Abstract
2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a carcinogenic heterocyclic aromatic amine formed in cooked meats, is metabolically activated to electrophilic intermediates that form covalent adducts with DNA and protein. We previously identified an adduct of PhIP formed at the Cys34 residue of human serum albumin following reaction of albumin with the genotoxic metabolite 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP). The major adducted peptide recovered from a tryptic/chymotryptic digest was identified as the missed-cleavage peptide LQQC*[SO2PhIP]PFEDHVK, a [Cysteine-S-yl-PhIP]-S-dioxide linked adduct. In this investigation, we have characterized the albumin adduction products of N-sulfooxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-sulfooxy-PhIP), which is thought to be a major genotoxic metabolite of PhIP formed in vivo. Targeted and data-dependent scanning methods showed that N-sulfooxy-PhIP adducted to the Cys34 of albumin in human plasma to form LQQC*[SO2PhIP]PFEDHVK at levels that were 8 to 10-fold greater than the adduct levels formed with N-(acetyloxy)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-acetoxy-PhIP) or HONH-PhIP. We also discovered that N-sulfooxy-PhIP forms an adduct at the sole tryptophan (Trp214) residue of albumin in the sequence AW*[PhIP]AVAR. However, stable adducts of PhIP with albumin were not detected in human hepatocytes. Instead, PhIP and 2-amino-1-methyl-6-(5-hydroxy)-phenylimidazo[4,5-b]pyridine (5-HO-PhIP), a solvolysis product of the proposed nitrenium ion of PhIP, were recovered during the proteolysis, suggesting a labile sulfenamide linkage had formed between an N-oxidized intermediate of PhIP and Cys34 of albumin. A stable adduct was formed at the Tyr411 residue of albumin in hepatocytes, and identified as a deaminated product of PhIP, Y*[desaminoPhIP]TK, where the 4-HO-tyrosine group bound to the C-2 imidazole atom of PhIP.
Keywords: Carcinogens, heterocyclic aromatic amines, serum albumin adducts
INTRODUCTION
2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is a carcinogenic heterocyclic aromatic amine (HAA) formed in cooked meat.1–4 PhIP undergoes metabolic activation by cytochrome P450s, to form 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP).5,6 HONH-PhIP undergoes esterification by sulfotransferase (SULTs) or N-acetyltransferase (NATs) to form the corresponding esters7 which undergo heterolytic cleavage to produce the presumed nitrenium ion that binds to DNA.8,9 The major DNA adduction product of N-oxidized metabolites of PhIP has been characterized as is N-(deoxyguanosin-8-y1)-PhIP (dG-C8-PhIP).10,11 The dG-C8-PhIP adduct has been employed as a biomarker in human studies.12,13
The measurement of DNA adducts in human biospecimens remains an analytical challenge because many DNA lesions are repaired and the trace levels of DNA adducts present in tissues may be difficult to measure, even by sensitive LC/MS-based methods.9,14 The same reactive metabolites of HAAs (or other procarcinogens) also may bind to proteins to form stable covalent adducts.15,16 The employment of protein carcinogen adducts as biomarkers is a promising approach to assess exposure to hazardous chemicals because stable covalent protein adducts are not repaired and expected to accumulate during chronic exposure and follow the kinetics of the lifetime of the protein in vivo.17,18
Hemoglobin (Hb) and serum albumin are the two most abundant proteins in blood and have long life times. The chemistry of reaction of Hb and albumin with many electrophilic genotoxicants and toxicants has been investigated.16,19–21 Like structurally related arylamines, N-oxidation is a major pathway of metabolism of PhIP in humans.22–24 Hb adducts of aromatic amines are formed by a co-oxidation reaction of the N-hydroxylated arylamine metabolites with oxy-Hb to form the arylnitroso intermediate, which selectively bind at the Hb-Cys93β residue and form arylsulfinamide adducts.18,25,26 Arylsulfinamide adducts are labile and undergo hydrolysis under acidic or alkaline pH to produce the parent arylamine. The labile nature of the Hb arylsulfinamide bond has been exploited to monitor occupational and environmental exposures to a number of primary arylamines.27 In the case of 4-aminobiphenyl (4-ABP), the structure of the Hb sulfinamide linkage was proven by X-ray crystallography.28 However, a study with radiolabelled 14C-PhIP showed that PhIP binding to Hb was very low in humans and Hb-PhIP adducts do not appear to be promising candidate biomarkers.29 In contrast, the binding of PhIP to albumin occurred at levels up to several percent of the ingested dose,29,30 and may be sufficient to biomonitor albumin-PhIP adducts in humans by LC/MS-based methods. However, the structure(s) of the albumin-PhIP adduct(s) formed in humans is unknown.
Human albumin contains only one reduced Cys residue.31 The Cys34 of albumin is a strong nucleophile and it also serves as a powerful antioxidant and scavenger of reactive oxygen species.21,32 The Cys34 residues of rodent and human albumin trap reactive electrophiles of HAAs, e.g. 2-amino-3-methylimidazo[4,5-f]quinoline,33 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline34 and PhIP,35–39 and other toxic electrophiles.21 We have shown that human albumin forms adducts with HONH-PhIP and 2-nitroso-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO-PhIP) at the Cys34 via a sulfinamide linkage.37,39 N-(Acetyloxy)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-acetoxy-PhIP), a metabolite of PhIP that forms covalent DNA adducts,10,40 reacts with albumin to form an unstable sulfenamide linkage at Cys34.38 We also identified several adducts formed between human albumin and 2-nitro-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO2-PhIP).38
The formation of covalent adducts of carcinogens to proteins such as albumin is thought to be driven not only by the abundance and relative nucleophilicities of the different amino acid residues available for reaction, but also by non-covalent interactions of the reactive metabolite with different receptor regions of proteins, which can direct the sites of carcinogen adduct formation.41 For example, the major adduct formed between the activated hydroxamic acid of 4-ABP and rat albumin occurred at the sole tryptophan residue, which is situated in a fatty acid binding site in a relatively hydrophobic position deep within the native protein.42 Studies employing genetically engineered mammalian cell lines expressing human NATs showed that HONH-PhIP is poorly bioactivated by NAT1 and NAT2.43–45 In comparison, cell lines expressing SULT1A1 were highly effective in the bioactivation of HONH-PhIP, to form the presumed reactive metabolite N-sulfooxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-sulfooxy-PhIP), which adducted to DNA.46,47 Thus, the contribution of N-sulfooxy-PhIP to the chemical modification protein (or DNA) may be much greater than N-acetoxy-PhIP. N-sulfooxy-PhIP is appreciably more polar than HONH-PhIP and N-acetoxy PhIP, and it exists as a negatively charged species at physiological pH. Therefore, the sites of reactivity of N-sulfooxy-PhIP with albumin may be different from other N-oxidized PhIP metabolites.37–39 To our knowledge, studies on the covalent binding of N-sulfooxy-PhIP with protein or DNA have not been reported.
Our goal is to comprehensively characterize the adduction products of N-oxidized metabolites of PhIP with human albumin and develop albumin-PhIP adducts as biomarkers for molecular epidemiology studies that seek to understand the role of PhIP in human cancer. In this study, we report our findings on the relative reactivity of human albumin with HONH-PhIP, N-acetoxy-PhIP, and N-sulfooxy-PhIP, all of which produce the proposed PhIP nitrenium ion (Fig. 1).8
Figure 1.
Structures of PhIP and N-oxidized metabolites
MATERIALS AND METHODS
Caution
PhIP is a carcinogen and should be handled in a well-ventilated fume hood with the appropriate protective clothing.
Chemicals and Materials
PhIP was purchased from Toronto Research Chemicals (Toronto, ON, Canada). 2-Amino-1-methyl-6-[2H5]-phenylimidazo[4,5-b]pyridine ([2H5]-PhIP, 99% isotopic purity) was a gift from Dr. Mark Knize and Dr. Kristen Kulp (Lawrence Livermore National Laboratory, Livermore, CA). Human serum albumin, trypsin, chymotrypsin, pronase E, leucine aminopeptidase, prolidase, meta-chloroperoxybenzoic acid, tert-butyl hydroperoxide, dimethyldioxirane and sodium periodate, and LC-MS grade formic were purchased from Sigma-Aldrich (St. Louis, MO). LC-MS grade solvents were purchased from Fisher Scientific (Pittsburgh, PA). All other chemicals were ACS grade, and purchased from Sigma-Aldrich unless stated. Isolute C18 solid-phase extraction (SPE) columns (25 mg) were from Biotage (Charlotte, NC). Pierce Albumin Depletion Kit was from Thermo Fisher (Rockford, IL). Amicon Ultra centrifugal filter units (10,000 mw cutoff) were from Millipore (Billerica, MA). Human plasma was purchased from Bioreclamation LLC (Hicksville, NY).
Synthesis of HONH-PhIP, N-Acetoxy-PhIP, and 2-Amino-1-methyl-6-(5-hydroxy)-phenylimidazo[4,5-b]pyridine (5-HO-PhIP), N-sulfooxy-PhIP, and N-(deoxyguanosin-8-y1)-PhIP (dG-C8-PhIP)
The syntheses of HONH-PhIP and N-acetoxy-PhIP were previously reported.10,38,48 5-HO-PhIP was obtained by the decomposition of N-acetoxy-PhIP in phosphate buffer (pH 7.4).35,49 Two methods were employed for the synthesis of N-sulfooxy-PhIP. The first method employed sulfur trioxide/pyridine.50 HONH-PhIP (5 µg, 21 nmol) was reacted with a 1.3 molar excess of sulfur trioxide in 50 µL anhydrous pyridine at room temperature for 18 h. The second method was described by Beland and Miller for the synthesis of 2-acetylaminfluorene N-sulfate.51 A 5-fold molar excess of N,N'-dicyclohexylcarbodiimide (DCCI) dissolved in DMF previously dried over 3Å molecular sieves was added to HONH-PhIP (5 µg, 21 nmol), followed by a 1.2x molar ratio of H2SO4 to HONH-PhIP. The mixture was purged with argon, and then mixed for 6 h at room temperature. Thereafter, the mixtures were vacuum centrifuged to dryness using a high vacuum CentriVap Cold Trap (Labconco, MO). The N-sulfooxy-PhIP residues were dissolved in 1:1 H2O:CH3CN, sonicated for 1 min, and then immediately subjected to infusion into the MS source, or reacted with DNA or albumin. The UV spectra of HONH-PhIP, N-acetoxy-PhIP and PhIP were acquired in CH3OH, employing an Agilent 8453 spectrophotometer (Agilent Technologies, Santa Clara, CA) (Supporting Information Fig. S-1). The spectral data are consistent with previous results.37 The ESI-MSn product ion spectra support the proposed structure of N-sulfooxy-PhIP ([M+H]+ m/z 321.1→[M+H-SO3]+ m/z 241.1; [M-H]− m/z 319.1→[M-H-SO3]− m/z 239.1). Further mass spectral data are provided (vide infra). dG-C8-PhIP and its internal standard [13C10]-dG-C8-PhIP were synthesized as described.10,11
Trapping N-sulfooxy-PhIP, N-Acetoxy-PhIP or HONH-PhIP with 2′-Deoxyguanosine (dG)
The reaction condition of N-acetoxy-PhIP with dG to form dG-C8-PhIP was previously described10,11 and employed to trap other electrophiles of PhIP in this study. The N-sulfooxy-PhIP (13.4 µg, 42 nmol, in 20 µL 50% CH3CN), N-acetoxy-PhIP (11.8 µg, 42 nmol, in 20 µL CH3OH) or HONH-PhIP (10.1 µg, 42 nmol, in 20 µL C2H5OH) were added to a solution of dG (1 mg/mL in 50 mM potassium phosphate buffer, pH 8.0). The reaction mixture was agitated at 37 °C for 1 h. Reaction products were applied to an Isolute C18 SPE column, followed by washing the resin with 10% CH3OH, and eluting dG-C8-PhIP with 100% CH3OH.11
Modification of Human Serum Albumin and Plasma with HONH-PhIP, N-Acetoxy-PhIP, and N-sulfooxy-PhIP
Mixed disulfides formed at Cys34 of albumin were reduced by treatment with βME.33 Reduced serum albumin (1 nmol in 300 µL 100 mM potassium phosphate buffer, pH 7.4) was reacted with HONH-PhIP (3 nmol/10 µL C2H5OH), N-acetoxy-PhIP (3 nmol/10 µL CH3OH) or N-sulfooxy-PhIP (3 nmol/20 µL 50% CH3CN). The mixtures were incubated at 37 °C for 12 h (HONH-PhIP), or 3 h (N-acetoxy-PhIP and N-sulfooxy-PhIP) with agitation. The PhIP products unbound to albumin were removed by solvent extraction with 2x volume of ethyl acetate, twice. The albumin solution underwent vacuum centrifugation for 5 min to remove the residual ethyl acetate, and the solution was subjected to a buffer exchange (450 µL of 100 mM potassium phosphate buffer, pH 7.4, 2-times) to remove residual unbound PhIP using centrifugal filters (10,000 mw cutoff).
Human plasma (5 µL) was diluted with 300 µL 100 mM potassium phosphate buffer, pH 7.4, followed by addition of HONH-PhIP, N-acetoxy-PhIP, N-sulfooxy-PhIP at a molar ratio of 1:3 (albumin: carcinogen). The mixture was incubated at 37 °C for 12 h, with constant agitation. The albumin was processed as described above. The albumin was purified with Albumin Depletion Kit (Pierce Biotechnology, Rockford, IL) employing chromatographic conditions provided by the supplier. The protein concentration was estimated by measuring the UV absorption at 280 nm (extinction coefficient, 35,218 M−1 cm−1).69
PhIP DNA and Albumin Adduct Formation in Human Hepatocytes
Human liver samples were obtained from patients undergoing liver resection for primary or secondary hepatomas through the Centre de Ressources Biologiques (CRB)-Santé of Rennes (http://www.crbsante-rennes.com, CHRU Pontchaillou, Rennes, France). The research protocol was conducted under French legal guidelines and the local institutional ethics committee. Hepatocytes were isolated by a two-step collagenase perfusion procedure, and parenchymal cells were seeded in Petri dishes at a density of 3 ×106 viable cells/19.5-cm2 dish in 3 mL of Williams’ modified medium except that fetal calf serum was replaced with human albumin pre-reduced with βME (1 g/L).52 The cells were then incubated with PhIP (50 µM) or PhIP:[2H5]-PhIP (1:1, 50 µM) for 24 h. The cells were retrieved from the Petri dishes and pelleted by centrifugation.
PhIP DNA Adduct Measurements in Human Hepatocytes by UPLC-ESI/MS3
DNA was isolated by chloroform phenol extraction, and dG-C8-PhIP was quantitated by UPLC-ESI/MS3 at the MS3 scan stage.11,52 [13C10]-dG-C8-PhIP was employed as an internal standard and spiked into DNA prior to enzymatic digestion at a level of 1 adduct per 106 DNA bases. The analyses were conducted with a Waters NanoAcquity UPLC system (Waters Corp., New Milford, MA) equipped with a Waters Symmetry trap column (180 µm × 20 mm, 5 µm particle size), a Michrom C18 AQ column (0.3 × 150 mm, 3 µm particle size) and a Michrom CaptiveSpray™ source interfaced to a linear quadrupole ion-trap mass spectrometer (LTQ Velos, Thermo Fisher, San Jose, CA), employing chromatographic conditions and MS data acquisition parameters as described.52
Albumin Isolation and Proteolytic Digestion
The albumin in the cell culture media of hepatocytes was extracted twice with 2 vol of ethyl acetate to remove unmetabolized PhIP, and then the aqueous phase was precipitated with 2 vol of chilled ethanol. The precipitated albumin was centrifuged, and resuspended in 0.3 mL 100 mM postassium phosphate buffer (pH 7.4). The protein was then isolated by the Albumin Depletion Kit as described above, followed by desalting using Millipore centrifugal filters (10,000 mw cutoff).
Trypsin/Chymotrypsin Digestion
Albumin (50 µg in 200 µL 50 mM ammonium bicarbonate buffer, pH 8.5) was digested with trypsin (1/50, w/w, protease/protein) and chymotrypsin (1/25, w/w, protease/protein), which were dissolved in 1 mM HCl containing 2 mM CaCl2. The mixture was incubated at 37 °C for 20 h. For denaturation of albumin, the protein (50 µg) was resuspended in 0.25 M Tris Buffer (pH 7.4) containing 8 M urea. Dithiothreitol (DTT) (1.5 mg, 20 mM) was added to the solution, and the mixture was incubated at 55 °C for 1 h, followed by addition of iodoacetamide (IAA) (6 mg, 65 mM). The mixture was incubated in dark at 22 °C for 1 h. Excess DTT and IAA were removed using centrifugal filters. The albumin was dissolved in 200 µL 50 mM ammonium bicarbonate buffer, pH 8.5, and subjected to proteolytic digestion as described above. After digestion, the mixture was diluted with 10x volume of distilled water, and applied to an Isolute C18 SPE column. Polar peptides were removed with 10% CH3OH (2 mL) and adducts were eluted with CH3OH (1 mL), followed by concentration to dryness by vacuum centrifugation.
Trypsin Digestion
Denatured or non-denatured PhIP-modified albumin (50 µg in 200 µL 50 mM ammonium bicarbonate buffer, pH 8.5) was digested with trypsin (1/50, w/w, protease/protein). The mixture was incubated at 37 °C for 20 h, followed by SPE purification described as above.
Pronase E/Leucine Aminopeptidase/Prolidase Digestion
Unmodified or PhIP-modified albumin (50 µg in 200 µL 50 mM ammonium bicarbonate buffer, pH 8.5) was digested with pronase E (1/2, w/w, protease/protein), leucine aminopeptidase (1/30, w/w, protease/protein), and prolidase (1/8, w/w, protease/protein). The mixture was incubated at 37 °C for 20 h, followed by SPE purification described as above.
Mass Spectral Measurements of PhIP Metabolites and Ultraperformance Liquid Chromatography-Electrospray Ionization-Multistage Mass Spectrometry (UPLC-ESI/MSn) for albumin PhIP Adducts
Mass spectral data of metabolites and synthetic peptides LQQCPF (New England Inc., Gardner, MA) were acquired by infusion with an Orbitrap Elite™ hydrid ion trap-Orbitrap mass spectrometer (Thermo Fisher, San Jose, CA) and a Michrom Advance CaptiveSpray™ source (Auburn, CA). Typical instrument tuning parameters were as follows: capillary temperature, 270 °C; source spray voltage, 2 kV; S-lens RF level, 69%; skimmer offset voltage, 0 V; source fragmentation, 5 V. Helium, set at 1 millitorr, was used as the collision and damping gas in the ion trap. There was no sheath or auxiliary gas. All analyses were conducted in the positive ionization mode except for the characterization of the N-sulfooxy-PhIP. The isolation width was set at m/z 1 for the MS2 and MS3 scan modes, the activation Q was set at 0.35, and the activation time was 10 ms.
The UPLC-ESI/MSn system consists of a Dionex Ultimate 3000 LC (Thermo Scientific, Waltham, MA) and an Orbitrap EliteTM Hydrid Ion Trap-Orbitrap mass spectrometer. The peptides of the albumin digest were resolved with a Magic C18AQ column (0.3 mm x 150 mm, Micrhom Bioresource Inc., Auburn, CA) with a 25 min (targeted MS/MS) or 60 min (data-dependent scanning) gradient starting from 100% A solvent (5% CH3CN with 0.01% HCO2H) to 100% B solvent (95% CH3CN with 0.01% HCO2H) at a flow rate of 5 µL/min. Chromeleon 7.2 Chromatography Data System was used for the HPLC management. The Advance CaptiveSpray was employed as the ion source and parameters were set as follows: capillary temperature, 270 °C; ionization voltage, 2 kV for positive ion mode; in-source fragmentation, 5 V; 1 µscan; maximum injection time, 10 ms for MS and 50 ms for MSn; MS fragmentation, a normalized collision energy of 35%; no auxiliary and sheath gases were used. The isolation width was set at m/z 1 for both MS2 and MS3 scan modes, and the activation Q was set at 0.35. AGC (automated gain control) was set 30,000 for ion trap (IT) MS and 10,000 for IT MSn, 106 for Orbitrap (FT) MS, and 50,000 for FT The Orbitrap was routinely calibrated in positive and negative ion modes using Pierce LTQ Velos ESI Positive Ion Calibration Solution (2 µg/mL caffeine, 1 µg/mL MRFA, 0.001% Ultramark 1621 and 0.00005% n-butylamine) and Pierce ESI Negative Ion Calibration Solution (2.9 µg/mL sodium dodecyl sulfate, 5.4 µg/mL sodium taurocholate and 0.001% Ultramark 1621) (Pierce Biotechnology, Rockford, IL).
For the mass tag triggered data-dependent acquisition method, a full MS scan (mass range, 100–1800 Dalton) was obtained, followed by 5 data dependent scans (mass range, 100–1800 Da), which were triggered by a pattern of the unlabeled PhIP to [2H5]-PhIP adducts at a partner intensity ratio of 65–100%, by enabling the mass tags option (m/z 5, 2.5 or 1.67 for singly, doubly or triply charged ions).38 The MS/MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 180 s with a repeat of 3 and a repeat duration of 60 s. CID was selected for ion fragmentation with a normalized collision energy of 35%. The maximum injection time of MS/MS was set at 50 ms and the isolation width was set as 2 m/z.
Data Analysis
Xcalibur version 2.2 was employed for data acquisition and data processing. Peptide adducts were identified manually and facilitated by MyriMatch (version 2.1.140)53,54 using a 31 protein subset database containing albumin from an initial search against the RefSeq human protein database, version 37.3. MyriMatch was configured to have Cys to contain carbamidomethyl (+57.0 Da) for IAA treatment, or (+32.0 or 48.0 Da) for oxidation of Cys to the sulfinic or sulfonic acids, and to allow for the possibility of oxidation (+16.0 Da) on methionines and deamidation (−17.0 Da) of N-terminal glutamines. Peptides modified with HONH-PhIP and HONH-[2H5]-PhIP were allowed to have dynamic modifications at [C,K,Y,S,T,W,H] of 222.1 (HONH-PhIP - H2O) and 227.1 Da (HONH-[2H5]-PhIP - H2O), or dynamic modifications at [C] of 238.1 and 243.1 Da for adduction with nitroso-PhIP (sulfinamide adducts) and 254.1 and 259.1 Da (sulfonamide adducts). Candidate peptides were allowed to have trypsin cleavages or protein termini at one or both termini (semitryptic search), and up to two missed cleavages were permitted. The precursor error was set at 1.25 m/z, but fragment ions were required to match within 0.5 m/z. Analyses of modified albumin were also performed with trypsin/chymotrypsin digests, employing the same configurations. The IDPicker algorithm v3.0.634 filtered the identifications for each spectrum with a 2% identification false discovery rate at the peptide spectrum match level.
RESULTS
Characterization of HONH-PhIP, N-acetoxy-PhIP, N-sulfooxy-PhIP
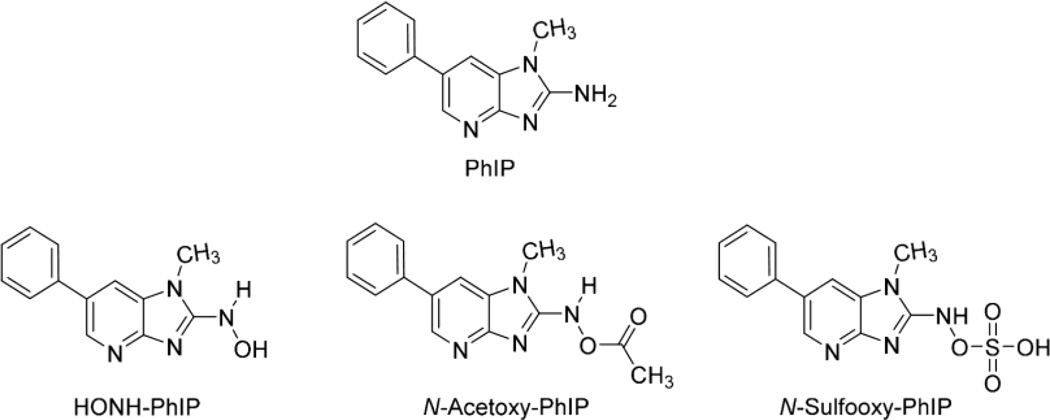
The UV spectra of PhIP, HONH-PhIP, and N-acetoxy-PhIP are provided in Supporting Information Fig. S-1. Because of its instability, the N-sulfooxy-PhIP could not be isolated and characterized by UV or online UPLC/MS analysis. Therefore, the yield of synthesis could not be determined. N-Sulfooxy-PhIP underwent rapid hydrolysis once dissolved in aqueous solution, and only HONH-PhIP was detected by HPLC. The sulfate ester of N-hydroxy-N-acetyl-2-aminofluorene was also reported to be highly unstable in aqueous solution.51 In contrast, N-acetoxy-PhIP had sufficient stability to assay by UPLC/MS and UV measurement.38 The full scan mass spectra of HONH-PhIP, N-acetoxy-PhIP, and N-sulfooxy-PhIP, acquired by infusion, are shown in Figs 2A, 2B and 2C. The protonated ions [M+H]+ are observed for HONH-PhIP (m/z observed 241.1081 vs. calculated 241.1084) and N-acetoxy-PhIP (m/z observed 283.1192 vs. calculated 283.1190); however, the N-sulfooxy-PhIP underwent extensive hydrolysis and the protonated ion was present at extremely low abundance ([M+H]+ at m/z 321.0647 vs. calculated 321.0652). The base peak of N-sulfooxy-PhIP was observed at m/z 223.0979 (calculated 223.0978) in the full scan mass spectrum (Figure 2A) and assigned as the nitrenium ion ([M+H-H2SO4]+. The relative abundance of the 223.0979 ion in Fig. 2A was considerably greater than that observed in the full scan mass spectra of N-acetoxy-PhIP and HONH-PhIP (Figures 2B and 2C), an observation consistent with the labile nature of the N-sulfooxy linkage of N-sulfooxy-PhIP. The minor ion present in the full scan spectrum of N-sulfooxy-PhIP at m/z 208.0866 (m/z calculated 208.0869), is consistent with in source fragmentation and loss of hydroxylamine-O-sulfonic acid [NH2OSO3H]. Further fragmentation of the nitrenium ion (m/z 223.0979) produces the ions at m/z 196.0866 (m/z calculated 196.0869) [HCN], and m/z 179.0601 (m/z calculated 179.0604) [HCN+NH3] (Figure 2A).
Figure 2.
(A) ESI-MS full scan mass spectra of (A) N-sulfooxy-PhIP(m/z 321.0652), (B) N-acetoxy-PhIP (m/z 283.1190), and (C) HONH-PhIP (m/z 241.1084) in the positive ion mode. The ion observed at m/z 225.11130 is attributed to PhIP and occurs by in-source reduction of HONH-PhIP. (D) Product ion spectrum of N-sulfooxy-PhIP ([M-H]m/z 319.0506 acquired at 0 collision energy, isolation width m/z 1). (E) Product ion spectra of N-sulfooxy-PhIP ion at m/z 319.1; and (F) product ion spectra of N-sulfooxy-PhIP at the MS3 scan stage (319.0 > 239.1 >) in the negative ion mode. The cluster of ions designated X between m/z 255 – 263 in the full scan mass spectrum of N-acetoxy-PhIP (B) are background ions present in the reaction medium.
The product ion spectra of N-acetoxy-PhIP and HONH-PhIP are consistent with the spectral data in the literature.38 N-Acetoxy-PhIP (m/z observed 283.1192) undergoes CID with losses of C2H3O2 (m/z observed 224.1057 vs. calculated 224.1057) to form the PhIP radical cation, and C2H4O2 (m/z observed 223.0981 vs. calculated 223.0978) to form PhIP nitrenium ion (Supporting Information, Fig. S-2A). CID studies with HONH-PhIP and 1-[2H3C]-HONH-PhIP have shown that nitrenium ion formation in the gas phase occurs primarily by homolytic cleavages at the hydrogen atom bonded to the C1 methyl atom and the HO-N2 bond of HONH-PhIP.55 This species may rearrange to form a pyrazine ring,56 which undergoes fragmentation to produce ions at m/z 206.0713, 196.0870 and 179.0605 as described in the Supporting Information. These fragment ions are observed in the product ion spectra of N-acetoxy-PhIP (Supporting Information, Fig. S-2B) and HONH-PhIP (data not shown).
The deprotonated ion of N-sulfooxy-PhIP [M-H]− was observed in the negative ion mode at m/z 319.0499 (calculated 319.0506) (Figure 2D), and undergoes fragmentation to form the HONH-PhIP [M-H-SO3]− at m/z 239.0939 (calculated 239.0938) (Figure 2E). The product ion spectrum of the N-sulfooxy-PhIP at the MS3 scan stage (319. 1 > 239.1 > ) displays a radical anion at m/z 224.0703 (calculated 224.0704), attributed to a loss of CH3· from HONH-PhIP (Figure 2F). These mass spectral data support the proposed structure as N-sulfooxy-PhIP.
Trapping Reactive N-Oxidized Electrophiles of PhIP with dG
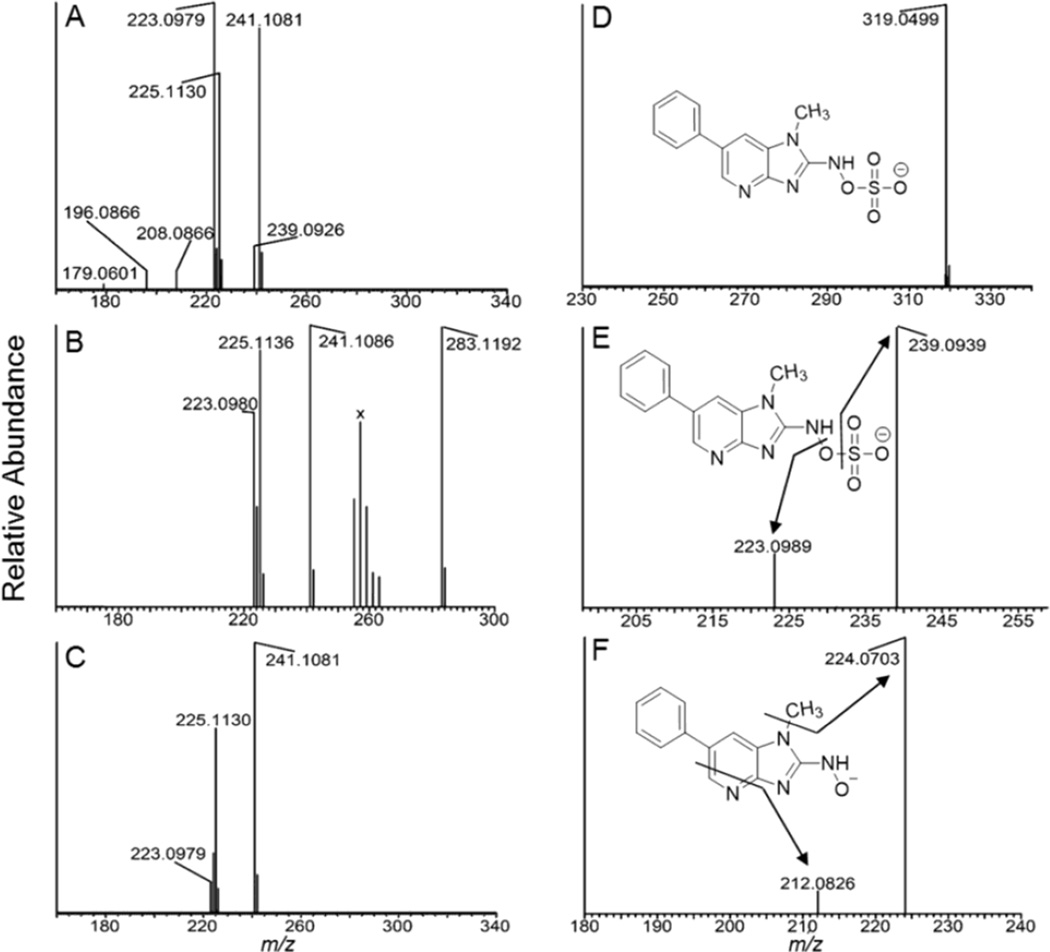
dG was used to trap the electrophilic N-oxidized metabolites of PhIP, by formation of dG-C8-PhIP, which was monitored by UPLC-ESI/MSn.11,52 The relative amounts of dG-C8 adduct formation were based on the total ion counts of dG-C8-PhIP adduct acquired at the MS3 scan stage. Representative chromatograms and the product ion spectrum are shown in Supporting Information (Fig. S-3). The spectrum is in excellent agreement to our previous data.11 The greatest amount of dG-C8-PhIP formation occurred with N-acetoxy-PhIP, followed by N-sulfooxy-PhIP, and lastly HONH-PhIP (Fig. 3). There was no significant difference between the peak area ion counts of dG-C8-PhIP obtained by the reaction of dG with the two different synthetic preparations of N-sulfooxy-PhIP (p=0.633, t-test). The dG-C8-PhIP adduct level obtained with N-sulfooxy-PhIP was 10–13 times greater than the adduct level formed with HONH-PhIP (p<0.005, t-test), providing further evidence for the existence of the reactive N-sulfooxy-PhIP electrophile. The lower level of dG-C8-PhIP obtained by reaction of dG with N-sulfooxy-PhIP than N-acetoxy-PhIP could be explained by the unstable nature of N-sulfooxy-PhIP, which undergoes extensive solvolysis during the reaction with dG. We could not measure N-sulfooxy-PhIP and assumed that the yield of the synthesis was quantitative. However, the yield of N-sulfooxy-PhIP may have been considerably lower and resulted in lower amounts of dG-C8-PhIP formed. Although, as reported below, the level of albumin Cys34 adduct formation was greater with N-sulfooxy-PhIP than N-acetoxy-PhIP.
Figure 3.
Peak area counts of dG-C8-PhIP adducts at the MS3 scan stage (m/z 490.1 > 374.3 >) from the reaction of dG with HONH-PhIP, N-acetoxy-PhIP, N-sulfooxy-PhIP (pyridine-SO3), or N-sulfooxy-PhIP (DMF-H2SO4). Data are three independent measurements (Mean ± SD, n=3).
Data-Dependent and Targeted Mass Spectrometric Characterization of Albumin Cys34 Adducts with N-Oxidized Metabolites of PhIP and [2H5]-PhIP
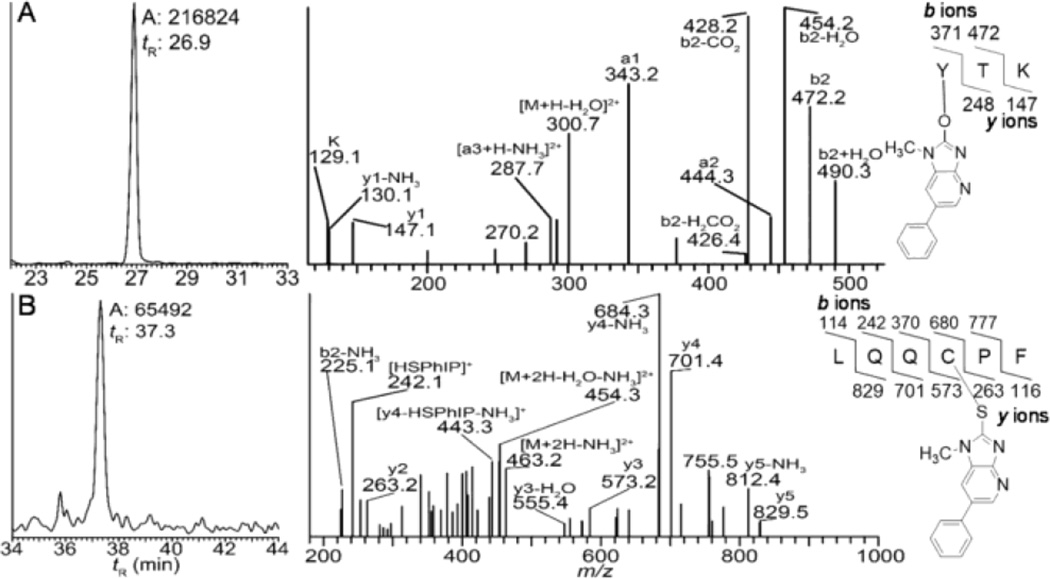
Adducts of PhIP formed with commercial albumin and albumin in human plasma modified with a 3-fold molar excess of N-sulfooxy-PhIP:N-sulfooxy-[2H5]-PhIP, N-acetoxy-PhIP:N-acetoxy-[2H5]-PhIP, or HONH-PhIP:HONH-[2H5]-PhIP (all at 1:1 molar ratio), following digestion with trypsin and chymotrypsin were characterized by data-dependent scanning.38,39 The Xcalibur software was programmed to switch from the full survey scan MS to the MS/MS scan mode, which was triggered by the characteristic isotopic pattern of unlabeled PhIP/[2H5]-PhIP at a partner intensity ratio of 65–100%.38 This wide tolerance for the partner intensity was employed because the deuterium isotope effect caused the [2H5]-PhIP-peptide adducts to elute about 2 s earlier than the unlabeled adducts in some instances, resulting in a distortion of the 1:1 ratio of unlabeled PhIP/[2H5]-PhIP adducts.38 The MyriMatch search program and manual verification of the isotope tags resulted in the identification of three Cys34 peptide adducts previously characterized with HONH-PhIP and N-acetoxy-PhIP: the single-missed cleavage sulfinamide LQQC*[SOPhIP]PFEDHVK ([M+3H]3+ at m/z 527.1) and single-missed cleavage sulfonamide, LQQC*[SO2PhIP]PFEDHVK ([M+3H]3+ at m/z 533.2) and the fully digested sulfinamide LQQC*[SOPhIP]PF ([M+2H]2+ at m/z 487.2) (Table 1).38 LQQC*[SOPhIP]PFEDHVK elutes at ~28 min, followed by LQQC*[SO2PhIP]PFEDHVK (tR 31 min) and LQQC*[SO2PhIP]PF (tR 34 min). The mass tag data-dependent acquisition method searched by MyriMatch or manual searching did not identify other adducts in the tryptic/chymotryptic digests of albumin modified with N-sulfooxy-PhIP or other N-oxidized PhIP metabolites. Thus, similar to the results previously obtained by reaction of HONH-PhIP or N-acetoxy-PhIP with human albumin.38 Cys34 is the major nucleophilic amino acid residue of albumin that reacts with N-sulfooxy-PhIP.
Table 1.
A list of identified peptide adducts from N-sulfooxy-PhIP and N-sulfooxy-[2H5]-PhIP, N-acetoxy-PhIP and N-acetoxy-[2H5]-PhIP, or HONH-PhIP and HONH-[2H5]-PhIP modified albumin using mass tag triggered data dependent scanning method.
| Proteolytic Enzymes | Peptides Adducts | Modified sites |
|---|---|---|
| Trypsin/ chymotrypsin |
LQQC*[SOPhIP]PFEDHVK | Cys34 |
| LQQC*[SO2PhIP]PFEDHVK | Cys34 | |
| LQQC*[SOPhIP]PF | Cys34 | |
| Pronase E/ leucine aminopeptidas/ prolidase |
C[SO2PhIP] | Cys34 |
| C*[SO2PhIP]PF | Cys34 | |
| W[PhIP] | Trp214 | |
| H[PhIP] | His |
The His residues of SA that reacted with N-oxidized PhIP are uncertain. There are 16 His residues in SA and the His containing PhIP adduct(s) was identified as a mono amino acid adduct following pronase digestion.
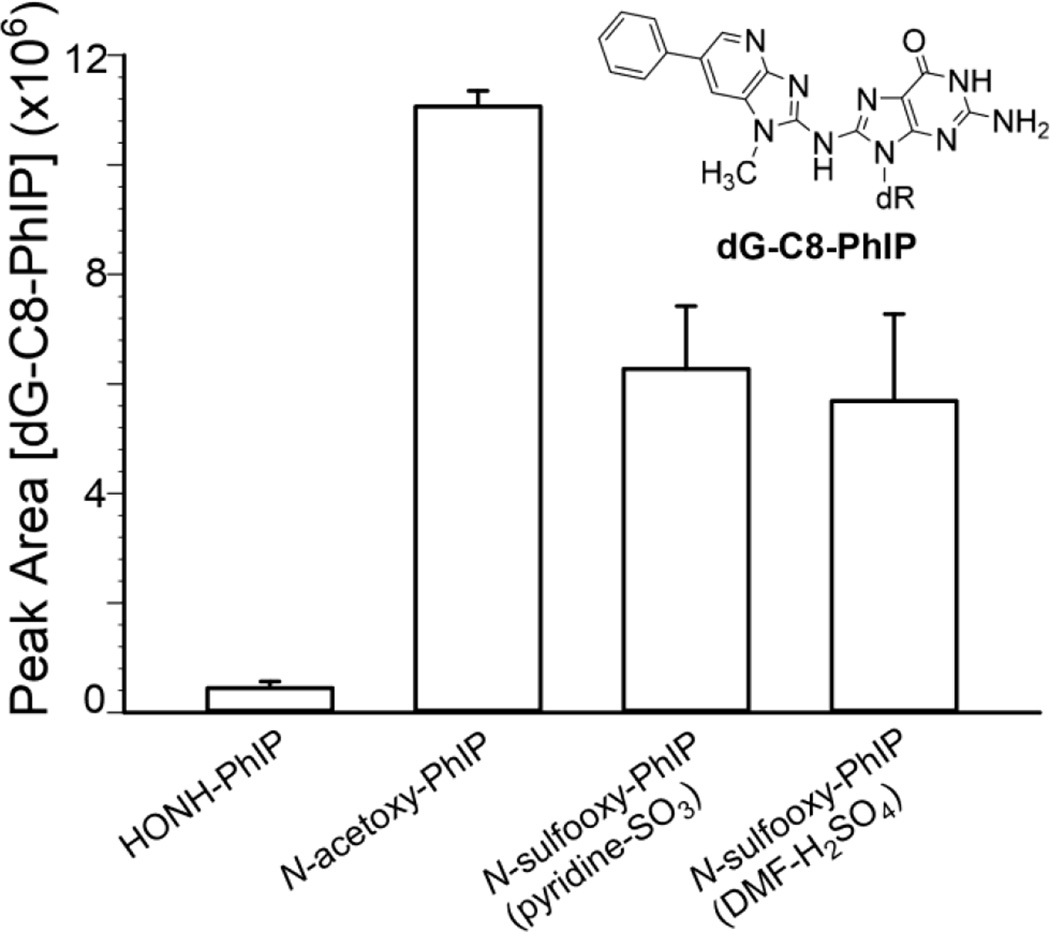
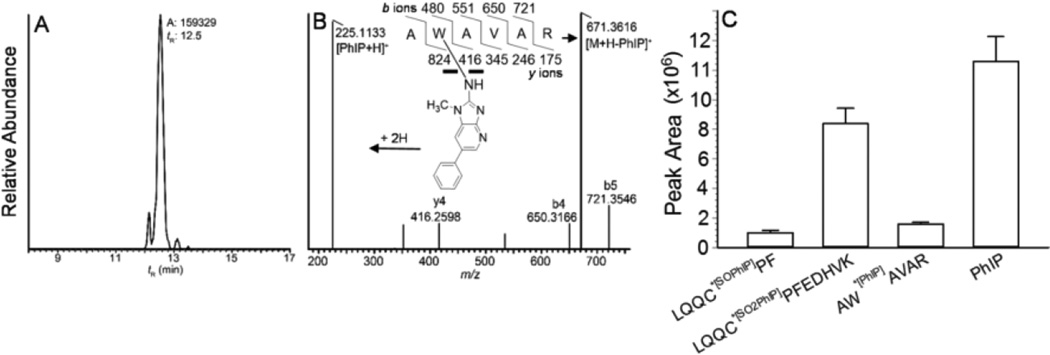
A representative mass chromatogram of albumin-Cys34 peptide adducts formed with N-sulfooxy-PhIP is shown in Figure 4 along with the higher energy collision dissociation (HCD) product ion spectra (LQQC*[SOPhIP]PF, [M+2H]2+ at m/z 487.2; LQQC*[SOPhIP]PFEDHVK, [M+3H]3+ at m/z 527.9, and LQQC*[SO2PhIP]PFEDHVK, [M+3H]3+ at m/z 533.2). The product ion spectrum of the sulfinamide LQQC*[SOPhIP]PF contains two major ions, PhIP (m/z 225.1) and [M+H-PhIP]+ (m/z 749.1) (Fig. 4A). The protonated ion of PhIP is also the base peak in the product ion spectrum of LQQC*[SOPhIP]PFEDHVK (Fig. 4B). The sulfonamide linked adduct is considerably more stable towards HCD and CID, and a majority of -b and -y ions are seen in both HCD- and CID- product ion spectra; protonated PhIP (m/z 225.1) is the base peak in the HCD-product ion spectrum of LQQC*[SO2PhIP]PFEDHVK (Fig. 4C), but a minor ion in CID product ion spectrum.38
Figure 4.
Reconstructed mass chromatograms (left panel) and product ion spectra (right panel) of Cys34 adducts from N-sulfooxy-PhIP treated human plasma (left panel) of (A) LQQC*[SOPhIP]PF ([M+2H]2+ at m/z 487.2 > 225.1), (B) LQQC*[SOPhIP]PFEDHVK ([M+3H]3+, m/z 527.9 > 225.1), and (C) LQQC*[SO2PhIP]PFEDHVK ([M+3H]3+ at m/z 533.2 > 225.1) and corresponding HCD product ion spectra (right panel), following tryptic/chymotryptic digestion of albumin. tR, Retention time; A, area ion counts of the peak.
The relative amounts of LQQC*[SOPhIP]PF and LQQC*[SO2PhIP]PFEDHVK adducts formed by reaction of human albumin or human plasma with N-sulfooxy-PhIP, N-acetoxy-PhIP, or HONH-PhIP were determined by targeted UPLC-ESI/MS2. There was no statistically significant difference between the level of the LQQC*[SOPhIP]PF formed by N-sulfooxy-PhIP, N-acetoxy-PhIP or HONH-PhIP in either albumin in human plasma samples or commercial human albumin (p=0.231, one-way ANOVA) (Unpublished results, Y. Wang). However, the amount of LQQC*[SO2PhIP]PFEDHVK recovered from N-sulfooxy-PhIP modified human plasma was 2x greater than the amounts formed with N-acetoxy-PhIP and HONH-PhIP modified samples (p<0.05, t-test).There was no significant difference between LQQC*[SO2PhIP]PFEDHVK levels produced by N-acetoxy-PhIP and HONH-PhIP (p>0.05, t-test) (unpublished results, Y. Wang).
Identification of Albumin Trp214 Adduct of PhIP at AW*[PhIP]AVAR
The sole tryptophan residue of rat albumin, Trp214, was reported to bind to the genotoxic metabolite N-sulfonyloxy-N-acetyl-4-aminobiphenyl.42 Human albumin also contains one sole Trp, which resides at the same sequence location as for rat albumin.31 We sought to determine whether the Trp214 of human albumin had reacted with the N-sulfooxy-PhIP to form an adduct that had escaped detection by mass tag triggered data-dependent acquisition method. Human albumin and human plasma modified with N-sulfooxy-PhIP were monitored for AW*[PhIP]AVAR ([M+2H]2+ at m/z 448.2), by targeted UPLC-ESI/MS2, following digestion of albumin with trypsin. The mass chromatogram and product ion spectrum of the adduct from plasma albumin, obtained by accurate mass measurements with the Orbitrap, are shown in Figs 5A and 5B. The product ion spectrum of AW*[PhIP]AVAR ([M+2H]2+ at m/z 448.2) contains predominant ions assigned to protonated PhIP (m/z 225.1) and [M+H-PhIP]+ (m/z 671.4), with minor ions assigned as -b ions [m/z 650.3 (b4), m/z 721.4 (b5)], and -y ion at m/z 416.3 (y4). The ion attributed to protonated PhIP and the –b and -y ions were within 2.3 ppm of the calculated values.
Figure 5.
(A) Reconstructed mass chromatograms obtained by CID of AW*[PhIP]AVAR ([M+2H]2+ m/z 448.2385 > 225.1135, 671.3624; mass tolerance, 2 ppm) of N-sulfooxy-PhIP modified albumin digested with trypsin, (B) product ion spectrum AW*[PhIP]AVAR, and (C) relative peak area ion counts of peptide adducts of albumin modified with N-sulfooxy-PhIP: AW*[PhIP]AVAR recovered from trypsin digest; LQQC*[SOPhIP]PF ([M+2H]2+ at m/z 487.2 > 225.1, 749.4) and PhIP ([M+H]+ at m/z 225.1 > 210.0) recovered from trypsin/chymotrypsin digest; and LQQC*[SO2PhIP]PFEDHVK ([M+3H]3+ at m/z 533.2 > 670.2, 679.0) recovered from trypsin/chymotrypsin digestion following m-CPBA oxidation of albumin.
The plasma PhIP albumin peptide adducts recovered from optimized enzyme digestion conditions are depicted in Fig. 5C. In the absence of stable, isotopically labelled internal standards, quantitative measurements of the relative amounts of adducts formed cannot be determined and only relative ion abundances are reported. The AW*[PhIP]AVAR recovered from a tryptic digest is comparable to the signal of LQQ*C[SO]PF recovered from the tryptic/chymotryptic digest, but the response is only ~ 25% of the signal of LQQC*[SO2PhIP]PFEDHVK recovered from tryptic/chymotryptic digest (following m-CPBA oxidation and stabilization of the Cys34 S-N linked PhIP adducts).39,54,57 The peak area ion count of PhIP, recovered from the tryptic/chymotryptic digest, is 10-fold greater than AW*[PhIP]AVAR (Fig. 5C), indicating that a significant proportion of the N-sulfooxy-PhIP bound to albumin formed labile Cys34 S-N linked adducts that underwent hydrolysis during proteolytic digestion.
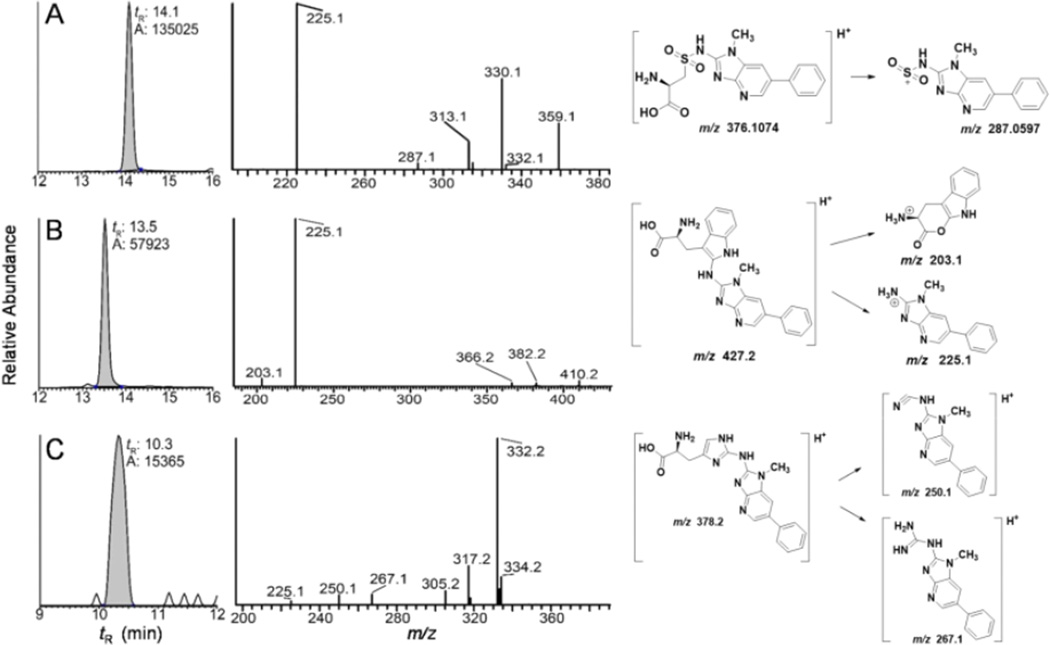
Identification of Albumin Adducts of PhIP formed with Cys, Trp, and His
PhIP-modified albumin was screened for amino acid adducts in albumin or human plasma reacted with N-oxidized metabolites of PhIP:[2H5]-PhIP, following digestion with pronase E, leucine aminopeptidase and prolidase. The C[SO2PhIP], C*[SO2PhIP]PF, W[PhIP], and H[PhIP] from albumin were identified, by the mass tag triggered data dependent scan method (Table 1). Subsequently, these adducts were assayed by targeted UPLC-ESI/MS2. The mass chromatograms of the adducts formed by reaction of albumin in plasma with N-sulfooxy-PhIP are shown in Figure 6. The product ion spectrum of C[SO2PhIP] ([M+H]+ at m/z 376.1) displays a base peak attributed to protonated PhIP (m/z 225.1), an ion at m/z 287.1 due to the charged PhIP-SO2 moiety ([M+H-C3H7NO2]+); other fragment ions are observed at m/z 313.1 ([M+H-NH3-H2CO2]+), m/z 330.1 ([M+H-H2CO2]+), and m/z 359.1 [M+H-NH3]+ (Fig. 6A). The W[PhIP] adduct ([M+H]+ at m/z 427.1) undergoes CID to produce a minor fragment ion at m/z 203.1 due to the loss of PhIP ([M+H-224]+, the base peak in the spectrum is protonated PhIP at m/z 225.1. Other fragment ions are observed at m/z 366.2 ([M+H-CO2-NH3]+, m/z 382.2 [M+H-·HCO2]+, and m/z 410.2 [M+H-NH3]+ (Fig. 6B). The mass spectrum is compatible with a structure of an adduct formed between the exocyclic amine group of PhIP and the C-2 atom of the indole group of Trp. The product ion spectra of the H[PhIP] adduct ([M+H]+ at m/z 378.1) is shown in Fig. 6C. Fragment ions at occurring at m/z 225.1, 250.1 and 267.1 are consistent with a proposed adduct structure containing a linkage between the exocyclic amine group of PhIP and the C-2 atom of the imidazole ring of His. The product ion spectra of the [2H5]-PhIP amino acid adducts displayed the same pattern of fragmentation with a shift in mass/charge of 5 m/z (data not shown).
Figure 6.
Reconstructed mass chromatograms (left pane) of (A) C[SO2PhIP] ([M+H]+ at m/z 376.1 > 225.1), (B) W[PhIP] ([M+H]+ at m/z, 421.1 > 225.1), and (C) H[PhIP] ([M+H]+ at m/z 378.1 > 225.1, 332.2) from N-sulfooxy-PhIP modified albumin following digestion with pronase E, leucine aminopeptidase, and prolidase. The product ion mass spectra and proposed diagnostic product ions are depicted (right panel).
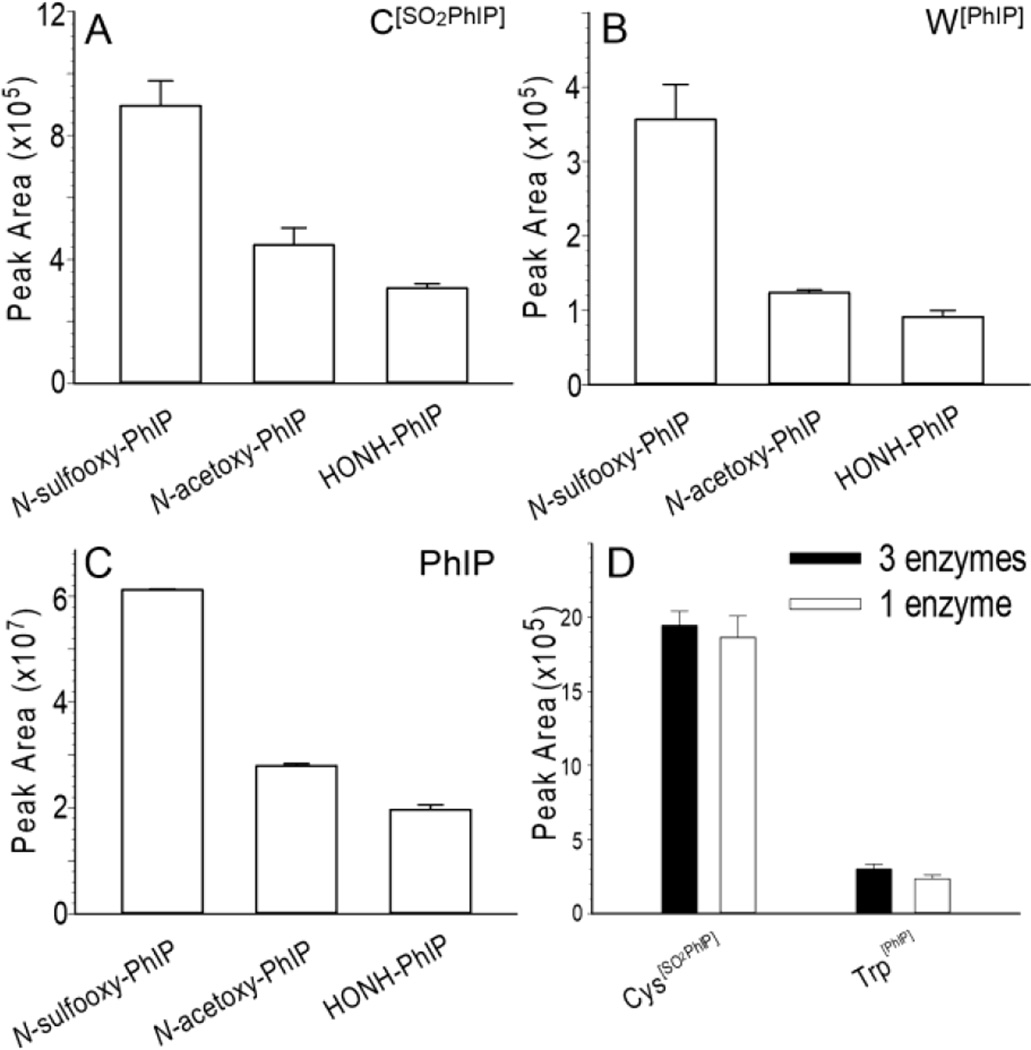
The level of W[PhIP] adduct, based on ion counts, is about half the signal of C[SO2PhIP]; however, the ion count of PhIP, formed by hydrolysis of unstable Cys S-N linked adducts, is more than100x greater than the ion counts of either C [SO2PhIP] or W[PhIP] recovered from albumin digested with the 3-enzyme mixture (Fig. 7). The levels of Cys[SO2PhIP] and Trp[PhIP] are 2x and 3x greater in N-sulfooxy-PhIP treated than N-acetoxy-PhIP or HONH-PhIP treated human plasma (Figs 7A and 7B). The amount of PhIP recovered from N-sulfooxy-PhIP modified albumin sample is also ~3x greater than from N-acetoxy-PhIP and HONH-PhIP modified human plasma. The high levels of PhIP recovered from N-sulfooxy-PhIP treated albumin in plasma is indicative of the existence of unstable Cys34 S-N linked PhIP adducts (Fig. 7C). The ion counts of H[PhIP] is 10-fold lower than the ion counts of C [SO2PhIP] and 5-folder lower than the counts of W[PhIP] adducts in N-sulfooxy-PhIP modified albumin (Unpublished observations, Y. Wang).
Figure 7.
(A) Peak area ion count estimates of the UPLC-ESI/MS2 analysis of (A) C[SO2PhIP] ([M+H]+ at m/z 376.1 > 225.1), (B) W[PhIP] ([M+H]+m/z, 421.1 > 225.1), and (C) PhIP ([M+H]+ at m/z 225.1 > 210.0) recovered from albumin in human plasma modified with N-sulfooxy-PhIP following by protein digestion with pronase E, leucine aminopeptidase and prolidase. (D) Peak area ion count estimates of the UPLC-ESI/MS2 analysis of C[SO2PhIP] ([M+H]+m/z, 376.1 > 225.1) and W[PhIP] ([M+H]+, m/z, 421.1 > 225.1) recovered from N-sulfooxy-PhIP modified commercial albumin following digestion with pronase E, leucine aminopeptidase and prolidase (black bar), or pronase E alone (white bar). Albumin digest (200 ng) was injected on column for all samples (Mean ± SD, n=3).
The proficiency of proteolytic digestion of N-sulfooxy-PhIP modified albumin by 3 enzymes (pronase E, leucine aminopeptidase and prolidase) and 1 enzyme (pronase E) on the recovery of C[SO2PhIP] and W[PhIP] was examined. In contrast to our previous findings, where C[SO2PhIP] was recovered primarily as the tripeptide C[SO2PhIP]PF from PhIP-modified commercial albumin digested with this 3 enzyme mixture, we saw no significant difference between the amino acid adduct C[SO2PhIP] or W[PhIP] levels, when PhIP modified albumin was digested with 3 enzymes or pronase E alone (p>0.05, t-test) (Fig. 7D), and the levels of incompletely digested dipeptide or tripeptide adducts of PhIP were negligible.
Characterization of Albumin PhIP Adducts and Cys34SO3H in Human Hepatocytes
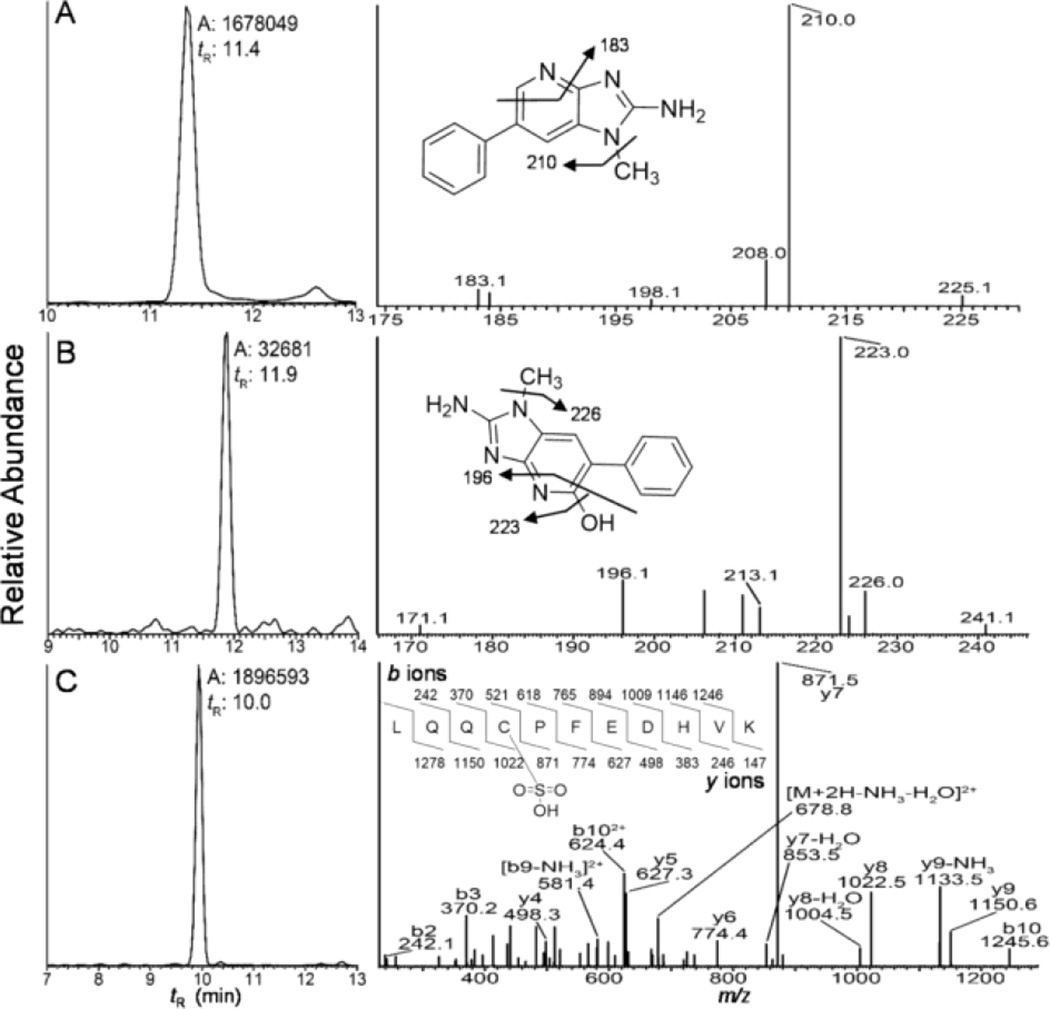
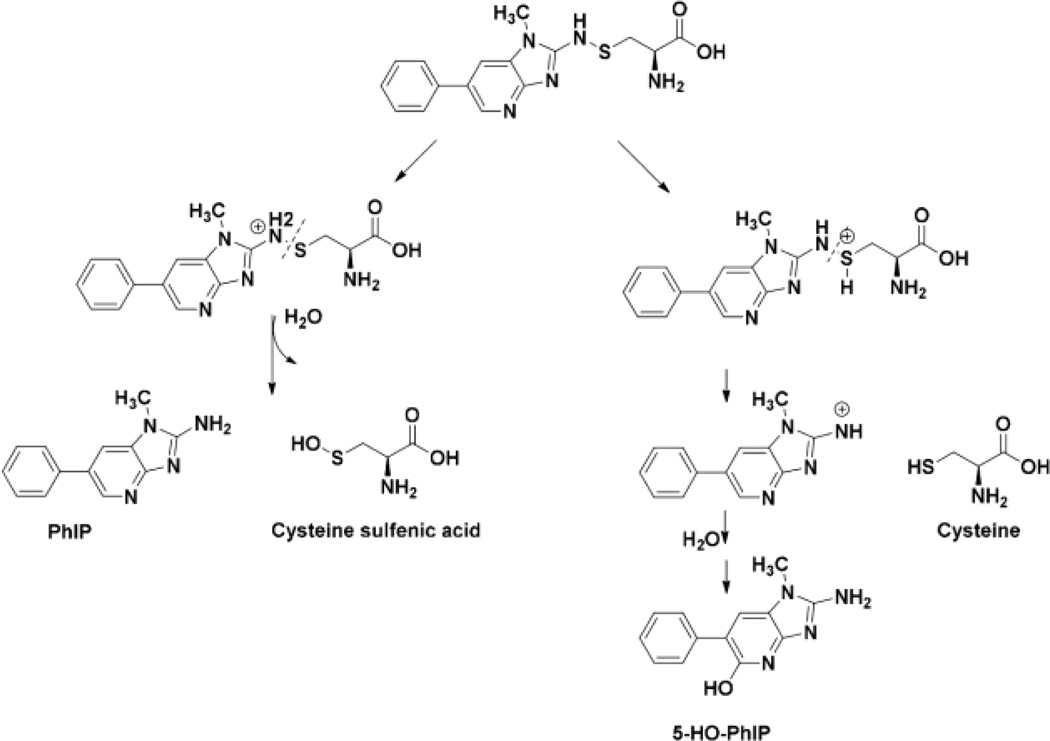
Human hepatocytes were incubated with PhIP (50 µM) or PhIP:[2H5]-PhIP (50 µM, 1:1 ratio) for 24 h and examined, respectively, for dG-C8-PhIP and albumin adduct formation. High levels of dG-C8-PhIP adducts were formed: the levels were 10.4 ± 0.3 adducts per 106 bases (Mean ± SD, 3 independent measurements), confirming PhIP had undergone extensive bioactivation by P450-mediated N-oxidation in hepatocytes (Fig. S-4).52 However, the mass tag triggered data-dependent acquisition method failed to detect albumin-PhIP:[2H5]-PhIP adducts. Targeted MS2 scanning also failed to detect albumin-PhIP peptide adducts at either Cys34 or Trp214 residues following tryptic, tryptic/chymotyptic digestion, or digestion of albumin with the three enzyme mixture. Instead, PhIP and 5-HO-PhIP were detected in the proteolytic digests (Figs. 8A and 8B). Spiking experiments of PhIP in the hepatocyte media, followed by solvent extraction, proteolytic digestion, and UPLC-ESI/MS2 targeting for PhIP showed that all of the spiked PhIP had been removed during the albumin workup (Unpublished observations, Y. Wang). These findings suggest the presence of labile Cys34 S-N linked PhIP adducts, which underwent hydrolysis during proteolytic digestion to form PhIP and 5-HO-PhIP (Scheme I). The albumin isolated from hepatocytes was oxidized with m-CPBA to convert labile Cys34 S-N linked adducts of PhIP to the stable sulfonamide adducts;39,54 however, targeted MS2 scanning failed to detect LQQC*[SO2PhIP]PFEDHVK. The signal of the PhIP sulfonamide adduct is more than 10-fold weaker in response than the signal for PhIP under ESI/MS2 conditions.54 We surmise that the level of the sulfonamide adduct is below the limit of detection under our current UPLC-ESI/MS conditions.
Figure 8.
Reconstructed mass chromatograms (left panel) of (A) PhIP ([M+H]+ at m/z 225.1 > 210.0), (B) 5-HO-PhIP ([M+H]+ at m/z 242.1 > 223.1), and (C) LQQC*[SO3]PFEDHVK ([M+2H]2+ at m/z 696.3 > 871.4, 1022.3) from hepatocytes treated with PhIP, and the corresponding mass spectra (right panel).
Scheme 1.
Proposed mechanism of hydrolysis of Cys34 PhIP-sulfenamide to produce PhIP and 5-HO-PhIP
HONH-PhIP undergoes oxidation to form nitroso-PhIP, and a redox cycling mechanism with the generation ROS can occur.58,59 In this study, we discovered that the level of LQQC*[SO3]PFEDHVK is increased by 6 fold in PhIP-treated hepatocytes compared to the control (p<0.001, t-test) (Unpublished results, Y. Wang). The UPLC/MS chromatogram and product ion spectrum of LQQC*[SO3]PFEDHVK are shown in Figure 8C. Thus, in addition to the formation of macromolecular adducts, N-oxidized PhIP metabolites induce oxidative stress in human hepatocytes, and Cys34 of albumin is acting as a scavenger of ROS.
We previously characterized several albumin adduction products formed between commercial human albumin and NO2-PhIP.37 The NO2 moiety of NO2-PhIP undergoes nucleophilic displacement by the HS group of Cys34 and HO group of Tyr411 of albumin to form, respectively, LQQC*[desaminoPhIP]PF and Y*[desaminoPhIP]TK, following tryptic/chymotryptic digestion.37 We screened albumin in human hepatocytes treated with PhIP for Y*[desaminoPhIP]TK and LQQC*[desaminoPhIP]PF and both adducts were identified (Figure 9).
Figure 9.
Reconstructed mass chromatograms (left panel) of (A) Y*[desaminoPhIP]TK ([M+2H]2+ at m/z 309.7 > 343.2, 428.3) and (B) LQQC*[desaminoPhIP]PF ([M+2H]2+ at m/z 471.7 > 684.3, 701.3) from human hepatocytes treated with PhIP (50 µM), and the corresponding mass spectra (right panel).
DISCUSSION
PhIP undergoes extensive bioactivation through P450 mediated N-oxidation to form HONH-PhIP as a major pathway of metabolism in humans.22–24 In our goal to establish albumin adducts of PhIP as biomarkers for human studies, we have characterized the adduction products of the major N-oxidized metabolites of PhIP, including HONH-PhIP, NO-PhIP, N-acetoxy-PhIP, and N-sulfooxy-PhIP with albumin.37–39,54 Our findings show that Cys34 is the principal nucleophile of albumin to react with all of these N-oxidized metabolites of PhIP, to form a putative sulfenamide, and more stable sulfinamide and sulfonamide linked adducts.37–39 Crystallography and NMR studies with human albumin have shown that the Cys34 resides in a shallow crevice of the protein,60–62 but it is accessible to solvent. Moreover, the Cys34 residue of albumin has an unusually low pKa value, 6.5 as compared to about 8.0 to 8.5 in many other proteins or peptides,20,63 and it predominantly exists as the thiolate anion at physiological pH, which explains its reactivity towards N-oxidized PhIP metabolites and many other electrophiles.21
The Trp214 was identified as a secondary binding site of albumin for N-oxidized metabolites of PhIP, and the hexapeptide adduct AW*[PhIP]AVAR was recovered from a tryptic digest. Trp214 is not located on the surface of albumin, but it resides in a hydrophobic cavity of subdomain IIA, which is the principal region of ligand binding of human albumin.61,64 Surprisingly, the recovery of AW*[PhIP]AVAR, by tryptic digestion, was not increased by denaturation of albumin prior to proteolysis. A previous study in rats dosed with 4-ABP, identified Trp214 of albumin as a major site of adduction of an activated form of the hydroxamic acid of N-acetyl-4-ABP.42 The synthetic sulfate esters of N-hydroxy-N-acetyl-4-ABP, N-hydroxy-N-acetyl-2-aminofluorene, and N-hydroxy-N,N'–diacetylbenzidine were also found to bind to Trp214 of commercial human albumin and recovered as AW*AV following pronase digestion.26 Most recently, an activated metabolite of Nevirapine, a non-nucleoside reverse transcriptase inhibitor, was reported to bind to Trp214 of human albumin.65 Our data show that various N-oxidized metabolites of PhIP react with Trp214 of albumin. The potential of Trp214 of albumin to serve as a target site of genotoxicants and electrophiles for human biomonitoring is unexplored.
We extended our findings on the reactivity of DNA and albumin with N-oxidized metabolites of PhIP in solution to studies in human hepatocytes. PhIP undergoes bioactivation in hepatocytes and high levels of dG-C8-PhIP are formed (10.4 ± 0.3 adducts per 106 DNA bases); however, Cys34 or Trp214 adducts were below the limit of detection. Instead, PhIP and 5-HO-PhIP were recovered from the proteolytic digest of albumin. PhIP and 5-HO-PhIP were also recovered from pronase digests of rat albumin modified with N-acetoxy-PhIP.35 These data suggest that a labile Cys S-N linked sulfenamide adduct of PhIP had formed with albumin and underwent hydrolysis during proteolytic digestion (Scheme 1).39 The oxidation of PhIP-modified albumin by m-CPBA converts labile Cys S-N linked adducts to the sulfonamide linkage, which is stable towards proteolytic digestion.39 We oxidized the albumin isolated from human hepatocytes with m-CPBA prior to enzyme digestion, and the level of PhIP and 5-HO-PhIP liberated during proteolysis did decrease by several fold. However, LQQC*[SO2PhIP]PFEDHVK was not detected, suggesting that the level of sulfonamide adduct is below the detection limit or the efficacy of oxidation by m-CPBA was somehow inhibited by components co-extracted with albumin from human hepatocytes.
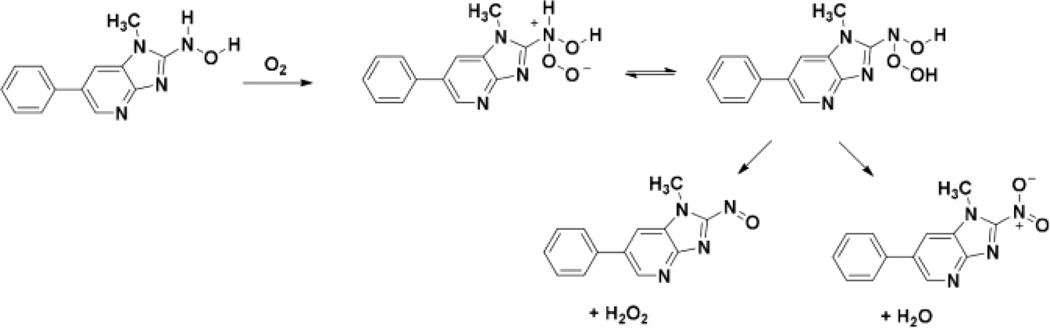
We did however detect albumin-PhIP peptide adducts formed with Tyr411 (Y*[desaminoPhIP]TK) and Cys34 (LQQC*[desaminoPhIP]PF), where the exocyclic amino moiety of PhIP was displaced by the nucleophilic HO group of Tyr and HS group of Cys. We previously reported that NO2-PhIP undergoes nucleophilic substitution by Tyr411 and Cys34 residues of human albumin in vitro.38 The Y*[desaminoPhIP]TK and LQQC*[desaminoPhIP]PF formed in hepatocytes most likely arise from the reaction of NO2-PhIP with albumin. Arylhydroxylamines undergo oxidation to form their nitroso and nitro derivatives under aerobic conditions.66 HONH-PhIP also undergoes oxidation under these conditions to form NO2-PhIP (R. Turesky, unpublished observations). A proposed mechanism for NO2-PhIP formation is shown in Scheme 2. Consistent with our findings, a desamino glutathione PhIP conjugate was characterized in hepatocytes and bile of rats pretreated with polychlorinated biphenyls.67
Scheme 2.
A proposed reactive N-hydroperoxy intermediate of HONH-PhIP which collapses to form NO2-PhIP.66
The response of the ESI/MS signal for the PhIP-DNA adduct, dG-C8-PhIP, is far greater than the signals obtained for albumin-PhIP peptide adducts. However, the amount of DNA and albumin in hepatocyte culture differ, and relative reactivity of N-oxidized PhIP with DNA and albumin is difficult to determine, particularly, in the absence of internal standards for quantitative measurements of PhIP-albumin adducts. Also, the relative abundances of these macromolecules differ. There is approximately 15 µg of nuclear DNA (45 nmol DNA bases) per 106 hepatocytes, whereas only ~1 µg of albumin (15 pmol albumin) is secreted into the media per day per 106 hepatocytes.68 In addition, the cell media contain 1 mg/mL of exogenously added albumin, and thus the albumin secreted into the media is diluted by ~1,000-fold. Currently, we do not know if PhIP-albumin adduct formation occurs primarily by reaction of N-oxidized PhIP intermediates with newly synthesized albumin within the hepatocyte or with albumin residing in the extracellular media.
In addition to macromolecular adduct formation, N-oxidized PhIP metabolites form reactive oxygen species (ROS) that can oxidize protein and induce toxicity.58,59 HONH-PhIP can undergo oxidation by P450, transition metals, or by oxygen to form NO-PhIP and ROS, and a redox cycling mechanism catalyzed by NADPH-P450 reductase can regenerate HONH-PhIP.58 There was a 6-fold increase in the levels of the sulfonic acid at Cys34 of albumin, and LQQC*[SO3]PFEDHVK was detected in hepatocytes treated with PhIP. Thus, Cys34 of albumin serves as an antioxidant and scavenger of free radicals induced by metabolites of PhIP in human hepatocytes.
In a summary, amino acid and peptide adducts of Cys34, Trp214, LQQC*[SOPhIP]PF, LQQC**[SOPhIP]PFEDHVK, LQQC**[SO2PhIP]PFEDHVK, AW*[PhIP]AVAR, Y411*[desaminoPhIP]TK), and LQQC*[desaminoPhIP]PF were identified in human albumin reacted with N-oxidized metabolites of PhIP. We plan to determine whether these albumin peptide adducts may be candidate protein biomarkers of PhIP in humans.
Supplementary Material
ACKNOWLEDGEMENT
We would like to thank the Dr. David Tabb Laboratory at the Department of Biomedical Informatics, and the Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, for the introduction to and use of the MyriMatch software program. The authors acknowledge the Centre de Ressources Biologiques (CRB)-Santé of Rennes for managing the patient samples.
Funding Sources.
This research was supported by Grant 2R01 CA122320 (R.J.T.), and in part by National Cancer Institute Cancer Center Support grant no. CA-77598 (R.J.T.), and the PNREST Anses, Cancer TMOI AVIESAN, 2013/1/166 (S.L.).
ABBREVIATIONS
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- 5-HO-PhIP
2-amino-1-methyl-6-(5-hydroxy)-phenylimidazo[4,5-b]pyridine
- N-acetoxy-PhIP
N-(acetyloxy)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- HONH-PhIP
2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- NO-PhIP
2-nitroso-1-methyl-6-phenylimidazo[4,5-b]pyridine
- NO2-PhIP
2-nitro-1-methyl-6-phenylimidazo[4,5-b]pyridine
- N-sulfooxy-PhIP
N-sulfooxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- Hb
hemoglobin
- HAA
heterocyclic aromatic amine
- HCD
higher-energy collisional dissociation
- m-CPBA
meta-chloroperoxybenzoic acid
- UPLC-ESI/MS/MS
ultraperformance liquid chromatography tandem mass spectrometry
- β-mercaptoethanol
βME
- SPE
solid phase extraction
- dG
deoxyguanosine
- dR
deoxyribose
Footnotes
UV spectra of PhIP, HONH-PhIP and N-acetoxy-PhIP; product ion spectra of N-acetoxy-PhIP and fragmentation mechanism; mass chromatogram and product ion spectra of dG-C8-PhIP obtained by the reaction of dG with N-sulfooxy-PhIP and human hepatocytes treated with PhIP. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felton JS, Jagerstad M, Knize MG, Skog K, Wakabayashi K. Contents in foods, beverages and tobacco. In: Nagao M, Sugimura T, editors. Food Borne Carcinogens Heterocyclic Amines. Chichester, England: John Wiley & Sons Ltd; 2000. pp. 31–71. [Google Scholar]
- 3.Knize MG, Dolbeare FA, Carroll KL, Moore DH, Felton JS. Effect of cooking time and temperature on the heterocyclic amine content of fried beef patties. Food Chem. Toxicol. 1994;32:595–603. doi: 10.1016/0278-6915(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 4.Sinha R, Rothman N, Brown ED, Salmon CP, Knize MG, Swanson CS, Rossi SC, Mark SD, Levander OA, Felton JS. High concentrations of the carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) occur in chicken but are dependent on the cooking method. Cancer Res. 1995;55:4516–4519. [PubMed] [Google Scholar]
- 5.Zhao K, Murray S, Davies DS, Boobis AR, Gooderham NJ. Metabolism of the food derived mutagen and carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) by human liver microsomes. Carcinogenesis. 1994;15:1285–1288. doi: 10.1093/carcin/15.6.1285. [DOI] [PubMed] [Google Scholar]
- 6.Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem. Res. Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- 7.Kato R. Metabolic activation of mutagenic heterocyclic aromatic amines from protein pyrolysates. CRC Crit. Rev. Toxicol. 1986;16:307–348. doi: 10.3109/10408448609037466. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen TM, Novak M. Synthesis and decomposition of an ester derivative of the procarcinogen and promutagen, PhIP, 2-amino-1-methyl-6-phenyl-1H–imidazo[4,5-b]pyridine: unusual nitrenium ion chemistry. J.Org.Chem. 2007;72:4698–4706. doi: 10.1021/jo070306p. [DOI] [PubMed] [Google Scholar]
- 9.Turesky RJ, Le Marchand L. Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem. Res. Toxicol. 2011;24:1169–1214. doi: 10.1021/tx200135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO, Jr, Kadlubar FF. Identification of N-(deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem. Res. Toxicol. 1992;5:691–697. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 11.Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2’-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem. Res. Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu D, Turesky RJ, Tao Y, Langouet SA, Nauwelaers GC, Yuan JM, Yee D, Yu MC. DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 4-aminobiphenyl are infrequently detected in human mammary tissue by liquid chromatography/tandem mass spectrometry. Carcinogenesis. 2012;33:124–130. doi: 10.1093/carcin/bgr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, Turesky RJ. Identification of carcinogen DNA adducts in human saliva by linear quadrupole ion trap/multistage tandem mass spectrometry. Chem. Res. Toxicol. 2010;23:1234–1244. doi: 10.1021/tx100098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tretyakova N, Goggin M, Sangaraju D, Janis G. Quantitation of DNA adducts by stable isotope dilution mass spectrometry. Chem Res.Toxicol. 2012;25:2007–2035. doi: 10.1021/tx3002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JA. Carcinogenesis by chemicals: an overview--G. H. A. Clowes memorial lecture. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- 16.Rubino FM, Pitton M, Di FD, Colombi A. Toward an “omic” physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom. Rev. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- 17.Tornqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, Rydberg P. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J. Chromatogr.B Analyt.Technol.Biomed. Life Sci. 2002;778:279–308. doi: 10.1016/s1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 18.Skipper PL, Tannenbaum SR. Protein adducts in the molecular dosimetry of chemical carcinogens. Carcinogenesis. 1990;11:507–518. doi: 10.1093/carcin/11.4.507. [DOI] [PubMed] [Google Scholar]
- 19.Liebler DC. Proteomic approaches to characterize protein modifications: new tools to study the effects of environmental exposures. Environ. Health Perspect. 2002;110(Suppl 1):3–9. doi: 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aldini G, Regazzoni L, Orioli M, Rimoldi I, Facino RM, Carini M. A tandem MS precursor-ion scan approach to identify variable covalent modification of albumin Cys34: a new tool for studying vascular carbonylation. J. Mass Spectrom. 2008;43:1470–1481. doi: 10.1002/jms.1419. [DOI] [PubMed] [Google Scholar]
- 21.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol. Lett. 2012;213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malfatti MA, Dingley KH, Nowell-Kadlubar S, Ubick EA, Mulakken N, Nelson D, Lang NP, Felton JS, Turteltaub KW. The urinary metabolite profile of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine is predictive of colon DNA adducts after a low-dose exposure in humans. Cancer Res. 2006;66:10541–10547. doi: 10.1158/0008-5472.CAN-06-1573. [DOI] [PubMed] [Google Scholar]
- 23.Walters DG, Young PJ, Agus C, Knize MG, Boobis AR, Gooderham NJ, Lake BG. Cruciferous vegetable consumption alters the metabolism of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Carcinogenesis. 2004;25:1659–1669. doi: 10.1093/carcin/bgh164. [DOI] [PubMed] [Google Scholar]
- 24.Gu D, McNaughton L, LeMaster D, Lake BG, Gooderham NJ, Kadlubar FF, Turesky RJ. A comprehensive approach to the profiling of the cooked meat carcinogens 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, and their metabolites in human urine. Chem. Res. Toxicol. 2010;23:788–801. doi: 10.1021/tx900436m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiese M. The biochemical production of ferrihemoglobin-forming derivatives from aromatic amines, and mechanisms of ferrihemoglobin formation. Pharmacol.Rev. 1966;18:1091–1161. [PubMed] [Google Scholar]
- 26.Tannenbaum SR, Skipper PL, Wishnok JS, Stillwell WG, Day BW, Taghizadeh K. Characterization of various classes of protein adducts. Environ. Health Perspect. 1993;99:51–55. doi: 10.1289/ehp.939951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu MC, Skipper PL, Tannenbaum SR, Chan KK, Ross RK. Arylamine exposures and bladder cancer risk. Mutat. Res. 2002;506–507:21–28. doi: 10.1016/s0027-5107(02)00148-3. [DOI] [PubMed] [Google Scholar]
- 28.Ringe D, Turesky RJ, Skipper PL, Tannenbaum SR. Structure of the single stable hemoglobin adduct formed by 4-aminobiphenyl in vivo . Chem. Res. Toxicol. 1988;1:22–24. doi: 10.1021/tx00001a003. [DOI] [PubMed] [Google Scholar]
- 29.Dingley KH, Curtis KD, Nowell S, Felton JS, Lang NP, Turteltaub KW. DNA and protein adduct formation in the colon and blood of humans after exposure to a dietary-relevant dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Epidemiol. Biomarkers Prev. 1999;8:507–512. [PubMed] [Google Scholar]
- 30.Garner RC, Lightfoot TJ, Cupid BC, Russell D, Coxhead JM, Kutschera W, Priller A, Rom W, Steier P, Alexander DJ, Leveson SH, Dingley KH, Mauthe RJ, Turteltaub KW. Comparative biotransformation studies of MeIQx and PhIP in animal models and humans. Cancer Lett. 1999;143:161–165. doi: 10.1016/s0304-3835(99)00118-4. [DOI] [PubMed] [Google Scholar]
- 31.Peters T., Jr Serum albumin. Adv.Protein Chem. 1985;37:161–245. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- 32.Colombo G, Clerici M, Giustarini D, Rossi R, Milzani A, Dalle-Donne I. Redox albuminomics: oxidized albumin in human diseases. Antioxidants & redox signaling. 2012;17:1515–1527. doi: 10.1089/ars.2012.4702. [DOI] [PubMed] [Google Scholar]
- 33.Turesky RJ, Skipper PL, Tannenbaum SR. Binding of 2-amino-3-methylimidazo[4,5-f]quinoline to hemoglobin and albumin in vivo in the rat. Identification of an adduct suitable for dosimetry. Carcinogenesis. 1987;8:1537–1542. doi: 10.1093/carcin/8.10.1537. [DOI] [PubMed] [Google Scholar]
- 34.Lynch AM, Murray S, Boobis AR, Davies DS, Gooderham NJ. The measurement of MeIQx adducts with mouse haemoglobin in vitro and in vivo: implications for human dosimetry. Carcinogenesis. 1991;12:1067–1072. doi: 10.1093/carcin/12.6.1067. [DOI] [PubMed] [Google Scholar]
- 35.Reistad R, Frandsen H, Grivas S, Alexander J. In vitro formation and degradation of 2-amino-1-methyl-6- phenylimidazo[4,5-b]pyridine (PhIP) protein adducts. Carcinogenesis. 1994;15:2547–2552. doi: 10.1093/carcin/15.11.2547. [DOI] [PubMed] [Google Scholar]
- 36.Chepanoske CL, Brown K, Turteltaub KW, Dingley KH. Characterization of a peptide adduct formed by N-acetoxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a reactive intermediate of the food carcinogen PhIP. Food Chem. Toxicol. 2004;42:1367–1372. doi: 10.1016/j.fct.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Peng L, Turesky RJ. Mass spectrometric characterization of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine N-oxidized metabolites bound at Cys34 of human serum albumin. Chem Res.Toxicol. 2011;24:2004–2017. doi: 10.1021/tx2003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng L, Dasari S, Tabb DL, Turesky RJ. Mapping serum albumin adducts of the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by data-dependent tandem mass spectrometry. Chem. Res. Toxicol. 2012;25:2179–2193. doi: 10.1021/tx300253j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng L, Turesky RJ. Capturing labile sulfenamide and sulfinamide serum albumin adducts of carcinogenic arylamines by chemical oxidation. Anal. Chem. 2013;85:1065–1072. doi: 10.1021/ac3028273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frandsen H, Grivas S, Andersson R, Dragsted L, Larsen JC. Reaction of the N 2 -acetoxy derivative of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) with 2’-deoxyguanosine and DNA. Synthesis and identification of N 2 -(2’-deoxyguanosin-8-yl)-PhIP. Carcinogenesis. 1992;13:629–635. doi: 10.1093/carcin/13.4.629. [DOI] [PubMed] [Google Scholar]
- 41.Skipper PL. Influence of tertiary structure on nucleophilic substitution reactions of proteins. Chem Res.Toxicol. 1996;9:918–923. doi: 10.1021/tx960028h. [DOI] [PubMed] [Google Scholar]
- 42.Skipper PL, Obiedzinski MW, Tannenbaum SR, Miller DW, Mitchum RK, Kadlubar FF. Identification of the major serum albumin adduct formed by 4-aminobiphenyl in vivo in rats. Cancer Res. 1985;45:5122–5127. [PubMed] [Google Scholar]
- 43.Wu RW, Tucker JD, Sorensen KJ, Thompson LH, Felton JS. Differential effect of acetyltransferase expression on the genotoxicity of heterocyclic amines in CHO cells. Mutat. Res. 1997;390:93–103. doi: 10.1016/s0165-1218(97)00005-0. [DOI] [PubMed] [Google Scholar]
- 44.Wu RW, Panteleakos FN, Kadkhodayan S, Bolton-Grob R, McManus ME, Felton JS. Genetically modified Chinese hamster ovary cells for investigating sulfotransferase-mediated cytotoxicity and mutation by 2-amino-1-methyl-6- phenylimidazo[4,5-b]pyridine. Environ. Mol. Mutagen. 2000;35:57–65. [PubMed] [Google Scholar]
- 45.Metry KJ, Zhao S, Neale JR, Doll MA, States JC, McGregor WG, Pierce WM, Jr, Hein DW. 2-Amino-1-methyl-6-phenylimidazo [4,5-b] pyridine-induced DNA adducts and genotoxicity in Chinese hamster ovary (CHO) cells expressing human CYP1A2 and rapid or slow acetylator N-acetyltransferase 2. Mol.Carcinog. 2007;46:553–563. doi: 10.1002/mc.20302. [DOI] [PubMed] [Google Scholar]
- 46.Nowell S, Ambrosone CB, Ozawa S, MacLeod SL, Mrackova G, Williams S, Plaxco J, Kadlubar FF, Lang NP. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000;10:789–797. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 47.Dobbernack G, Meinl W, Schade N, Florian S, Wend K, Voigt I, Himmelbauer H, Gross M, Liehr T, Glatt H. Altered tissue distribution of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-DNA adducts in mice transgenic for human sulfotransferases 1A1 and 1A2. Carcinogenesis. 2011;32:1734–1740. doi: 10.1093/carcin/bgr204. [DOI] [PubMed] [Google Scholar]
- 48.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 49.Langou‰t S, Paehler A, Welti DH, Kerriguy N, Guillouzo A, Turesky RJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in rat and human hepatocytes. Carcinogenesis. 2002;23:115–122. doi: 10.1093/carcin/23.1.115. [DOI] [PubMed] [Google Scholar]
- 50.Attaluri S, Iden CR, Bonala RR, Johnson F. Total synthesis of the aristolochic acids, their major metabolites, and related compounds. Chem. Res. Toxicol. 2014;27:1236–1242. doi: 10.1021/tx500122x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beland FA, Miller DW, Mitchum RK. Synthesis of the ultimate hepatocarcinogen 2-acetylaminofluorene N -sulphate. J.Chem.Soc.Chem.Commun. 1983:30–31. [Google Scholar]
- 52.Nauwelaers G, Bessette EE, Gu D, Tang Y, Rageul J, Fessard V, Yuan JM, Yu MC, Langouet S, Turesky RJ. DNA adduct formation of 4-aminobiphenyl and heterocyclic aromatic amines in human hepatocytes. Chem Res.Toxicol. 2011;24:913–925. doi: 10.1021/tx200091y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J.Proteome.Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng L, Turesky RJ. Optimizing proteolytic digestion conditions for the analysis of serum albumin adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, a potential human carcinogen formed in cooked meat. J.Proteomics. 2014;30:267–278. doi: 10.1016/j.jprot.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fede JM, Thakur AP, Gooderham NJ, Turesky RJ. Biomonitoring of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and its carcinogenic metabolites in urine. Chem. Res. Toxicol. 2009;22:1096–1105. doi: 10.1021/tx900052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen C, Ma X, Malfatti MA, Krausz KW, Kimura S, Felton JS, Idle JR, Gonzalez FJ. A comprehensive investigation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) metabolism in the mouse using a multivariate data analysis approach. Chem Res.Toxicol. 2007;20:531–542. doi: 10.1021/tx600320w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gan LS, Skipper PL, Peng XC, Groopman JD, Chen JS, Wogan GN, Tannenbaum SR. Serum albumin adducts in the molecular epidemiology of aflatoxin carcinogenesis: correlation with aflatoxin B1 intake and urinary excretion of aflatoxin M1. Carcinogenesis. 1988;9:1323–1325. doi: 10.1093/carcin/9.7.1323. [DOI] [PubMed] [Google Scholar]
- 58.Kim D, Kadlubar FF, Teitel CH, Guengerich FP. Formation and reduction of aryl and heterocyclic nitroso compounds and significance in the flux of hydroxylamines. Chem. Res. Toxicol. 2004;17:529–536. doi: 10.1021/tx034267y. [DOI] [PubMed] [Google Scholar]
- 59.Murata M, Kawanishi S. Mechanisms of oxidative DNA damage induced by carcinogenic arylamines. Front Biosci (Landmark Ed) 2011;16:1132–1143. doi: 10.2741/3739. [DOI] [PubMed] [Google Scholar]
- 60.Thomas AT, Stewart BJ, Ognibene TJ, Turteltaub KW, Bench G. Directly coupled high-performance liquid chromatography-accelerator mass spectrometry measurement of chemically modified protein and peptides. Anal. Chem. 2013;85:3644–3650. doi: 10.1021/ac303609n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carter DC, Ho JX. Structure of serum albumin. Adv.Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 62.Christodoulou J, Sadler PJ. 1H NMR of albumin in human blood plasma: drug binding and redox reactions at Cys34. FEBS Lett. 1995;376:1–5. doi: 10.1016/0014-5793(95)01231-2. [DOI] [PubMed] [Google Scholar]
- 63.Stewart AJ, Blindauer CA, Berezenko S, Sleep D, Tooth D, Sadler PJ. Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. FEBS J. 2005;272:353–362. doi: 10.1111/j.1742-4658.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- 64.He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 65.Antunes AM, Godinho AL, Martins IL, Oliveira MC, Gomes RA, Coelho AV, Beland FA, Marques MM. Protein adducts as prospective biomarkers of nevirapine toxicity. Chem. Res. Toxicol. 2010;23:1714–1725. doi: 10.1021/tx100186t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Becker AR, Sternson LA. Oxidation of phenylhydroxylamine in aqueous solution: a model for study of the carcinogenic effect of primary aromatic amines. Proc. Natl. Acad. Sci. U. S. A. 1981;78:2003–2007. doi: 10.1073/pnas.78.4.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexander J, Wallin H, Rossland OJ, Solberg KE, Holme JA, Becher G, Andersson R, Grivas S. Formation of a glutathione conjugate and a semistable transportable glucuronide conjugate of N 2 -oxidized species of 2-amino-1-methyl-6-phenylimidazo[4,5- b ]pyridine (PhIP) in rat liver. Carcinogenesis. 1991;12:2239–2245. doi: 10.1093/carcin/12.12.2239. [DOI] [PubMed] [Google Scholar]
- 68.Kostadinova R, Boess F, Applegate D, Suter L, Weiser T, Singer T, Naughton B, Roth A. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol. Appl. Pharmacol. 2013;268:1–16. doi: 10.1016/j.taap.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 69.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficent of a protein. Protein Science: a publication of the Protein Society. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.