indicate two individual transgenic lines of PIF3 promoter with all three EIN3 biding motifs mutated.
See also Figure S2.
The similar kinetics between the induction of PIF3 and that of EBF2 or ERF1 suggested that PIF3 might be a direct target gene of EIN3 (Figure 2B and S2A). Previous studies showed that EIN3 binds to specific promoter elements named EIN3 binding sites (EBS) to regulate downstream gene expression [15, 16]. Bioinformatics analysis identified several putative EIN3 binding sites (EBS) in the PIF3 promoter (Figure S2D). We thus divided the PIF3 promoter sequence into four regions, i.e. A to D, each containing at least one putative EBS element (Figure 2D). Chromatin immunoprecipitation (ChIP) of light-grown iE/qm seedlings followed by PCR assay showed that EIN3 preferentially binds to regions A, B and C of the PIF3 promoter in vivo (Figure 2D and S2C). Subsequent yeast-one-hybrid assay revealed that EIN3 binds specifically to region B in yeast cells (Figure S2E), implying additional factors might be required to enable EIN3 association with region A and C in planta. Further yeast-one-hybrid assays using mutated EBS elements showed that one of the three putative EBS elements in region B (CTCTGC) primarily mediates EIN3 binding in yeast (Figure S2D and S2E). Electrophoretic mobility shift assay (EMSA) indicated that EIN3 was able to directly bind to the three EBS elements in the region B in vitro (Figure 2E and 2F). Moreover, the ACC induction of PIF3p: GUS activity was greatly decreased when all the three EBS elements in the region B of the PIF3 promoter were mutated (Figure 2H), further confirming the essential role of these EBS elements in mediating ethylene-activated PIF3 gene expression. Taken together, EIN3 directly binds to the specific EBS elements in the PIF3 promoter to activate its transcription.
Ethylene activates a PIF3-dependent growth-promotion pathway that is progressively fortified with light-induced PIFs degradation
Interestingly, we found that, as in light condition, ethylene also induced PIF3 gene expression in darkness in an EIN3/EIL1-dependent manner (Figure S3A). We then examined whether PIF3 contributes to ethylene-regulated hypocotyl elongation in dark-grown seedlings. In contrast to their pronounced phenotypes in light, neither pif3 mutant nor PIF3OX showed any obvious difference from WT with or without ACC treatment of etiolated seedlings (Figure 3A). The finding that PIF3 seems to lack a role on hypocotyl growth in dark could be attributed to functional redundancy among multiple PIFs. Previous studies showed that PIF1, 3, 4 and 5 are highly abundant and act redundantly to promote hypocotyl elongation in dark, as none of the single, double or triple mutants but only the pif1 pif3 pif4 pif5 quadruple mutant (pifQm) displays significantly reduced hypocotyls [10, 11](Figure S3B). Our results demonstrated that all of the PIF1, PIF3, PIF4 and PIF5 proteins were accumulated in darkness and dramatically declined upon light irradiation (Figure S3C). It is noticed that with ACC treatment, light-induced degradation of the constitutively expressing PIF3-MYC protein is a little different from on MS (Figure S3C), suggesting a possible role of ethylene in affecting PIF3 protein stability. However, light predominantly degrades PIFs protein (Figure S3C). It is thus likely that PIF functions have reached a saturated level in etiolated seedlings and the ethylene induction of PIF3 or overexpression of PIF3 by genetic means cannot exert an additive effect on promoting hypocotyl elongation in dark. Nevertheless, when grown in light, most PIF proteins are degraded, leading to low abundance of PIF proteins (Figure S3C) [10, 12]. Under this condition, the action of individual PIFs becomes discernible, as evidenced by the findings that the pif3, pif4 and pif5 monogenic mutant each displayed shorter hypocotyls than WT and ethylene-promoted hypocotyl elongation was eliminated by the pif3 single mutant (Figure S3D).
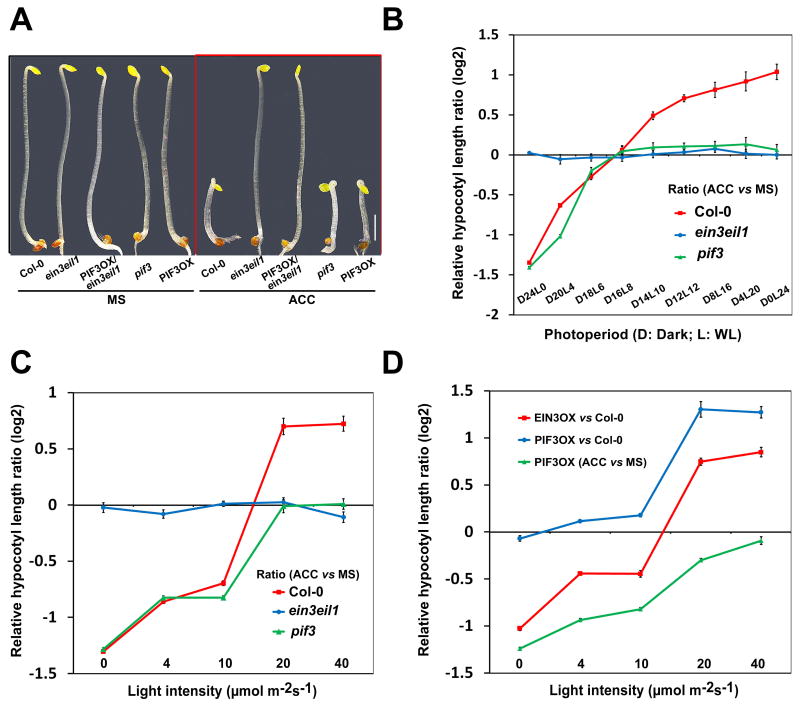
Figure 3. Ethylene-activated PIF3 pathway is progressively fortified with light-induced PIFs protein degradation.
(A) Hypocotyl phenotypes of 3-day-old seedlings grown on MS±10μM ACC.
(B-D) The ratios (in log2) of hypocotyl lengths of 5-day-old seedlings under increasing photoperiods (130 μmol m-2s-1) (B) or intensities of continuous light (C, D) on MS±10μM ACC. Mean ±s.d., n>20.
See also Figure S3.
To address this scenario in more details, we measured the effect of ethylene on hypocotyl elongation in a series of photoperiods starting from complete darkness to continuous light. In WT seedlings, ethylene dramatically repressed hypocotyl elongation in dark, but it gradually turned into a stimulatory signal with increasing light duration (Figure 3B). Expectedly, ein3 eil1 completely abolished any response to ethylene treatment regardless of the photoperiodic conditions (Figure 3B). However, pif3 was found to behave normally as WT in the short photoperiodic conditions under which ethylene was repressive, but entirely failed to respond to ethylene in the long photoperiodic conditions where ethylene was stimulatory for hypocotyl elongation (Figure 3B). The influence of light intensities on ethylene-regulated hypocotyl elongation was also examined. Similarly, ethylene's effect on hypocotyl elongation was progressively switched from repressive to stimulatory with increasing intensity of continuous light (Figure 3C). Likewise, pif3 acted as WT under weak light intensities (<10 μmol m-2s-1), but behaved like ein3 eil1 under strong light intensities (>20 μmol m-2s-1) (Figure 3C). Therefore, EIN3/EIL1 are required for the dual actions of ethylene in hypocotyl elongation: repressive in dark and stimulatory in light, while PIF3 only mediates ethylene's stimulatory function in light. Further support of this notion came from the observations that, with increasing light intensities, the effect of EIN3OX on hypocotyl elongation was gradually reversed from repression to stimulation, while PIF3OX had little effect in dark but became progressively effective on hypocotyl elongation (Figure 3D). Therefore, with increasing light illumination, PIFs redundancy is gradually dampening, and ethylene-induced PIF3 becomes progressively effective in modulating hypocotyl elongation.
Ethylene activates an ERF1-depedent growth-inhibition pathway that is gradually saturated by light
Ethylene application further decreased the hypocotyl length of the pifQm mutant in dark (Figure S3B), implying that ethylene inhibits dark-grown hypocotyl elongation through a separate pathway independent of PIFs. Furthermore, the hypocotyl of PIF3OX was notably inhibited by ACC treatment in dark and dim light, and such ACC-evoked inhibition was progressively attenuated with increasing light intensity (Figure 3D). These results suggest the existence of a PIF3-independent pathway operating to mediate ethylene inhibition of hypocotyl elongation, even the function of PIFs is saturated.
We next sought to further characterize the molecular composition of the ethylene-induced growth-inhibition pathway. Of those ethylene-regulated genes, Ethylene Response Factor 1 (ERF1), an AP2-type transcription factor, represents a good candidate as it was previously reported to be a direct target of EIN3 and suppress hypocotyl elongation [15]. We found that ethylene activated ERF1 gene expression in both dark and light conditions in an EIN3/EIL1-dependent manner (Figure S2A and S4A). Moreover, transgenic overexpression of ERF1 (ERF1OX) led to marked inhibition of hypocotyl elongation in dark (Figure S4B), mimicking the effect of ethylene. However, such inhibition was gradually diminishing with the increase of light intensity, and eventually disappeared in strong light conditions (Figure 4A and 4B), reminiscent of the inhibitory effect of ethylene on PIF3OX (Figure 3D). Therefore, ERF1 mediates an ethylene-activated growth-inhibition pathway that operates effectively in dark and dim light, but hardly in strong light conditions.
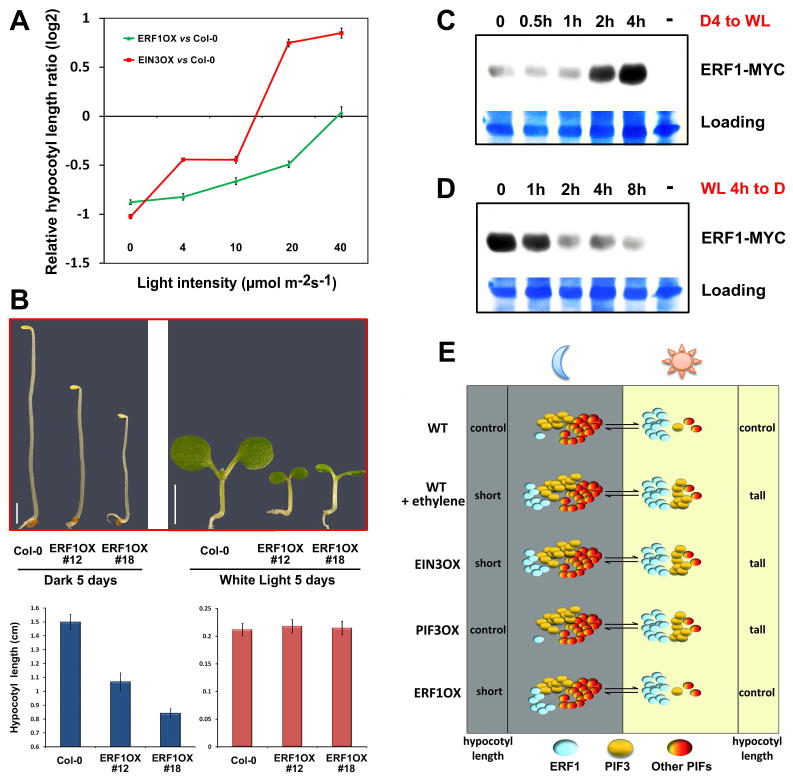
Figure 4. Ethylene activates an ERF1-depedent growth-inhibition pathway that is progressively attenuated by light-stabilized ERF1 protein accumulation.
(A) The ratios (in log2) of hypocotyl lengths of 5-day-old seedlings under increasing intensities of continuous light. Mean ±s.d., n>20.
(B) Phenotype and hypocotyl lengths of 5-day-old dark or continuous light (40μmol m-2s-1) grown seedlings on MS±1μM ACC. Mean ±s.d., n>20.
(C-D) Immunoblot assay of ERF1 protein (by anti-MYC antibody) in 4-day-old etiolated seedlings constitutively over-expressing ERF1-MYC on MS treated with light irradiation (C), or first illuminated for 4 hr then returned to darkness (D).
(E) A schematic diagram illustrating the regulation of PIF and ERF1 proteins by light and ethylene. Light induces PIFs protein degradation but stabilizes ERF1 protein so that PIFs activity is redundant in darkness while ERF1 activity is redundant in light.
See also Figure S4.
Light de-stabilizes PIF3 but stabilizes ERF1 to control the output of ethylene's effect on hypocotyl elongation
We then investigated how light modulates the function of ERF1. In sharp contrast to PIF proteins, we found that ERF1 protein was evidently stabilized when transferring the dark-growth seedlings into light exposure (Figure 4C). Conversely, ERF1 protein became unstable when light-irradiated seedlings were put back to darkness (Figure 4D). ERF1 belongs to a large family of ERFs, most of which contain a highly conserved GCC-box binding domain and some ERFs (e.g. ERF2) are over 50% identical with ERF1 at amino acid level [17]. Consistent with the existence of functional redundancy among multiple ERF proteins, the erf1 and other erf single mutants showed no visibly phenotype compared with WT in light (Figure S4C). Therefore, we conclude that light increasingly stabilizes ERF1 (and probably other ERF proteins) so that the ERFs-mediated hypocotyl inhibition is saturated in strong light condition, providing an explanation for why ethylene-induced ERF1 or transgenic overexpression of ERF1 functions in dark but not in strong light.
Light is previously reported to degrade PIFs by activating phytochromes to directly interact with PIFs, and enhance the accumulation of HY5 by removing COP1/SPA's repression [10, 18]. Our study reveals that ERF1 is dramatically stabilized by light irradiation, suggesting that it might represent a new pathway in mediating light-regulated plant development. The relatively slow kinetics of protein accumulation upon light irradiation (about 2 hr and longer) compared to PIFs (less than 1 hr) implies that photoreceptors are not likely to directly regulate ERF1. Therefore, further studies are worthy to examine whether the COP1/SPA complexes modulate ERF1 protein stability, and whether the light-induced stabilization of ERF1 and other ERF proteins contributes to the establishment of photomorhpogenesis.
Collectively, we propose a model illustrating how light reverses the function of ethylene in hypocotyl elongation (Figure 4E and S4D). Ethylene constantly activates a promoting pathway dependent on PIF3, and an inhibiting pathway mediated by ERF1. Light progressively destabilizes PIF3 and other PIF proteins but stabilizes ERF1 protein. As a result, PIF proteins become limited in light but saturated in dark, whereas ERF1 protein is inadequate in dark but abundant in light. Therefore, ethylene-activated PIF3 accumulation is primarily manifested in light to promote hypocotyl elongation, whereas ethylene-induced ERF1 accumulation is mainly functional in dark to repress hypocotyl elongation. Accordingly, overexpression of EIN3 precisely mimics the ethylene effect in both conditions, but overexpression of PIF3 has an effect merely in light while overexpression of ERF1 is only effective in dark (Figure 4E).
Interestingly, ethylene was also reported to activate two functionally opposite transcription factors in rice plants to help survive different types of flooding: ethylene induces SUB1A transcription to restrain rice growth for energy saving during flash flood [19], whereas under deepwater flood, ethylene stimulates SNORKEL1/2 transcription to promote stem elongation for keeping leaves above water [20]. It is thus fascinating to explore whether analogous strategies are universally utilized by plants to flexibly and effectively respond to various environmental changes.
Experimental Procedures
Supplementary Material
Acknowledgments
We thank Ning Wei for critical reading of the manuscript; Chao Cheng, Mingqiu Dai, Yu Zhang, Tianying Shi, Mantong Zhao, Qiong Zhao and Zheng Wang for experiment assistance; and Fengying An for providing the iE/qm seed [13]. This work was supported by grants from National Natural Science Foundation (91017010), Ministry of Agriculture (2010ZX08010-002), and Ministry of Science and Technology (2009CB119101) of China to H.G; grants from NIH (GM-47850) and NSF (MCB- 0929100) to X.W.D; S.Z. was a Monsanto fellow, and both S.Z. and H.S. are also supported by China Scholarship Council.
Footnotes
Supplemental Information: Supplemental Information includes extended experimental procedures, 4 figures, 1 table and their legends.
References
- 1.Spartz AK, Gray WM. Plant hormone receptors: new perceptions. Genes & Development. 2008;22:2139–2148. doi: 10.1101/gad.1693208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen YF, Etheridge N, Schaller GE. Ethylene signal transduction. Annals of Botany. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casal JJ, Fankhauser C, Coupland G, Blazquez MA. Signalling for developmental plasticity. Trends in Plant Science. 2004;9:309–314. doi: 10.1016/j.tplants.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 5.Ecker JR. THE ETHYLENE SIGNAL-TRANSDUCTION PATHWAY IN PLANTS. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- 6.Smalle J, Haegman M, Kurepa J, VanMontagu M, VanderStraeten D. Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2756–2761. doi: 10.1073/pnas.94.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 8.Alonso JM, Stepanova AN. The ethylene signaling pathway. Science. 2004;306:1513–1515. doi: 10.1126/science.1104812. [DOI] [PubMed] [Google Scholar]
- 9.Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2010 doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. Ethylene-Induced Stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 Is Mediated by Proteasomal Degradation of EIN3 Binding F-Box 1 and 2 That Requires EIN2 in Arabidopsis. Plant Cell. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008;55:821–831. doi: 10.1111/j.1365-313X.2008.03551.x. [DOI] [PubMed] [Google Scholar]
- 15.Solano R, Stepanova A, Chao QM, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes & Development. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosugi S, Ohashi Y. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 2000;28:960–967. doi: 10.1093/nar/28.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau OS, Deng XW. Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol. 2010 doi: 10.1016/j.pbi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Xu K, Xu X, Fukao T, Canias P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 20.Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.