Abstract
The high-mobility group (HMG) domain containing proteins regulate transcription, DNA replication and recombination. They adopt L-shaped folds and are structure-specific DNA binding motifs. Here, I define the L-motif super-family that consists of DNA-binding HMG-box proteins and the L-motif of the histone mRNA binding domain of Stem-Loop Binding Protein (SLBP). The SLBP L-motif and HMG-box domains adopt similar L-shaped folds with three α-helices and two or three small hydrophobic cores that stabilize the overall fold, but have very different and distinct modes of nucleic acid recognition. A comparison of the structure, dynamics, protein-protein and nucleic acid interactions, and regulation by PTMs of the SLBP and the HMG-box L-motifs reveals the versatile and diverse modes by which L-motifs utilize their surfaces for structure-specific recognition of nucleic acids to regulate gene expression.
Keywords: HMG-box domain, Stem-Loop Binding Protein (SLBP), RNA binding domain, phosphorylation, DNA binding domain, protein-nucleic acid interaction
1. INTRODUCTION
Nucleic acid binding motifs in proteins generally recognize specific sequences in the cognate RNA or DNA using modular domains. While the structural basis for sequence-specific recognition of DNA and RNA is easier to understand, it is quite challenging to understand the mechanism of structure-specific recognition or “shape readout” of both DNA and RNA (1). Tertiary interactions between secondary structure elements in RNAs such as the tetraloop-receptor interaction (2), A-minor interactions (3), kissing hairpin loops (4), pseudoknots (5), ribose zippers (2), adenosine wedges (6) etc can result in a infinite number of unique RNA folds (7–10). Noncanonical DNA structures are also present as intermediates in DNA recombination, repair, and replication and are frequently stabilized by proteins (11–15). Nucleic acid binding proteins that are structure-specific either recognize a pre-formed structure in RNA and DNA, or can induce DNA bending, DNA looping, helical distortions and unfolding of RNA (16). One functional consequence of structure-specific nucleic acid recognition is to facilitate the formation of higher-order multi-protein DNA/RNA complexes whereby binding of one protein triggers conformational changes in the nucleic acid that are recognized by another protein in the macromolecular complex.
Here I highlight the structure and mechanism of RNA recognition of a new RNA binding fold, the L-motif, that is present in the histone mRNA specific processing factor, Stem-Loop Binding Protein (SLBP) (17–19), also known as Hairpin Binding Protein (HBP) (20) and compare it to the canonical L-motif observed in HMG-box domains (21) that bind DNA. The high mobility group (HMG) proteins are abundant non-histone nuclear proteins that associate with chromatin, as has been summarized in some excellent recent reviews (22–24). In 1990, R. Tjian and colleagues proposed that the HMG-box motif from Upstream Binding Factor or UBF is a novel DNA binding motif (25). Since then, the HMG-box domain has been identified in plants, yeast and vertebrates and is involved in transcription, DNA replication, recombination and repair (22, 26). The L-shaped scaffold is quite versatile and can bind and bend DNA in the minor groove. It also plays a role in DNA looping (27). These boxes have high affinity (Kds ~10−9 M) towards four-way junction DNA (28), cruciform DNA (29–31), cisplatin-modified DNA (32–34) as well as tRNA (30), double-stranded RNA (35, 36) and single-stranded RNA (37). There are two broad subfamilies of HMG-box containing proteins: those that bind bent or distorted DNA in a non-sequence-specific manner and have two or more tandemly arranged HMG-box domains followed by an acidic tail that has a regulatory function; and a second class of sequence-specific single HMG-box containing transcription factors usually with no acidic tail (38, 39). HMG-box proteins from the former class such as HMG1 (40), HMG2 (41), HMGD (42), and NHP6a (43) bind pre-bent DNA such as Holliday junctions or cisplatin-modified DNA (34, 44). HMG-box proteins from the second class, such as lymphoid enhancer-binding factors TCF-1 and LEF-1 (45–47), sex-determining region Y (SRY) (48), and the SRY-related HMG box (Sox) family (49), are observed frequently in transcription factors that bend specific DNA sequences. The HMG domains are highly selective but in a subtle fashion such that it is difficult to predict DNA binding preferences. For example, the HMG1 box A binds four-way junction DNA with a Kd of 200 nM, but the UBF box has low affinity (Kd 1.5 µM) towards the same DNA (50). The sequences and structures of HMG-box domains from both subgroups are remarkably similar.
Interestingly, recent crystal structures of the histone mRNA specific RNA processing factor, Stem-Loop Binding Protein (SLBP) (18) bound to histone mRNA stem-loop and the exonuclease 3’hExo/ERI1 reveal that its RNA binding domain (RBD) is structurally related to the HMG-box domain. SLBP recognizes the structure of the A-form RNA hairpin and distorts and unfolds the RNA tetraloop. There is no sequence similarity between the SLBP L-motif and HMG-box domains, indicating they are evolutionarily distant. However the overall folding topologies and their architectural functional roles are similar. The similarities in structure, dynamics, and regulation by posttranslational modifications of the SLBP RNA binding L-motif and the DNA-binding HMG-box domains lead to new hypotheses. Do HMG-box proteins bind RNA, and do they play a role in RNA processing? There is some experimental evidence in the literature that this may be a plausible idea. Does SLBP play a direct role in DNA replication? No functional roles for SLBP besides its role in histone mRNA metabolism have been described. Herein I compare the SLBP L-motif with HMG-box domains and discuss their distinct modes of structure-specific recognition of RNA and DNA, respectively.
2. THE HMG-BOX L-MOTIF FOLD
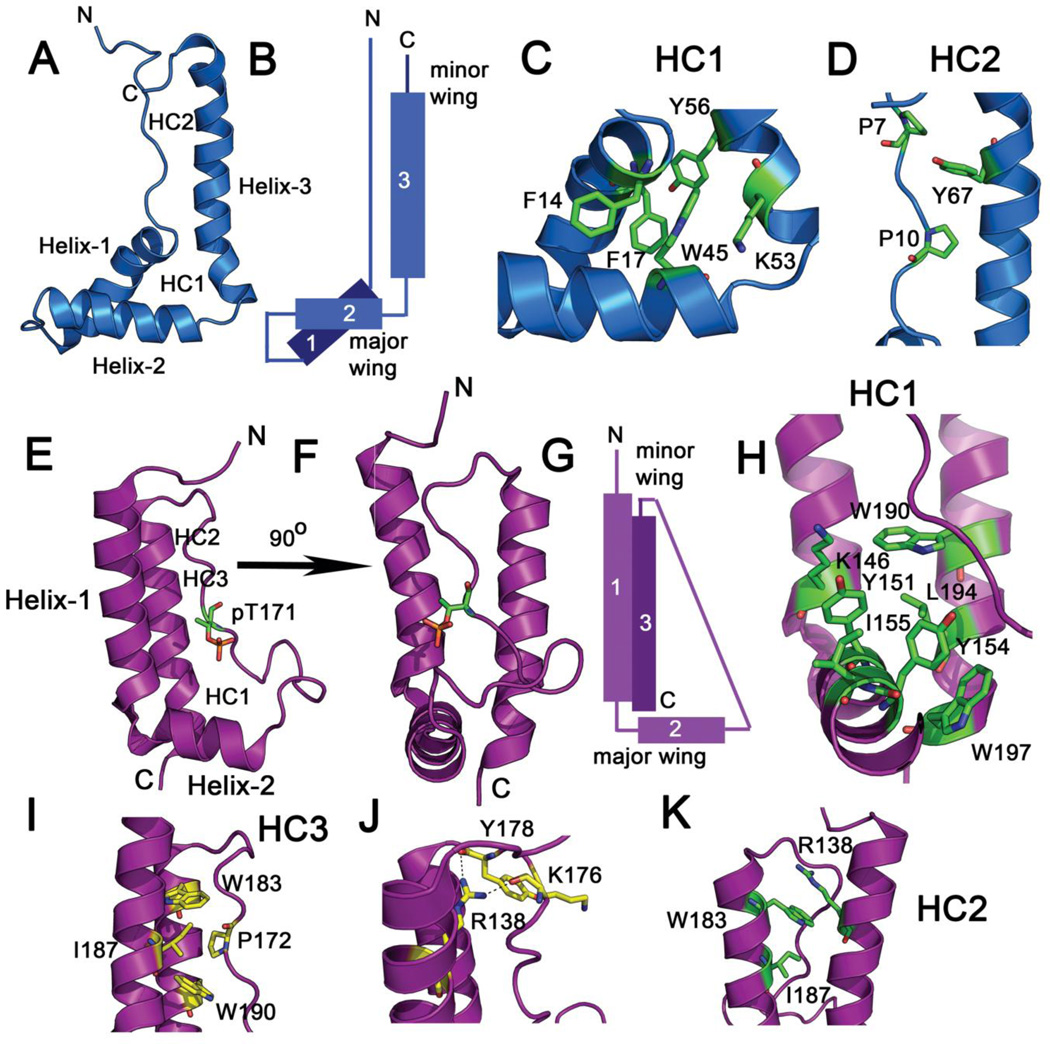
High-resolution structures of several HMG-box domains have been solved by NMR spectroscopy (21, 51–58) and X-ray crystallography (59, 60). The HMG-box domain is an L-shaped DNA-binding motif (Figure 1A, 1B) consisting of ~75–80 amino acids and three α-helices (21). The angle between the two arms (helix-2 and helix-3 or the major wing) of the “L” shape is ~80° and can show ~20° variation between different HMG-box structures (61). The long arm of the “L” (also known as the minor wing) consists of helix-3 and the extended N-terminus, whereas the short arm is made up of helix-1 and helix-2. An extended N-terminal strand packs against helix-3. The orientation of helix-1 differs slightly in many HMG-box structures. The loops that connect the helices can vary in length. The position of helix-2 and helix-3 to form the L-shape is maintained by interactions between conserved aromatic and aliphatic residues that form a compact hydrophobic core (Figure 1C, 1D). The solution NMR structure of the B domain of HMG1 (PDB code 1HME) (21) shows stacking interactions between Phe14, Phe17, Trp45, Lys53, and Tyr56 side chains to form the major tightly packed hydrophobic cluster (HC1) (Figure 1C). In addition, Pro7 and Pro10 form a second hydrophobic core (HC2) in the N-terminal extension that stack against Tyr67 of helix-3, thereby stabilizing the overall fold (Figure 1D).
Figure 1.
(A) Structure of the HMG-box fold from HMG1A (PDB code 1HME) is shown. The location of the two hydrophobic cores, the major core 1 (HC1) and minor core 2 (HC2) are labeled. α-Helices 1 and 2 form the major wing and Helix-3 along with the N-terminal strand forms the minor wing. (B) Schematic of the L-shape fold of the HMG-box domain is depicted. (C) Interaction between hydrophobic side chains in HC1 of HMG1A located in the major wing. (D) Interaction between Y67, P7, and P10 in HC2 of HMG1A is depicted. (E) Structure of the L-motif of SLBP from the structure of the T171 phosphorylated SLBP-histone mRNA stem-loop-3’hExo ternary complex (PDB code 4QOZ) is shown. The phosphothreonine is shown in green stick. The location of the three hydrophobic cores, HC1, HC2, and HC3 is labeled. The front view is shown in (E) and the side view is shown in (F). (G) Schematic of the L-motif fold of SLBP is depicted. (H) Interaction between hydrophobic side chains in HC1 of SLBP located in the major wing. Interaction between hydrophobic side chains in HC2 and HC3 of SLBP located in the minor wing is shown in (I) and (K). (J) A stabilizing salt bridge interaction made by the side chain of R138 with the backbone carbonyl oxygens of Y178 and K176 locks the turn in place in the minor wing.
Proteins belonging to the non-sequence-specific subclass i.e. HMG1A (51, 62), HMG1B (21, 52), and HMG-D (53) have structures that are quite stable in the absence of DNA. In contrast, members of the sequence-specific subclass such as the HMG-box domains of LEF-1 (63), Sox-4 (57), and Sox-5 (58, 64) show significant conformational freedom, and are either disordered or partially ordered in the absence of DNA. These HMG-box domains undergo a disorder-to-order transition upon DNA binding. NMR studies of the Sox-4 HMG-box (57) indicate that in the absence of DNA, the N-terminal strand is disordered and does not pack against helix-3 via HC2. The major hydrophobic core, HC1, is well defined, but HC2 is absent in the structure of free Sox-4 (57, 65). DNA binding leads to packing of HC2 and an ordered complex. The solution NMR structure of Sox-5 (58) shows increased flexibility of helix-3 and a dynamic equilibrium between partially unfolded and folded states. Thermodynamic analysis of Sox-5 unfolding (64) demonstrates that the HMG-box domain unfolds in two separate transitions. The first melting transition is assigned to HC2 (Tm = 34°C) and the second cooperative transition to the melting of HC1 (Tm = 46°C). The HMG-box of LEF-1 is also unstable in the absence of DNA and tends to self-associate as a function of temperature (63). Only partial NMR assignments were obtained for the LEF-1 HMG-box due to exchange broadening of a significant number of resonances (63). Cis-trans isomerization of Pro68 that lies towards the C-terminus of helix-3 and close to HC2, leads to doubling of backbone resonances for several residues and is at least in part responsible for the observed conformational heterogeneity (63). Pro68 adopts a well-ordered trans configuration in the LEF-1/DNA complex. It has been suggested that the increased conformational mobility of the sequence-specific class may be required to bind, distort, and bend a sequence of DNA via a induced fit mechanism in which both the protein and the DNA co-fold to form a stable, low energy complex.
3. THE L-MOTIF OF SLBP
The L-motif of human SLBP (Figure 1E, 1F, G) (PDB codes 4QOZ and 4L8R) has an overall fold that is remarkably similar to that of the L-shaped HMG-box domain (18). The SLBP L-motif is ~73 amino acids across species and consists of three α-helices. In the crystal structures of the human SLBP L-motif bound to the histone mRNA stem-loop and the exonuclease 3’hExo (PDB codes 4QOZ and 4L8R) (18), the L-motif is made up of α-helix-1 and α-helix-2 that lie orthogonal to each other to form the L-shape (Figure 1E). Helix-2 is connected to helix-3 by a long loop of 20 amino acids (Val158-Tyr178) that is constitutively phosphorylated at Thr171 (17, 66). Helix-3 is the RNA recognition helix and is slightly bent when bound to RNA. The orientation of helix-3 is parallel to helix-1. In the presence of RNA, the L-shape is stabilized by a major hydrophobic core (HC1) at the junction of the long and short arms of the “L”, similar to HMG-box domains (Figure 1H). The aromatic and aliphatic side chains of Lys146, Tyr151, Tyr154, Ile155, Trp190 Leu194, and Trp197 pack against each other at the junction of the two arms to stabilize the fold (Figure 1H). In addition, several van der Waals and salt bridge interactions are observed between helix-1, helix-3 and the loop connecting helices 2 and 3 that maintain the orientation of the helices. The Pro172 ring, which is in the trans configuration in the crystal structure, shows van der Waals interactions with Trp183, Ile187, and Trp190 to form a hydrophobic core (HC3) (Figure 1I). The guanidinium side chain of Arg138 in helix-1 hydrogen bonds to the backbone carbonyl oxygens of Tyr178 and Lys176 in the loop locking the turn in place thereby orienting helix-3 (Figure 1J). The side chain of Trp183 packs between the aliphatic side chains of Arg138 and Ile187 to form a second minor hydrophobic core (HC2) (Figure 1K).
In spite of these extensive interactions in the crystal structure of the RNA bound complex, similar to the sequence-specific family of HMG-box domains, the free SLBP L-motif is not a stable fold in solution (17, 67, 68), and is disordered in the absence of RNA. The domain has a hydrodynamic radius that is characteristic of a pre-molten globule (67) and although it has helical secondary structure by circular dichroism, it lacks a stable tertiary fold by NMR (67). NMR studies also show that the free SLBP L-motif exhibits cis-trans proline isomerization about the Thr171-Pro172 bond (17). Isomerization of Pro172 from the trans configuration, to the cis-configuration would disrupt the hydrophobic core of the L-motif, thereby destabilizing the domain in the absence of RNA. Consistent with this, mutation of Pro172 to Gly gives one major conformation of the free SLBP L-motif in NMR studies (17). However the source of severe destabilization of the fold in the RNA-free state is unclear. Unlike globular domains that are stabilized by a major central hydrophobic core, SLBP is stabilized by three small hydrophobic core elements, which may be one reason for the observed instability. Similar to LEF-1, Sox-4, and Sox-5, the SLBP L-motif undergoes a disorder-to-order transition upon binding RNA (17). As discussed below, there are significant conformational rearrangements in the histone mRNA tetraloop in the bound complex (18). Therefore co-folding of SLBP and histone mRNA stem-loop via an induced fit mechanism is necessary to form a stable and specific complex.
4. RECOGNITION OF DNA AND RNA BY L-MOTIF SCAFFOLDS
(A) DNA RECOGNITION BY HMG-BOX TYPE L-MOTIFs
HMG-box domains from both sub-classes have been shown to bind DNA in the minor groove using the concave face of the L-shape with a pincher like hold on the DNA (Figure 2A). A hydrophobic amino acid is intercalated between bases in the minor groove and extensive non-sequence-specific protein-DNA contacts are made with a hydrophobic interface of the HMG-box, as well as hydrogen bonding interactions with the phosphate backbone. The DNA is bent and underwound with positive roll angles, resulting in a shallow and wide minor groove and a compressed major groove. The determinants for DNA sequence-specificity are generally believed to lie in the minor wing of the HMG-box consisting of helix-3 and the N-terminal strand (61, 69).
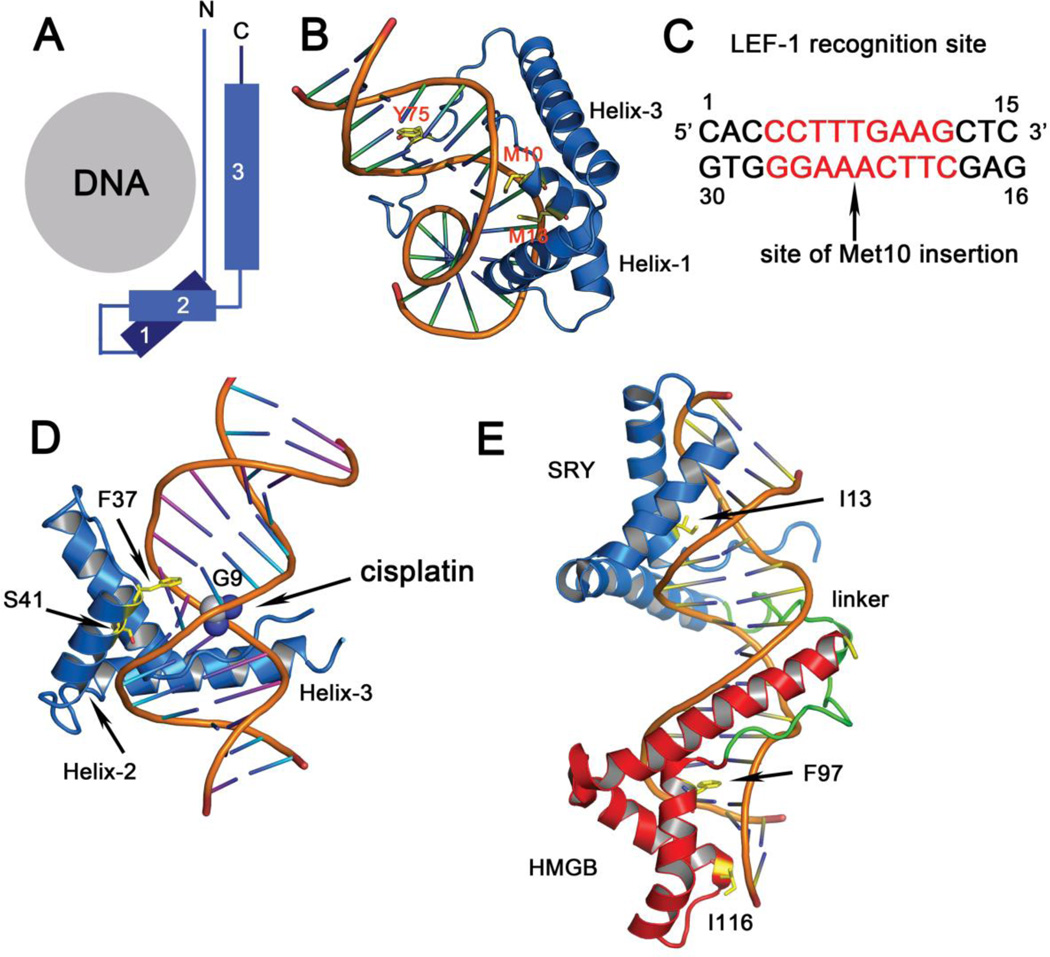
Figure 2.
Structures of HMG-box domains bound to DNA. (A) Summary schematic of the binding interface of DNA and the L-motif of HMG-box domains. All HMG-box domains bind DNA in the minor groove using the concave face of the L-shape. (B) Mode of interaction of the HMG domain from LEF-1 complexed to 15 bp DNA oligonucleotide observed in the solution NMR structure (PDB code 2LEF). Three side chains that intercalate between DNA bases are shown. The HMG domain wraps around the DNA minor groove resulting in a significant conformational change in the DNA. (C) Sequence of the 15 bp DNA oligonucleotide showing the site of Met10 intercalation. (D) Structure of HMG1A bound to a 16 base pair DNA duplex modified with cisplatin (PDB code 1CKT) is shown. The cisplatin is shown in cpk. (E) Structure of the SRY.B tandem HMG boxes bound to 16 bp DNA (PDB code 2GZK) is depicted. Only the hydrophobic side chains that intercalate and distort the DNA are depicted in stick. The linker connecting the two boxes is in green.
An example of a structure from the sequence-specific class of HMG-box proteins is the solution NMR structure of the LEF-1 HMG-box domain bound to a 15 bp oligonucleotide from the TCRα enhancer (Figure 2B, 2C) (PDB code 2LEF). The structure reveals that the DNA is bent by ~117° towards the major groove. A tyrosine residue (Tyr75) anchors the protein by inserting into one end of the minor groove. DNA bending results in formation of a wide and shallow minor groove in a 9 bp recognition site, with an overall geometry similar to A-DNA, that accommodates helices 1 and 2 of LEF-1. The side chain of Met10, which is located near the N-terminus of helix-1, is partially inserted between the A23 and A24 bases in the center of the DNA recognition element, disrupting base stacking (but not base pairing) in this region. Met13 contacts the A24 base and the side chain of Phe9 packs against the T8 ribose on the opposite strand (Figure 2B). The interactions of Met10, Met13, and Phe9 stabilize the minor groove of the bent DNA. The average minor groove width 5’ from the site of intercalation between A23 and A24 nt is 11 Å, with a depth of only 1 Å, and becomes narrower, with an average width of 5 Å and a depth of 4.6 Å on the 3’ end (Figure 2C). In addition to extensive interactions with the minor groove, the basic C-terminus of the HMG-box interacts with the phosphates that lie in the major groove. It is proposed (61) that an important difference between LEF-1 and HMG-box domains from the non-sequence-specific family lies in the length of helix-3, which is kinked at Pro67, and this kink appears to be important for orienting the C-terminal basic region so that it can make extensive contacts with the phosphate backbone.
An example of a structure from the non-sequence-specific sub-class of HMG-box domains consists of the 2.5 Å crystal structure of the complex of HMG1A bound to a 16 base pair DNA duplex modified with cisplatin (PDB code 1CKT) (70). Similar to LEF-1, the HMG1A binds with the concave face facing toward the minor groove of DNA. The DNA is bent by 61° and the minor groove resembles A-DNA. The aromatic side chain of Phe37 inserts into the G8-G9 base pair step (formed by cisplatin) and is located at the N-terminus of helix-2. Small conformational changes are observed in the HMG domain around Phe37. A noteworthy difference between HMG1A and LEF-1 is the location of the DNA bend, which lies two base pairs away from the HMG1A binding site. The register of Phe37 is also different from Met10 of LEF-1, which is located at the N-terminus of Helix-1 of the HMG domain. This mode of binding is confirmed in solution, as the Phe37Ala mutant is severely impaired in its ability to bind cisplatin-modified DNA (70). Position 37 is always conserved as a hydrophobic amino acid in the non-sequence-specific sub-class of HMG domains, but is a polar residue in the sequence-specific sub-family. The loop preceding Phe37 (residues 28–37) is dynamic in solution and remains flexible in the crystal in the presence of DNA, as can be ascertained by high B-factors and weak electron density in this region. Cisplatin modification destacks G8-G9, bending the DNA and exposing these bases into the minor groove. The role of Phe37 appears to be to act as a wedge by stacking onto the solvent exposed face of G9. Extensive non-specific contacts to DNA are observed with the N-terminal strand, helix-1 and helix-2 of the HMG1A box. Helix-1 contacts the sugar-phosphate backbone of the unmodified strand and helix-2 contacts the backbone of the cisplatin-modified strand.
Several HMG-box containing proteins, such as human UBF and HMGB1 have more than one copy of the HMG-box domain that are arranged in tandem. How do multiple boxes influence DNA bending? The solution NMR structure of two tandem HMG boxes, i.e. the HMG box of SRY linked to the HMGB box from HMGB1 complexed to DNA provides insight into the mode of DNA recognition via multiple HMG-boxes (PDB code 2GZK) (Figure 2E). HMGB1 has two HMG-box domains (A and B boxes) with an intervening linker that binds non-sequence specifically to DNA. Since non-sequence-specific binding leads to heterogeneity of the NMR sample, box-A was replaced with the HMG-box of SRY to give a hybrid molecule SRY-HMGB (SRY.B) and a stable, well-defined complex with a 16 bp DNA duplex. The SRY.B box binds a 119 bp DNA with a Kd of ~10 nM in EMSA assays, which is ~2–3 fold higher than observed for the single SRY box/DNA complex. The Kd for interaction of SRY.B with the shorter 16 bp DNA used for NMR studies was determined to be ~120–140 nM.
The structure of the DNA in the complex is bent by 101.5 ± 9.1° with each HMG-box contributing equally to the overall bend. The DNA is underwound and is between A-DNA and B-DNA. The minor groove is shallow and wide and the major groove is compressed as is observed in the structure of the single HMG-box/DNA complex. The average twist angle is 31.4 ± 0.4° compared to a twist angle in B-DNA of 35.8° and a positive roll angle of 7.75 ± 0.7° is observed, compared to a roll angle of −3.65° typically observed in B-DNA. The DNA bend in the SRY portion is due to intercalation of Ile13 between two adenine bases causing a positive roll at this base step (A12 and A13). Phe12 also packs against the deoxyribose and base of T20 and T21 similar to that observed in the SRY/DNA complex (71). Extensive hydrogen bonding and electrostatic interactions are observed with the backbone phosphates and the deoxyribose groups. In the HMGB-box region there are two sites of intercalation made by Phe97 and Ile116 (Figure 2E). Phe97 inserts into the G3-C30/A4-T29 base step and contacts all four bases. Ile116 intercalates into G1-C32/G2-C31, also contacting all four bases. The overall structures of the HMG-boxes are similar to those observed in the single domain/DNA complexes (21, 51, 52, 72). The r.m.s.d. between the structure of free SRY and the DNA bound protein in SRY.B is 1.7 Å, and that between the HMGB-box portion (of protein and DNA backbone) and the B-box/DNA structure is 2.5Å. The linker between the two boxes lies in the minor groove and is anchored into place by a specific base contact between Thr79 and A10. The structure explains how tandem HMG-boxes can bind longer DNA stretches to structurally distort the DNA backbone. While the HMG-boxes act independently to bend DNA, the interactions of the individual domains are enhanced by extensive contacts between the linker and the minor groove of DNA.
(B) HISTONE mRNA RECOGNITION BY THE SLBP L-MOTIF
The crystal structures of the SLBP L-motif bound histone stem-loop RNA reveal three features that are shared with the mode of DNA recognition exhibited by sequence-specific HMG-box domains, such as Sox-4, Sox-5, SRY, and LEF-1. First, as discussed earlier, the HMG-boxes recognize DNA in a structure and sequence-specific manner and are capable of distorting and bending DNA. SLBP also distorts the RNA structure, particularly in the tetraloop. Second, many HMG-box proteins are partially disordered especially in the minor wing of the L-motif. Similar to SLBP, these proteins undergo a disorder-to-order transition upon binding DNA. Third, both SLBP and the HMG-box domains make sequence-specific contacts in the minor wing of the L-motif in addition to interaction of a hydrophobic residue from the major wing with the minor groove of DNA. Other than these observations, the mode of RNA recognition by SLBP is quite different compared to the HMG-box/DNA complexes.
SLBP forms a specific complex with a 16 nt stem-loop that is highly conserved in the 3’ untranslated region (3’ UTR) of replication-dependent histone mRNAs. Human, Drosophila, and Xenopus SLBP proteins bind histone mRNA stem-loops with a Kd ~1–10 nM (17, 73–75). The stem is capped by a UYUM tetraloop that is unique to histone mRNAs. Solution NMR structures of the stem-loop from the histone H4–12 gene has been solved in the absence of SLBP (74, 76). The six base pair RNA stem is A-form and starts with a G-C base pair at the base and ends in a U-A closing base pair. The tetraloop is well ordered in both structural ensembles, however the uridine at position 12 adopts different conformations. In one NMR structure, U12 is stacked onto the other uridines of the loop (76), and in the second structure (74) it is flipped out to solvent. Therefore unlike tetraloops of the GNRA, UNCG, and CUYG sub-families (1), the loop is conformationally flexible and sensitive to solution conditions and the environment.
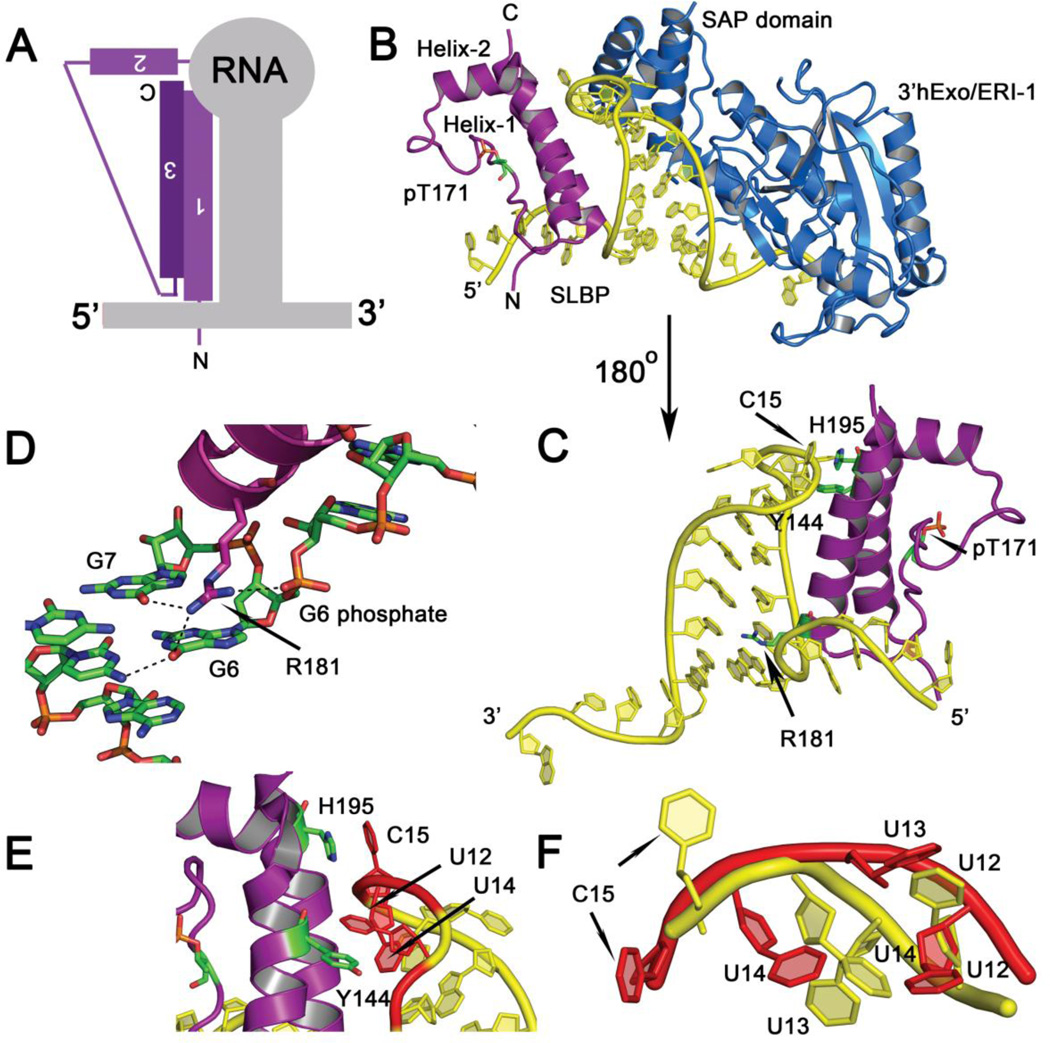
In the crystal structures of human SLBP in a ternary complex with histone mRNA stem-loop and 3’hExo (18), SLBP binds the stem-loop in an inverted L-shape via the outer surface of the SLBP L-motif (Figure 3A, 3B). Almost all RNA contacts are made by helices 1 and 3. Helix-3 is the RNA recognition helix and approaches the major groove, extending from the base of the stem to the tetraloop (Figure 3C). No contacts are made in the minor groove of the RNA. The only residue that makes sequence specific contacts, Arg181, is located in the minor wing of the L-motif in helix-3 (Figure 3C, 3D). The guanidinium group of Arg181 is involved in three hydrogen bonds: to the O6 of guanines G6 and G7 at the base of the RNA helix, and the G6 phosphate (Figure 3D). These hydrogen bonds anchor the minor wing of the SLBP L-motif to the base of the stem, positioning the major wing and the N-terminal end of helices 1 and 3 near the tetraloop. In addition, there are extensive nonspecific interactions made by helices 1 and 3 with the RNA backbone. Tyr144 of helix-1 forms π-π stacking interactions with U12 of the RNA tetraloop, and shows a T-shaped stacking interaction with U14 in the tetraloop (Figure 3E). Tyr144 likely triggers a conformational change in the tetraloop causing it to distort and unfold. Intriguingly, C.elegans SLBP has an arginine at position 144 and position 12 in the tetraloop is a cytosine instead of a uridine (75), suggesting a different mode of recognition of the tetraloop for C.elegans SLBP as compared to human SLBP. It has been shown that C.elegans SLBP is very specific for the C.elegans histone mRNA and will not bind the human histone H4.12 stem-loop mRNA in EMSA assays (75). In addition, His-195 from helix-3 stacks against C15 in the tetraloop (Figure 3E). While histidine is conserved at position 195 in human, xenopus, sea urchin, ciona, and Drosophila SLBPs, it is a tyrosine in C. elegans SLBP. These stacking interactions alter the structure of the RNA and stabilize a new unfolded tetraloop conformation (Figure 3F). The distorted tetraloop contacts the SAP domain of 3’hExo/ERI1 in this structure, thereby facilitating formation of a higher order complex.
Figure 3.
(A) Summary schematic showing the mode of histone mRNA stem-loop recognition by the SLBP L-motif. (B) Crystal structure of the Thr171 phosphorylated SLBP-histone mRNA stem-loop-3’hExo/ERI-1 ternary complex (PDB code 4QOZ). SLBP is in purple, the RNA in yellow, and the exonuclease 3’hExo in blue. The SAP domain of 3’hExo contacts the tetraloop on the opposing face from SLBP. The orientation shown is from the 5’ end of the RNA to the 3’ end. (C) The back view (180° rotation from (B)) of the SLBP RNA complex is shown. For clarity, 3’hExo is not displayed. The three side chains (R181, Y144, and H195) that contact RNA bases or make stacking interactions are shown in green. (D) Hydrogen bonds mediated by the guanidinium group of R181 to the O6 of G6 and G7 at the base of the stem and the phosphate of G6 is shown. (E) Base stacking interactions with U12 and U14 of the RNA tetraloop are depicted. (F) Superposition of the tetraloop structure of free RNA tetraloop (PDB code 1JWC) (in red) and the tetraloop in the RNA complex (4QOZ) (in yellow). The uridines are flipped out and the tetraloop structure unfolded in the protein bound state.
A unique feature of the SLBP L-motif is that it is phosphorylated at a conserved threonine (Thr171 in human SLBP and Thr230 in Drosophila SLBP) in its L-motif, and also at several sites C-terminal to the L-motif (discussed in more detail in the next section). Thr171 is located in the concave face of the L-motif, in the flexible 20 amino acid loop that links helices 2 and 3. Phosphorylation at this site increases the affinity of the SLBP L-motif for the histone stem-loop by 7–11 fold (17, 66) when measured by nitrocellulose filter binding assays or SPR. Although the overall effect on the Kd is small, mutation of Thr230 to alanine or dephosphorylation increases the off-rate for the RNA by ~100-fold while increasing the on-rate (of the more basic L-motif) by 10-fold (17). Therefore, the Thr171/Thr230 dephosphorylated form is characterized by rapid on-off kinetics, yielding an unstable complex. Comparison of the crystal structures of T171 phosphorylated and dephosphorylated SLBP bound to histone mRNA (PDB codes 4QOZ and 4L8R) (18) shows that phosphorylation stabilizes helix-2 and the loop via a network of five hydrogen bonds that helps stabilize the complex. The T171 phosphate stabilizes that protein-RNA complex in solution and the phosphate moiety has a depressed pKa (< 4.5) measured by 31P NMR (77), consistent with its involvement in a network of electrostatic interactions. 31P NMR studies also indicate that the phosphate at this position experiences torsional strain in solution (77). This is a likely consequence of the phosphate bringing together secondary structure elements to stabilize the flexible L-motif fold.
The role of the flexible loop between helix 2 and 3 in modulating RNA binding affinity remains unclear, and the biological data on loop mutants cannot be rationalized solely based on the crystal structures. For example, the amino acid sequence following the 171TPNK is 175FKKY in human and xenopus SLBPs and in particular Lys177 is conserved in most SLBPs, except sea urchin and C.elegans SLBPs, where the sequence is FQVT and LINE, respectively. Mutagenesis studies have shown that replacing this sequence in both sea urchin and C.elegans SLBPs with FKKY results in a dramatic improvement in histone mRNA binding (75, 78), however surprisingly no contacts are made with the RNA by this sequence. In addition, although one conformation is observed for the loop in the crystal, NMR studies suggest that the loop, especially around the phosphate, may fluctuate between two slowly interconverting conformers (77). Future structural studies on cognate and non-cognate SLBP-RNA complexes should provide greater mechanistic insight into the precise mode of RNA recognition by the SLBP L-motif.
5. MODULATION OF L-MOTIF FUNCTION BY POST-TRANSLATIONAL MODIFICATIONS
Several L-motifs, especially those of HMGB1 and HMGB2 are posttranslationally modified by serine/threonine phosphorylation, lysine methylation, lysine acetylation, glycosylation, and poly (ADP)-ribosylation (for reviews see (22, 23, 79) ) (Figure 4). These posttranslational modifications (PTMs) may play important functional roles in regulating subcellular localization, DNA binding, as well as protein-protein interactions, although the biological roles are just beginning to be understood.
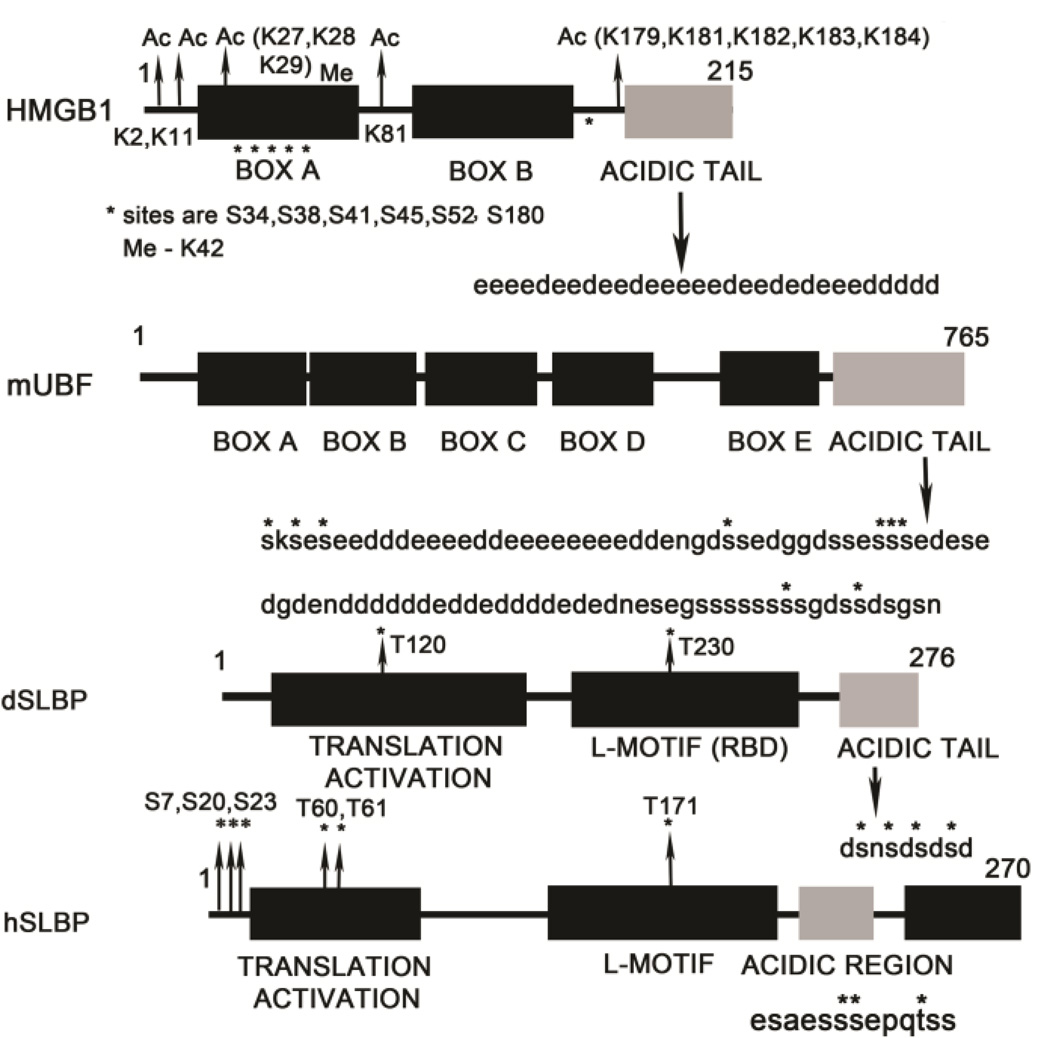
Figure 4.
Structural organization of L-motif containing proteins that have C-terminal acidic tails is shown as a schematic. The sequence of the acidic tail is depicted. Asterisks highlight serines and threonines that are phosphorylated in vivo, lysines that are acetylated in HMGB1 are designated “Ac”, and lysines that are methylated are designated “Me”.
HMGA1a and HMGB1 are extensively acetylated on lysine residues in Box A and close to the acidic tail (Figure 4) by the histone acetyltransferases CBP (CREB-binding protein) and PCAF (p300) in vitro as well as in vivo (80, 81). Remarkably, 17 out of 43 lysines of HMGB1 are acetylated (82). Constitutive acetylation of Lys2 in the N-terminus of HMGB1 enhances the ability of the L-motif to bind bent DNA substrates such as cisplatinated DNA (83), DNA ends (84), and also enhances HMGB1 mediated stimulation of enzymatic ligation of DNA fragments (85, 86). Acetylation of both Lys2 and Lys11 has been reported to enhance binding of HMGB1 to four-way junctions and DNA minicircles (87). Acetylation of HMGB proteins also enhances protein-protein interactions with the nucleosome as well as the SWI/SNF chromatin remodelling complex (88). Acetylation of Sox2 at Lys75 is an export signal and promotes translocation of Sox2 from the nucleus to the cytoplasm (89).
In contrast to acetylation, methylation of HMGB1 at Lys42 weakens its affinity for DNA (90) and also alters its subcellular localization by promoting its export into the cytoplasm (90). The HMGA proteins are also methylated on lysines and arginines and increased levels of arginine methylation are observed in leukemias and prostate tumors (91).
Phosphorylation of the HMGB proteins occurs in the Box A L-motif at Ser34, Ser38, Ser41, Ser45, and Ser52. These sites lie close to two nuclear localization signals and may regulate nucleocytoplasmic shuttling of HMGB proteins. Although the kinases that phosphorylate the specific serines have not been identified, calcium/phospholipid-dependent protein kinase can phosphorylate HMGB proteins in vitro (92). As mentioned in section 4, the SLBP L-motif phosphorylated at Thr171/Thr230 (human/Drosophila SLBP) and phosphorylation at this site regulates RNA binding. Thr171 phosphorylation is also important for subcellular localization of SLBP into the nucleus in both human (93) and Drosophila SLBPs (94) and regulates histone pre-mRNA processing in Drosophila (94) and mRNA decay of human SLBP (93) by regulating the stability of the SLBP-histone mRNA complex (93). It is also a binding site for the prolyl isomerase Pin1 which catalyzes dissociation of the SLBP L-motif from the histone stem-loop by promoting trans-to-cis isomerisation of the phospho-Thr-Pro bond in the TPNK sequence (93). The effects of Pin1 on mRNA stability in mammalian cells have been well characterized on a genome-wide scale (95). Besides phosphorylation, no other PTMs have been reported for the SLBP L-motif.
HMGB1 has also been reported to undergo glycosylation and poly (ADP)-ribosylation (PAR) (96). The physiological role of glycosylation is not clear. PARylation of HMGB1 has been linked to its subcellular localization during necrosis but the functional relevance in normal cellular function is unclear (97).
6. PHOSPHORYLATION OF THE ACIDIC TAIL
Some HMG-box containing proteins like human HMG4L, HMG1–3, HMG1L10, SP100-HMG, UBF, SSRP-1(26), Drosophila HMG-D (98) and plant HMGB, have HMG-box domains that are followed by a short basic linker and a ~20–30 residue unstructured acidic tail that is rich in Asp and Glu residues and is also phosphorylated (Figure 4). The structural and functional consequences of phosphorylation of the tail have been particularly well studied. The acidic tail has been shown to lower the affinity of HMGB1 for DNA by directly interacting with the DNA binding interface on the HMG-boxes, thereby auto-inhibiting the protein (99–104). This acidic region has diverse functional roles in the abundant non-histone HMG proteins. In the case of HMGB1, the acidic tail is required for transcriptional activation, and for preferential binding to DNA minicircles and four-way junctions as compared to linear DNA (31, 105). It interacts with other proteins such as the basic N-terminus of histone H1, thereby increasing the affinity of HMGB1 for DNA while lowering the affinity of H1 (106, 107). This is a potential mechanism that allows HMGB1 access to certain promoters. The acidic tail also facilitates a chaperone function of HMG proteins via protein-protein interactions with transcription factors such as p53 (108).
Maximal transcription activation of UBF proteins also requires its acidic C-terminal tail. Phosphorylation of UBF by CKII (109) in its C-terminal tail stimulates Pol1 transcription by stabilizing the interaction of UBF with the TATA-binding protein complex SL1 (110, 111). SL1 is a complex composed of TBP and three TBP-associated factors namely TAF148, TAF163, and TAF1110. Besides participating in protein-protein interactions, the acidic tail of xenopus UBF interacts directly with the N-terminal HMG-box of xUBF (112). This interaction is required for folding of the promoter DNA promoting negative DNA supercoiling, DNA looping, and the formation of a disc like structure called the enhancesome that is optimal for recruitment of the TATA-box protein complex (112).
Drosophila and human SLBP proteins also have an acidic region that is separated from the L-motif by a short (9–15 amino acid) linker (Figure 4). In Drosophila SLBP, the acidic region is phosphorylated at four serines and phosphorylation of the C-terminal tail is important for RNA processing (113). It is also important for increasing binding of dSLBP to the stem-loop RNA by ~30-fold (17, 67) although the mechanism by which this occurs in not clear. Comparison of the kinetics of binding by Surface Plasmon Resonance (SPR) of the C-terminally truncated L-motif that lacks the four phosphorylated serines (but is phosphorylated at Thr230), to that of the full length phosphorylated dSLBP protein shows that the effect is not on the on-rate but on the off-rate of binding (17). This is consistent with previous stability measurements (67) that showed that dSLBP phosphorylated in the C-terminus forms a more stable complex with RNA. It is possible, therefore, that the phosphorylated C-terminal tail may interact with the L-motif to form a higher affinity complex with the RNA. In human SLBP, three phosphorylation sites have been mapped in the acidic region at Ser221, Ser222, and Thr226 in vivo by mass spectrometry (93). However the functional role of phosphorylation at these sites, its effect on RNA binding, or pre-mRNA processing are currently unknown.
7. PROTEIN-PROTEIN INTERACTIONS MEDIATED BY L-MOTIFs
As expected from their diverse functional roles in the cell, HMG-box containing proteins interact with a plethora of proteins that regulate transcription such as transcription factors and chromatin. For example, the HMG boxes of HMGB1 interact with the C-terminal domain of the progesterone receptor to promote the receptor binding to DNA (114). HMG-box containing proteins have also been shown to be part of the chromatin remodelling complexes SWI/SNF, BAF, and FACT (115, 116), but the roles of the L-motifs in these interactions are unclear.
The SLBP L-motif has been proposed to interact with proteins involved in histone pre-mRNA processing, mRNA decay, and mRNA translation. The best-characterized interaction of the SLBP L-motif is with the prolyl isomerase Pin1 (93). NMR chemical shift mapping experiments show that Pin1 interacts only with the phospho-Thr171 form of the SLBP-L-motif and not with the unphosphorylated protein (93). The interaction requires the N-terminal WW domain of Pin1 as well as the prolyl isomerase domain. Pin1 acts with the phosphatase PP2A to dephosphorylate SLBP in vivo at a number of sites, including T171. Chemical inhibition of Pin1 by the inhibitor PiB in vivo showed significant (2–3 fold) phosphate enrichment of human SLBP at seven sites (Ser7, Ser20, Ser23, Thr171, Ser221 Ser222, Thr226) by mass spectrometry (93). The interaction of the SLBP L-motif with Pin1 is also important for SLBP ubiquitination, particularly via the Ser20/Ser23 phosphodegron sequence in the N-terminus (93). The SLBP L-motif has also been reported to directly associate in vivo and in vitro with the Lsm4 subunit of the Lsm-1–7 complex that is important for mRNA degradation of all eurkaryotic mRNAs (117). The SLBP L-motif also interacts directly with the h subunit of eIF3 in directed yeast two-hybrid assays (118), although the functional role of this interaction in stimulating translation of histone mRNAs is uncharacterized.
8. FUNCTIONAL ANALOGIES BETWEEN HMG DOMAIN PROTEINS AND SLBP
There is no obvious sequence similarity between the SLBP L-motif and HMG domains indicating they are evolutionarily unrelated. However, they share common structural features and function. These are structural or architectural proteins that bend and distort the nucleic acid to facilitate formation of higher order nucleic acid-protein complexes. The “L” shape is particularly suited in this regard as it offers different modes of nucleic acid recognition. The inner concave face has a large surface area so it can wrap around the minor groove of DNA. The long arm of the “L” can act as a molecular ruler to anchor to the base of an RNA A-helix and distort the loop. Another common feature is that at least a subset of L-motif containing proteins are regulated by posttranslational modifications. Phosphorylation of the L-motif and in an acidic domain C-terminal to the L-motif modulates affinity of the L-motif for the nucleic acid, and in turn regulates nucleic acid-protein assembly.
9. OTHER DNA AND RNA BINDING PROTEINS (DRBPs)
There are many examples of multifunctional transcription factors or their sub-domains that can bind both DNA and RNA (DRBPs) (for reviews see (119, 120)). Proteins such as the zinc finger containing protein TFIIIA from Xenopus (121, 122), human p53 (123, 124), the Wilms tumor protein (WT1) (125–127), the Bicoid homeodomain (128, 129), and others are known to bind DNA and RNA. However, double-stranded B-form DNA and A-form RNA have very different structures, and not surprisingly, few proteins recognize both ds-DNA and ds-RNA. On the other hand, there are several examples of proteins that recognize single-stranded DNA and RNA as well as Z-DNA and Z-RNA. Such interactions may be particularly relevant during transcription, when DRBP transcription factors bind DNA and may also associate with long non-coding RNAs or the nascent mRNA transcript for pre-mRNA processing.
Crystal structures of the Zα–domain of RNA adenosine deaminase 1 (ADAR1) bound to left-handed Z-DNA (130) and Z-RNA (131) reveal a similar mode of interaction of the Z–domain with the phosphate backbone in a sequence-independent manner. The Z–domain of ADAR1 has also recently been reported to bind the G-quadruplex of the c-Myc promoter (132) and NMR studies indicate that the mode of interaction is likely to be similar to the interaction of the Zα-domain of Z-DNA and Z-RNA.
Although the RNA-recognition motif (RRM) is very specific for binding single-stranded RNA (133) and may also be involved in protein-protein interactions (133), the RRMs of TBP-associated factor 15 (TAF15)(134), and TAR DNA-binding protein 43 (TDP43)(135, 136) can bind both single-stranded DNA and single-stranded RNA. Crystal structures of RRM3 of TDP43 bound to ssDNA and ssRNA show similar π–stacking interactions of aromatic residues of TDP43 with RNA and DNA bases (135, 136). These structural studies reveal that several RNA and DNA binding proteins may have dual selectivity towards distinct conformations of both DNA and RNA.
10. SUMMARY
The goal of this review was to highlight the varied roles of L-motifs in DNA and RNA metabolism. The functional roles of HMG domain containing proteins in transcription and in other nuclear processes such as DNA replication and repair are well established. However, a possible role in RNA processing has not been investigated. In view of the structural comparison between the SLBP L-motif and HMG domains presented here, and the ability of HMG-box domains to recognize A-form DNA, a testable hypothesis is that HMG domains could also bind RNA with high affinity, although they may utilize a different recognition mode for RNA recognition compared to their mode of DNA association. Consistent with this hypothesis, there are several reports that HMG-box containing proteins can bind RNA substrates in vitro. UBF has been demonstrated to bind tRNA in EMSA assays (30). HMGB1 can associate with branched RNA substrates such as 5S rRNA and the L-3 ribozyme in vitro with high affinity (36). Binding of HMGB1 to the L-3 ribozyme also impaired its RNA cleavage activity (36). HMG-D has been shown to bind the double-stranded HIV-1 TAR and RBE RNA substrates (35). In these studies, it is not clear whether the observed association with RNA is non-specific or if HMG-box domains have functional roles in RNA metabolism in vivo. Another intriguing question is: does SLBP play a direct role in DNA metabolism, especially in DNA replication? Although SLBP does not bind B-form DNA (137), it is conceivable that it may recognize branched non-B DNA intermediates during replication. Such a speculative model is intriguing because SLBP is expressed only during S-phase of the cell cycle, and is one of two cell cycle regulated factors involved in histone mRNA metabolism whose expression is correlated with that of histone mRNA (138). The only other protein that is cell cycle regulated during S-phase is CstF64 (139), a component of the heat-labile factor (HLF) complex (composed of CPSF, CsfF64 and symplekin) (140). The signals that coordinate histone protein biogenesis and histone mRNA expression with DNA replication are not understood. In summary, I suggest the possibility that L-motifs, by nature of their structure, may act as double agents in both RNA and DNA metabolism. Future studies may unveil additional roles of L-motifs in binding structured nucleic acids and in regulating gene expression.
HIGHLIGHTS.
L-motifs are a family of structure-specific nucleic acid binding modules
L-motifs bind and distort the structure of DNA and RNA
L-motifs are found in architectural proteins such as SLBP and HMG-box proteins
They are important for formation of higher order nucleic-acid/protein assemblies
L-motifs are regulated by post-translational modifications such as phosphorylation
ACKNOWLEDGEMENTS
R.T was supported by the National Institutes of Health (NIH) grants 1RO1-GM076660 and 1RO1-GM076660 ARRA (Phosphorylation-dependent recognition of a histone mRNA stem-loop by SLBP).
ABBREVIATIONS
- 3’ UTR
3’ untranslated region
- PTMs
posttranslational modifications
- r.m.s.d
root-mean-square deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thapar R, Denmon AP, Nikonowicz EP. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip Rev RNA. 2014;5:49–67. doi: 10.1002/wrna.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 3.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulyanov NB, Mujeeb A, Du Z, Tonelli M, Parslow TG, James TL. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J Biol Chem. 2006;281:16168–16177. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 5.Ferre-D'Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 6.Gagnon MG, Steinberg SV. The adenosine wedge: a new structural motif in ribosomal RNA. RNA. 2010;16:375–381. doi: 10.1261/rna.1550310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 9.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 10.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 11.Lilley DM. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980;77:6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panayotatos N, Wells RD. Cruciform structures in supercoiled DNA. Nature. 1981;289:466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- 13.Wells RD, Goodman TC, Hillen W, Horn GT, Klein RD, Larson JE, Muller UR, Neuendorf SK, Panayotatos N, Stirdivant SM. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- 14.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 15.Svozil D, Kalina J, Omelka M, Schneider B. DNA conformations and their sequence preferences. Nucleic Acids Res. 2008;36:3690–3706. doi: 10.1093/nar/gkn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu Rev Biophys Biomol Struct. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Lam TT, Tonelli M, Marzluff WF, Thapar R. Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP-RNA complex by phosphorylation and proline isomerization. Biochemistry. 2012;51:3215–3231. doi: 10.1021/bi2018255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan D, Marzluff WF, Dominski Z, Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3'hExo ternary complex. Science. 2013;339:318–321. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3' end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 20.Martin F, Schaller A, Eglite S, Schumperli D, Muller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir HM, Kraulis PJ, Hill CS, Raine AR, Laue ED, Thomas JO. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Malarkey CS, Churchill ME. The high mobility group box: the ultimate utility player of a cell. Trends Biochem Sci. 2012;37:553–562. doi: 10.1016/j.tibs.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 25.Jantzen HM, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 26.Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilley DM. DNA--protein interactions. HMG has DNA wrapped up. Nature. 1992;357:282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- 28.Webb M, Thomas JO. Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain. J Mol Biol. 1999;294:373–387. doi: 10.1006/jmbi.1999.3150. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn A, Voit R, Stefanovsky V, Evers R, Bianchi M, Grummt I. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 1994;13:416–424. doi: 10.1002/j.1460-2075.1994.tb06276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Copenhaver GP, Putnam CD, Denton ML, Pikaard CS. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994;22:2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 32.Treiber DK, Zhai X, Jantzen HM, Essigmann JM. Cisplatin-DNA adducts are molecular decoys for the ribosomal RNA transcription factor hUBF (human upstream binding factor) Proc Natl Acad Sci U S A. 1994;91:5672–5676. doi: 10.1073/pnas.91.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong B, Masse JE, Yen YM, Giannikopoulos P, Feigon J, Johnson RC. Binding to cisplatin-modified DNA by the Saccharomyces cerevisiae HMGB protein Nhp6A. Biochemistry. 2002;41:5404–5414. doi: 10.1021/bi012077l. [DOI] [PubMed] [Google Scholar]
- 34.Pil PM, Lippard SJ. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 35.Arimondo PB, Gelus N, Hamy F, Payet D, Travers A, Bailly C. The chromosomal protein HMG-D binds to the TAR and RBE RNA of HIV-1. FEBS Lett. 2000;485:47–52. doi: 10.1016/s0014-5793(00)02183-9. [DOI] [PubMed] [Google Scholar]
- 36.Bell AJ, Jr, Chauhan S, Woodson SA, Kallenbach NR. Interactions of recombinant HMGB proteins with branched RNA substrates. Biochem Biophys Res Commun. 2008;377:262–267. doi: 10.1016/j.bbrc.2008.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu T, King DL, LaBonne C, Kafatos FC. A Drosophila single-strand DNA/RNA-binding factor contains a high-mobility-group box and is enriched in the nucleolus. Proc Natl Acad Sci U S A. 1993;90:6488–6492. doi: 10.1073/pnas.90.14.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi ME, Falciola L, Ferrari S, Lilley DM. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992;11:1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas JO, Travers AA. HMG1 and 2, and related 'architectural' DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 42.Churchill ME, Jones DN, Glaser T, Hefner H, Searles MA, Travers AA. HMG-D is an architecture-specific protein that preferentially binds to DNA containing the dinucleotide TG. EMBO J. 1995;14:1264–1275. doi: 10.1002/j.1460-2075.1995.tb07110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolodrubetz D, Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J Biol Chem. 1990;265:3234–3239. [PubMed] [Google Scholar]
- 44.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 45.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 47.Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 48.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 49.Prior HM, Walter MA. SOX genes: architects of development. Mol Med. 1996;2:405–412. [PMC free article] [PubMed] [Google Scholar]
- 50.JR Po, Norman DG, Bramham J, Bianchi ME, Lilley DM. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hardman CH, Broadhurst RW, Raine AR, Grasser KD, Thomas JO, Laue ED. Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1995;34:16596–16607. doi: 10.1021/bi00051a007. [DOI] [PubMed] [Google Scholar]
- 52.Read CM, Cary PD, Crane-Robinson C, Driscoll PC, Norman DG. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993;21:3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones DN, Searles MA, Shaw GL, Churchill ME, Ner SS, Keeler J, Travers AA, Neuhaus D. The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster. Structure. 1994;2:609–627. doi: 10.1016/s0969-2126(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 54.Allain FH, Yen YM, Masse JE, Schultze P, Dieckmann T, Johnson RC, Feigon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Yang W, Wu J, Shi Y. Solution structure of the first HMG box domain in human upstream binding factor. Biochemistry. 2002;41:5415–5420. doi: 10.1021/bi015977a. [DOI] [PubMed] [Google Scholar]
- 56.Yang W, Xu Y, Wu J, Zeng W, Shi Y. Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor. Biochemistry. 2003;42:1930–1938. doi: 10.1021/bi026372x. [DOI] [PubMed] [Google Scholar]
- 57.van Houte LP, Chuprina VP, van der Wetering M, Boelens R, Kaptein R, Clevers H. Solution structure of the sequence-specific HMG box of the lymphocyte transcriptional activator Sox-4. J Biol Chem. 1995;270:30516–30524. doi: 10.1074/jbc.270.51.30516. [DOI] [PubMed] [Google Scholar]
- 58.Cary PD, Read CM, Davis B, Driscoll PC, Crane-Robinson C. Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5. Protein Sci. 2001;10:83–98. doi: 10.1110/ps.32801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rong H, Li Y, Shi X, Zhang X, Gao Y, Dai H, Teng M, Niu L, Liu Q, Hao Q. Structure of human upstream binding factor HMG box 5 and site for binding of the cell-cycle regulatory factor TAF1. Acta Crystallogr D Biol Crystallogr. 2007;63:730–737. doi: 10.1107/S0907444907017027. [DOI] [PubMed] [Google Scholar]
- 60.Gao N, Jiang W, Gao H, Cheng Z, Qian H, Si S, Xie Y. Structural basis of human transcription factor Sry-related box 17 binding to DNA. Protein Pept Lett. 2013;20:481–488. [PubMed] [Google Scholar]
- 61.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 62.Broadhurst RW, Hardman CH, Thomas JO, Laue ED. Backbone dynamics of the A-domain of HMG1 as studied by 15N NMR spectroscopy. Biochemistry. 1995;34:16608–16617. doi: 10.1021/bi00051a008. [DOI] [PubMed] [Google Scholar]
- 63.Love JJ, Li X, Chung J, Dyson HJ, Wright PE. The LEF-1 high-mobility group domain undergoes a disorder-to-order transition upon formation of a complex with cognate DNA. Biochemistry. 2004;43:8725–8734. doi: 10.1021/bi049591m. [DOI] [PubMed] [Google Scholar]
- 64.Crane-Robinson C, Read CM, Cary PD, Driscoll PC, Dragan AI, Privalov PL. The energetics of HMG box interactions with DNA. Thermodynamic description of the box from mouse Sox-5. J Mol Biol. 1998;281:705–717. doi: 10.1006/jmbi.1998.1895. [DOI] [PubMed] [Google Scholar]
- 65.Weiss MA. Floppy SOX: mutual induced fit in hmg (high-mobility group) box-DNA recognition. Mol Endocrinol. 2001;15:353–362. doi: 10.1210/mend.15.3.0617. [DOI] [PubMed] [Google Scholar]
- 66.Borchers CH, Thapar R, Petrotchenko EV, Torres MP, Speir JP, Easterling M, Dominski Z, Marzluff WF. Combined top-down and bottom-up proteomics identifies a phosphorylation site in stem-loop-binding proteins that contributes to high-affinity RNA binding. Proc Natl Acad Sci U S A. 2006;103:3094–3099. doi: 10.1073/pnas.0511289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thapar R, Marzluff WF, Redinbo MR. Electrostatic contribution of serine phosphorylation to the Drosophila SLBP--histone mRNA complex. Biochemistry. 2004;43:9401–9412. doi: 10.1021/bi036315j. [DOI] [PubMed] [Google Scholar]
- 68.Bansal N, Zhang M, Bhaskar A, Itotia P, Lee E, Shlyakhtenko LS, Lam TT, Fritz A, Berezney R, Lyubchenko YL, Stafford WF, Thapar R. Assembly of the SLIP1-SLBP complex on histone mRNA requires heterodimerization and sequential binding of SLBP followed by SLIP1. Biochemistry. 2013;52:520–536. doi: 10.1021/bi301074r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Read CM, Cary PD, Preston NS, Lnenicek-Allen M, Crane-Robinson C. The DNA sequence specificity of HMG boxes lies in the minor wing of the structure. EMBO J. 1994;13:5639–5646. doi: 10.1002/j.1460-2075.1994.tb06902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 71.Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 72.Murphy EC, Zhurkin VB, Louis JM, Cornilescu G, Clore GM. Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J Mol Biol. 2001;312:481–499. doi: 10.1006/jmbi.2001.4977. [DOI] [PubMed] [Google Scholar]
- 73.Battle DJ, Doudna JA. The stem-loop binding protein forms a highly stable and specific complex with the 3' stem-loop of histone mRNAs. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zanier K, Luyten I, Crombie C, Muller B, Schumperli D, Linge JP, Nilges M, Sattler M. Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression. RNA. 2002;8:29–46. doi: 10.1017/s1355838202014061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Michel F, Schumperli D, Muller B. Specificities of Caenorhabditis elegans and human hairpin binding proteins for the first nucleotide in the histone mRNA hairpin loop. RNA. 2000;6:1539–1550. doi: 10.1017/s135583820000056x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DeJong ES, Marzluff WF, Nikonowicz EP. NMR structure and dynamics of the RNA-binding site for the histone mRNA stem-loop binding protein. RNA. 2002;8:83–96. doi: 10.1017/s1355838202013869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thapar R. Contribution of protein phosphorylation to binding-induced folding of the SLBP-histone mRNA complex probed by phosphorus-31 NMR. FEBS Open Bio. 2014;4:853–857. doi: 10.1016/j.fob.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robertson AJ, Howard JT, Dominski Z, Schnackenberg BJ, Sumerel JL, McCarthy JJ, Coffman JA, Marzluff WF. The sea urchin stem-loop-binding protein: a maternally expressed protein that probably functions in expression of multiple classes of histone mRNA. Nucleic Acids Res. 2004;32:811–818. doi: 10.1093/nar/gkh193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Q, Wang Y. High mobility group proteins and their post-translational modifications. Biochim Biophys Acta. 2008;1784:1159–1166. doi: 10.1016/j.bbapap.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 81.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 82.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ugrinova I, Pasheva EA, Armengaud J, Pashev IG. In vivo acetylation of HMG1 protein enhances its binding affinity to distorted DNA structures. Biochemistry. 2001;40:14655–14660. doi: 10.1021/bi0113364. [DOI] [PubMed] [Google Scholar]
- 84.Ugrinova I, Mitkova E, Moskalenko C, Pashev I, Pasheva E. DNA bending versus DNA end joining activity of HMGB1 protein is modulated in vitro by acetylation. Biochemistry. 2007;46:2111–2117. doi: 10.1021/bi0614479. [DOI] [PubMed] [Google Scholar]
- 85.Stros M, Cherny D, Jovin TM. HMG1 protein stimulates DNA end joining by promoting association of DNA molecules via their ends. Eur J Biochem. 2000;267:4088–4097. doi: 10.1046/j.1432-1327.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- 86.Nagaki S, Yamamoto M, Yumoto Y, Shirakawa H, Yoshida M, Teraoka H. Non-histone chromosomal proteins HMG1 and 2 enhance ligation reaction of DNA double-strand breaks. Biochem Biophys Res Commun. 1998;246:137–141. doi: 10.1006/bbrc.1998.8589. [DOI] [PubMed] [Google Scholar]
- 87.Assenberg R, Webb M, Connolly E, Stott K, Watson M, Hobbs J, Thomas JO. A critical role in structure-specific DNA binding for the acetylatable lysine residues in HMGB1. Biochem J. 2008;411:553–561. doi: 10.1042/BJ20071613. [DOI] [PubMed] [Google Scholar]
- 88.Ugrinova I, Pashev IG, Pasheva EA. Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry. 2009;48:6502–6507. doi: 10.1021/bi9004304. [DOI] [PubMed] [Google Scholar]
- 89.Baltus GA, Kowalski MP, Zhai H, Tutter AV, Quinn D, Wall D, Kadam S. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 2009;27:2175–2184. doi: 10.1002/stem.168. [DOI] [PubMed] [Google Scholar]
- 90.Ito I, Fukazawa J, Yoshida M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J Biol Chem. 2007;282:16336–16344. doi: 10.1074/jbc.M608467200. [DOI] [PubMed] [Google Scholar]
- 91.Sgarra R, Diana F, Bellarosa C, Dekleva V, Rustighi A, Toller M, Manfioletti G, Giancotti V. During apoptosis of tumor cells HMGA1a protein undergoes methylation: identification of the modification site by mass spectrometry. Biochemistry. 2003;42:3575–3585. doi: 10.1021/bi027338l. [DOI] [PubMed] [Google Scholar]
- 92.Ramachandran C, Yau P, Bradbury EM, Shyamala G, Yasuda H, Walsh DA. Phosphorylation of high-mobility-group proteins by the calcium-phospholipid-dependent protein kinase and the cyclic AMP-dependent protein kinase. J Biol Chem. 1984;259:13495–13503. [PubMed] [Google Scholar]
- 93.Krishnan N, Lam TT, Fritz A, Rempinski D, O'Loughlin K, Minderman H, Berezney R, Marzluff WF, Thapar R. The Prolyl Isomerase Pin1 Targets Stem-Loop Binding Protein (SLBP) To Dissociate the SLBP-Histone mRNA Complex Linking Histone mRNA Decay with SLBP Ubiquitination. Mol Cell Biol. 2012;32:4306–4322. doi: 10.1128/MCB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lanzotti DJ, Kupsco JM, Yang XC, Dominski Z, Marzluff WF, Duronio RJ. Drosophila stem-loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol Biol Cell. 2004;15:1112–1123. doi: 10.1091/mbc.E03-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krishnan N, Titus MA, Thapar R. The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins. PLoS One. 2014;9:e85427. doi: 10.1371/journal.pone.0085427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reeves R, Chang D, Chung SC. Carbohydrate modifications of the high mobility group proteins. Proc Natl Acad Sci U S A. 1981;78:6704–6708. doi: 10.1073/pnas.78.11.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ditsworth D, Zong WX, Thompson CB. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J Biol Chem. 2007;282:17845–17854. doi: 10.1074/jbc.M701465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Payet D, Travers A. The acidic tail of the high mobility group protein HMG-D modulates the structural selectivity of DNA binding. J Mol Biol. 1997;266:66–75. doi: 10.1006/jmbi.1996.0782. [DOI] [PubMed] [Google Scholar]
- 99.Stros M, Stokrova J, Thomas JO. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994;22:1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sheflin LG, Fucile NW, Spaulding SW. The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry. 1993;32:3238–3248. doi: 10.1021/bi00064a005. [DOI] [PubMed] [Google Scholar]
- 101.Watson M, Stott K, Thomas JO. Mapping intramolecular interactions between domains in HMGB1 using a tail-truncation approach. J Mol Biol. 2007;374:1286–1297. doi: 10.1016/j.jmb.2007.09.075. [DOI] [PubMed] [Google Scholar]
- 102.Stott K, Watson M, Howe FS, Grossmann JG, Thomas JO. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J Mol Biol. 2010;403:706–722. doi: 10.1016/j.jmb.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 103.Knapp S, Muller S, Digilio G, Bonaldi T, Bianchi ME, Musco G. The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry. 2004;43:11992–11997. doi: 10.1021/bi049364k. [DOI] [PubMed] [Google Scholar]
- 104.Muller S, Bianchi ME, Knapp S. Thermodynamics of HMGB1 interaction with duplex DNA. Biochemistry. 2001;40:10254–10261. doi: 10.1021/bi0100900. [DOI] [PubMed] [Google Scholar]
- 105.Webb M, Payet D, Lee KB, Travers AA, Thomas JO. Structural requirements for cooperative binding of HMG1 to DNA minicircles. J Mol Biol. 2001;309:79–88. doi: 10.1006/jmbi.2001.4667. [DOI] [PubMed] [Google Scholar]
- 106.Ueda T, Chou H, Kawase T, Shirakawa H, Yoshida M. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry. 2004;43:9901–9908. doi: 10.1021/bi035975l. [DOI] [PubMed] [Google Scholar]
- 107.Cato L, Stott K, Watson M, Thomas JO. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol. 2008;384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 108.Rowell JP, Simpson KL, Stott K, Watson M, Thomas JO. HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure. 2012;20:2014–2024. doi: 10.1016/j.str.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 109.Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg HG, Grummt I. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 1992;11:2211–2218. doi: 10.1002/j.1460-2075.1992.tb05280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jantzen HM, Chow AM, King DS, Tjian R. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 1992;6:1950–1963. doi: 10.1101/gad.6.10.1950. [DOI] [PubMed] [Google Scholar]
- 111.Lin CY, Navarro S, Reddy S, Comai L. CK2-mediated stimulation of Pol I transcription by stabilization of UBF-SL1 interaction. Nucleic Acids Res. 2006;34:4752–4766. doi: 10.1093/nar/gkl581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 113.Dominski Z, Yang XC, Raska CS, Santiago C, Borchers CH, Duronio RJ, Marzluff WF. 3' end processing of Drosophila melanogaster histone pre-mRNAs: requirement for phosphorylated Drosophila stem-loop binding protein and coevolution of the histone pre-mRNA processing system. Mol Cell Biol. 2002;22:6648–6660. doi: 10.1128/MCB.22.18.6648-6660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roemer SC, Adelman J, Churchill ME, Edwards DP. Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding. Nucleic Acids Res. 2008;36:3655–3666. doi: 10.1093/nar/gkn249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1799:175–180. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lyons SM, Ricciardi AS, Guo AY, Kambach C, Marzluff WF. The C-terminal extension of Lsm4 interacts directly with the 3' end of the histone mRNP and is required for efficient histone mRNA degradation. RNA. 2014;20:88–102. doi: 10.1261/rna.042531.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gorgoni B, Andrews S, Schaller A, Schumperli D, Gray NK, Muller B. The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA. 2005;11:1030–1042. doi: 10.1261/rna.7281305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol. 2014;15:749–760. doi: 10.1038/nrm3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nolte RT, Conlin RM, Harrison SC, Brown RS. Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc Natl Acad Sci U S A. 1998;95:2938–2943. doi: 10.1073/pnas.95.6.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Churchill ME, Tullius TD, Klug A. Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc Natl Acad Sci U S A. 1990;87:5528–5532. doi: 10.1073/pnas.87.14.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 124.Nedbal W, Frey M, Willemann B, Zentgraf H, Sczakiel G. Mechanistic insights into p53-promoted RNA-RNA annealing. J Mol Biol. 1997;266:677–687. doi: 10.1006/jmbi.1996.0813. [DOI] [PubMed] [Google Scholar]
- 125.Stoll R, Lee BM, Debler EW, Laity JH, Wilson IA, Dyson HJ, Wright PE. Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA. J Mol Biol. 2007;372:1227–1245. doi: 10.1016/j.jmb.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 126.Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 2014;28:2304–2313. doi: 10.1101/gad.250746.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhai G, Iskandar M, Barilla K, Romaniuk PJ. Characterization of RNA aptamer binding by the Wilms' tumor suppressor protein WT1. Biochemistry. 2001;40:2032–2040. doi: 10.1021/bi001941r. [DOI] [PubMed] [Google Scholar]
- 128.Baird-Titus JM, Clark-Baldwin K, Dave V, Caperelli CA, Ma J, Rance M. The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site. J Mol Biol. 2006;356:1137–1151. doi: 10.1016/j.jmb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 129.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 130.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 131.Placido D, Brown BA, Lowenhaupt K, 2nd, Rich A, Athanasiadis A. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kang HJ, Le TV, Kim K, Hur J, Kim KK, Park HJ. Novel interaction of the Z-DNA binding domain of human ADAR1 with the oncogenic c-Myc promoter G-quadruplex. J Mol Biol. 2014;426:2594–2604. doi: 10.1016/j.jmb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 133.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 134.Schwartz JC, Cech TR, Parker RR. Biochemical Properties and Biological Functions of FET Proteins. Annu Rev Biochem. 2014 doi: 10.1146/annurev-biochem-060614-034325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kuo PH, Chiang CH, Wang YT, Doudeva LG, Yuan HS. The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids. Nucleic Acids Res. 2014;42:4712–4722. doi: 10.1093/nar/gkt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, Allain FH. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- 137.Kupsco JM, Wu MJ, Marzluff WF, Thapar R, Duronio RJ. Genetic and biochemical characterization of Drosophila Snipper: A promiscuous member of the metazoan 3'hExo/ERI-1 family of 3' to 5' exonucleases. RNA. 2006;12:2103–2117. doi: 10.1261/rna.186706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3' end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Romeo V, Griesbach E, Schumperli D. CstF64: Cell Cycle Regulation and Functional Role in 3' End Processing of Replication-Dependent Histone mRNAs. Mol Cell Biol. 2014;34:4272–4284. doi: 10.1128/MCB.00791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Luscher B, Schumperli D. RNA 3' processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]