Abstract
Wnt signaling pathways and microRNAs (miRNAs) are critical regulators of development. Aberrant Wnt signaling pathways and miRNA levels lead to developmental defects and diverse human pathologies including but not limited to cancer. Wnt signaling pathways regulate a plethora of cellular processes during embryonic development and maintain homeostasis of adult tissues. A majority of Wnt signaling components are regulated by miRNAs which are small noncoding RNAs that are expressed in both animals and plants. In animal cells, miRNAs fine tune gene expression by pairing primarily to the 3′untranslated region of protein coding mRNAs to repress target mRNA translation and/or induce target degradation. miRNA-mediated regulation of signaling transduction pathways is important in modulating dose-sensitive response of cells to signaling molecules. This review discusses components of the Wnt signaling pathways that are regulated by miRNAs in the context of development and diseases. A fundamental understanding of miRNA functions in Wnt signaling transduction pathways may yield new insight into crosstalks of regulatory mechanisms essential for development and disease pathophysiology leading to novel therapeutics.
Keywords: Post-transcriptional regulation, Regulatory network, β-catenin, Non-canonical Wnt signaling, Cancer
1. Introduction
The Wnt signaling pathways are highly conserved throughout evolution. Wnt signaling can be classified as (a) canonical β-catenin-dependent Wnt pathway and (b) non-canonical β-catenin-independent Wnt/Planar cell polarity (Wnt/PCP) and Wnt/calcium (Ca2+) pathways (Fig. 1A). The Wnt signaling pathways are employed in a plethora of cellular processes during embryonic development and in adult animals. Wnt signaling pathways are tightly regulated at all levels from transcriptional regulation to post-translational modification [1]. Inappropriate Wnt pathway activity results in developmental disorders and diseases, including cancer, skeletal disorders, neuronal diseases, and cardiovascular diseases [1–6]. Recently microRNAs (miRNAs) are found to regulate components of the Wnt signaling pathways (Table 1) [7,8]. In addition, the Wnt signaling cascades have been linked to the production and activity of miRNAs (reviewed in [8]). Thus, both miRNAs and Wnt signaling pathways interact to regulate various biological processes in the developing embryo. This review is divided into four main sections. The first section discusses the current knowledge on Wnt signaling pathways in the context of development. The second section discusses the biogenesis and function of microRNAs. The third and fourth sections of the review summarize miRNAs that are known to target the Wnt pathways in the context of development and disease, respectively.
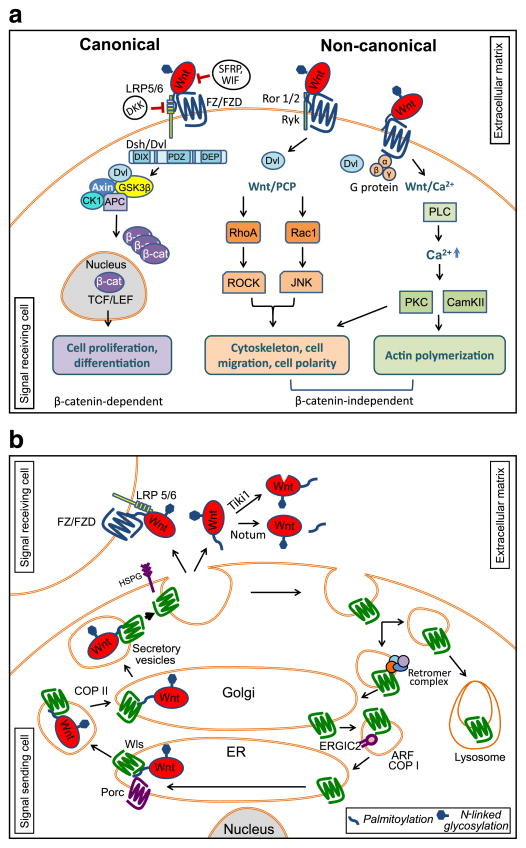
Fig. 1.
Wnt signaling pathways. a) In canonical Wnt pathway, β-catenin is normally degraded by the destruction complex composed of Axin, APC, CK1, and GSK3β. Upon Wnt ligand binding to the frizzled (FZ/FDZ) and LRP5/6 co-receptor, disheveled (Dsh/Dvl) disassembles the β-catenin destruction complex, resulting in an accumulation of β-catenin that can enter the nucleus. In conjunction with TCF/LEF, β-catenin transcriptionally activates target genes. In non-canonical Wnt/Planar cell polarity pathway (Wnt/PCP), its activation is mediated by Wnt ligand binding to the FZ/FDZ and co-receptors Ror1, Ror2, or Ryk. Wnt/PCP is mediated by small GTPases RhoA and Rac1 to activate JNK and ROCK pathways, respectively, resulting in actin remodeling events. In Wnt/Ca2+ pathway, the activation of FZ/FDZ is mediated by disheveled and heterotrimetic G-proteins. Calcium release activates PKC, calcineurin, and CamKII that activates cell migration. b) Wnt proteins are N-glycosylated and lipid modified by the oligosacchary transferase complex and acyltransferase Porcupine within the endoplasmic reticulum (ER), respectively. Wnts are then engaged by Wntless (Wls) for secretory pathway transit and extracellular secretion. Wnt-Wls complex is transferred from ER to Golgi in COP II regulated vesicles, and from the Golgi to extracellular matrix in secretory vesicles [351,352]. Secretion mechanisms of Wnts are not completely understood; however, it has been reported that Heparan Sulfate Proteoglycans (HSPGs) are involved in Wnt stabilization in extracellular matrix [353]. After secretion, the Wnt ligand may be enzymatically inactivated by Tiki1 or Notum. Tiki1 is a protease that cleaves the amino terminal residues of Wnt [141], and Notum removes an essential palmitoleate moiety from Wnt [143]. Functional Wnts are then targeted to their receptors (FZ/FZD) via one of the proposed models mentioned in the text. Following extracellular release of Wnts, Wls is routed to multicellular vesicles for lysosomal degradation or shuttled back to the Golgi by retromer mediated targeting [354–356]. Golgi targeted Wls is transported to the ER by ADP-Ribosylation Factor family proteins (ARF) regulated COP I vesicles and Endoplasmic Reticulum-Golgi Intermediate Compartment protein 2 (ERGIC2) for subsequent rounds of loading ([357]; EMS and SST, unpublished).
Table 1.
miRNAs regulate components of Wnt signaling pathway in various cancers.
| microRNA | Target gene | Type of cancer | Effect | Reference |
|---|---|---|---|---|
| miR-499 | Wnt1, Inducible Signaling Pathway Protein 2 (WISP-2) | Adrenocortical tumors | Tumor suppressor | [326] |
| miR-21 | TGFβR2 | Bladder cancer | Oncogene | [296] |
| miR-144 | EZH2 | Bladder cancer | Oncogene Inhibits SFRP-1, APC |
[327] |
| miR-182-5p | Smad4 | Bladder cancer | Oncogene | [328] |
| miR-493 | FZ4, RhoC | Bladder cancer cell lines | Tumor suppressor Inhibits cell motility |
[329] |
| miR-1862 | CTNNB1, MEK | Bladder cancer | Tumor suppressor | [330,331] |
| miR-200b | PKCα | Bronchial epithelial cell cancer | Tumor suppressor Inhibits Rac1 mediated motility | [332] |
| miR-26a | GSK3β | Cholangiocarcinoma | Oncogene | [333] |
| miR-148a | Wnt10B | Endometrial cancer | Tumor suppressor | [334] |
| miR-141 | Sox-17 | Esophageal cancer | Oncogene | [335] |
| miR-27 | APC | Gastric cancer | Oncogene | [336] |
| miR-218 | LEF1 | Glioblastoma | Tumor suppressor | [337] |
| miR-577 | LRP6, β-catenin | Glioblastoma | Tumor suppressor | [338] |
| miR-155 | CK1α | Liposarcoma | Oncogene | [339] |
| miR-31 | SFRP-4 | Lung cancer | Oncogene | [317] |
| miR-708 | TMEM88 | Lung adenocarcinoma | Oncogene | [340] |
| miR-200a | β-catenin | Meningiomas, nasopharyngeal carcinoma, gastric adenocarcinoma | Tumor suppressor | [276,341,342] |
| miR-200a | ZEB1/SIP1 | Meningiomas | Tumor suppressor | [276] |
| miR-155 | APC | Papillary thyroid carcinoma tumor | Oncogene | [343] |
| miR-15a | Wnt3a | Prostate cancer | Tumor suppressor | [344] |
| miR-16-1 | Wnt3a | Prostate cancer | Tumor suppressor | [344] |
| miR-34a | LEF1 | Prostate cancer | Tumor suppressor | [345] |
| miR-34b/c | β-catenin | Prostate cancer | Tumor suppressor | [346] |
| miR-221 | DVL2 | Prostate cancer | Tumor suppressor Inhibits migration |
[347] |
| miR-320 | β-catenin | Prostate cancer | Tumor suppressor | [348] |
| miR-490 (5p and 3p) | DAAM1 | Thrombocythemia platelets | Tumor suppressor Interacts with DVL that activates Wnt/PCP pathway | [349] |
| miR-483-3p | β-catenin | Urological cancer | Tumor suppressor | [350] |
| miR-1862 | β-catenin | Urological cancer | Tumor suppressor | [350] |
2. Wnt signaling pathways in development
In 1982, Nusse and Varmus identified the mouse proto-oncogene Wnt1 (Int1) in Wnt signaling [9]. Inappropriate expression of Wnt1 induces a transformed phenotype in mammary epithelial cells [10] and in transgenic mice [11]. Injection of Wnt1 mRNA into Xenopus embryos led to duplication of the embryonic axis, revealing its role in the canonical Wnt pathway [12]. In efforts to identify homologs of Wnt1, Moon and coworkers identified Wnt5a in Xenopus. However, when Wnt5a mRNA was injected into Xenopus embryos, it led to developmental defects of the head and tail as a result of cellular movement perturbation which are different than the defects induced by Wnt1. Ectopic expression of the Xenopus Wnt5a in zebrafish embryos caused increased intracellular calcium concentration and the stimulation of calcium signaling phenocopied that of Wnt5a signaling, indicating that Wnt5a is one of the major ligands responsible for non-canonical Wnt signaling [13]. These seminal publications elucidated the critical roles that Wnt signaling pathways play in development.
2.1. The canonical β-catenin-dependent Wnt pathway
Canonical Wnt signaling has been shown to control diverse biological processes and functions, including cellular proliferation and differentiation [14], survival [15], cell fate decisions [16], stem cell maintenance and somatic cell reprogramming [17]. Furthermore, canonical Wnt signaling has been shown to be critical in important embryological events, including axis specification [18] and gastrulation [19], as well as in organogenesis, including development of the breast [20], limb [21], heart [22], central nervous system [23], and bone [24].
In canonical Wnt pathway, the absence of the Wnt ligand leads to rapid phosphorylation of cytoplasmic β-catenin by glycogen synthase kinase 3β (GSK3β) at Ser33, Ser37 and Thr41 [25] and by casein kinase Ia (CK Ia) at Ser45 [26]. GSK3β is found as a part of the destruction complex, which includes Axin and adenomatous polyposis coli (APC) [4] (Fig. 1). The post-translational phosphorylation of β-catenin targets it for ubiquitinylation and subsequent proteasomal degradation [27], preventing its nuclear accumulation. However, binding of canonical Wnt ligand to its corresponding transmembrane receptor frizzled (FZ/FZD) as well as co-receptor LRP5/6 [28] results in recruitment of cytoplasmic protein disheveled (Dsh/Dvl) to the cell membrane. Dsh/Dvl transduces Wnt ligand activation of both canonical Wnt/β-catenin and the non-canonical Wnt signaling pathways [29]. Dsh/Dvl protein has three highly conserved domains, an N-terminal DIX (disheveled, Axin) domain, a central PDZ (postsynaptic density 95, discs large, zonula occludens-1) domain, and a C-terminal DEP (Dvl, Egl-10, Pleckstrin) domain [29]. The Dsh/Dvl DIX domain and its proximal region are important for Dsh/Dvl oligomerization which is required for relay of signal and subsequent stabilization of β-catenin [30]. The binding of FZ to the activated Dsh/Dvl recruits Axin and GSK3β to cell membrane, thereby, dismantling the destruction complex and inhibiting phosphorylation of β-catenin [31]. This results in increased stability of β-catenin, facilitating its nuclear accumulation to one pole of the embryo. Asymmetric localization of nuclear β-catenin is conserved in invertebrate sea urchin species [32,33], starfish Asterina pectinifera [34], ascidian Halocynthia roretzi [35], and vertebrate Xenopus laevis [36]. Together with the Tcf/Lef family of transcription factors, β-catenin activates transcription of several genes involved in diverse biological processes, such as cellular proliferation, apoptosis, and differentiation [14,17].
Besides functioning as a transcriptional co-activator, β-catenin is also a component of the adherens junction [37]. β-catenin links the cadherin molecules to the α-catenin, leading to strong cadherin-mediated cell adhesion [38]. The level of β-catenin in the adhesion complex at the plasma membrane affects the availability of β-catenin functioning as a transcription co-activator in the nucleus [38]. This is demonstrated with experiments in which perturbation of cadherin complexes has an effect on Wnt/β-catenin regulated processes. For example, overexpression of cadherins in Xenopus embryos results in inhibited dorsal axis formation because binding of cadherin to endogenous β-catenin antagonizes β-catenin’s role as a nuclear transcription co-activator [39,40]. Similarly, overexpression of sea urchin cadherin results in depletion of nuclear β-catenin, abrogating endomesodermal cell types [32,41].
Wnt/β-catenin signaling is regulated at many levels, including by secreted proteins that antagonize the Wnt ligand [3]. Among these are secreted frizzled-related proteins (SFRPs) and Wnt inhibitory protein (WIF) that bind to Wnt ligands, preventing interaction between the Wnt ligand with the frizzled receptor [42]. Other Wnt inhibitors include Dikkopf (DKK) and WISE/SOST protein families that bind to LRP5/6 in inhibiting the Wnt signaling pathway [43,44].
In summary, the canonical Wnt/β-catenin signaling pathway is highly conserved for anterior–posterior axis formation, endodermal specification, and organ formation in most animals [39,41,45–48].
2.2. The non-canonical Wnt pathways
The non-canonical Wnt pathways or the β-catenin independent Wnt signaling includes the Wnt/Planar cell polarity (PCP) pathway and the Wnt/calcium (Ca2+) pathway (Fig. 1A). The non-canonical Wnt signaling pathways regulate convergence and extension of the body axis during embryogenesis in Xenopus [49,50], zebrafish [51], and mice [52,53]. In addition, the non-canonical Wnt pathways regulate cell motility which is critical for many cell type establishment and organ development, such as kidney ureteric branching [54], neural crest cell migration [55], neural axon guidance [56,57], neuroectoderm formation [23,58], female gonadal development [59], skeletogenesis [60], and osteogenesis [61].
In the Wnt/PCP signaling pathway, the Wnt ligand binds to non-canonical FZ and its co-receptors to activate two parallel pathways stimulating small GTPases RhoA and Rac1 [62,63]. This signaling is transduced through the PDZ and DEP domains of Dsh/Dvl that directly interact with FZ at the cell membrane [31] and indirectly activate RhoA [64]. Activation of RhoA GTPase turns on the Rho-associated kinase (ROCK) cascade, which in turn results in dynamic cytoskeletal rearrangement, leading to formation of lamellopodia and filopodia-like structures associated with the leading edge during cell migration [62]. Further, ROCK phosphorylates Myosin Light Chain (MLC) phosphatase, which associates with acto-myosin filaments [65]. Wnt/PCP signaling has been shown to be important for the initiation of sea urchin primitive gut (archenteron) invagination by activating RhoA [66,67]. Inhibition of RhoA results in failure to initiate invagination movements; whereas, constitutively active RhoA induced precocious invagination of the archenteron [66]. Similarly, inhibition of ROCK also results in blocking sea urchin gut invagination [66].
The second branch of Wnt/PCP signaling activates the Rac GTPase which in turn stimulates c-Jun N-terminal kinase (JNK) activity, resulting in modification of actin cytoskeleton [68]. Moreover, JNK activation which occurs downstream of RhoA, but independent of ROCK, has been shown to be essential during Xenopus convergent extension movements [69]. JNK phosphorylates focal adhesion adaptor protein paxillin to promote cellular migration. Paxillin and its interacting proteins, such as focal adhesion kinase, c-Src, vinculin, Erk, p38 MAPK, MEK, have all been implicated in cell migration [70].
In the Wnt/Ca2+ pathway, binding of non-canonical Wnt to its cognate receptor FZ/FZD results in activation of heterotrimeric G proteins that regulate phospholipase C (PLC), PDZ, and p38 [71]. This activation of PLC is coupled with short-lived increase in concentration of intracellular second messengers such as inositol 1,4,5-triphosphate (IP3), 1,2-diacylglycerol (DAG), and Ca2+ with the consequences of rapid alteration of the cellular functions [72,73]. Elevated levels of intracellular Ca2+ is sufficient for activation of calcium-sensitive enzymes such as calmodulin dependent protein kinase II (CaMKII), protein kinase C (PKC) [74], calcineurin (Cn) [75] and calpain (a non-lysosomal cysteine protease) [76]. PKC activates small GTPase Cdc42 which remodels actin cytoskeleton and shape cell polarity [77–79]. Activated CaMKII, PKC as well as Cn, mediate gene transcription via NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells), CREB (cAMP response element-binding protein) and NFAT (nuclear factor of activated T cells) transcription factors [62]. NFAT has been shown to regulate transcription of heavy chains of several isoforms of motor protein Myosin (Delling et al., 2000), implicating its role in regulation cellular movements by controlling actomyosin contractility [80]. Using Xenopus embryos, it was shown that expression of truncated Dsh/Dvl possessing only PDZ and DEP domain relayed Wnt signaling predominantly via Wnt/Ca2+ cascade as seen in elevated levels of intracellular Ca2+ and activation of Ca2+-dependent enzymes such as PKC and calcineurin [62]. An overexpression of Dsh-DEP domain in sea urchin embryos manifests a dominant negative effect over endogenous Dsh/Dvl and blocks specifically non-canonical signaling pathways with developmental defects, including a lack of endodermal markers and organized skeleton [60].
Co-receptors for the non-canonical Wnt/PCP pathway include the receptor tyrosine kinases of Ryk and Ror families (Ror1 and Ror2), which are orphan receptors that have been shown to mediate polarized cell migration [81,82]. In development, Ryk (receptor tyrosine kinase-related tyrosine kinase) together with Wnt5 mediate growth cone repulsion in Drosophila commissural axons [83]. In Xenopus, Ryk cooperates with FZ7 and Wnt11 during gastrulation and convergent extension movements [84,85]. As in Drosophila, mammalian Ryk is required for axon guidance and neurite outgrowth [86–90]. Further, Ryk-deficient mice exhibit defects similar to Wnt/PCP mutants, such as the misorientation of stereocilia in the inner ear [91]. In glioma-derived cells, Ryk is important for the Wnt5a-dependent induction of matrix metalloproteinase (MMP-2) that degrades extracellular matrix to facilitate cancer cell motility and increase invasive activity [92]. Thus, Ryk is a pharmacological target for human glioma.
Studies have shown that Ror1 and Ror2 interact genetically to regulate skeletal and cardiovascular systems [93]. Ror2 mutants exhibit similar phenotypes as the Wnt5a mutant mice in that they show dwarfism, facial anomalies, short limbs and tails, and respiratory dysfunction that lead to neonatal lethality [94]. In contrast to Ror2 mutant mice, Ror1 mutant mice do not have apparent morphological abnormalities, yet they die neonatally due to respiratory dysfunction and cyanosis, similar to Wnt5a and Ror2 mutants [93]. Ror1 and Ror2 double mutant mice exhibit enhanced Ror2 mutant phenotypes with skeletal defects, as well as complete transposition of great arteries [93]. In humans, mutation of Ror2 co-receptors results in skeletal disorders dominant brachydactyly type B and recessive Robinow Syndrome [95,96]. In addition, overexpression of Ror1 and Ror2 results in various cancers [97–104]. Ror2 expression is observed in squamous cell carcinoma of the oral cavity, osteosarcoma cell lines, melanoma, renal cell carcinoma, leiomyosarcoma, and gastrointestinal stromal tumor. Ror1 overexpression is involved in hematopoietic malignancies of B cell lineage in humans, neuroblastoma, breast cancer, renal cancer, and lung adenocarcinoma. Because of their role in activating Wnt/PCP pathway to promote cell migration, small molecules and antibody-based therapies against Ror1 and Ror2 are potential targets for cancer therapy [105].
In summary, the non-canonical Wnt/PCP pathway regulates cell polarity in morphogenetic processes [60,106–108], and the Wnt/Ca2+ pathway regulates the transcription of genes controlling cell fate and cell migration [62]. Both branches of the non-canonical Wnt pathways have the ability to mediate actin polymerization in morphogenetic movements which are critical for proper development.
2.3. Processing of Wnt ligand and its importance in development
While signaling molecules within receiving cells are essential for activation of the various Wnt dependent signaling pathways, Wnt processing within ligand producing cells also significantly influences Wnt signaling. The synthesis and movement of Wnt ligands are strictly regulated in the signal producing cells. Wnts, cysteine rich signaling ligands, are synthesized in the rough endoplasmic reticulum (ER) and simultaneously undergo N-glycosylation by oligosaccharyl transferase complex (OST) and disulfide bond formation facilitated by an ER-resident protein GRP78/BiP [109–113]. Within the ER, Wnts (with the exception of the fly-specific WntD) are also palmitoylated by the membrane-bound acyltransferase Porcupine (Porc) [109,114–117]. This post-translational lipid modification renders Wnts hydrophobic and prevents them from moving within the cellular secretory pathway as a soluble protein [118,119]. Modified Wnts are escorted through the secretory pathway to the plasma membrane for extracellular release by a dedicated chaperone protein, Wntless (Wls) [120–122]. The post-translational processing of Wnt ligands is essential, as all 19 human Wnts require palmitoylation by Porcupine and interaction with Wls in order to ensure efficient secretion [123]. The exact mechanism of Wnt extracellular release and targeting to receiving cells has not yet been fully elucidated. However, Wnts have been detected in the extracellular space in a variety of different forms that would enable targeting of otherwise hydrophobic Wnts to receiving cells. These include packaging of Wnts into exosomes and lipoprotein particles, delivery of membrane bound Wnts on membrane processes known as cytonemes, and interaction with extracellular proteins that mask Wnt hydrophobic domains. Indeed, the modality of Wnt extracellular travel may also influence specificity of Wnt pathway activation in the receiving cells [124–131].
Changes in the expression of the factors required for Wnt maturation would be expected to have pleiotropic effects, as they will affect all Wnts. Hence, some specific developmental defects and human diseases have been attributed to disrupted expression of Wnt processing factors. Mutations in human Porcupine result in focal dermal hypoplasia (FDH) that is characterized by skin deformities and various ocular and dental malformations [132,133]. Knockout mutations in porcupine block Wnt signaling, resulting in early embryonic lethality in mice due to gastrulation failure [134,135]. Similarly, the knockdown of porcupine or treatment with C59 (specific inhibitor of Porcupine) results in defective sea urchin endodermal gene regulatory network and apical neurogenic domain [136]. Thus, Wnt processing by Porcupine is requisite for proper Wnt signaling.
Wnt–Wls interaction is crucial for proper development. Disruption of mammalian WLS (Gpr 177) by a lacZ insertion exhibits defective body axis formation [137]. While conditional deletion of WLS caused pancreatic hypoplasia in pancreatic precursor cells, germline deletion of WLS in mice results in failed anterior-posterior axis formation, similar to β-catenin deletion phenotypes [138]. In addition to regulating proper embryonic axis formation, Wnt and Wls levels are also important for maintaining normal tissue homeostasis. Elevated Wnt and Wls protein levels have been observed in patient colorectal cancers, as compared to matched normal colon tissue [139]. Furthermore, knockdown of WNT and WLS in colon cancer cell lines with activating mutations in β-catenin and APC show decreased Wnt reporter activity, suggesting the presence of ligand with its chaperone protein further increases Wnt pathway activation [139].
Secreted Wnt ligand can be inactivated by the catalytic activity of extracellular Wnt inhibitors, such as Tiki1 and Notum [140,141]. Tiki1 is a protease that cleaves amino terminal residues of Wnt proteins [141], whereas Notum has recently been shown to cleave the acyl group from Wnts [142,143]. Both changes prevent Wnt binding to the FZ receptor and downstream signaling. Loss of Notum in Drosophila leads to increased Wnt signaling activity, resulting in abnormal wing growth [140,144]. The critical role of Notum in head development is conserved from planarians to vertebrate [145,146]. In Xenopus embryos, Notum is critical for neural and head induction by acting within the presumptive neuroectoderm [142]. Inactivation of extracellular WNT inhibitors, Tiki1 and Notum, may lead to the development of therapeutic treatments for degenerative diseases caused by WNT deficiency [147].
2.4. Cross regulation of the Wnt signaling pathways
How Wnt signaling achieves specificity remains unclear. The same Wnt ligands or frizzled receptors can activate either the canonical Wnt/β-catenin or the non-canonical Wnt pathways, such as in the case of Wnt 2 [148–150], 3 [151–153], 4 [154,155], 5 [156,157] and FZ4 [156,158], 7 [156,159–162]. Recent studies have shown that one way Wnt signal transduction achieves specificity is by selective activation of the type of Wnt pathway depending on the concentration of Wnt ligands. For example, in primary human articular chondrocytes, high concentration of Wnt3A activates the Wnt/β-catenin pathway, whereas low concentration of Wnt3A activates the Wnt/Ca2+ pathway [163]. Thus, Wnt3 can activate both canonical Wnt/β-catenin and non-canonical signaling in the same cells and turns on different target genes, depending on the level of Wnt3. In the Xenopus embryo, Wnt11 ligand is enriched on the dorsal side of the embryo and present in lower concentrations on the ventral side of the embryo [164]. This Wnt11 gradient results in Wnt11 activating CamKII of the non-canonical Wnt/Ca2+ pathway on the ventral side of the embryo [165], and Wnt11 activating the Wnt/β-catenin pathway for dorsal axis formation [166]. Further, the canonical and non-canonical Wnt pathways are highly interconnected and they cross-regulate each other. For example, the Wnt/Ca2+ pathway has been found to inhibit the Wnt/β-catenin pathway [167–169]. Thus, pattern formation and organogenesis during embryogenesis is achieved by the diversity of Wnt mediated cellular effects, depending on gradients of Wnt ligands and cross regulation of canonical and non-canonical Wnt signaling pathways.
3. microRNAs biogenesis and function in development
miRNAs are non-coding regulatory RNA molecules that are approximately 21–23 nucleotides in length [170]. The first microRNA (miRNA) genes identified were lin-4 and let-7 that control developmental timing in nematodes by modulating the expression of lin-28 and lin-41, respectively, at the post-transcriptional level [171,172]. miRNAs are found in viruses, fungi, plants, and animals that are capable of modulating a large fraction of an organism’s transcriptome [173–176]. miRNAs are capable of regulating thousands of genes that are involved in diverse biological processes, including cellular differentiation and development [177]. Because a single miRNA regulates numerous gene targets, dysregulation of miRNAs is associated with various pathologies such as cancer [178], autoimmune [179], developmental disorders [180], and many others. The miRNA field has grown tremendously and miRNAs are now recognized as critical regulators of gene expression.
3.1. Synthesis and regulatory mechanism of miRNAs
Most miRNAs in the genome are located in the regions that are distant from previously annotated genes, suggesting that they are in an independent transcription unit with a promoter of their own [170]. A minority of miRNAs are derived from the intronic regions of the pre-mRNAs transcribed from the protein coding genomic sequences, suggesting that these miRNAs are dependent on the promoter region of the associated gene and the mRNA splicing mechanisms [181]. Furthermore, some miRNA genes are also found clustered in the genome, indicating their transcription as a multi-cistronic primary transcript [182].
Majority of miRNAs are transcribed as primary miRNA (pri-miRNA) transcripts by the RNA polymerase II, although some are transcribed by RNA polymerase III as well (Bartel, 2004). Most pri-miRNAs are initially processed by Drosha and its cofactor DGCR8 [183,184] in the nucleus to form the pre-miRNAs which are exported into the cytoplasm. Here, the pre-miRNAs are further processed by Dicer, leading to formation of double-stranded RNA fragments (20–25 bp) [185,186]. One strand of the resulting miRNA:miRNA* duplex, also known as the guide strand, is then loaded on to the RNA-induced silencing (RISC) complex [187], forming mature miRNAs that are able to recognize and bind their target mRNAs. The mechanism of strand selection had been reported to be dependent upon the relative free energies of the duplex ends, as the small RNA whose 5′-end exhibits the less stable end is preferentially maintained in the mature silencing complex [187,188]. The passenger strand denoted with an asterisk (miRNA*) is either degraded or in some cases, may become functional miRNA that can target different mRNA [189]. miRNA* species are bona fide trans-regulatory RNA molecules with demonstrable endogenous regulatory effects in D. melanogaster [189]. It was also shown that the miRNA* species are often present at physiologically relevant levels and can associate with Argonaute proteins, similar to their complementary miRNA guide strand. Moreover, using functional assays, the inhibitory activity of miRNA* species in both cultured cells and transgenic animals also validated their functions (Okamura et al., 2008).
A subset of intronic miRNAs, known as mirtrons, is processed independent of Drosha [190–193]. Mirtrons lack the lower hairpin and the flanking single-stranded regions. Instead, they are processed by small nuclear RNAs of the splicing machinery. After debranching of the lariat intron, the spliced introns fold back into a stem-loop structure that obviates the Drosha-mediated processing.
In animals, binding of miRNA to the target mRNA is facilitated by the “seed” or the “core” sequence. The seed sequence is a 6–8 nucleotide stretch present at the 5′ end of the miRNA from positions 2 to 8 and is generally considered necessary and sufficient for miRNA function [194–199]. The miRNA “anchor” sequence (nucleotides 13–16), or 3′-supplementary pairing, plays a modest role in target recognition [173, 200]. Further, a single target mRNA (3′untranslated region in particular) can have more than one different miRNA binding sites, and a single miRNA can also regulate multiple target mRNAs by binding to its corresponding binding site in the 3′UTRs of target mRNAs [195–197,201]. Target seed sites are particularly conserved in the 3′UTR of target mRNAs, suggesting that the predominant regulatory functions of miRNAs are exerted from the 3′UTR [202]. However, recent studies indicate that miRNA binding sites can also occur in the coding sequence [203] as well as in the promoter region of the target gene [204,205].
3.2. miRNAs in developmental processes
Results from various studies have implicated the role of miRNAs in animal development [206–209]. Loss of dicer1, a key enzyme in the processing of miRNAs, is lethal to early mouse development and defects become apparent as early as embryonic day 7.5 [210]. Knockdown of maternal and zygotic Dicer in zebrafish results in aberrant morphogenesis during gastrulation, brain formation, somitogenesis, and heart development [211]. A global depletion of miRNAs by knockdown of Dicer and Drosha in the sea urchin embryo results in early developmental defects, including gastrulation failure and embryonic lethality [212].
miRNAs have also been established as key regulators of cell fate and differentiation [213]. miRNAs may function both as a switch and a fine-tuner of the gene regulatory network [214]. Importance of miRNA lin-4 as a developmental switch during normal temporal regulation of post-embryonic developmental events in Caenorhabditis elegans is well established [171,172]. miRNA lin-4 mediates translational repression of its target mRNA lin-14 which facilitates switching of C. elegans larva from stage L1 to L2 and then to L3.
Studies have also shown that miRNAs play an important role in regulation of cell proliferation and apoptosis during development [215]. Drosophila melanogaster miRNA dme-miR-bantam promotes tissue growth by targeting the apoptotic gene hid, and the dme-miR-2 family targets the proapoptotic genes Grim and Reaper [216].
During embryonic development, cells within the embryos are exposed to numerous ligands and morphogens of several signaling pathways (such as TGFβ, Wnt, Hedgehog, Notch, Hippo, EGF, VEGF, PDGF) to allow tissue induction coordination, patterning, growth and morphogenesis [214]. miRNAs may sharpen morphogen gradients in the developing embryo. For example, miR-15/16 targets Nodal receptor activating receptor type 2A to allow selective expression of Nodal in the most dorsal and anterior part of the embryo in forming the Spemann’s organizer in X. laevis [217,218]. miRNAs can also serve as a positive regulator by amplifying signal strength in duration to allow cell responsiveness to sub-threshold stimuli [7]. This is the case in the RAS-RAF-mitogen activated protein kinase (MAPK) and phosphatidylinositide 3-kinase-AKT cascades where miR-21 enhances in RTK signaling by inhibiting PTEN and Sprouty [219,220] and miR-126 sustains VEGF signaling by suppressing SPRED1 and PIK3R2 mRNAs [221,222].
In summary, during developmental transitions where a tissue-specific miRNA is upregulated and its pool of target genes are downregulated, the effective concentration of the miRNA could greatly increase [214]. The transcription factors of the developmental gene regulatory network, whose protein exerts a switch-like response, may be suppressed in the first place through molecular repression by its regulatory miRNA. In this manner, miRNAs may precisely and promptly fine-tune gene expression thresholds which are an important feature of cell fate decisions [7,214].
4. microRNA regulation of Wnt signaling pathways in the context of development
4.1. Embryonic development
miR-34 has been shown to directly regulate several Wnt/β-catenin components, including β-catenin, Wnt1, Wnt3, LRP6, and LEF1, affecting Xenopus body axis polarity and downstream Wnt-dependent patterning genes [223]. The embryonic axis in early Xenopus embryos is controlled by Wnt/β-catenin signaling [39]. Injection of β-catenin mRNA lacking its UTR into the Xenopus ventral blastomeres resulted in axis duplication, while injection of β-catenin constructs with 790 bp of its 3′UTR (containing miR-34 regulatory sites) exhibited significantly less axial defects [223]. This indicates that miRNA repression of β-catenin is critical for establishing embryonic axis.
In the sea urchin, β-catenin is regulated by at least two miRNAs [224], one of which has the exact same sequence as the mammalian miR-25 which targets mammalian β-catenin [225]. Blocking endogenous miRNA regulation of sea urchin β-catenin resulted in increased transcript levels of Wnt responsive endodermal regulatory genes and affected development of the larval gut and the structure of circumesophageal musculature [224]. Thus, regulation of β-catenin by miRNAs is a conserved regulatory mechanism utilized by developing vertebrate and invertebrate embryos.
4.2. Bone development: osteoblast differentiation
Recently several studies have reported the role of miRNAs in mediating differentiation of mesenchymal stem cells (MSCs) into osteoblasts. MSCs have the ability to self-renewal and differentiate into multiple cell lineages including osteoblasts, chondrocytes, adipocytes, cartilage, tendons, muscle and skin. miRNAs have the potential to drive differentiation from one to the other lineage by targeting TGFβ, BMP, Wnt signaling pathways that are involved in osteoblast differentiation [226–230]. Several miRNAs have been identified to activate the Wnt signaling pathway which promotes osteoblast differentiation. miR-27, miR-142-3p, and miR-135a-5p target APC which leads to accumulation of β-catenin and activation of Wnt signaling to promote osteoblast differentiation in human fetal osteoblastic 1.19 (hFOB1.19) and 3T3-L1 cells [231–233]. miR-29 participates in the fine-tuning of Wnt signaling such that the canonical Wnt signaling induces miR-29a transcription which down regulates inhibitors of canonical Wnt signaling such as DKK1, Kremen2, and SFRP2 to promote osteoblast differentiation in hFOB1.19 cells [234]. DKK1 is also down regulated by miR-335-5p in murine osteoblast-like cells (MC3T3-E1 cells) [235]. miR-218 is another miRNA that directly suppresses SFRP2 and DKK2 which result in enhanced Wnt/β-catenin signaling activity that promote osteogenic differentiation in human adipose-derived stem cells [236]. Further, miR-218 is a part of a feed-forward regulatory circuit such that Wnt/β-catenin signaling activity increases the expression of miR-218 [236]. Previous studies using 3T3-L1 preadipocyte cells and human mesenchymal stem cells demonstrated that miR-709, miR-344, and miR-346 directly inhibit 3′UTR GSK3β, leading to increased β-catenin which represses adipogenesis [237–239].
In contrast, miR-30e inhibits Wnt signaling by inhibiting LPR6 co-receptor of Wnt ligands, promoting adipocyte differentiation in mouse bone marrow stromal cell, mesenchymal cell line, C3H10T1/2, and mouse preadipocyte 3T3-L1 cells [240]. A large scale microarray experiment identified miR-210 to target TCF712 in HEK-293FT cells to promote adipogenesis by repressing Wnt signaling [241]. Similarly, the Drosophila miR-8 (homologous to the mammalian miR-200c/141 or miR200b/a429 cluster) directly inhibits TCF and Wls, resulting in antagonizing Wnt and promotes bone marrow stromal cells to differentiate into adipocytes [242]. An accumulating literature indicates that miRNAs maintain the balance between osteoblast versus adipocyte differentiation as well as osteoblast versus osteoclast differentiation [243]. Thus, miRNAs can potentially be used as a therapeutic treatment of metabolic bone diseases such as osteoporosis.
4.3. Cardiac development
Heart formation and function require precise cardiac gene expression for myogenesis, morphogenesis, and contractility that are controlled by complex regulatory networks at the transcriptional and post-transcriptional levels [244]. miRNAs are important regulators of cardiovascular development, and dysregulation of their expression has been linked to cardiac diseases [245–247]. miR-19b overexpression study revealed its regulation of the left-right symmetry and cardiac development in zebrafish embryo by directly suppressing β-catenin (CTNNB1) of the Wnt signaling pathway [248,249]. The deformed cardiac phenotype was partially rescued by lithium chloride treatment which inhibits GSK3β, leading to an accumulation of β-catenin [248]. miR-499 in rat bone marrow-derived mesenchymal stem cells was able to drive cardiac differentiation as shown by increased expression of cardiac-specific genes (Nkx2.5, GATA4, MEF2C, and cTnl) through Wnt/β-catenin signaling pathway. miR-499’s activation of the Wnt/β-catenin signaling pathway was demonstrated by decreased ratio of phosphorylated/dephosphorylated β-catenin, although its targets within the Wnt pathway have not been directly identified [250]. miR-1 is a key muscle-specific miRNA expressed in cardiac and smooth muscle cells [251,252]. miR-1 promotes differentiation of cardiomyocytes by suppressing the activities of Wnt and FGF signaling pathways [253]. Specifically, miR-1 directly inhibits FZ7 to repress Wnt signaling [253]. Because miRNAs are able to regulate all stages of cardiac development, certain miRNA expression signatures are used for clinical diagnosis of human heart disease [245]. miRNA-based therapeutics for cardiac disease are also in the near future.
4.4. Other organ development
4.4.1. Brain
Increasing evidence indicates the essential role of miRNAs in brain development [254–256]. A conditional knockout using Wnt-1 Cre-mediated loss of Dicer resulted in malformation of midbrain and cerebellum and a failure of neural crest and dopaminergic differentiation [257]. miR-296, -124, and -101 were identified to modulate the GSK3β levels which inhibit the Wnt signaling pathway, using luciferase assays, mutagenesis of miRNA binding sites, and β-catenin activity assays in 293 T cells [257]. Further, the size of the midbrain dopaminergic progenitor cells is regulated by miR-135a2, inhibiting the Wnt1 morphogen in the embryonic midbrain [258]. miR-135a2 suppresses Lmx1b, Ccnd1, GSK3β, and TCF7l2 in an autoregulatory negative feedback loop to control the size of the dopamine progenitor region [258]. In addition to playing a critical role in various aspects of normal brain development, miRNAs are also important for neuronal repair. Previous study indicates that the intact Dicer-dependent miRNA pathway is necessary for functional recovery after peripheral nerve injury in adult mouse [259]. Subsequently, miR-431 was identified to stimulate regenerative axon growth by silencing Kremen1 (DKK1), which is a negative regulator of the Wnt/β-catenin signaling pathway [260].
4.4.2. Testis
Wnt signaling plays an important role in sex determination during gonadal development [261,262]. Perturbation of the Wnt pathway with Drosophila miR-310 to miR-313 cluster deletion antagonizes Wnt signaling pathway activity, resulting in aberrant germ and somatic cell differentiation in the testis [263]. miR-310/313 is the human ortholog hsa-miR-25 that suppresses the Wnt/β-catenin pathway by directly targeting Drosophila β-catenin and TCF in vivo [263].
4.4.3. Lung
Alveolar epithelial cell (AEC) trans-differentiation is a process where AECII trans-differentiates into AECI during lung recovery after injuries. miR-375 overexpression inhibited alveolar epithelial trans-differentiation by targeting the FZ8 of the Wnt/β-catenin pathway [264].
5. microRNA regulation of Wnt signaling pathways in the context of disease
5.1. Heart disease
miRNAs that regulate Wnt/β-catenin signaling pathway have been implicated in contributing to not only normal heart development, but also cardiac dysfunction in disease development. During Coxsackie viral infection, virally induced ERK1/2 activation leads to phosphorylation of ETS-1/2 transcription factors, which induce miR-126 expression [265]. miR-126 inhibits SPRED1, LRP6, WRCH1, to mediate crosstalks between ERK1/2 and Wnt/β-catenin pathways. During Coxsackie viral infection, an upregulation of miR-126 promotes cyto-pathogenicity by stimulating GSK3β and suppressing Wnt/β-catenin signaling, sensitizing the cells to virus-induced cell death and viral progeny release [265]. Also, in patients with Duchenne muscular dystrophy with dystrophin deficiency, miR-199a-5p is elevated in the dystrophic muscle [266]. miR-199a-5p suppresses FZ4, JAG1, and Wnt2 that leads to inhibition of Wnt signaling factors that regulate myogenesis [266].
5.2. Bladder pain syndrome
Human bladder pain syndrome (BPS) is characterized with defects in the proteoglycan core proteins and the tight junction proteins that cause increased urothelial permeability [267,268]. miR-199-5p is highly expressed in the bladder smooth muscle that has previously been found to regulate components of the intercellular junctions [269]. Recently miR-199a-5p was found to directly inhibit Wnt2 or Wnt-dependent cell proliferation and simultaneously activate myocardin and MRTF-A-dependent smooth muscle differentiation [270]. Attenuation of miR-199a-5p causes a significant reduction of the bladder smooth muscle cell size and increased cell proliferation. Thus, the ability of miR199a-5p to inhibit cell proliferation and activate muscle differentiation makes it important for smooth muscle hypertrophy and fibrosis [270].
5.3. Diabetes
Central to the pathophysiology of diabetic nephropathy is mesangial cells (MCs) that undergo phenotypic transition into myofibroblasts [271,272]. Profibrotic growth factor TGF-β1 responds to high glucose levels in glomerular mesangial cells and regulates MC phenotypic changes during diabetic nephropathy progression [273]. miR-215 directly regulates Catenin-β Interacting Protein (CTNNBIP1), which suppresses Wnt/β-catenin signaling that leads to activation of α-SMA and fibronectin expression in TGF-β1 treated mouse mesangial cells (MMCs) [274]. Thus, miR-215 suppresses CTNNBIP1 that promotes the TGF-β1 induced pathogenesis of diabetic nephropathy.
5.4. Various cancers
Both Wnt signaling pathways and miRNAs play essential roles in embryonic development as well as in tissue homeostasis maintenance in adults. Recently microRNAs have been found to regulate tumor-related pathways (reviewed by [275]). As crucial biological regulators, miRNAs suppress targets of the signaling pathway that can act to promote or suppress cancer progression in a context-dependent or target-dependent way. We will discuss examples of miRNA regulation of Wnt signaling pathways that lead to specific types of cancers. Additional miRNAs that regulate Wnt pathways components in other cancers not mentioned in the text are summarized in Table 2.
5.4.1. Brain cancer
miRNAs regulate several types of brain cancer. In meningiomas, one of the most common human brain tumors, miR-200a was found to directly target β-catenin to inhibit cell proliferation [276]. miR-200a also acts as a multifunctional tumor suppressor miRNA that promotes cell adhesion and differentiation by directly inhibiting ZEB and SIP1, leading to increased E-cadherin and cell adhesion [276].
Glioma is a tumor that originates from glial cells in the brain or the spine. It makes up approximately 30% of all brain and central nervous system and 80% of all malignant brain tumors [277]. The majority of known miRNAs that regulate the Wnt pathway in glioma are oncogenic. From miRNA expression profiling of tumor biopsy specimens of human astrocytic gliomas, miR-328 was found to be upregulated in invading glioma cells in vivo and directly suppress SFRP1 [278]. Oncogenic miR-603 directly inhibits WIF1 and CTNNBIP1 and promotes glioma growth [279]. miR-96 was identified to indirectly regulate the Wnt/β-catenin pathway by suppressing HMG-box transcription factor protein 1 (HBP-1) that leads to increased glioma cell proliferation. Importantly, the tumorigenicity of U-87 MG cells was reduced by miR-96 silencing [280]. Oncogenic miR-24 inhibits ST7L tumor suppressor gene that negatively regulates the Wnt/β-catenin pathway [281]. miR-222 and miR-92b target DKK-2 and DKK-3, respectively, that lead to constitutive activation of β-catenin in glioma cells [282,283]. Inhibition of miR-222 leads to decreased tumorigenesis in vivo and serves as a potential target in glioma therapy. In addition, miR-30a-5p directly inhibits PRDM1, a transcriptional regulator that suppresses the proliferation and invasion by glioma cells by inhibiting Wnt/β-catenin signaling [284].
In medulloblastoma cells, overexpression of miR-193a and miR-224 induced highest Wnt pathway specific upregulation, although their specific targets of the Wnt signaling pathway have not been identified [285].
5.4.2. Colorectal cancer
Colorectal cancer is the second leading cause of cancer-related mortality in Western countries [286]. p53 function and activation of canonical Wnt signaling cascades are often dysregulated in cancer. p53 was found to transactivate miR-34 which suppresses the transcriptional activity of β-catenin and TCF/LEF complexes [223]. Loss of p53 or miR-34 contributes to neoplastic progression in colorectal cancer cells. miR-34-mediated suppression of Axin2 results in increased nuclear GSK3β and decreased Snail in colorectal cancer [287]. Leading to a similar effect, tumor suppressor miR-23b directly inhibits FZ7 to halt tumor progression [288]. Tumor suppressor miR-29b down regulates coactivators of β-catenin, including TCF7L2, Snail, and BCL9L, in SW480 cells [289]. Further, ectopic expression of miR-29b inhibits anchorage independent cell growth and promoted EMT in vitro. Targeting the non-canonical Wnt/Ca2+ pathway, miR-224 inhibits Cdc42, resulting in suppression of cell migration [290]. Dependent on tumor grade, miR-101 is differentially expressed such that its expression suppresses cell growth, hypoxic survival and invasive potential [291]. miR-101 was found to repress activation of Wnt/β-catenin signaling pathway and EMT to hamper the aggressive behavior of colorectal cancer in vivo. miR-93 directly targets Smad7 which is important for the nuclear accumulation of β-catenin. Indirectly, miR-93 induces downregulation of β-catenin, Axin, c-Myc, and CyclinD [292]. Furthermore, forced expression of miR-93 resulted in reduced colon cancer invasion, migration, and proliferation. A novel regulatory mechanism was discovered in which tumor suppressor miR-145 directly inhibits catenin δ-1 that results in aberrant translocation of β-catenin through defective nuclear shuttling with p21-activated kinase 4 (PAK4) in colon cancer cells [293]. The inability of β-catenin translocation into the nucleus leads to downregulation of key β-catenin activated genes that promote cell proliferation, such as c-Myc and CyclinD1.
miR135a/b is an oncogenic miRNA that suppresses APC directly to promote β-catenin pathway activation in colorectal cancer [294]. Other oncogenic miRs indirectly target various negative regulators of the Wnt pathways. For example, miR-574-5p suppresses Qki6/7, a RNA family protein, leading to increased expression of β-catenin and p27 [295]. This results in increased proliferation, migration and invasion, and decreased differentiation and cell cycle exit of mouse and human colorectal cancer cells. Importantly, inhibition of miR-574-5p suppresses the growth of tumors in the nude mice [295]. miR-21 enhances colon cancer by directly inhibiting a negative regulator of the Wnt/β-catenin pathway, TGFβ-R2, which indirectly causes decreased levels of β-catenin, c-Myc, and CyclinD1 [296]. Another example is oncogenic miR-7 that inhibits Ying Yang 1 transcription factor, which is a negative regulator of Wnt/β-catenin signaling that down regulates SFRP-1 and APC [297].
5.4.3. Liver cancer
Dysregulation of the Wnt/β-catenin signaling pathway is observed in hepatocellular carcinoma (HCC). miR-214 directly targets β-catenin and the enhancer of zeste homologue2 (EZH2), which also regulates β-catenin [298,299]. miR-214 is a tumor suppressor miRNA that directly and indirectly targets β-catenin to inhibit cell growth by downregulating c-Myc, Cyclin D1, TCF-1, and LEF1 in HCC [299]. Upregulation of miR-139 was observed in HCC cells and demonstrated to directly inhibit TCF4 via luciferase assay [300]. In addition, overexpression of miR-139 suppressed β-catenin signaling and inhibits cell proliferation and invasion, suggesting it as a potential therapeutic for treating HCC patients. miR-122 and miR-148b directly inhibit Wnt1, resulting in reduced β-catenin and c-Myc and prevent HCC cell growth [301,302]. miR-142-3p is found to directly target Rac1, a downstream effector of the non-canonical Wnt/PCP pathway and to inhibit migration and invasion of HCC [303]. Tumor suppressor miR-612 directly inhibits AKT2 accompanied with inhibited epithelial-mesenchymal transition and upregulation of E-cadherin that sequesters β-catenin in the cytoplasm [304,305]. Further, miR-612 level is negatively correlated with TCF/LEF transcriptional activity and transcript levels CyclinD and c-Myc (downstream genes of β-catenin signaling), indicating that miR-612 indirectly inhibits the Wnt/β-catenin pathway [304]. Another tumor suppressor miR-145 targets IRS-1 and IRS-2 in the insulin-like growth factor pathway [306], which is known to cross regulate the Wnt/β-catenin pathway [307,308]. Increased miR-145 expression leads to the reduction of β-catenin protein levels, thus exhibiting its tumor suppressor function in HCC.
miRNAs can also enhance HCC. For example, oncogenic miR-106b and miR-155 were upregulated in hepatoma cells and both directly inhibit APC, resulting in an accumulation of β-catenin that promotes the proliferation of human hepatoma cells [309,310]. Although not directly targeting the Wnt pathway, upregulated oncogenic miR-130a directly inhibits tumor suppressor Runx3 expression which results in activation of Wnt/β-catenin signaling and HCC resistance to cisplatin, a commonly used chemotherapeutic drug used for treatment [311]. Recently, oncogenic miR-153 was identified to regulate a tumor suppressor WWOX, inhibitor of the Wnt/β-catenin signaling pathway [312]. Overexpression of miR-153 leads to increased proliferation in HCC in a murine liver cancer model.
5.4.4. Breast cancer
In addition to regulating HCC, miR-145 targets Mucin, Fascin-1, c-Myc, SMAD2/3, and IGF-1R to inhibit breast cancer tumor cell invasion and indirectly causes downregulation of β-catenin and cadherin11 [313, 314]. miR-31 acts as a tumor suppressor by targeting RhoA to inhibit cell motility [315,316]; however, it also acts as an oncogenic miR by targeting Wnt antagonists DKK1 and indirectly decreases SFRP1, SFRP4, and WIF1 levels [317]. Moreover, tumor suppressor miR-24 inhibits Wnt4, resulting in inhibition of cell motility and EMT [318].
Oncogenic miR-374a is overexpressed in breast cancer cell lines and it promotes epithelial to mesenchymal transition (EMT) and metastasis [319]. miR-374a directly inhibits multiple negative regulators of the Wnt/β-catenin signaling cascade, including Wnt5A (ligand for non-canonical Wnt), PTEN, and WIF1 [319]. Previously oncogenic miR-142 and miR-150 were increased in human breast cancer stem cells (BCSCs) [320]. miR-142 suppresses APC directly and transactivates miR-150 which induces cell growth. Further, knockdown of miR-142 reduced the clonogenicity of BCSCs in vitro and tumor growth in vivo. Interestingly, metastatic breast cancer cells express many osteoblast related genes (osteomimicry) that promote its metastasis to the bone [321]. miRNAs play an important role in regulating osteoblast differentiation during normal bone development [322–324]. For example, miR-218 is significantly upregulated during osteoblast differentiation and targets multiple inhibitors of Wnt signaling [325]. Oncogenic miR-218 found in metastatic breast cancer cells is a potent activator of Wnt signaling that promotes the osteomimicry to facilitate breast cancer cells that metastasize to the bone.
6. Concluding remarks
Increasing evidence indicates the importance of miRNAs in modulating signal transduction pathways (reviewed in [7]). The highly dose-sensitive nature of developmental signaling pathways is under intense miRNA regulation. Current studies indicate that miRNAs participate in signaling networks, both as additional layer of transcriptional control and as feed-forward or feedback devices that result in robustness to the output of cell signaling. Deciphering miRNA function in the context of a developing embryo where crosstalks of miRNAs, signaling pathways and regulatory complexities are taken into account will be critical in revealing the physiological functions of miRNAs.
Many human diseases result from disruption of developmental signaling pathways where miRNAs play a key role in promoting or inhibiting disease progression. Thus, a thorough understanding of miRNA function and miRNA interaction and crosstalk with various signaling pathways will be critical in understanding normal regulation of development and maintenance of tissue homeostasis. A fundamental understanding of miRNA regulation of signal transduction pathways may lead to discovery of novel anti-miRNA-based diagnostics and therapeutics for cancer and other human diseases.
Acknowledgments
We thank feedback from the anonymous reviewers. This project is supported by United States National Institutes of Health NIGMS 5P20GM103653-02 to JLS.
Abbreviations
- Wls
wntless
- Porc
porcupine
- HSPG
heparin sulfate proteoglycans
- ARF
ADP-Ribosylation Factor
- Dsh/Dvl
disheveled
- DIX
disheveled Axin domain
- PDZ
postsynaptic density 95, discslarge, zonulaoccludens-1
- DEP
Dvl, Egl-10, Pleckstrin
- LRP5/6
low-density lipoproteinco-receptor 5 and 6
- FZ/FZD
frizzled receptor
- APC
adenomatous polyposis coli
- LEF1
lymphoid enhancer binding factor 1
- GSK3β
glycogen synthase kinase3B
- SFRPs
secreted frizzled-related proteins
- WIF
Wnt inhibitory protein
- DKK
dikkopf
- ROR
receptorty rosine kinase-like orphan receptor
- Ryk
receptorty rosine kinase-related tyrosine kinase
- MMP-2
matrix metalloproteinase
- Wnt/PCP
Wnt Planar cell polarity
- WGEF
weak similarity GEF
- PLC
phospho-lipase C
- IP3
inositol 1,4,5-triphosphate
- DAG
1,2-diacylglycerol
- PKC
protein kinase C
- CamKII
calmodulin dependent protein kinase II
- Cn
calcineurin
- NFκβ
nuclear factor kappa-light-chain-enhancer of activated B cells
- CREB
cAMP response element-binding protein
- NFAT
nuclear factor of activated T cells
- EMT
epithelial to mesenchymal transition
- DGCR8
DiGeorge syndrome critical region 8
- RISC
RNA induced silencing complex
- miRNA
microRNA
- 3′UTR
3′untranslated region
Footnotes
Conflict of interest statement
The authors declare no conflict of interests.
References
- 1.Clevers H. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. Nat Rev Genet. 2004;5(9):691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H, Nusse R. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald BT, Tamai K, He X. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim W, Kim M, Jho EH. Biochem J. 2013;450(1):9–21. doi: 10.1042/BJ20121284. [DOI] [PubMed] [Google Scholar]
- 6.Anastas JN, Moon RT. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 7.Inui M, Martello G, Piccolo S. Nat Rev Mol Cell Biol. 2010;11(4):252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 8.Schepeler T. Crit Rev Oncog. 2013;18(4):357–371. doi: 10.1615/critrevoncog.2013006128. [DOI] [PubMed] [Google Scholar]
- 9.Nusse R, Varmus HE. Cell. 1982;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 10.Rijsewijk F, van Deemter L, Wagenaar E, Sonnenberg A, Nusse R. EMBO J. 1987;6(1):127–131. doi: 10.1002/j.1460-2075.1987.tb04729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Cell. 1988;55(4):619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 12.Sokol S, Christian JL, Moon RT, Melton DA. Cell. 1991;67(4):741–752. doi: 10.1016/0092-8674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 13.Slusarski DC, Yang-Snyder J, Busa WB, Moon RT. Dev Biol. 1997;182(1):114–120. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- 14.Teo JL, Kahn M. Adv Drug Deliv Rev. 2010;62(12):1149–1155. doi: 10.1016/j.addr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Lin CL, Wang JY, Huang YT, Kuo YH, Surendran K, Wang FS. J Am Soc Nephrol. 2006;17(10):2812–2820. doi: 10.1681/ASN.2005121355. [DOI] [PubMed] [Google Scholar]
- 16.Martin BL, Kimelman D. Dev Cell. 2012;22(1):223–232. doi: 10.1016/j.devcel.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miki T, Yasuda SY, Kahn M. Stem Cell Rev. 2011;7(4):836–846. doi: 10.1007/s12015-011-9275-1. [DOI] [PubMed] [Google Scholar]
- 18.Hikasa H, Sokol SY. Cold Spring Harb Perspect Biol. 2013;5(1):a007955. doi: 10.1101/cshperspect.a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JY, Marston DJ, Walston T, Hardin J, Halberstadt A, Goldstein B. Curr Biol. 2006;16(20):1986–1997. doi: 10.1016/j.cub.2006.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boras-Granic K, Wysolmerski JJ. Organogenesis. 2008;4(2):116–122. doi: 10.4161/org.4.2.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao B, Yang Y. Curr Opin Genet Dev. 2013;23(4):438–444. doi: 10.1016/j.gde.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gessert S, Kuhl M. Circ Res. 2010;107(2):186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- 23.Mulligan KA, Cheyette BN. J Neuroimmune Pharmacol. 2012;7(4):774–787. doi: 10.1007/s11481-012-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day TF, Yang Y. J Bone Joint Surg Am. 2008;90(Suppl 1):19–24. doi: 10.2106/JBJS.G.01174. [DOI] [PubMed] [Google Scholar]
- 25.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. Genes Dev. 1996;10(12):1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 26.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Cell. 2002;108(6):837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 27.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, et al. Curr Biol. 1999;9(4):207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 28.Cadigan KM, Liu YI. J Cell Sci. 2006;119(Pt 3):395–402. doi: 10.1242/jcs.02826. [DOI] [PubMed] [Google Scholar]
- 29.Gao C, Chen YG. Cell Signal. 2010;22(5):717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz-Romond T, Metcalfe C, Bienz M. J Cell Sci. 2007;120(Pt 14):2402–2412. doi: 10.1242/jcs.002956. [DOI] [PubMed] [Google Scholar]
- 31.Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, et al. Mol Cell. 2003;12(5):1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Development. 1999;126(2):345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 33.Weitzel HE, Illies MR, Byrum CA, Xu R, Wikramanayake AH, Ettensohn CA. Development. 2004;131(12):2947–2956. doi: 10.1242/dev.01152. [DOI] [PubMed] [Google Scholar]
- 34.Miyawaki K, Yamamoto M, Saito K, Saito S, Kobayashi N, Matsuda S. Dev Growth Differ. 2003;45(2):121–128. doi: 10.1034/j.1600-0854.2004.00681.x. [DOI] [PubMed] [Google Scholar]
- 35.Kawai N, Iida Y, Kumano G, Nishida H. Dev Dyn. 2007;236(6):1570–1582. doi: 10.1002/dvdy.21169. [DOI] [PubMed] [Google Scholar]
- 36.Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, et al. J Cell Biol. 1997;136(5):1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. J Cell Sci. 1999;112(Pt 8):1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 38.Heuberger J, Birchmeier W. Cold Spring Harb Perspect Biol. 2010;2(2):a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, et al. Cell. 1994;79(5):791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 40.Fagotto F, Funayama N, Gluck U, Gumbiner BM. J Cell Biol. 1996;132(6):1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wikramanayake AH, Huang L, Klein WH. Proc Natl Acad Sci U S A. 1998;95(16):9343–9348. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. J Cell Sci. 2008;121:737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 43.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Nature. 1998;391(6665):357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 44.Semenov M, Tamia K, He X. J Biol Chem. 2005;280:26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 45.Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, et al. Development. 1996;122(10):2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- 46.Kelly GM, Erezyilmaz DF, Moon RT. Mech Dev. 1995;53(2):261–273. doi: 10.1016/0925-4773(95)00442-4. [DOI] [PubMed] [Google Scholar]
- 47.Pelegri F, Maischein HM. Mech Dev. 1998;77(1):63–74. doi: 10.1016/s0925-4773(98)00132-4. [DOI] [PubMed] [Google Scholar]
- 48.Imai K, Takada N, Satoh N, Satou Y. Development. 2000;127(14):3009–3020. doi: 10.1242/dev.127.14.3009. [DOI] [PubMed] [Google Scholar]
- 49.Tada M, Smith JC. Development. 2000;127(10):2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- 50.Wallingford JB, Rowning BA, Vogeli KM, Rothbacher U, Fraser SE, Harland RM. Nature. 2000;405(6782):81–85. doi: 10.1038/35011077. [DOI] [PubMed] [Google Scholar]
- 51.Heisenberg CP, Tada M, Rauch GJ, Saude L, Concha ML, Geisler R, et al. Nature. 2000;405(6782):76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- 52.Kibar Z, Vogan KJ, Groulx N, Justice MJ, Underhill DA, Gros P. Nat Genet. 2001;28(3):251–255. doi: 10.1038/90081. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Hamblet NS, Mark S, Dickinson ME, Brinkman BC, Segil N, et al. Development. 2006;133(9):1767–1778. doi: 10.1242/dev.02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merkel CE, Karner CM, Carroll TJ. Pediatr Nephrol. 2007;22(11):1825–1838. doi: 10.1007/s00467-007-0504-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthews HK, Marchant L, Carmona-Fontaine C, Kuriyama S, Larrain J, Holt MR, et al. Development. 2008;135(10):1771–1780. doi: 10.1242/dev.017350. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Thekdi N, Smallwood PM, Macke JP, Nathans J. J Neurosci. 2002;22(19):8563–8573. doi: 10.1523/JNEUROSCI.22-19-08563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyuksyutova AI, Lu CC, Milanesio N, King LA, Guo N, Wang Y, et al. Science. 2003;302(5652):1984–1988. doi: 10.1126/science.1089610. [DOI] [PubMed] [Google Scholar]
- 58.Range RC, Angerer RC, Angerer LM. PLoS Biol. 2013;11(1):e1001467. doi: 10.1371/journal.pbio.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jordan BK, Shen JH, Olaso R, Ingraham HA, Vilain E. Proc Natl Acad Sci U S A. 2003;100(19):10866–10871. doi: 10.1073/pnas.1834480100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Byrum CA, Xu R, Bince JM, McClay DR, Wikramanayake AH. Dev Dyn. 2009;238(7):1649–1665. doi: 10.1002/dvdy.21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. J Biol Chem. 2007;282(19):14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 62.Komiya Y, Habas R. Organogenesis. 2008;4(2):68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugimura R, Li L. Birth Defects Res C Embryo Today. 2010;90(4):243–256. doi: 10.1002/bdrc.20195. [DOI] [PubMed] [Google Scholar]
- 64.Habas R, Kato Y, He X. Cell. 2001;107(7):843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 65.Heng YW, Koh CG. Int J Biochem Cell Biol. 2010;42(10):1622–1633. doi: 10.1016/j.biocel.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 66.Beane WS, Gross JM, McClay DR. Dev Biol. 2006;292(1):213–225. doi: 10.1016/j.ydbio.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 67.Croce J, Duloquin L, Lhomond G, McClay DR, Gache C. Development. 2006;133(3):547–557. doi: 10.1242/dev.02218. [DOI] [PubMed] [Google Scholar]
- 68.Tada M, Concha ML, Heisenberg CP. Semin Cell Dev Biol. 2002;13(3):251–260. doi: 10.1016/s1084-9521(02)00052-6. [DOI] [PubMed] [Google Scholar]
- 69.Kim GH, Han JK. Dev Dyn. 2005;232(4):958–968. doi: 10.1002/dvdy.20262. [DOI] [PubMed] [Google Scholar]
- 70.Mierke CT. Phys Biol. 2013;10(6):065005. doi: 10.1088/1478-3975/10/6/065005. [DOI] [PubMed] [Google Scholar]
- 71.Ahumada A, Slusarski DC, Liu X, Moon RT, Malbon CC, Wang HY. Science. 2002;298(5600):2006–2010. doi: 10.1126/science.1073776. [DOI] [PubMed] [Google Scholar]
- 72.Kohn AD, Moon RT. Cell Calcium. 2005;38(3–4):439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 73.Ma L, Wang HY. J Biol Chem. 2007;282(39):28980–28990. doi: 10.1074/jbc.M702840200. [DOI] [PubMed] [Google Scholar]
- 74.Sheldahl LC, Slusarski DC, Pandur P, Miller JR, Kuhl M, Moon RT. J Cell Biol. 2003;161(4):769–777. doi: 10.1083/jcb.200211094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rao TP, Kuhl M. Circ Res. 2010;106(12):1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 76.De A. Acta Biochim Biophys Sin (Shanghai) 2011;43(10):745–756. doi: 10.1093/abbs/gmr079. [DOI] [PubMed] [Google Scholar]
- 77.Habas R, Dawid IB, He X. Genes Dev. 2003;17(2):295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlessinger K, McManus EJ, Hall A. J Cell Biol. 2007;178(3):355–361. doi: 10.1083/jcb.200701083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlessinger K, Hall A, Tolwinski N. Genes Dev. 2009;23(3):265–277. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- 80.Rao A, Luo C, Hogan PG. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 81.Minami Y, Oishi I, Endo M, Nishita M. Dev Dyn. 2010;239(1):1–15. doi: 10.1002/dvdy.21991. [DOI] [PubMed] [Google Scholar]
- 82.Green J, Nusse R, van Amerongen R. Cold Spring Harb Perspect Biol. 2014;6(2) doi: 10.1101/cshperspect.a009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshikawa S, McKinnon RD, Kokel M, Thomas JB. Nature. 2003;422(6932):583–588. doi: 10.1038/nature01522. [DOI] [PubMed] [Google Scholar]
- 84.Kim GH, Her JH, Han JK. J Cell Biol. 2008;182(6):1073–1082. doi: 10.1083/jcb.200710188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin S, Baye LM, Westfall TA, Slusarski DC. J Cell Biol. 2010;190(2):263–278. doi: 10.1083/jcb.200912128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y, Shi J, Lu CC, Wang ZB, Lyuksyutova AI, Song XJ, et al. Nat Neurosci. 2005;8(9):1151–1159. doi: 10.1038/nn1520. [DOI] [PubMed] [Google Scholar]
- 87.Keeble TR, Cooper HM. Int J Biochem Cell Biol. 2006;38(12):2011–2017. doi: 10.1016/j.biocel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 88.Lu W, Yamamoto V, Ortega B, Baltimore D. Cell. 2004;119(1):97–108. doi: 10.1016/j.cell.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 89.Keeble TR, Halford MM, Seaman C, Kee N, Macheda M, Anderson RB, et al. J Neurosci. 2006;26(21):5840–5848. doi: 10.1523/JNEUROSCI.1175-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li L, Hutchins BI, Kalil K. J Neurosci. 2009;29(18):5873–5883. doi: 10.1523/JNEUROSCI.0183-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Macheda ML, Sun WW, Kugathasan K, Hogan BM, Bower NI, Halford MM, et al. J Biol Chem. 2012;287(35):29312–29323. doi: 10.1074/jbc.M112.362681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Habu M, Koyama H, Kishida M, Kamino M, Iijima M, Fuchigami T, et al. J Biochem. 2014;156(1):29–38. doi: 10.1093/jb/mvu015. [DOI] [PubMed] [Google Scholar]
- 93.Nomi M, Oishi I, Kani S, Suzuki H, Matsuda T, Yoda A, et al. Mol Cell Biol. 2001;21(24):8329–8335. doi: 10.1128/MCB.21.24.8329-8335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.DeChiara TM, Kimble RB, Poueymirou WT, Rojas J, Masiakowski P, Valenzuela DM, et al. Nat Genet. 2000;24(3):271–274. doi: 10.1038/73488. [DOI] [PubMed] [Google Scholar]
- 95.Oldridge M, Fortuna AM, Maringa M, Propping P, Mansour S, Pollitt C, et al. Nat Genet. 2000;24(3):275–278. doi: 10.1038/73495. [DOI] [PubMed] [Google Scholar]
- 96.Schwabe GC, Trepczik B, Suring K, Brieske N, Tucker AS, Sharpe PT, et al. Dev Dyn. 2004;229(2):400–410. doi: 10.1002/dvdy.10466. [DOI] [PubMed] [Google Scholar]
- 97.Baskar S, Kwong KY, Hofer T, Levy JM, Kennedy MG, Lee E, et al. Clin Cancer Res. 2008;14(2):396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 98.Shabani M, Asgarian-Omran H, Vossough P, Sharifian RA, Faranoush M, Ghragozlou S, et al. Leuk Lymphoma. 2008;49(7):1360–1367. doi: 10.1080/10428190802124000. [DOI] [PubMed] [Google Scholar]
- 99.Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J, et al. PLoS ONE. 2012;7(3):e31127. doi: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rabbani H, Ostadkarampour M, Danesh Manesh AH, Basiri A, Jeddi-Tehrani M, Forouzesh F. Iran Biomed J. 2010;14(3):77–82. [PMC free article] [PubMed] [Google Scholar]
- 101.Wright TM, Brannon AR, Gordan JD, Mikels AJ, Mitchell C, Chen S, et al. Oncogene. 2009;28(27):2513–2523. doi: 10.1038/onc.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Edris B, Espinosa I, Muhlenberg T, Mikels A, Lee CH, Steigen SE, et al. J Pathol. 2012;227(2):223–233. doi: 10.1002/path.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kobayashi M, Shibuya Y, Takeuchi J, Murata M, Suzuki H, Yokoo S, et al. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107(3):398–406. doi: 10.1016/j.tripleo.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 104.Bicocca VT, Chang BH, Masouleh BK, Muschen M, Loriaux MM, Druker BJ, et al. Cancer Cell. 2012;22(5):656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rebagay G, Yan S, Liu C, Cheung NK. Front Oncol. 2012;2:34. doi: 10.3389/fonc.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Axelrod JD. Semin Cell Dev Biol. 2009;20(8):964–971. doi: 10.1016/j.semcdb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 107.Simons M, Mlodzik M. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kikuchi A, Yamamoto H, Sato A, Matsumoto S. Int Rev Cell Mol Biol. 2011;291:21–71. doi: 10.1016/B978-0-12-386035-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 109.Tanaka K, Kitagawa Y, Kadowaki T. J Biol Chem. 2002;277(15):12816–12823. doi: 10.1074/jbc.M200187200. [DOI] [PubMed] [Google Scholar]
- 110.Burrus LW, McMahon AP. Exp Cell Res. 1995;220(2):363–373. doi: 10.1006/excr.1995.1327. [DOI] [PubMed] [Google Scholar]
- 111.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.MacDonald BT, Hien A, Zhang X, Iranloye O, Virshup DM, Waterman ML, et al. J Biol Chem. 2014;289(26):18122–18136. doi: 10.1074/jbc.M114.575027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verras M, Papandreou I, Lim AL, Denko NC. Mol Cell Biol. 2008;28(23):7212–7224. doi: 10.1128/MCB.00947-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, et al. Dev Cell. 2006;11(6):791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 115.Zhai L, Chaturvedi D, Cumberledge S. J Biol Chem. 2004;279(32):33220–33227. doi: 10.1074/jbc.M403407200. [DOI] [PubMed] [Google Scholar]
- 116.Ching W, Hang HC, Nusse R. J Biol Chem. 2008;283(25):17092–17098. doi: 10.1074/jbc.M802059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Komekado H, Yamamoto H, Chiba T, Kikuchi A. Genes Cells. 2007;12(4):521–534. doi: 10.1111/j.1365-2443.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 118.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, et al. Nature. 2003;423(6938):448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 119.Herr P, Basler K. Dev Biol. 2012;361(2):392–402. doi: 10.1016/j.ydbio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 120.Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Cell. 2006;125(3):509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 121.Bartscherer K, Pelte N, Ingelfinger D, Boutros M. Cell. 2006;125(3):523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 122.Goodman RM, Thombre S, Firtina Z, Gray D, Betts D, Roebuck J, et al. Development. 2006;133(24):4901–4911. doi: 10.1242/dev.02674. [DOI] [PubMed] [Google Scholar]
- 123.Najdi R, Proffitt K, Sprowl S, Kaur S, Yu J, Covey TM, et al. Differentiation. 2012;84(2):203–213. doi: 10.1016/j.diff.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gross JC, Chaudhary V, Bartscherer K, Boutros M. Nat Cell Biol. 2012;14(10):1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 125.Koles K, Nunnari J, Korkut C, Barria R, Brewer C, Li Y, et al. J Biol Chem. 2012;287(20):16820–16834. doi: 10.1074/jbc.M112.342667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mulligan KA, Fuerer C, Ching W, Fish M, Willert K, Nusse R. Proc Natl Acad Sci U S A. 2012;109(2):370–377. doi: 10.1073/pnas.1119197109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Neumann S, Coudreuse DY, van der Westhuyzen DR, Eckhardt ER, Korswagen HC, Schmitz G, et al. Traffic. 2009;10(3):334–343. doi: 10.1111/j.1600-0854.2008.00872.x. [DOI] [PubMed] [Google Scholar]
- 128.Greco V, Hannus M, Eaton S. Cell. 2001;106(5):633–645. doi: 10.1016/s0092-8674(01)00484-6. [DOI] [PubMed] [Google Scholar]
- 129.Hsiung F, Ramirez-Weber FA, Iwaki DD, Kornberg TB. Nature. 2005;437(7058):560–563. doi: 10.1038/nature03951. [DOI] [PubMed] [Google Scholar]
- 130.Korkut C, Ataman B, Ramachandran P, Ashley J, Barria R, Gherbesi N, et al. Cell. 2009;139(2):393–404. doi: 10.1016/j.cell.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Nature. 2005;435(7038):58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 132.Grzeschik KH, Bornholdt D, Oeffner F, Konig A, del Carmen Boente M, Enders H, et al. Nat Genet. 2007;39(7):833–835. doi: 10.1038/ng2052. [DOI] [PubMed] [Google Scholar]
- 133.Wang X, Reid Sutton V, Omar Peraza-Llanes J, Yu Z, Rosetta R, Kou YC, et al. Nat Genet. 2007;39(7):836–838. doi: 10.1038/ng2057. [DOI] [PubMed] [Google Scholar]
- 134.Barrott JJ, Cash GM, Smith AP, Barrow JR, Murtaugh LC. Proc Natl Acad Sci U S A. 2011;108(31):12752–12757. doi: 10.1073/pnas.1006437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Biechele S, Cox BJ, Rossant J. Dev Biol. 2011;355(2):275–285. doi: 10.1016/j.ydbio.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 136.Cui M, Siriwon N, Li E, Davidson EH, Peter IS. Proc Natl Acad Sci U S A. 2014;111(47):E5029–E5038. doi: 10.1073/pnas.1419141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fu J, Jiang M, Mirando AJ, Yu HM, Hsu W. Proc Natl Acad Sci U S A. 2009;106(44):18598–18603. doi: 10.1073/pnas.0904894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carpenter AC, Rao S, Wells JM, Campbell K, Lang RA. Genesis. 2010;48(9):554–558. doi: 10.1002/dvg.20651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, et al. Nat Commun. 2013;4:2610. doi: 10.1038/ncomms3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Giraldez AJ, Copley RR, Cohen SM. Dev Cell. 2002;2(5):667–676. doi: 10.1016/s1534-5807(02)00180-6. [DOI] [PubMed] [Google Scholar]
- 141.Zhang X, Abreu JG, Yokota C, MacDonald BT, Singh S, Coburn KL, et al. Cell. 2012;149(7):1565–1577. doi: 10.1016/j.cell.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang X, Cheong SM, Amado NG, Reis AH, MacDonald BT, Zebisch M, et al. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kakugawa S, Langton PF, Zebisch M, Howell SA, Chang TH, Liu Y, et al. Nature. 2015;519(7542):187–192. doi: 10.1038/nature14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gerlitz O, Nellen D, Ottiger M, Basler K. Int J Dev Biol. 2002;46(1):173–176. [PubMed] [Google Scholar]
- 145.Flowers GP, Topczewska JM, Topczewski J. Development. 2012;139(13):2416–2425. doi: 10.1242/dev.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Petersen CP, Reddien PW. Science. 2011;332(6031):852–855. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nusse R. Nature. 2015;519(7542):163–164. doi: 10.1038/nature14208. [DOI] [PubMed] [Google Scholar]
- 148.Teh MT, Blaydon D, Ghali LR, Briggs V, Edmunds S, Pantazi E, et al. J Cell Sci. 2007;120(Pt 2):330–339. doi: 10.1242/jcs.03329. [DOI] [PubMed] [Google Scholar]
- 149.Onizuka T, Yuasa S, Kusumoto D, Shimoji K, Egashira T, Ohno Y, et al. J Mol Cell Cardiol. 2012;52(3):650–659. doi: 10.1016/j.yjmcc.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 150.Karasawa T, Yokokura H, Kitajewski J, Lombroso PJ. J Biol Chem. 2002;277(40):37479–37486. doi: 10.1074/jbc.M205658200. [DOI] [PubMed] [Google Scholar]
- 151.Endo Y, Beauchamp E, Woods D, Taylor WG, Toretsky JA, Uren A, et al. Mol Cell Biol. 2008;28(7):2368–2379. doi: 10.1128/MCB.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Galli LM, Munji RN, Chapman SC, Easton A, Li L, Onguka O, et al. Dev Dyn. 2014;243(6):833–843. doi: 10.1002/dvdy.24123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Varela-Nallar L, Grabowski CP, Alfaro IE, Alvarez AR, Inestrosa NC. Neural Dev. 2009;4:41. doi: 10.1186/1749-8104-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lyons JP, Mueller UW, Ji H, Everett C, Fang X, Hsieh JC, et al. Exp Cell Res. 2004;298(2):369–387. doi: 10.1016/j.yexcr.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 155.Heinonen KM, Vanegas JR, Lew D, Krosl J, Perreault C. PLoS ONE. 2011;6(4):e19279. doi: 10.1371/journal.pone.0019279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Umbhauer M, Djiane A, Goisset C, Penzo-Mendez A, Riou JF, Boucaut JC, et al. EMBO J. 2000;19(18):4944–4954. doi: 10.1093/emboj/19.18.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, et al. Science. 2003;301(5638):1391–1394. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 158.Robitaille J, MacDonald ML, Kaykas A, Sheldahl LC, Zeisler J, Dube MP, et al. Nat Genet. 2002;32(2):326–330. doi: 10.1038/ng957. [DOI] [PubMed] [Google Scholar]
- 159.Hering H, Sheng M. FEBS Lett. 2002;521(1–3):185–189. doi: 10.1016/s0014-5793(02)02831-4. [DOI] [PubMed] [Google Scholar]
- 160.Medina A, Reintsch W, Steinbeisser H. Mech Dev. 2000;92(2):227–237. doi: 10.1016/s0925-4773(00)00240-9. [DOI] [PubMed] [Google Scholar]
- 161.Le Grand F, Jones AE, Seale V, Scime A, Rudnicki MA. Cell Stem Cell. 2009;4(6):535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Development. 2005;132(11):2587–2597. doi: 10.1242/dev.01857. [DOI] [PubMed] [Google Scholar]
- 163.Nalesso G, Sherwood J, Bertrand J, Pap T, Ramachandran M, De Bari C, et al. J Cell Biol. 2011;193(3):551–564. doi: 10.1083/jcb.201011051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schroeder KE, Condic ML, Eisenberg LM, Yost HJ. Dev Biol. 1999;214(2):288–297. doi: 10.1006/dbio.1999.9426. [DOI] [PubMed] [Google Scholar]
- 165.Kuhl M, Sheldahl LC, Malbon CC, Moon RT. J Biol Chem. 2000;275(17):12701–12711. doi: 10.1074/jbc.275.17.12701. [DOI] [PubMed] [Google Scholar]
- 166.Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, et al. Cell. 2005;120(6):857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 167.Slusarski DC, Corces VG, Moon RT. Nature. 1997;390(6658):410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- 168.Kestler HA, Kuhl M. J Cell Biol. 2011;193(3):431–433. doi: 10.1083/jcb.201103167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, et al. Mol Cell Biol. 2003;23(1):131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bartel DP. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 171.Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 172.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 173.Bartel DP. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 175.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Science. 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 176.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 177.He L, Hannon GJ. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 178.Iorio MV, Croce CM. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhu S, Pan W, Qian Y. J Mol Med (Berl) 2013;91(9):1039–1050. doi: 10.1007/s00109-013-1043-z. [DOI] [PubMed] [Google Scholar]
- 180.Bian S, Sun T. Mol Neurobiol. 2011;44(3):359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim VN, Nam JW. Trends Genet. 2006;22(3):165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 182.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 183.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 184.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 185.Lund E, Dahlberg JE. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 186.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. EMBO J. 2002;21(21):5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 188.Khvorova A, Reynolds A, Jayasena SD. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 189.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. Nat Struct Mol Biol. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ruby JG, Jan CH, Bartel DP. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. Cell. 2007;130(1):89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mol Cell. 2007;28(2):328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kim YK, Kim VN. EMBO J. 2007;26(3):775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 195.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 196.Brennecke J, Stark A, Russell RB, Cohen SM. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 198.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, et al. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lai EC. Nat Genet. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 200.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Friedman RC, Farh KK, Burge CB, Bartel DP. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 204.Chekulaeva M, Filipowicz W. Curr Opin Cell Biol. 2009;21(3):452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 205.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. Proc Natl Acad Sci U S A. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Alvarez-Garcia I, Miska EA. Development. 2005;132(21):4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 207.Bushati N, Cohen SM. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 208.Ambros V. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 209.Wienholds E, Plasterk RH. FEBS Lett. 2005;579(26):5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 210.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 211.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 212.Song JL, Stoeckius M, Maaskola J, Friedlander M, Stepicheva N, Juliano C, et al. Dev Biol. 2012;362(1):104–113. doi: 10.1016/j.ydbio.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ivey KN, Srivastava D. Cell Stem Cell. 2010;7(1):36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 214.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. Nat Genet. 2011;43(9):854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jovanovic M, Hengartner MO. Oncogene. 2006;25(46):6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 216.Stark A, Brennecke J, Russell RB, Cohen SM. PLoS Biol. 2003;1(3):E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, et al. Nature. 2007;449(7159):183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 218.Niehrs C. Nat Rev Genet. 2004;5(6):425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- 219.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 221.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. Dev Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, et al. Development. 2008;135(24):3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 223.Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, et al. Sci Signal. 2011;4(197):ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Stepicheva N, Nigam PA, Siddam A, Peng CF, Song JL. Dev Biol. 2015 doi: 10.1016/j.ydbio.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Anton R, Chatterjee SS, Simundza J, Cowin P, Dasgupta R. PLoS ONE. 2011;6(10):e26257. doi: 10.1371/journal.pone.0026257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bianco P, Riminucci M, Gronthos S, Robey PG. Stem Cells. 2001;19(3):180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 227.Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, et al. Proc Natl Acad Sci U S A. 2009;106(49):20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kang H, Hata A. BMB Rep. 2014 doi: 10.5483/BMBRep.2015.48.6.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lin GL, Hankenson KD. J Cell Biochem. 2011;112(12):3491–3501. doi: 10.1002/jcb.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Vimalraj S, Selvamurugan N. Curr Issues Mol Biol. 2012;15(1):7–18. [PubMed] [Google Scholar]
- 231.Wang T, Xu Z. Biochem Biophys Res Commun. 2010;402(2):186–189. doi: 10.1016/j.bbrc.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 232.Hu W, Ye Y, Zhang W, Wang J, Chen A, Guo F. Mol Med Rep. 2013;7(2):689–693. doi: 10.3892/mmr.2012.1207. [DOI] [PubMed] [Google Scholar]
- 233.Chen C, Peng Y, Peng J, Jiang S. J Mol Endocrinol. 2014;52(3):311–320. doi: 10.1530/JME-14-0013. [DOI] [PubMed] [Google Scholar]
- 234.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. J Biol Chem. 2010;285(33):25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, et al. J Bone Miner Res. 2011;26(8):1953–1963. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhang WB, Zhong WJ, Wang L. Bone. 2014;58:59–66. doi: 10.1016/j.bone.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 237.Chen H, Mo D, Li M, Zhang Y, Chen L, Zhang X, et al. Cell Signal. 2014;26(11):2583–2589. doi: 10.1016/j.cellsig.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 238.Chen H, Wang S, Chen L, Chen Y, Wu M, Zhang Y, et al. FEBS Lett. 2014;588(3):429–435. doi: 10.1016/j.febslet.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 239.Wang Q, Cai J, Cai XH, Chen L. PLoS ONE. 2013;8(9):e72266. doi: 10.1371/journal.pone.0072266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H. J Biol Chem. 2012;287(10):7503–7511. doi: 10.1074/jbc.M111.292722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Qin L, Chen Y, Niu Y, Chen W, Wang Q, Xiao S, et al. BMC Genomics. 2010;11:320. doi: 10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kennell JA, Gerin I, MacDougald OA, Cadigan KM. Proc Natl Acad Sci U S A. 2008;105(40):15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Arfat Y, Xiao WZ, Ahmad M, Zhao F, Li DJ, Sun YL, et al. Curr Med Chem. 2014 doi: 10.2174/0929867321999141106121227. [DOI] [PubMed] [Google Scholar]
- 244.Olson EN. Science. 2006;313(5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Chen J, Wang DZ. J Mol Cell Cardiol. 2012;52(5):949–957. doi: 10.1016/j.yjmcc.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 247.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, et al. Circ Res. 2009;105(6):585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Li M, Hu X, Zhu J, Zhu C, Zhu S, Liu X, et al. Cell Physiol Biochem. 2014;33(6):1988–2002. doi: 10.1159/000362975. [DOI] [PubMed] [Google Scholar]
- 249.Qin DN, Qian L, Hu DL, Yu ZB, Han SP, Zhu C, et al. Cell Biochem Biophys. 2013;66(3):709–722. doi: 10.1007/s12013-013-9516-9. [DOI] [PubMed] [Google Scholar]
- 250.Zhang LL, Liu JJ, Liu F, Liu WH, Wang YS, Zhu B, et al. Biochem Biophys Res Commun. 2012;420(4):875–881. doi: 10.1016/j.bbrc.2012.03.092. [DOI] [PubMed] [Google Scholar]
- 251.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kwon C, Han Z, Olson EN, Srivastava D. Proc Natl Acad Sci U S A. 2005;102(52):18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lu TY, Lin B, Li Y, Arora A, Han L, Cui C, et al. J Mol Cell Cardiol. 2013;63C:146–154. doi: 10.1016/j.yjmcc.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sun E, Shi Y. Exp Neurol. 2014 doi: 10.1016/j.expneurol.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 255.Follert P, Cremer H, Beclin C. Front Mol Neurosci. 2014;7:5. doi: 10.3389/fnmol.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Petri R, Malmevik J, Fasching L, Akerblom M, Jakobsson J. Exp Cell Res. 2014;321(1):84–89. doi: 10.1016/j.yexcr.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 257.Liu Y, Huang T, Zhao X, Cheng L. Biochem Biophys Res Commun. 2011;408(2):259–264. doi: 10.1016/j.bbrc.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 258.Anderegg A, Lin HP, Chen JA, Caronia-Brown G, Cherepanova N, Yun B, et al. PLoS Genet. 2013;9(12):e1003973. doi: 10.1371/journal.pgen.1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wu D, Raafat A, Pak E, Clemens S, Murashov AK. Exp Neurol. 2012;233(1):555–565. doi: 10.1016/j.expneurol.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wu D, Murashov AK. Front Mol Neurosci. 2013;6:35. doi: 10.3389/fnmol.2013.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kim Y, Capel B. Dev Dyn. 2006;235(9):2292–2300. doi: 10.1002/dvdy.20894. [DOI] [PubMed] [Google Scholar]
- 262.Bernard P, Harley VR. Int J Biochem Cell Biol. 2007;39(1):31–43. doi: 10.1016/j.biocel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 263.Pancratov R, Peng F, Smibert P, Yang S, Jr, Olson ER, Guha-Gilford C, et al. Development. 2013;140(14):2904–2916. doi: 10.1242/dev.092817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang Y, Huang C, Reddy Chintagari N, Bhaskaran M, Weng T, Guo Y, et al. Nucleic Acids Res. 2013;41(6):3833–3844. doi: 10.1093/nar/gks1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ye X, Hemida MG, Qiu Y, Hanson PJ, Zhang HM, Yang D. Cell Mol Life Sci. 2013;70(23):4631–4644. doi: 10.1007/s00018-013-1411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Alexander MS, Kawahara G, Motohashi N, Casar JC, Eisenberg I, Myers JA, et al. Cell Death Differ. 2013;20(9):1194–1208. doi: 10.1038/cdd.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Slobodov G, Feloney M, Gran C, Kyker KD, Hurst RE, Culkin DJ. J Urol. 2004;171(4):1554–1558. doi: 10.1097/01.ju.0000118938.09119.a5. [DOI] [PubMed] [Google Scholar]
- 268.Sanchez Freire V, Burkhard FC, Kessler TM, Kuhn A, Draeger A, Monastyrskaya K. Am J Pathol. 2010;176(1):288–303. doi: 10.2353/ajpath.2010.090552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Monastyrskaya K, Sanchez-Freire V, Hashemi Gheinani A, Klumpp DJ, Babiychuk EB, Draeger A, et al. Am J Pathol. 2013;182(2):431–448. doi: 10.1016/j.ajpath.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 270.Hashemi Gheinani A, Burkhard FC, Rehrauer H, Aquino Fournier C, Monastyrskaya K. J Biol Chem. 2015 doi: 10.1074/jbc.M114.618694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Simonson MS. Kidney Int. 2007;71(9):846–854. doi: 10.1038/sj.ki.5002180. [DOI] [PubMed] [Google Scholar]
- 272.Mishra R, Cool BL, Laderoute KR, Foretz M, Viollet B, Simonson MS. J Biol Chem. 2008;283(16):10461–10469. doi: 10.1074/jbc.M800902200. [DOI] [PubMed] [Google Scholar]
- 273.Mishra R, Simonson MS. Arterioscler Thromb Vasc Biol. 2008;28(3):541–547. doi: 10.1161/ATVBAHA.107.157339. [DOI] [PubMed] [Google Scholar]
- 274.Mu J, Pang Q, Guo YH, Chen JG, Zeng W, Huang YJ, et al. PLoS ONE. 2013;8(3):e58622. doi: 10.1371/journal.pone.0058622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sun X, He Y, Huang C, Ma TT, Li J. Cell Signal. 2013;25(12):2805–2811. doi: 10.1016/j.cellsig.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 276.Saydam O, Shen Y, Wurdinger T, Senol O, Boke E, James MF, et al. Mol Cell Biol. 2009;29(21):5923–5940. doi: 10.1128/MCB.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Goodenberger ML, Jenkins RB. Cancer Genet. 2012;205(12):613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 278.Delic S, Lottmann N, Stelzl A, Liesenberg F, Wolter M, Gotze S, et al. Neuro Oncol. 2014;16(2):179–190. doi: 10.1093/neuonc/not164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Guo M, Zhang X, Wang G, Sun J, Jiang Z, Khadarian K, et al. Cancer Lett. 2015 doi: 10.1016/j.canlet.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 280.Yan Z, Wang J, Wang C, Jiao Y, Qi W, Che S. FEBS Lett. 2014;588(17):3038–3046. doi: 10.1016/j.febslet.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 281.Chen L, Zhang A, Li Y, Zhang K, Han L, Du W, et al. Cancer Lett. 2013;329(2):174–180. doi: 10.1016/j.canlet.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 282.Li Q, Shen K, Zhao Y, He X, Ma C, Wang L, et al. FEBS Lett. 2013;587(12):1742–1748. doi: 10.1016/j.febslet.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 283.Li Q, Shen K, Zhao Y, Ma C, Liu J, Ma J. J Transl Med. 2013;11:302. doi: 10.1186/1479-5876-11-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Wang X, Wang K, Han L, Zhang A, Shi Z, Zhang K, et al. Cancer Lett. 2013;331(2):211–219. doi: 10.1016/j.canlet.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 285.Gokhale A, Kunder R, Goel A, Sarin R, Moiyadi A, Shenoy A, et al. J Cancer Res Ther. 2010;6(4):521–529. doi: 10.4103/0973-1482.77072. [DOI] [PubMed] [Google Scholar]
- 286.Available from: http://www.cancer.org/acs/groups/content/documents/document/acspc-042280.pdf2014.
- 287.Kim NH, Cha YH, Kang SE, Lee Y, Lee I, Cha SY, et al. Cell Cycle. 2013;12(10):1578–1587. doi: 10.4161/cc.24739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, et al. Nat Commun. 2011;2:554. doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 289.Subramanian M, Rao SR, Thacker P, Chatterjee S, Karunagaran D. J Cell Biochem. 2014;115(11):1974–1984. doi: 10.1002/jcb.24869. [DOI] [PubMed] [Google Scholar]
- 290.Ke TW, Hsu HL, Wu YH, Chen WT, Cheng YW, Cheng CW. Dis Markers. 2014;2014:617150. doi: 10.1155/2014/617150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L, et al. J Pathol. 2013;229(3):379–389. doi: 10.1002/path.4097. [DOI] [PubMed] [Google Scholar]
- 292.Tang Q, Zou Z, Zou C, Zhang Q, Huang R, Guan X, et al. Tumour Biol. 2014 doi: 10.1007/s13277-014-2771-6. [DOI] [PubMed] [Google Scholar]
- 293.Yamada N, Noguchi S, Mori T, Naoe T, Maruo K, Akao Y. Cancer Lett. 2013;335(2):332–342. doi: 10.1016/j.canlet.2013.02.060. [DOI] [PubMed] [Google Scholar]
- 294.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, et al. Cancer Res. 2008;68(14):5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 295.Ji S, Ye G, Zhang J, Wang L, Wang T, Wang Z, et al. Gut. 2013;62(5):716–726. doi: 10.1136/gutjnl-2011-301083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, et al. Carcinogenesis. 2012;33(1):68–76. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT, et al. Oncogene. 2013;32(42):5078–5088. doi: 10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]
- 298.Xia H, Ooi LL, Hui KM. PLoS ONE. 2012;7(9):e44206. doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Wang X, Chen J, Li F, Lin Y, Zhang X, Lv Z, et al. Biochem Biophys Res Commun. 2012;428(4):525–531. doi: 10.1016/j.bbrc.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 300.Gu W, Li X, Wang J. Oncol Rep. 2014;31(1):397–404. doi: 10.3892/or.2013.2831. [DOI] [PubMed] [Google Scholar]
- 301.Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, et al. Liver Int. 2012;32(5):752–760. doi: 10.1111/j.1478-3231.2011.02750.x. [DOI] [PubMed] [Google Scholar]
- 302.Zhang JG, Shi Y, Hong DF, Song M, Huang D, Wang CY, et al. Sci Rep. 2015;5:8087. doi: 10.1038/srep08087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Wu L, Cai C, Wang X, Liu M, Li X, Tang H. FEBS Lett. 2011;585(9):1322–1330. doi: 10.1016/j.febslet.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 304.Tang J, Tao ZH, Wen D, Wan JL, Liu DL, Zhang S, et al. Biochem Biophys Res Commun. 2014;447(1):210–215. doi: 10.1016/j.bbrc.2014.03.135. [DOI] [PubMed] [Google Scholar]
- 305.Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, et al. J Exp Med. 2013;210(4):789–803. doi: 10.1084/jem.20120153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Law PT, Ching AK, Chan AW, Wong QW, Wong CK, To KF, et al. Carcinogenesis. 2012;33(6):1134–1141. doi: 10.1093/carcin/bgs130. [DOI] [PubMed] [Google Scholar]
- 307.Dearth RK, Cui X, Kim HJ, Kuiatse I, Lawrence NA, Zhang X, et al. Mol Cell Biol. 2006;26(24):9302–9314. doi: 10.1128/MCB.00260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Longato L, de la Monte S, Kuzushita N, Horimoto M, Rogers AB, Slagle BL, et al. Hepatology. 2009;49(6):1935–1943. doi: 10.1002/hep.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Shen G, Jia H, Tai Q, Li Y, Chen D. Carcinogenesis. 2013;34(1):211–219. doi: 10.1093/carcin/bgs320. [DOI] [PubMed] [Google Scholar]
- 310.Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, et al. Hepatology. 2012;56(5):1631–1640. doi: 10.1002/hep.25849. [DOI] [PubMed] [Google Scholar]
- 311.Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q, et al. Biochem Biophys Res Commun. 2012;425(2):468–472. doi: 10.1016/j.bbrc.2012.07.127. [DOI] [PubMed] [Google Scholar]
- 312.Hua HW, Jiang F, Huang Q, Liao Z, Ding G. Oncotarget. 2015 doi: 10.18632/oncotarget.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Sachdeva M, Mo YY. Cancer Res. 2010;70(1):378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW, Nam SJ, et al. J Control Release. 2011;155(3):427–434. doi: 10.1016/j.jconrel.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 315.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 316.Valastyan S, Weinberg RA. Cell Cycle. 2010;9(11) doi: 10.4161/cc.9.11.11843. [DOI] [PubMed] [Google Scholar]
- 317.Xi S, Yang M, Tao Y, Xu H, Shan J, Inchauste S, et al. PLoS ONE. 2010;5(10):e13764. doi: 10.1371/journal.pone.0013764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.De Menna M, D’Amato V, Ferraro A, Fusco A, Di Lauro R, Garbi C, et al. Oncogene. 2013;32(35):4110–4119. doi: 10.1038/onc.2012.419. [DOI] [PubMed] [Google Scholar]
- 319.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. J Clin Invest. 2013;123(2):566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Isobe T, Hisamori S, Hogan DJ, Zabala M, Hendrickson DG, Dalerba P, et al. Elife. 2014;3 doi: 10.7554/eLife.01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Weilbaecher KN, Guise TA, McCauley LK. Nat Rev Cancer. 2011;11(6):411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, et al. Proc Natl Acad Sci U S A. 2010;107(46):19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, et al. Nat Rev Endocrinol. 2012;8(4):212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, et al. Proc Natl Acad Sci U S A. 2008;105(37):13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, et al. J Biol Chem. 2012;287(50):42084–42092. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Iliopoulos D, Bimpaki EI, Nesterova M, Stratakis CA. Cancer Res. 2009;69(8):3278–3282. doi: 10.1158/0008-5472.CAN-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, et al. FEBS J. 2013;280(18):4531–4538. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 328.Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, et al. PLoS ONE. 2012;7(11):e51056. doi: 10.1371/journal.pone.0051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ueno K, Hirata H, Majid S, Yamamura S, Shahryari V, Tabatabai ZL, et al. Mol Cancer Ther. 2012;11(1):244–253. doi: 10.1158/1535-7163.MCT-11-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Hirata H, Hinoda Y, Ueno K, Nakajima K, Ishii N, Dahiya R. Carcinogenesis. 2012;33(3):501–508. doi: 10.1093/carcin/bgr302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Hirata H, Hinoda Y, Ueno K, Shahryari V, Tabatabai ZL, Dahiya R. Carcinogenesis. 2012;33(1):41–48. doi: 10.1093/carcin/bgr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Wang Z, Humphries B, Xiao H, Jiang Y, Yang C. J Biol Chem. 2014;289(26):18373–18386. doi: 10.1074/jbc.M114.554246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zhang J, Han C, Wu T. Gastroenterology. 2012;143(1):246–56 e8. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Aprelikova O, Palla J, Hibler B, Yu X, Greer YE, Yi M, et al. Oncogene. 2013;32(27):3246–3253. doi: 10.1038/onc.2012.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Jia Y, Yang Y, Zhan Q, Brock MV, Zheng X, Yu Y, et al. J Mol Diagn. 2012;14(6):577–585. doi: 10.1016/j.jmoldx.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 336.Zhang Z, Liu S, Shi R, Zhao G. Cancer Genet. 2011;204(9):486–491. doi: 10.1016/j.cancergen.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 337.Liu Y, Yan W, Zhang W, Chen L, You G, Bao Z, et al. Oncol Rep. 2012;28(3):1013–1021. doi: 10.3892/or.2012.1902. [DOI] [PubMed] [Google Scholar]
- 338.Zhang W, Shen C, Li C, Yang G, Liu H, Chen X, et al. Mol Carcinog. 2015 doi: 10.1002/mc.22304. [DOI] [PubMed] [Google Scholar]
- 339.Zhang P, Bill K, Liu J, Young E, Peng T, Bolshakov S, et al. Cancer Res. 2012;72(7):1751–1762. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H, Park CH, et al. Clin Cancer Res. 2012;18(13):3658–3667. doi: 10.1158/1078-0432.CCR-11-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Xia H, Ng SS, Jiang S, Cheung WK, Sze J, Bian XW, et al. Biochem Biophys Res Commun. 2010;39(1):535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 342.Cong N, Du P, Zhang A, Shen F, Su J, Pu P, et al. Oncol Rep. 2013;29(4):1579–1587. doi: 10.3892/or.2013.2267. [DOI] [PubMed] [Google Scholar]
- 343.Zhang X, Li M, Zuo K, Li D, Ye M, Ding L, et al. J Clin Endocrinol Metab. 2013;98(8):E1305–E1313. doi: 10.1210/jc.2012-3602. [DOI] [PubMed] [Google Scholar]
- 344.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 345.Liang J, Li Y, Daniels G, Sfanos K, De Marzo A, Wei J, et al. Mol Cancer Res. 2015 doi: 10.1158/1541-7786.MCR-14-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Liu H, Yin J, Wang H, Jiang G, Deng M, Zhang G, et al. Cell Signal. 2015 doi: 10.1016/j.cellsig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 347.Zheng C, Yinghao S, Li J. Med Oncol. 2012;29(2):815–822. doi: 10.1007/s12032-011-9934-8. [DOI] [PubMed] [Google Scholar]
- 348.Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, et al. Carcinogenesis. 2013;34(3):530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 349.Xu X, Gnatenko DV, Ju J, Hitchcock IS, Martin DW, Zhu W, et al. Blood. 2012;120(17):3575–3585. doi: 10.1182/blood-2012-02-411264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Veronese A, Visone R, Consiglio J, Acunzo M, Lupini L, Kim T, et al. Proc Natl Acad Sci U S A. 2011;108(12):4840–4845. doi: 10.1073/pnas.1101734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Buechling T, Chaudhary V, Spirohn K, Weiss M, Boutros M. EMBO Rep. 2011;12(12):1265–1272. doi: 10.1038/embor.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Port F, Hausmann G, Basler K. EMBO Rep. 2011;12(11):1144–1152. doi: 10.1038/embor.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Fuerer C, Habib SJ, Nusse R. Dev Dyn. 2010;239(1):184–190. doi: 10.1002/dvdy.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, et al. Dev Cell. 2008;14(1):120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 355.Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, Itasaki N, et al. Nat Cell Biol. 2008;10(2):170–177. doi: 10.1038/ncb1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Port F, Kuster M, Herr P, Furger E, Banziger C, Hausmann G, et al. Nat Cell Biol. 2008;10(2):178–185. doi: 10.1038/ncb1687. [DOI] [PubMed] [Google Scholar]
- 357.Yu J, Chia J, Canning CA, Jones CM, Bard FA, Virshup DM. Dev Cell. 2014;29(3):277–291. doi: 10.1016/j.devcel.2014.03.016. [DOI] [PubMed] [Google Scholar]