Abstract
The HCV-IRES sequence is vital for both protein translation and genome replication and serves as a potential target for anti-HCV therapy. We constructed a series of anti-HCV Group I introns (αHCV-GrpIs) to attack conserved target sites within the HCV IRES. These αHCV-GrpIs were designed to mediate a trans-splicing reaction that replaces the viral RNA genome downstream of the 5’ splice site with a 3’ exon that encodes an apoptosis-inducing gene. Pro-active forms of the apoptosis inducing genes BID, Caspase 3, Caspase 8, or tBax were modified by incorporation of the HCV NS5A/5B cleavage sequence in place of their respective endogenous cleavage sites to ensure that only HCV infected cells would undergo apoptosis following splicing and expression. Huh7.5 cells transfected with each intron were challenged at MOI 0.1 with HCV-Jc1FLAG2 which expresses a Gaussia Luciferase (GLuc) marker. Virus-containing supernatants were then assayed for GLuc expression as a measure of viral replication inhibition. Cellular extracts were analyzed for the presence of correct splice products by RT-PCR and DNA sequencing. We also measured levels of Caspase 3 activity as a means of quantifying apoptotic cell death. Each of these αHCV-GrpI introns was able to correctly splice their 3’ apoptotic exons onto the virus RNA genome at the targeted Uracil, and resulted in greater than 80% suppression of the GLuc marker. A more pronounced suppression effect was observed with TCID50 virus titrations, which demonstrated that these αHCV-GrpIs were able to suppress viral replication by more than 2 logs, or greater than 99%. Robust activation of the apoptotic factor within the challenged cells was evidenced by a significant increase of Caspase 3 activity upon viral infection compared to non-challenged cells. This novel genetic intervention tool may prove beneficial in certain HCV subjects.
Keywords: Hepatitis, Group I, Intron, Apoptosis, IRES
Introduction
Hepatitis C Virus (HCV) affects more than 170 million people worldwide, or 3% of the world population [1], with 4 million new cases and more than 300,000 deaths per year [2]. Clinical conditions of the disease range from an asymptomatic carrier state to persistent infections. Of those individuals infected, 70% will develop chronic HCV infections, and 20% of chronic infections will progress to cirrhosis and terminal hepatocellular carcinoma [3-6]. There are currently no vaccines available to the public to preven HCV due to the high genetic variability of the virus and its ability to escape host immune defenses [7].
The current standard of care (SOC) treatments may include a combination of pegylated interferon-α and ribavirin [8] and direct acting antivirals such as Sofosbuvir and Simeprevir [9]. The drug combination has unfavorable side effects and may ultimately lead to drug resistance and relapse. One of the main reasons may be attributed to the generation of quasispecies genome diversity common for HCV infections, a phenomenon that results in infection by a swarm of microvariants derived from a predominant “master sequence” within an individual host [10]. Quasispecies are more prominent in the setting of persistent infections and may be responsible for drug treatment failures [11,12]. Quasispecies result from the high error rate of the non-proofreading HCV RNA-dependent RNA polymerase (RdRp) leading to continuous production of mutated virus sequences which is one mechanism the virus employ to escape immune system defense [13]. This warrants a continued intensive search for alternative antiviral approaches to combating HCV.
HCV is a plus-strand RNA virus of the Hepacivirus genus, having a 9600 nt long genome encodin a single ORF flanked by highly conserved 5’ and 3’ untranslated regions (UTRs) [14]. The ORF encodes a single polyprotein that is modified post-translationally by both cellular and viral proteases to produce 3 structural (C, E1, E2) and 7 non-structural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins [15]. The 5’ UTR of the viral RNA contains an internal ribosome entry site (IRES) that is highly conserved among most known HCV quasispecies [16]. The 5’UTR of HCV facilitates viral replication and mediates cap-independent viral protein translation by acting as a scaffold and recruiting multiple protein factors during the initiation of translation upon early infection [17-19]. Because the IRES serves a crucial function for viral infection and propagation and is therefore highly conserved, it represents an ideal target for anti-HCV approaches employing nucleic acid homologies such as trans-splicing group I introns [20].
Trans-splicing group I introns derived from the cis-splicing group I intron of Tetrahymena thermophila mediate RNA splicing through two successive transesterification steps [21]. First, the intror recognizes a specific uracil on the target RNA during complementary base pairing with the surrounding sequence. The target RNA is then cleaved at that uracil, and the intron-attached 3’exon is cleaved from the group I intron and appended onto the cleaved target RNA to create a product RNA. If that product i capable of translation it will express a new protein encoded by the sequence of the 3’exon [22]. Group introns have been used successfully in a number of anti-viral applications including targeting of Dengue Fever virus [23], HCV [20], and HIV [24] genomes, and in post transcriptional gene manipulations including the restoration of wild-type p53 activity in three cancerous cell lines [25] and the repair of sickle β-globin mRNAs in erythrocyte precursors [26].
In this report we describe the construction and activity analysis of a series of anti-HCV Group I introns (αHCV-GrpIs). These αHCV-GrpIs were designed to be more effective than conventional group I introns by extending both the External Guide Sequence (EGS) to increase the target base pairing specificity, and the Internal Guide Sequence (IGS) to help stabilize the base pairing at the catalytic site [24]. Apoptosis-inducing gene sequences were incorporated as 3’exons to induce cell death upon successful splicing.
We verify the functional characteristics of two αHCV-GrpIs constructed to target conserved sequences within the IRES surrounding U329 of stem loop IIIf and U343 of stem loop IV. These αHCV-GrpIs mediate trans-splicing reactions that cleave the HCV RNA genome at the designated uracils and append a 3’ exon composed of sequences that complete the downstream region of the HCV IRES, start codon AUG, and N-terminal coding sequence of the HCV core protein, all of which are mandatory for efficient initiation of IRES-dependent translation. These reconstituted IRES sequences are followed by one of the apoptosis-inducing genes BID, Caspase 3, Caspase 8 or a truncated form of Bax called ΔN Bax (abbreviated as tBax in this publication). To prevent unintended expression of these apoptotic genes an to increase the specificity of activity for HCV infections, these genes are incorporated in their pro-active forms, replacing their respective endogenous cleavage sites with the HCV NS5A/5B cleavage sequence recognized by the virus protease NS3/4A [27]. Using this strategy, the functional “active” form of the pro-apoptotic gene is produced only upon correct trans-splicing and in the presence of the active HCV protease, ensuring that only infected cells can undergo apoptosis. This successful genetic approach may provide a useful tool for intervention strategies against HCV.
Materials & Methods
Target site selection
Genome sequences of most known HCV quasispecies were obtained from the Los Alamos Hepatitis C Sequence Database [28] and aligned in ClustalX [29]. The aligned sequences comprise the following GenBank Genlnfo identifiers: 22129792, 9843676, 21397075, 21397076, 21397077, 8926244, 221586, 2327072, 2327074, 2316097, 2327070, 329873, 329737, 221604, 221606, 20521945, 2943783, 3098638, 15529110, 12831192, 1944375, 14581919, 11559440, 11559442, 11559444, 11559446, 11559448, 11559450, 11559452, 11559454, 11559456, 11559458, 11559460, 11559462, 11559464, 11559466, 11559468, 1212741, 560788, 306286, 27544243, 7650221, 7650223, 7650225, 7650227, 7650229, 7650231, 7650233, 7650235, 7650237, 7650239, 7650241, 7650243, 650245, 7650247, 7650249, 7650251, 7650253, 7650255, 7650257, 7650259, 7650261, 7650263, 7650265, 7341102, 3098634, 3098636, 3098632, 2764397, 48237633, 471116, 38492204, 329739, 5420376, 4753718, 4753720, 1181831, 329770, 221610, 1030706, 1030705, 1030701, 10300702, 1030704, 1030703, 23957856, 960359, 385583, 62006146, 1160327, 59478, 221612, 221614, 5918928, 5918930, 5918964, 5918966, 5918932, 5918934, 5918936, 5918938, 5918940, 5918942, 5918944, 5918946, 5918948, 5918950, 5918952, 5918954, 5918956, 5918958, 5918960, 5918962, 567059, 437107, 46560633, 5738246, 19568932, 2443428, 329763, 464177, 15422182, 50235321, 6707281, 6707283, 7329200, 7329202, 7329204, 7329206, 7329208, 6707279, 6707285, 6010579, 53680893, 53680893, 221650, 13122263, 13122265, 13122267, 13122269, 13122271, 13122273, 13122261, 9757541, 7329210, 221608, 1483141, 6521008, 2895898, 558520, 514395, 633201, 676877, 1183032, 2252489, 2464303, 3660725, 60115454, 6011545, 60115457, 62362179, and 60115456. These sequences cover all major HCV genotypes including 1a, 1b, 1c, 2a, 2b, 2c, 2k, 3a, 3b, 3k, 4a, 5a, 6b, 6d, 6g, 6h, and 6k. The IRES is located in the 5’UTR of the virus genome and continues to 40 nucleotides of the core-coding sequence [30]. Target Uracils for Intron #19 (I19) targeting domain lllf and Intron #20 (I20) targeting domain IV are U329 and U343, respectively.
Addition of HCV NS3/4A serine protease recognition site to pro-apoptotic factors
Apoptotic factors BID and Caspase 3 were modified by replacing their endogenous cleavage sites (aa62 in BID and aa25 and 172 in Procaspase 3) with the NS3/NS4A recognition site NS5A/NS5B cleavage sequence: AEDVVCCCCSMSYS [27]. For Caspase 8, the 5’ end was truncated at aa186 in the Procaspase 8 and the NS5A/5B recognition sequence was inserted at aa211 and 375. These complete pro-apoptotic genes were synthesized by GeneArt (Life Technologies, USA) with nucleotide usage optimized preferably for mammalian expression.
Anti-HCV group I intron constructs
The sequence of group I intron splicing domain used in this study was derived from the catalytic core of the rRNA Tetrahymena thermophila on the pTT1A3-T7 plasmid (a kind gift from Dr. Thomas Cech, University of Colorado, Boulder). In the preliminary assay (Figure 1C), we constructed a set of introns attacking an artificial target that encoded the HCV IRES linked to a Fluc-reporter sequence. Cleavage of the HCV IRES sequence would result in a reduction of FLuc expression. Once we had determined the best attack site, we constructed our αHCV-GrpI introns based on the sequences surrounding that target site. The I19 and I20 were generated by PCR amplification using the following primer sets: I19 for: 5’GTTAACTTTTCTTTGAGGTTTAGGATTCGTGCTCATGCAGTCGGTCTGCGAGAAAAAGTTATCAGGCATGCACCT GGT3’; I19 rev: 5’ACCGGTTTTTCTTTGAGGTTTAGGATTCGTGCTCATGGTGCACGGTCTCGATTAGTACTCCAAAACTAATCAATAT ACTTTC3’; I20 for: 5’GTTAACTTTTCTTTGAGGTTTTCCTAAGGTGCTCGTGGTAAAAGTTATCAGGCATGCACCTGGT3’; and I20 rev: 5’ACCGGTTTTTCTTTGAGGTTTAGGATTCGTGCTCCGATTAGTACTCCAAAACTAATCAATATACTTTC3’. The forward primers contain EGS, IGS, and 5’ end of the intron splicing domain while the reverse primers contain 3’ end of the intron splicing domain, P10 helix, Loop Bulge (LB), reconstructed 3’IRES and an extended 30 nt-long core sequence (Table 1). Following PCR amplification and band isolation, the introns were restriction digested with HpaI and AgeI and inserted into a backbone vector pLRH [31]. The CMV promoter driving the expression of the intron was obtained by PCR amplification of the pCMV-DsRed-Express (Clontech, USA) using forward: 5’GGATCCTCAATATTGGCCATTAGCCATA3’ and reverse: 5’GTTAACCCTATAGTGAGTCGTATTAA3’ primers and was digested and inserted into the backbone vector at BamHI and HpaI sites. The 3’ apoptosis-inducing exon BID, Caspase 3, Caspase 8, or tBax was amplified by PCR amplification of the templates synthesized by MR. GENE using the following primers: BID for: 5’ACCGGTATGGACTGTGAGGTC3’: 5’ATGCATCTATTAGTCCATCCCATTTCTGG3’; Caspase 3 for: 5’ACCGGTATGGAGAACACTGAAAAC3’ and rev: 5’ATGCATCTATTAGTGATAAAAATAGAGTTCTTTTGTG3’; Caspase 8 for: 5’ACCGGTATGGGGGAGGAG3’ and rev: 5’ATGCATCTATTAATCAGAAGGGAAGACAAG3’; tBax for: 5’ACCGGTGCCCTTTTCTACTTTGCCAGCA3’ and rev: 5’ATGCATCTAGCCCATCTTCTTCCAGATGG3’. These apoptotic genes were inserted into the vector between AgeI and NsiI sites. The resulting introns were named I19BID, I19C3, I19C8, I19tBx, I20BID, I20C3, I20C8, and I20tBx. To create inactive control introns (ΔIntrons), the entire splicing domain was removed, leaving only the EGS, IGS, P10, LB and 3’exon to religate.
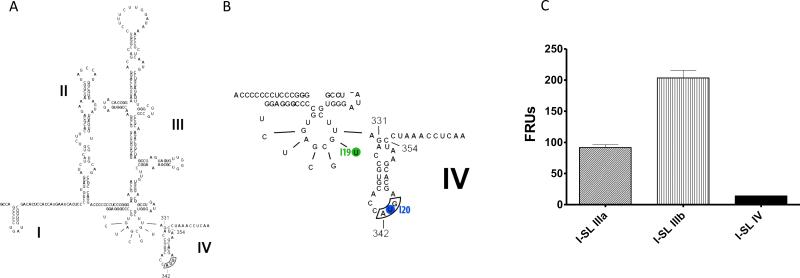
Figure 1.
Structure of HCV IRES. Adapted from Tumban E, 2009 [58]. (A) Complete IRES construct containing stem-loop I-IV. The AUG start codon of the core protein is bordered; (B) Stem-loop IIIf and IV enlarged to show target sites for I19 and I20 at U329 and U343 (circled), respectively; (C) FLuc assay demonstrating that the intron targeting stem loop IV (I-SL IV) inhibited a near-complete IRES translation compared to those targeting stem loop IIIa (I-SL IIIa) and IIIb (I-SL-IIIb).
Table 1.
Positions of target Uracils within the HCV IRES and sequences of group I intron domains. EGS, External Guide Sequence; IGS, Internal Guide Sequence; P10, P10 helix; LB, Loop Bulge.
| Version | Target U | EGS | IGS | P10 | LB | Reconstructed IRES+core |
|---|---|---|---|---|---|---|
| I19 | U329 | ttttctttgaggtttaggattcgtgctcatg | cggtctgcgaga | agaccg | cagt | tgcaccatgagcacgaatcctaaacctcaaagaaaa |
| I20 | U343 | ttttctttgaggttt | gtgctcgtggtg | gagcac | tcctaag | gaatcctaaacctcaaagaaaa |
Huh7.5 cell culture
All cell culture experiments were carried out in Huh7.5 human hepatocellular carcinoma cells (a kind gift from Dr. Charles Rice, the Rockefeller University, USA with written permission from Apath). Cells were routinely maintained in Dulbecco's Modified Eagle's Medium (DMEM; Sigma-Aldrich, USA) supplemented with 2 mM L-glutamine (Sigma-Aldrich, USA), 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, USA), and 1X non-essential amino acids (Gibco; Life Technologies, USA) and incubated at 37°C in a 5% CO2 atmosphere.
DNA transfection
Cells were plated in 6-well plates at 6×105 cells/well 24 h before transfection. We used Lipofectamine LTX + Plus reagent system (Life Technologies, USA) for all DNA transfection experiments. Transfection reaction was prepared by combining the following components: 1 μg intron plasmid DNA + 10 μl Plus reagent + 50 ng Cypridina plasmid DNA (as a normalizer; in the luciferase assay; NEB, USA) + 100 μl serum-free optiMEM (Life Technologies, USA). The reaction was incubated at RTfor 15 min followed by addition of 100 μl of 5% Lipofectamine LTX (5 μl LTX diluted in 95 μl serum-free optiMEM). The DNA:Lipofectamine LTX complex was allowed to form at RT for 30 min. DMEM was then added to the reaction to make up to 1 ml total volume. Cells were left in transfection media for 24 h at 37°C + 5% CO2 before replacing with fresh DMEM. Transfected cells were allowed to grow for another 24 h prior to virus inoculation.
JclFLAG2 HCV transcription
The Jc1FLAG2 plasmid (a kind gift from Dr. Charles Rice, the Rockefeller University, USA; [32]) encoding a full-length HCV was used to generate a transcription template as previously described [32]. Briefly, 10 μg of the plasmid was digested with XbaI for 3 h at 37°C and cleaned up using the Wizard SV Gel and PCR Clean-Up System (Promega,USA). The T7 RiboMAX Express Large Scale RNA Production System (Promega, USA) was used for a transcription reaction of 1 μg DNA template. The reaction was incubated for 30 min at 37°C. One U of supplied DNase was then added to eliminate the DNA template. The reaction was incubated for an additional 15 min at 37°C. The final RNA transcript was cleaned up and eluted in the RNA Storage Solution (Ambion; Life Technologies, USA) using the Qiagen RNeasy kit (Qiagen, USA). The purified RNA was aliquoted for single uses and stored frozen at −80°C.
Full-length infectious JclFLAG2 HCV preparation
Huh7.5 cells were washed twice before electroporation. In a multi-well electroporation plate (4 mm, 25 wells; BTX Harvard Apparatus, USA), 5×106 cells were mixed with 10 μg RNA, loaded onto each well (in total volume 400 μl) and electroporated using the BTX ElectroSquare Porator ECM 830 (BTX Harvard Apparatus, USA) with the following settings: 860 V, 5 pulses, 99 μs, 1.1 s interval, and high voltage. Electroporated cells were plated immediately after. Virus-infected cells were incubated at 37°C + 5% CO2. Virus-containing media was harvested every 4 h for up to 96 h, clarified by filtration, and concentrated using the Stirred Ultrafiltration Cell system (Model 8400; Millipore, USA). Concentrated virus was aliquoted into cryotubes for single uses and stored at −80°C. Virus titer was determined by TCID50 titration (see below).
HCV infection and sample collection
Cells were washed once with DMEM, followed by HCV inoculation at MOI 0.1. The inoculum were removed at 6 h. Cells were washed twice and replenished with 2 ml DMEM. Virus-containing supernatants were collected at 48 h (for apoptosis assay), 72 h (for luciferase assay), or 96 h (for TCID50) and filtered through a sterile 0.45 pm cellulose acetate syringe filter. Supernatants were stored frozen at −80°C until assayed. For Annexin V staining, cells were stained at 24 h post infection.
One-step RT-PCR
Splice products were detected by RT-PCR, followed by DNA sequencing to confirm the correct sequence of the splicing junctions. Total RNA was extracted from infected and uninfected cells using TRIZol Reagent (Life Technologies, USA) following the manufacturer's instructions without modification and eluted in 100 μl nuclease-free water. RNA quantity and purity was measured by Nanodrop (ThermoScientific, USA). Superscript III One-Step RT-PCR System with Platinum Taq High Fidelity kit (Life Technologies, USA) was used for all RT-PCR reactions. Briefly, the RNA was treated with TURBO DNA-free DNase (Ambion; Life Technologies, USA) for 30 min at 37°C to remove any DNA residues. To deactivate DNase, 0.1 volume of DNase Inactivating reagent was added to the reaction and incubated at RT for 5 min with periodic mixing. The RNA was collected after centrifugation at 10,000 × g for 1.5 min. Ten micrograms of DNA-free RNA was transferred to the RT-PCR reaction. The spliced product was detected using “splice primers” specific to the target (forward) and 3’ exon (reverse) that straddle the splice junction. The RT-PCR profile was as follows: 50°C 45 min for cDNA synthesis, 94°C 2 min, 40 cycles of 94°C 15 sec, 59°C 30 sec, and 68°C 1 min, and 68°C 7 min. PCR products were run on 0.9% agarose gel, followed by band visualization using the Gel Doc EZ Imager (Bio-Rad, USA) and band isolation using the Wizard SV Gel and PCR Clean-Up System (Promega,USA). Correct spliced products were confirmed by DNA sequencing (Genomics Core Facility, University of Notre Dame, USA).
Firefly luciferase assay (FLuc assay)
Transfected cells were washed twice with PBS before addition of 350 ul of 1X Lysis Buffer. Cells were incubated at RT for 5 min and transfererred to a microcentrifuge tube for storage at −80°C overnight. The next day, frozen cells were freezed and thawed repeatedly 3 times and centrifuged at 10,000 RPM for 2 min. Firefly Relative Units (FRUs) were read on 20 ul sample in a 96-well format after addition of 100 ul of Luciferase Assay Reagent (Promega, USA). Relative light units (RLUs) were measured with a spectramax plate reader (L MAXII 384, Molecular Devices, USA). Luminescence was integrated over 100 sec with a 3-second delay.
Gaussia luciferase assay (GLuc assay)
Collected supernatants were immediately mixed with an equal amount of 1X lysis buffer. Twenty μl of lysed sample was loaded on to a 96-well plate in triplicate. Gaussia luciferase (GLuc) activity was detected using the Renilla Luciferase Assay System (Promega, USA). Relative light units (RLUs) were measured with a spectramax plate reader (L MAXII 384, Molecular Devices, USA). Luminescence was integrated over 10 sec with a 2-second delay and measured upon injection of 50 μl of substrate reagent. For Cypridina luciferase (normalizer) measurement, we used BioLux Cypridina Luciferase Assay kit (NEB, USA) with identical settings to GLuc measurement except we set the integration time to 2 sec.
TCID50 assay
The titers of infectious HCV in infected cell culture supernatants were measured by immunohistochemistry staining in conjunction with an endpoint dilution assay as previously described [32]. Briefly, a 10-fold serial dilution of the virus-containing supernatants was performed and used to infect Huh7.5 cells in multiple wells of 96-well plates. Infection was allowed to incubate for 96 h at 37°C + 5% CO2. Infected cells were then washed with PBS and fixed with freezer-cold methanol for 30 min at −20°C. Cells were blocked in PBS + 1% BSA + 0.2% skim milk for 30 min, followed by staining with the NS5A-specific 9E10 primary antibody (diluted at 1:2000; a kind gift from Dr. Charles Rice, the Rockefeller University, USA; [32]) for 1 h. Endogenous peroxidase was quenched with 3% H2O2 for 5 min and cells were incubated with a secondary antibody from the ImmPRESS Anti-Rabbit Ig (peroxidase) Polymer Detection kit (diluted at 1:3; ImmPRESS; Vectorlab, USA) for 30 min. HCV-infected cells were visualized under light microscope after addition of peroxidase DAB substrate (diluted at 1 drop/ml of supplied buffer; DAB+; Dako, USA) for 1 h. The 50% tissue culture infectious dose (TCID50) was calculated using Reed & Muench's protocol [33].
Caspase 3 activity assay
At 48 h post infection, cells were trypsinized, pelleted, washed with PBS, and counted. The Caspase 3 activity was measured using the EnzChek Caspase-3 Assay Kit #2 (Life Technologies, USA) in accordance with the manufacturer's instructions. Briefly, cells were lysed on ice for 30 min in 50 μl cell lysis buffer. Following pelleting of cell debris at 5000 rpm for 5 min, 50 μl supernatant was combined with 50 ul of the 2X substrate working solution. The reaction was incubated in the dark for 30 min. The fluorescence was measured using a fluorescence plate reader (SPECTRAmax M2, Molecular Devices) with the following settings for excitation/emission: 496/520 nm. The RFUs readout for each sample was calculated on a per cell basis.
Annexin V staining
Apoptotic cells were visualized using the Annexin V-FITC Apoptosis Kit Plus (MBLI, USA). Briefly, cells were stained with 5 μl Annexin V-FITC diluted in 500 μl 1X Binding buffer for 10 min in the dark at RT. Apoptotic cells were visualized with a GFP filter. Total cells were counted and the percentage of positive cells were calculated.
Western blot analysis
At 96 h post infection, cell extracts were prepared in RIPA buffer supplemented with Halt Protease Inhibitor Cocktail (Thermo Scientific, USA) and phenylmethylsulfonyl fluoride (PMSF; Thermo Scientific, USA). The extracted proteins were sonicated for 15 sec twice at 50% pulse and then mixed with 1.25% β-mercaptoethanol and 1:3 volume LDS Sample Buffer (Thermo Scientific, USA) before boiling for 5 minutes. The protein concentrations in the samples were measured using NanoDrop and 12 μg of proteins were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes by electroblotting with an XCell SureLock unit (Life Technologies, USA). The protein blots were blocked overnight at 4°C with PBS+5% non-fat milk (NFM) and incubated with polyclonal anti-PKR (Santa Cruz, USA), monoclonal anti-pPKR (Abcam, USA), or polyclonal anti-β-actin (Thermo Scientific, USA) diluted at 1:1000 in PBS-T+0.5% NFM for 1 h at room temperature with agitation. After incubation, the membranes were washed for 5 minutes (x4) with PBS-T and incubated with a 1:1000 dilution of donkey anti-rabbit IgG-HRP (Santa Cruz, USA) in PBS-T +0.1% NFM for 1 hour at room temperature with agitation. The membranes were washed again for 5 minutes (x4) with PBS-T and developed using the SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific, USA) for 5 minutes at RT. Immunoreactive bands were detected by exposing the membranes to X-ray film. As a positive control, untransformed wild-type cells were infected and induced with Human Interferon Beta 1a (PBL Assay Science, USA) at 100U/ml concentration for 16 hours prior to protein extraction [34].
Statistical analysis
For reproducibility, each assay was performed in triplicates. Data were analyzed in Prism6 software (GraphPad, USA). Statistical significance (p-value < 0.05) between the intron-transfected cells vs. control cells were reported along with standard errors of the means.
Results
Target site selection
We retrieved all known HCV quasispecies from The Los Alamos HCV Sequence Database [28] and aligned them in ClustalX. Intron design focused to target the IRES sequence located in the 5’ region of the virus genome since this structure is highly conserved among HCV quasispecies (Figure 1A). Initially, we constructed and tested several group I introns targeting multiple sites throughout the IRES, but later narrowed down our focus to those that attack target sites at the very 3’ end of the IRES. Our initial results demonstrated that introns targeting stem loops IIIa and IIIb did not completely prohibit the function of the IRES to translate the firefly luciferase gene in the artificial target. In contrast, introns that were designed to target the very 3’ end of the IRES were able to knock down the IRES translation by 95% (Figure 1C). Twotarget sites, U329 and U343 (Figure 1B), were selected for further experiments. U329 is located in the conserved pseudoknot (stem loop IIIf) region which is part of the 40S binding domain crucial for viral translation [35, 36]. U343 is part of the AUG start codon (stem loop IV) of the core-coding sequence and is positioned at the P site during translation initiation [37].
Design of anti-HCV group I intron targeting conserved IRES sequences
The group I intron trans-splicing mechanism has been described extensively [23, 24]. Our optimized αHCV-GrpIs incorporate a more extensive complementary base-pairing between the EGS and IGS and the virus genomic RNA to improve both specificity and splicing activities at the targeted uracil (Figure 2: step A). The EGS can be of nearly any practical length from as short as 5 bases without compromising the splicing efficacy [38], although longer sequences may decrease the efficiency of the reaction due to internal interactions and/or non-specific binding to non-target RNAs [39]. The emphasis on the design and addition of the EGS reduce potential illegitimate binding of the intron to non-target RNA that may result in unintentional activation of the apoptosis in non-HCV infected cells. The ability of our introns to attack target single-stranded motifs in the secondary structure of the IRES was demonstrated by splice product detection.
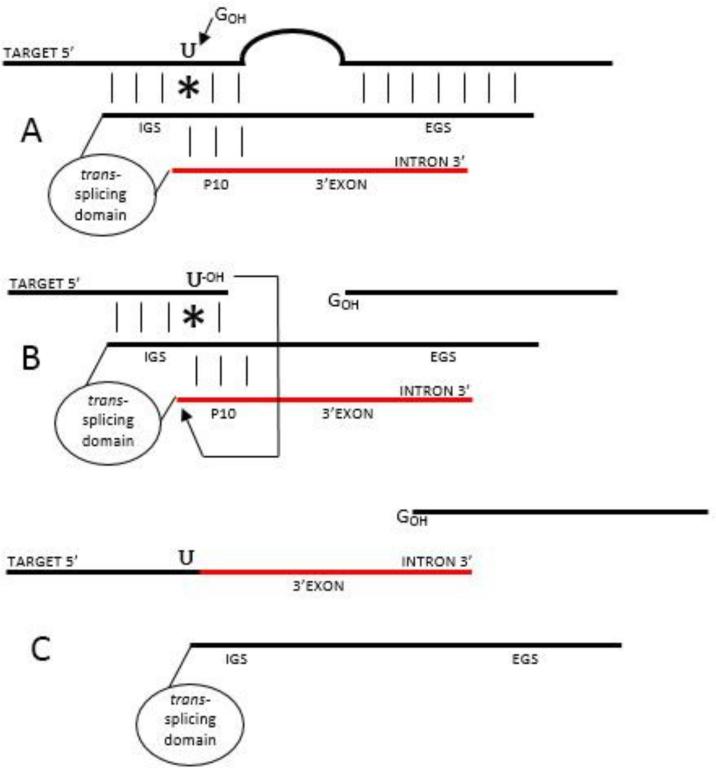
Figure 2.
Group I intron-mediated trans-splicing mechanism. Group I intron base pairs with the target through the IGS and EGS. A nucleophilic 3’-OH of an exogenous guanosine attacks the phosphate backbone at the 5’ splice site on the target RNA at the target Uracil and covalently binds to the excision product (A). While the P10 helix brings the intron-attached 3’ exon into a close proximity with the target Uracil, the free 3’-OH of the cleaved Uracil attacks the 3’ splice site (B) resulting in a seamless ligation of the target to the new 3’ exon (C).
The splicing reaction relies on the formation of the P1 and P10 helices between the target RNA and the group I intron. The IGS participates in the formation of the P1 helix that spans the reactive uracil required for proper trans-splicing mechanism [24]. Nucleophilic attack by the 3’-OH of a free exogenous guanosine cleaves the phosphate backbone of the target RNA molecule 3’ of the P1 helix-embedded uracil. The splicing reaction is facilitated allowing the intron to form a P10 helix that brings the 3’exon into proximity with the P1-attached 3’-OH, which later attacks the phosphate backbone upstream of the group I intron-attached 3’exon to complete the trans-splicing reaction (Figure 2: step B). To insure expression from the spliced RNA, we incorporated sequences that reconstituted the IRES sequence from the splice target in the HCV genome through the first 30 nt of the HCV core sequence. These sequences had been shown essential for effective expression from the HCV IRES [30]. The resulting spliced RNA has a reconstituted IRES sequence that is capable of translating the 3’exon sequence (Figure 2: step C).
Initial experiments were performed to select the most effectively targeted uracils in the upstream regions of the HCV IRES [U160 (stem loop IIIa), U194 (stem loop IIIb), U198 (stem loop IIIb), and U301 (stem loop IIIe)]. These initial intron constructs contained firefly luciferase (FLuc) as their 3’exon, and luciferase assays demonstrated significant levels of unintentional expression of FLuc in the absence of the target sequence. These results suggested that the incomplete HCV IRES sequence (up until stem loop IIIe) is still capable of initiating translation of the downstream coding sequence. The translation efficiency with such incomplete constructs may not have been at optimum levels but was enough to drive the expression of the 3’exon. Such background expression would not be acceptable in downstream applications using apoptosis-inducing genes, and this finding forced a re-design to target uracils at a different position, specifically more towards the 3’ end of the HCV IRES.
We subsequently tested introns designed to target U329 (stem loop IIIf) and U343 (stem loop IV) located at the very end of the IRES sequence (Table 1, Figure 3). Reconstituted IRES and downstream HCV core sequences from each of these uracils failed to generate any detectable background FLuc expression with our αHCV-GrpIs in the absence of target sequence. These targets became our preferred targets for αHCV-GrpI construction.
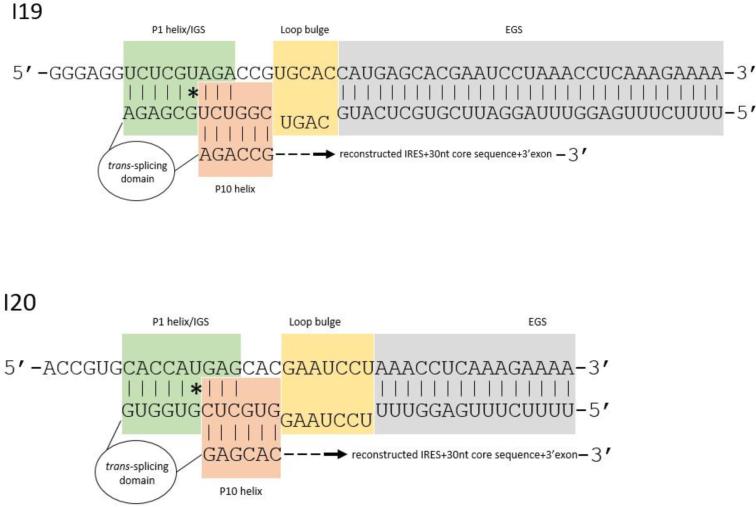
Figure 3.
Design of anti-HCV group I introns. Displaying I19 targeting U329 (top) and I20 targeting U343 (bottom). Target HCV and intron sequences are displayed in the 5’-3’ and 3’-5’direction, respectively. The intron contains the following components in 5’-3’ direction: EGS, LB, IGS, trans-splicing domain, P10, reconstructed IRES+30nt core sequence, and 3’exon.
The EGS of both introns were designed to base pair with the 5’ terminus of the HCV core sequence, immediately followed by a short stretch of a non-homologous loop bulge sequence (LB) to allow efficient formation of the P10 helix [40]. We improved the formation of the P1 helix in our introns by extending the IGS from 8 to 9 nucleotides and positioning the target uracil at nt 6 in this sequence. We also improved the formation of the P10 helix and the resulting splicing efficiency by making the last 3 nucleotides of the IGS complimentary to the first three nucleotides of the P10 helix sequence. Insertion of two stop codons, UAA and UAG, immediately 5’ of the intron UCG splice site insured translation of the 3’exon in the absence of splicing would not occur. Sequences that reconstructed the 3’ portion of the IRES from each of the target uracils along with the first 30 nt of the HCV core-coding sequence were inserted as the last elements of each αHCV-GrpI.
HCV NS3/4A serine protease recognition site
To prevent illegitimate expression of the apoptotic gene products from the 3’exon and to enhance the specificity of the apoptotic reactions for HCV infected cells, we modified the pro-apoptotic forms of BID, Caspase 3, and Caspase 8 by replacing their original cleavage sites with the NS5A/5B cleavage recognition site for HCV NS3/4A HCV serine protease. Inserting the sequence AEDVVCCCCSMSYS (NS5A/5B cleavage site) at aa62 of BID, 25 and 172 of Caspase 3, and 211 [27] and 375 of Caspase 8 generated “pro” forms of these apoptotic factors that could only be activated in the presence of the HCV protease in infected cells, and therefore resulted in the initiation of the apoptotic cascade. We chose to include the enzymes BID, Caspase 3, and Caspase 8 apoptotic genes in addition to the non-enzymatic tBax because their enzymatic properties enhance their ability to induce apoptosis without relying upon apoptotic protein buildup. The tBax has been reported to induce apoptosis more efficiently than full-length Bax in vitro through a caspase-independent mechanism [41, 42].
αHCV-GrpIs mediate correct trans-splicing reactions in HCV infected Huh7.5 cells
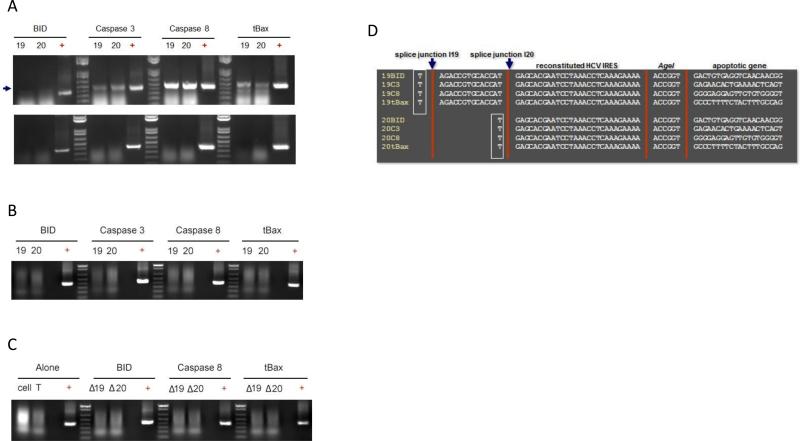
We examined each αHCV-GrpI for its ability to mediate the trans-splicing reaction in infected Huh7.5 cells by RT-PCR. The purpose of this assay was simply to prove that our introns are able to perform the splicing reaction against the viral RNA in a qualitative manner. The amount of unmodified HCV RNA species was not monitored because this assay was not intended to be quantitative. Cells were transfected with an intron-expressing plasmid (see Materials and Methods and Figure 3) and challenged 48 h post transfection with infectious CwmAG2 at a MOI 0.1. Total cellular RNA was extracted at 72 h post infection and analyzed for spliced products by one-step RT-PCR. The expected ~400 bp spliced products were evident for both I19 and I20 with each of the 3’exons; BID, Caspase 3, Caspase 8, or tBax, tested (Figure 4A: top). In contrast, spliced products were not detected in the absence of RT (Figure 4A: bottom) or from unchallenged cells expressing the αHCV-GrpIs (Figure 4B), confirming the specificity of the trans-splicing reactions for all the αHCV-GrpIs. In addition, control ΔIntrons (Materials and Methods), which lack the trans-splicing domain, failed to produce spliced products (Figure 4C), verifying that our αHCV-GrpIs produce splice product via the trans-splicing mechanism. DNA sequencing confirmed correct splice junctions on all splice products (Figure 4D).
Figure 4.
Splice product detection by RT-PCR. Total RNA was extracted from HCV infected cells at 48 h and the spliced products were detected by RT-PCR. (A) Top: introns cells, + reverse transcriptase (RT); bottom: -RT; (B) mock and uninfected intron cells; (C) ΔIntrons containing only guide sequences. Arrow indicates splice product; (D) DNA sequencing of the splice junction. Bordered indicates Uracil target.
αHCV-GrpIs suppress IRES-dependent translation from HCV reporter constructs in Huh7.5 cells
We utilized the Jc1FLAG2 construct containing the GLuc marker gene inserted between p7 and NS2, as previously described [43], for all our luciferase assays. Full-length Jc1FLAG2 viral RNA was generated by in vitro-transcription [43] and used to initiate infection in naïve Huh7.5 cells. The resulting infectious virus-containing media was collected, titrated, and used as a virus stock for downstream experiments. Huh7.5 cells were transfected with plasmids expressing each of the αHCV-GrpI or control plasmids at 48 h prior to HCV challenge at a MOI of 0.1. Supernatants were collected at 72 h post infection and the level of GLuc activity, which correlates directly to the total amount of virus in infected cells, was measured in each sample.
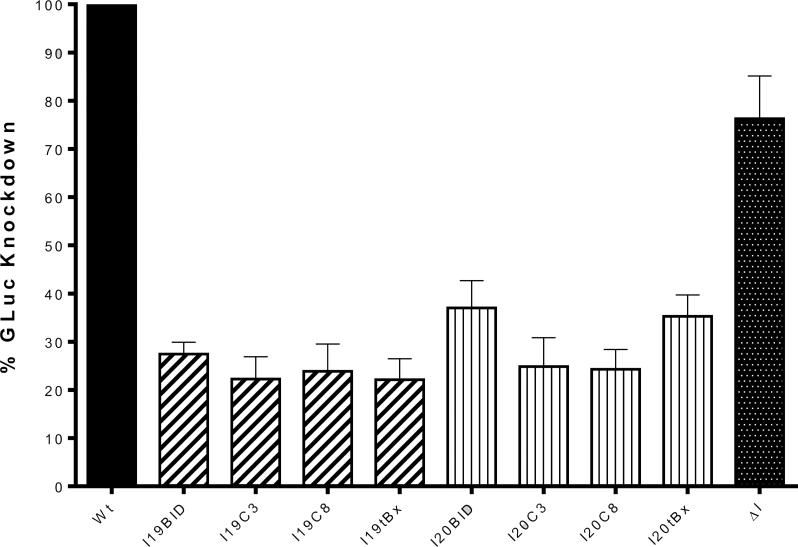
For these experiments we co-transfected a CMV promoted Cypridina luciferase expression plasmid along with the intron to allow normalization of the luciferase relative light unit (RLU) reads between samples. The amount of GLuc activity in the untransfected control infection was established as 100% for comparison purposes. As shown in Figure 5, our αHCV-GrpIs demonstrated varying levels of GLuc knockdown in Huh7.5 cells, with I19C3 and I19tBax being the most effective, reducing the level of GLuc activity to 20% of untransfected control cells, followed closely by I9BID, 19C8, I20C3, and I20C8. I20BID and I20tBx reduced GLuc to approximately 35% of untransfected controls. In contrast, the ΔIntron control demonstrated only a slight reduction in GLuc productivity to 80% of untransfected control cells. Overall, these results demonstrated that the transfected αHCV-GrpIs were able to effectively target HCV IRES RNA in infected Huh7.5 cells, knocking down the level of GLuc activity by as much as 80%.
Figure 5.
Luciferase assay. Huh7.5 cells were transfected with each intron 48 h prior to full-length, HCV-GLuc infection. Samples were collected at 72 h and measured for GLuc activity. GLuc measured for the control wild-type cells (Wt) was set to 100% for comparison with each intron. ΔI: intron lacking a trans-splicing domain containing only guide sequences. The error bars represent standard error of the mean computed frormfhree independent experiments.
Repression of infectious virus production in αHCV-GrpI intron-transfected cell cultures
Huh7.5 cell cultures were challenged at 48 hrs post transfection with HCV Jc1FLAG2 at MOI 0.1. Virus-containing cell supernatants were collected 96 h post challenge and virus titres were determined by IFA (Figure 6) in Huh7.5 cells. The anti-NS5A monoclonawntnbody 9E10 was used to stain infected cells [32]. Expression of each αHCV-GrpI in Huh7.5 cells was expected to reduce the yield of HCV following challenge with virus by eliminating productively infected cells through apoptotic cell death.
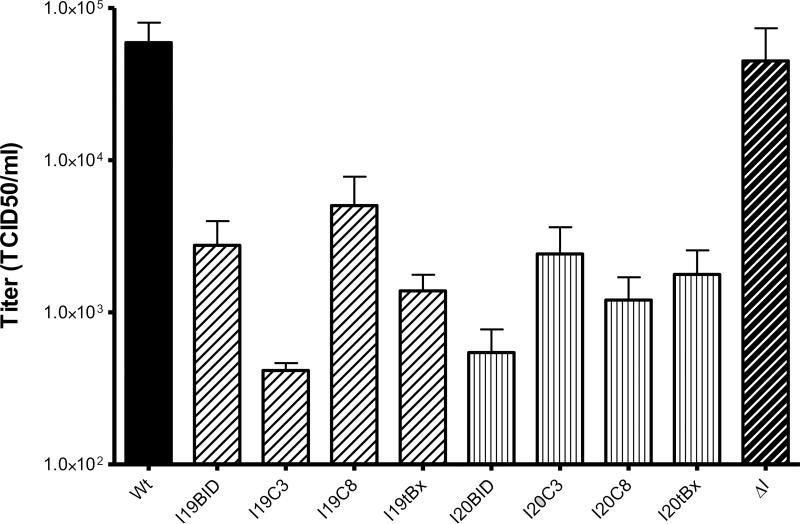
Figure 6.
TCID50 titration assay. Huh7.5 cells were transfected with each intron 48 h prior to full-length, HCV-GLuc infection. Samples were collected at 96 h and the viral titers were determined by TCID50. Wt: wild-type cells, ΔI: intron lacking a trans-splicing domain. Data represented in the scale of Log 10. The error bars represent standard error of the mean computed from three independent experiments.
In contrast to GLuc assays, the levels of virus suppression determined by TCID50 varied considerably between different introns. I19C3, the most active intron observed in the GLuc assay, and I20BID were the most effective in reducing the production of infectious virus from intron-transfected cells, suppressing the infectious virus production by more than 2 logs compared to the untransfected control cells (Figure 6). The I19tBax, which was as effective as I19C3 in GLuc reduction, appeared to be less effective as measured by the TCID50 assay, reducing the virus titer by slightly more than a log. The remaining αHCV-GrpIs, I20C8, I20tBax, I19BID, I19C8, and I20C3 demonstrated slightly lower suppression levels, reducing virus titers by approximately 1 log. In contrast, the ΔIntron lacking the trans-splicing domain did not demonstrate significant inhibition of virus production.
Cell-specific cytotoxicity induced in infected Huh7.5 cells upon trans-splicing
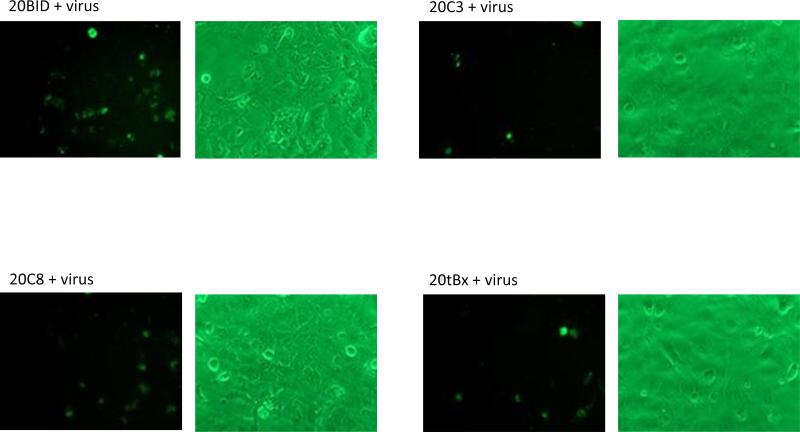
Huh 7.5 cells were transfected with each intron and subsequently challenged with Jc1FLAG2 at 48 h post transfection. Induction of apoptosis following HCV-challenge of αHCV-GrpI expressing cells was visualized by Annexin V-FITC staining at 48 h post infection. The percentage of apoptotic cells were reported based on the numbers of fluorescent positive cells counted from four separate fields divided by the total cell count.
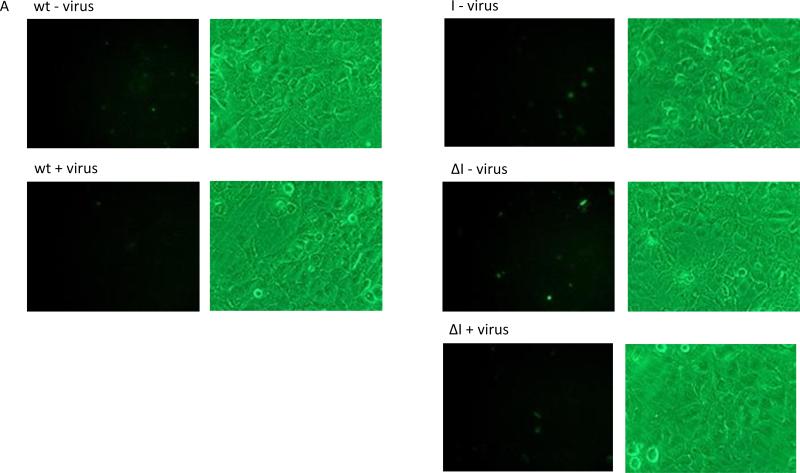
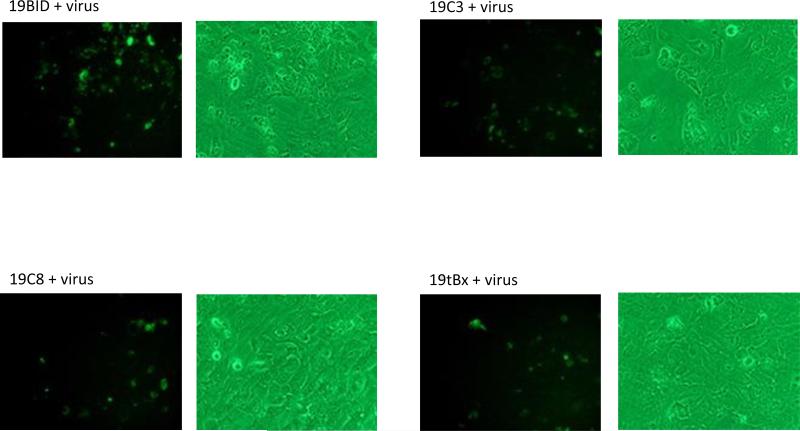
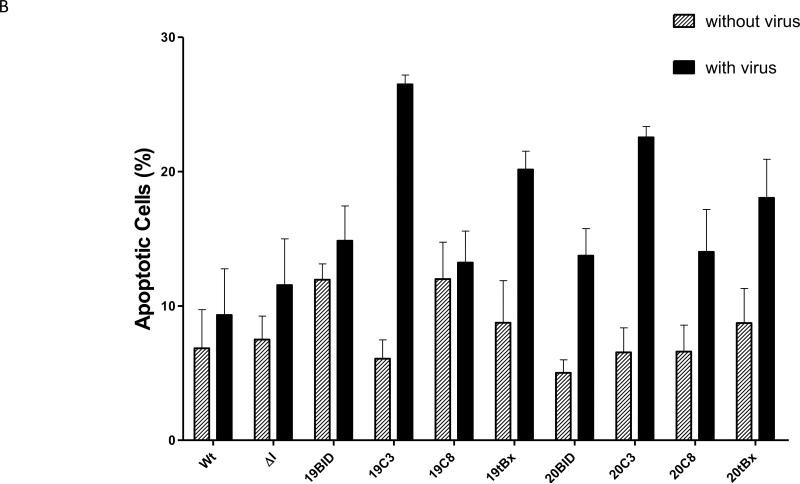
Cultures expressing the αHCV-GrpIs consistently exhibited greater numbers of fluorescing cells than the infected control culture when challenged with HCV (Figure 7A). These results suggested that apoptosis was in fact induced in the intron expressing cells upon virus infection. The ΔIntron lacking the trans-splicing domain exhibited a similar percentage of fluorescent cells as the infected control culture, indicating that the fluorescence observed in the αHCV-GrpI cultures was a result of targeted splicing by the αHCV-GrpIs and induction of apoptosis through translation of the splice product, and not some other non-specific effect such as antisense-mediated induction. Among the 3’exons tested, Caspase 3 and tBax were found to be the most effective in inducing apoptosis, bringing the total count of apoptotic cells significantly higher than that of the control cells without the virus infection (Figure 7B).
Figure 7.
Apoptotic cell death visualized by Annexin V staining. (A) HCV-infected cells were stained with Annexin V at 48 h post infection. Left: cells visualized under a fluorescent filter; Right: under a bright field; (B) Positive Annexin-V-stained cells were counted and divided by total cell count. For each sample, cells were counted manually from 4 separate fields; Wt: wild-type cells; ΔI: intron lacking a trans-splicing domain; + virus: with virus; - virus: without virus.
Elevated Caspase 3 activity in infected Huh7.5 cells upon trans-splicing
To insure cellular apoptosis upon intron trans-splicing to the HCV genome we designed the 3’exon to express the apoptotic-inducing genes BID, Caspase 3, Caspase 8, in their proactive form. As added insurance against apoptotic activity in the absence of HCV infection, we substiwfcd the native activation cleavage sequence for each of the proactive forms with the HCV NS3/4A protease cleavage sequence. As a control for apoptosis induction without proteolytic cleavage we also included the previously utilized, apoptotically active tBax coding sequence [44]. The BID and tBax proteins are involved in cytochrome-C release from the mitochondria while Caspase 3 and Caspase 8 cysteine proteases are involved in the execution phase of the cascade [45].
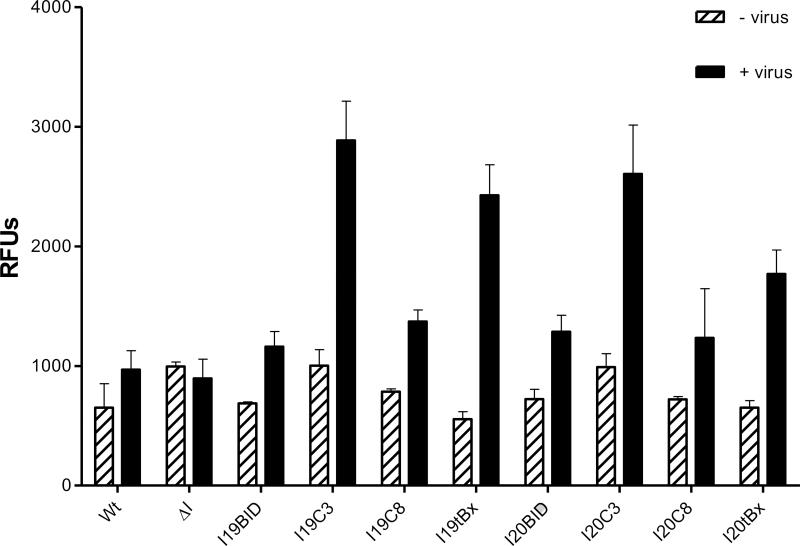
Huh7.5 cells were transfected with each intron and challenged at 48 h post transfection with HCV Jc1FLAG2, and the effectiveness of apoptosis induction was measured using a Caspase 3 assay [46]. As demonstrated in Figure 8, the level of Caspase 3 activity was significantly increased in the I19C3 and I20C3 cells (+ virus) compared to their respective negative controls (- virus). However, the Caspase 3 induction was most robust with I19tBax, which exhibited a difference of almost 6-fold between unchallenged and virus challenged cells. Surprisingly, the I-BID and I-Caspase 8 only induced Caspase 3 activity to a level comparable with the control wild-type cells. As expected, the ΔIntron did not exhibit a significant increase of Caspase 3 activity. The low amount of activated Caspase 3 observed in the ΔIntron and control cells may be attributed to cell death caused by DNA transfection procedures and/or the effect of a typical HCV infection. Together, these results indicated that Caspase 3 and tBax are the two best options as 3’ apoptotic exons among the four tested.
Figure 8.
Caspase 3 activity assay. Virus-containing medium from HCV-infected and uninfected Huh7.5 cells was collected at 48 h post infection and the level of total Caspase 3 was measured. Each bar represent the level of Caspase 3 without the virus (left) and with the virus (right). Wt: wild-type cells, ΔI: intron lacking a trans-splicing domain. The error bars represent standard error of the mean computed from three independent experiments.
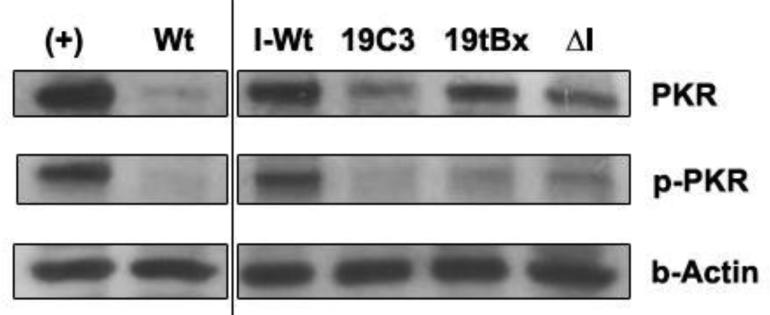
αHCV-GrpIs do not induce phosphoprylation of PKR
Double-stranded RNA formation through complementary base pairing of the ΔIntron EGS and the target viral RNA may trigger the activation of PKR activity resulting in the reduction of the viral protein synthesis [47]. We examined the possibility that the slight anti-HCV effect observed for ΔIntron transfected cells (Figure 5) might be due to activation of a specific PKR response. Cell lysates collected at 96 h post infection were assayed for PKR activity using Western blot analysis. However, we observed no significant difference in the level of PKR and phospho-PKR between the intron transfected cells and ΔIntron transfected cells (Figure 9). These results do not support the possibility that the slight knockdown of the GLuc level observed for ΔIntron (Figure 5) was caused by the activation of a PKR response, but rather by some inherent variation in the transfection procedure.
Figure 9.
Western blot analysis. Total proteins were extracted at 96 h post infection and immunoblotted for PKR, phospho-PKR, and β-actin. While untransformed cells infected with HCV (I-Wt) showed relatively high levels of both PKR and phospho-PKR, there was no significant difference evident in the concentration of PKR or phospho-PKR between cells transformed with 19C3, I19tBx, and ΔI group I introns. (+): IFN-induced positive control wild-type cells; Wt: untransformed, uninfected wild-type cells; I-Wt: HCV-infected untransformed wild-type cells, ΔI: intron lacking a trans-splicing domain.
Discussion
HCV is one of the leading causes of hepatocellular carcinoma, liver cirrhosis and liver transplantations in the United States [9, 48]. Novel strategies have been developed to combat the virus with different levels of success. Some of the approaches that are relevant to our research include the use of antisense [49-51], siRNA [52], aptamer [53, 54], hammerhead ribozyme [55] and group I intron [20]. Due to the high error-prone rate of the virus RdRp, target site selection is of utmost importance for these effector molecules to be effective against virus quasispecies. In our initial analysis of the virus 5’UTR, we observed that the sequence of the HCV IRES, one of the most characterized region on the virus genome, is well conserved among genotypes. Therefore, it represents a potential target region for RNA-based anti-viral inhibitor strategies. Since the IRES has a crucial function in both viral translation and replication processes required for viral maintenance and propagation, disrupting the IRES structure will result in interruption of both processes.
Group I introns have been used to target the HCV IRES successfully in liver cells using an in vitro-transcribed artificial target [20]. However, the previous study implemented the use of the Diphtheria toxin gene as their 3’exon which causes concern for the safety of such approach for human gene therapy. Our study differs in that we used naturally occurring apoptosis-inducing genes, eliminating the necessity for any type of bacterial toxin. In addition, our apoptotic genes may only be activated in the presence of active HCV virus, while uninfected cells will remain unaffected.
In this study we examine the effectiveness of two anti-viral group I introns against HCV. These introns, I19 and I20, target the highly conserved U329 of the domain IIIf and U343 of the domain IV in the IRES, respectively, both of which are required for proper translation initiation of the viral proteins. These introns are further modified from the original intron sequence to include an extended complimentary EGS, IGS, and P10 helix to increase the intron splicing efficacy [23]. The downstream sequence of the HCV IRES that follows the target site is also included to regenerate the complete IRES structure upon splicing.
The group I intron catalytic sequence itself is immediately followed by one of the cell death-inducing apoptotic genes, BID, Caspase 3, Caspase 8, or tBax, as the 3’exon. These apoptotic 3’exons are constructed in-frame with 30 nt of the HCV core-coding sequence. This sequence has been defined as essential for proper translation initiation from the HCV IRES sequence and is necessary for optimal expression of the 3’ exon encoded proteins [56].
When properly spliced, the resulting RNA encodes a fusion protein that induces cell death. The effect on viral suppression is therefore 2-tier; cleavage by the intron destroys the target virus genome, while splicing simultaneously induces cytotoxicity in the infected cells. This approach should have advantages over antisense oligonucleotide, siRNA, or simple target-cleaving ribozyme strategies where viral RNA is simply cleaved or blocked for translation.
Inappropriate initiation of translation or off-target splicing resulting in expression of these apoptotic factors is an undesired outcome for this strategy. Two stop codons are added in-frame immediately 3’ of the intron splicing domain and prior to the intron splice junction to prevent illegitimate translation of the 3’ exon. Further insurance is provided in the case of the BID, Caspase 3, and Caspase 8 constructs by incorporating them as modified proactive forms in which their endogenous cleavage sites are replaced with the HCV protease NS5A/5B cleavage site [27]. This strategy ensures that unintended splicing and illegitimate expression of the toxic 3’exon in the absence of the HCV infection is not possible.
Using RT-PCR with primers specific for the spliced product sequence, the targeting and trans-splicing capabilities of each intron is demonstrated against viral target RNA in transfected Huh7.5 cells, with splice junctions confirmed by sequence analysis. The negative control ΔIntron lacks any detectable splicing activity, and served as a control for the anti-viral interfering effect, if any, resulting from IGS and EGS homologous interactions with the viral IRES sequence. These experiments are designed to detect predicted splice products in HCV infected cells expressing functional introns but cannot detect potential off-target cleavage or splicing. However, based upon the extent of sequence homology required for the specific interactions that would lead to splicing we would expect off-target events to be extremely rare.
The ability of the introns to reduce the production of virus in infected cell cultures was assessed using the Jc1FLAG2 recombinant HCV that expresses the GLuc marker gene inserted between p7 and NS2 on the target viral genome. A decreased level of GLuc activity indicates reduced viral RNA production. The GLuc assay demonstrates virus knockdown for all our intron vs. non-intron (wild-type) cells especially for I19C3 and I19tBax where 80% knockdown was observed. The level of knockdown seen in our study is comparable to a previous aptamer study [57] but considerably higher than the previous group I intron study in which only 50% knockdown of luciferase was reported [20]. The insignificant, slight knockdown observed for the ΔIntron suggested that the presence of the intron in the cells may have had a small effect on virus production. This observation could not have resulted from illegitimate expression of the 3’ apoptotic exon because the ΔIntron is not capable of trans-splicing, as demonstrated by the RT-PCR results (Figure 4A-D). In addition, the knockdown does not appear to be related to activation of a PKR response since there was no significant difference in the PKR level observed between the I19C3, I19tBx intron and its corresponding ΔIntron. The most likely explanation for this result is inherent variation in the transfection procedure.
HCV-challenged intron expressing cells were also analyzed by TCID50 to determine the level of suppression of infectious virus production. The results among our introns are less uniform than the GLuc assay. We observed the level of suppression ranged between 1 and 3 logs, with I19C3 being the best effector among all introns tested, bringing down the infectious virus titer to almost 1000 fold (3 logs) compared to the non-intron cells. The ΔIntron control confirms our observation that expression of the intron constructs did affect infectious virus production.
The presence of cell death upon splicing is verified by Annexin V staining. Although the HCV-infected wild-type cells exhibit a small number of fluorescent cells, the effect is considered insignificant compared to what we observe in cells expressing the apoptosis-inducing introns, where cells undergoing apoptosis are observed in almost every field and with a much greater fluorescent intensity. When examined with bright field, wild-type cells maintain a healthy cell morphology and grow to confluence. The intron expressing cells, however, exhibit cell changes characteristic of apoptosis such as cell shrinkage and blebbing. There is no evidence of cell death in the ΔIntron expressing control cells indicating that apoptosis events in the intron expressing cells result from correct trans-splicing reactions.
We quantified the level of apoptosis by indirectly measuring the level of Caspase 3 activity in infected cells. To facilitate quantitative comparison, since the amount of cells was lower in the intron cells than wild-type, we corrected the fluctuation of the Caspase 3 between samples by calculating its activity on a per cell basis. Similar to the results shown in the Annexin V staining, an insignificant background level of Caspase 3 is evident in the control wild-type and ΔIntron cells, with or without the virus. The intron expressing cells, on the other hand, exhibit a significant increase in the level of Caspase 3, especially for I19C3 and I20C3 where the Caspase 3 levels are 3 times higher than their respective backgrounds without virus, or twice higher than the control wild-type cells. The 19tBx do not express the same level of total knockdown but display the most robust activation of Caspase 3 in these cells. In fact, the level of activated Caspase 3 is almost 6-fold higher than its respective background level without the virus. These results suggest that a robust induction of apoptotic cell death occurred within our intron expressing cells.
We have demonstrated the effectiveness of IRES-targeting αHCV-GrpIs in inhibiting viral replication in vitro. These results were obtained utilizing transient experiments where the intron constructs were introduced into the cells through chemical transfection. Based on our previous published data [23, 31], we expect to see a higher suppression effect when we have established clonal cell population of our introns. Due to the structural conservation in the IRES region among various HCV strains, our group I intron approach should be applicable to other HCV strains not tested in our study. The combination of more than one individual introns or a single chimeric intron simultaneously attacking different sites on the target virus may offer a different approach to better combat the virus. Alternatively, the intron may be designed to include an aptamer region to increase the splicing efficacy and specificity and to avoid viral escape mutants since aptamers rely on structural than sequence recognition of the target virus [53, 57]. Our current strategy may also prove to be useful when combined with other antiviral-based strategies and/or current and future drug treatment to completely eradicate the disease.
Highlights.
We constructed two anti-viral Group I introns targeting conserved regions of HCV IRES
Each intron included modified pro-apoptotic genes activated during HCV infection
Trans-splicing of the HCV genome was confirmed by RT-PCR and DNA sequencing
TCID50 and luciferase assays confirmed repression of virus in Huh7.5 cells
Elevated Caspase 3 and annexin V-positive cells indicated apoptosis of infected cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perz JF, et al. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–38. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142(6):1279–1287. e3. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Bradley DW. Studies of non-A, non-B hepatitis and characterization of the hepatitis C virus in chimpanzees. Curr Top Microbiol Immunol. 2000;242:1–23. doi: 10.1007/978-3-642-59605-6_1. [DOI] [PubMed] [Google Scholar]
- 4.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 5.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36(5 Suppl 1):S35–46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 6.Hoofnagle JH. Hepatitis C the clinical spectrum of disease. Hepatology. 1997;26(3 Suppl 1):15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 7.Di Lorenzo C, Angus AG, Patel AH. Hepatitis C virus evasion mechanisms from neutralizing antibodies. Viruses. 2011;3(11):2280–300. doi: 10.3390/v3112280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie JM, Chapman RW. Combination therapy for chronic hepatitis C interferon and ribavirin. Hosp Med. 1999;60(5):357–61. doi: 10.12968/hosp.1999.60.5.1116. [DOI] [PubMed] [Google Scholar]
- 9.Belousova V, Abd-Rabou AA, Mousa SA. Recent advances and future directions in the management of hepatitis C infections. Pharmacol Ther. 2014 doi: 10.1016/j.pharmthera.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15(1):41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 11.Farci P, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288(5464):339–44. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 12.Domingo E, Gomez J. Quasispecies and its impact on viral hepatitis. Virus Res. 2007;127(2):131–50. doi: 10.1016/j.virusres.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmichael GG. Medicine: silencing viruses with RNA Nature. 2002;418(6896):379–80. doi: 10.1038/418379a. [DOI] [PubMed] [Google Scholar]
- 14.Takamizawa A, et al. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65(3):1105–13. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauvelle C, et al. Hepatitis C virus vaccines - Progress and perspectives. Microb Pathog. 2013 doi: 10.1016/j.micpath.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Brown EA, et al. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20(19):5041–5. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg S. Recent advances in the molecular biology of hepatitis C virus. J Mol Biol. 2001;313(3):451–64. doi: 10.1006/jmbi.2001.5055. [DOI] [PubMed] [Google Scholar]
- 18.Kieft JS, et al. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292(3):513–29. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 19.Friebe P, Bartenschlager R. Genetic analysis of seguences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76(11):5326–38. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu KJ, Kim JH, Lee SW. Ribozyme-mediated selective induction of new gene activity in hepatitis C virus internal ribosome entry site-expressing cells by targeted trans-splicing. Mol Ther. 2003;7(3):386–95. doi: 10.1016/s1525-0016(02)00063-1. [DOI] [PubMed] [Google Scholar]
- 21.Cech TR. RNA editing: world's smallest introns? Cell. 1991;64(4):667–9. doi: 10.1016/0092-8674(91)90494-j. [DOI] [PubMed] [Google Scholar]
- 22.Long MB, et al. Ribozyme-mediated revision of RNA and DNA. J Clin Invest. 2003;112(3):312–8. doi: 10.1172/JCI19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter JR, et al. Targeting of highly conserved Dengue virus seguences with anti-Dengue virus trans-splicing group I introns. BMC Mol Biol. 2010;11:84. doi: 10.1186/1471-2199-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohler U, et al. Trans-splicing ribozymes for targeted gene delivery. J Mol Biol. 1999;285(5):1935–50. doi: 10.1006/jmbi.1998.2447. [DOI] [PubMed] [Google Scholar]
- 25.Shin KS, Sullenger BA, Lee SW. Ribozyme-mediated induction of apoptosis in human cancer cells by targeted repair of mutant p53 RNA. Mol Ther. 2004;10(2):365–72. doi: 10.1016/j.ymthe.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Lan N, et al. Ribozyme-mediated repair of sickele beta-globin mRNAs in erythrocyte precursors. Science. 1998;280(5369):1593–6. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 27.Hsu EC, et al. Modified apoptotic molecule (BID) reduces hepatitis C virus infection in mice with chimeric human livers. Nat Biotechnol. 2003;21(5):519–25. doi: 10.1038/nbt817. [DOI] [PubMed] [Google Scholar]
- 28.Kuiken C, et al. The Los Alamos hepatitis C sequence database. Bioinformatics. 2005;21(3):379–84. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds JE, et al. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14(23):6010–20. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nawtaisong P, et al. Effective suppression of Dengue fever virus in mosquito cell cultures using retroviral transduction of hammerhead ribozymes targeting the viral genome. Virol J. 2009;6:73. doi: 10.1186/1743-422X-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 33.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hygiene. 1938;27:493–497. [Google Scholar]
- 34.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6(6):513–22. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, et al. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1(5):526–37. [PMC free article] [PubMed] [Google Scholar]
- 36.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8(8):1045–55. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji H, et al. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101(49):16990–5. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byun J, et al. Efficient and specific repair of sickle beta-globin RNA by trans-splicing ribozymes. RNA. 2003;9(10):1254–63. doi: 10.1261/rna.5450203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ban G, Song MS, Lee SW. Cancer cell targeting with mouse TERT-specific group I intron of Tetrahymena thermophila. J Microbiol Biotechnol. 2009;19(9):1070–6. doi: 10.4014/jmb.0812.692. [DOI] [PubMed] [Google Scholar]
- 40.Bell MA, et al. Enhancing the second step of the trans excision-splicing reaction of a group I ribozyme by exploiting P9.0 and P10 for intermolecular recognition. Biochemistry. 2004;43(14):4323–31. doi: 10.1021/bi035874n. [DOI] [PubMed] [Google Scholar]
- 41.Usui K, et al. N-terminal deletion augments the cell-death-inducing activity of BAX in adenoviral gene delivery to nonsmall cell lung cancers. Oncogene. 2003;22(17):2655–63. doi: 10.1038/sj.onc.1206331. [DOI] [PubMed] [Google Scholar]
- 42.Carter JR, et al. Effective suppression of Dengue virus using a novel group-I intron that induces apoptotic cell death upon infection through conditional expression of the Bax C-terminal domain. Virol J. 2014;11(1):111. doi: 10.1186/1743-422X-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marukian S, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48(6):1843–50. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyota H, et al. Enforced expression of a truncated form of Bax-alpha (tBax) driven by human telomerase reverse transcriptase (hTERT) promoter sensitizes tumor cells to chemotherapeutic agents or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Anticancer Res. 2006;26(1A):99–105. [PubMed] [Google Scholar]
- 45.Antonsson B, Martinou JC. The Bcl-2 protein family. Exp Cell Res. 2000;256(1):50–7. doi: 10.1006/excr.2000.4839. [DOI] [PubMed] [Google Scholar]
- 46.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281(5381):13126. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 47.Qashqari H, et al. Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance. Infect Genet Evol. 2013;19:113–9. doi: 10.1016/j.meegid.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 48.Rosen HR. Clinical practice. Chronic hepatitis C infection. N Engl J Med. 2011;364(25):242938. doi: 10.1056/NEJMcp1006613. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Carmona MA, et al. Inhibition of hepatitis C virus gene expression by adenoviral vectors encoding antisense RNA in vitro and in vivo. J Hepatol. 2011;55(1):19–28. doi: 10.1016/j.jhep.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Jarczak D, et al. Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomicreplicons. FEBS J. 2005;272(22):5910–22. doi: 10.1111/j.1742-4658.2005.04986.x. [DOI] [PubMed] [Google Scholar]
- 51.Korf M, et al. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J Hepatol. 2005;43(2):225–34. doi: 10.1016/j.jhep.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 52.Zekri AR, et al. Consensus siRNA for inhibition of HCV genotype-4 replication. Virol J. 2009;6:13. doi: 10.1186/1743-422X-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikuchi K, et al. A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res. 2005;33(2):683–92. doi: 10.1093/nar/gki215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishikawa F, et al. Inhibition of HCV NS3 protease by RNA aptamers in cells. Nucleic Acids Res. 2003;31(7):1935–43. doi: 10.1093/nar/gkg291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonzalez-Carmona MA, et al. Hammerhead ribozymes with cleavage site specificity for NUH and NCH display significant anti-hepatitis C viral effect in vitro and in recombinant HepG2 and CCL13 cells. J Hepatol. 2006;44(6):1017–25. doi: 10.1016/j.jhep.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 56.Kohara M, et al. Expression and characterization of glycoprotein gp35 of hepatitis C virus using recombinant vaccinia virus. J Gen Virol. 1992;73(Pt 9):2313–8. doi: 10.1099/0022-1317-73-9-2313. [DOI] [PubMed] [Google Scholar]
- 57.Romero-Lopez C, et al. An engineered inhibitor RNA that efficiently interferes with hepatitis C virus translation and replication. Antiviral Res. 2012;94(2):131–8. doi: 10.1016/j.antiviral.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 58.Tumban E, Painter JM, Lott WB. Comparison between the HCU IRES domain IV RNA structure and the Iron Responsive Element. J Negat Results Biomed. 2009;8:4. doi: 10.1186/1477-5751-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]