Abstract
Epigenetic regulation of imprinted genes enables monoallelic expression according to parental origin, and its disruption is implicated in many cancers and developmental disorders. The expression of hormone receptors is significant in breast cancer as they are indicators of cancer cell growth rate and determine response to endocrine therapies. We investigated the frequency of aberrant events and variation in DNA methylation at nine imprinted sites in invasive breast cancer and examined the association with estrogen and progesterone receptor status. Breast tissue and blood from patients with invasive breast cancer (n=38) and benign breast disease (n=30) were compared to those from healthy individuals (n=36), matched to the cancer patients by age at diagnosis, ethnicity, BMI, menopausal status, and familial history of cancer. DNA methylation and allele-specific expression were analyzed by pyrosequencing. Tumor-specific methylation changes at IGF2 DMR2 were observed in 59% of cancer patients, IGF2 DMR0 in 38%, DIRAS3 DMR in 36%, GRB10 ICR in 23%, PEG3 DMR in 21%, MEST ICR in 19%, H19 ICR in 18%, KvDMR in 8%, and SNRPN/SNURF ICR in 4%. Variation of methylation was significantly greater in breast tissue from cancer patients than healthy individuals and benign breast disease. Aberrant methylation of three or more sites was significantly associated with negative estrogen-alpha (Fisher’s Exact Test, p=0.02) and progesterone-A (p=0.02) receptor status. Aberrant events and increased variation of imprinted gene DNA methylation therefore appear to be frequent in invasive breast cancer and are associated with negative estrogen and progesterone receptor status, without loss of monoallelic expression.
Keywords: DNA methylation, genomic imprinting, pyrosequencing, breast cancer, hormone receptor
Introduction
Genomic imprinting is the epigenetic regulation of genes to enable monoallelic expression according to parental origin, through differential methylation of regions labeled imprinting control regions (ICRs) when established in the germline, or differentially methylated regions (DMRs) when established post-fertilization. Loss of imprinting (LOI) is the loss of this monoallelic expression, and it is commonly the result of disruption of DNA methylation at ICRs and DMRs. LOI is associated with a range of disorders, such as the Beckwith-Wiedemann, Angelman and Prader-Willi syndromes1,2. Imprinted genes have also been implicated in a range of cancers, including those of the breast3 and ovaries4. Their expression has been associated with disease progression, including reduced survival in pancreatic cancer5 and aggressive prostate cancers6. Aberrant methylation of imprinted genes may be an early event involved in neoplastic transformation7, and it has been reported that 10% of apparently healthy individuals display LOI of IGF28.
DNA methylation can display stochastic variation, which may facilitate developmental plasticity and adaptation to environments, including that of malignant cells in the tumor microenvironment9. This variation can be gene-specific, such as that as observed in the placenta to enable adaptation to environmental challenges throughout pregnancy10. A significantly greater level of variation is observed in tumors, which often constitutes shifting in methylation ‘boundaries’ between CpG islands and shores11. Such events may occur early in tumorigenesis, demonstrated by the significantly increased variation observed in cervical cells of normal morphology12.
Breast cancers are classified according to the expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). ER-alpha and PR-A expression are determined by immunohistochemistry, with tumors in which more than 5–10% of cells stain positively classified as positive for expression. HER2 expression is determined by immunohistochemistry, with scores of 0–3 according to intensity of staining, and by fluorescence in situ hybridization (FISH) to detect amplification of the gene. Activation of these receptor signaling pathways results in cellular proliferation, and they are implicated in the progression of breast and gynecological cancers. Approximately two-thirds of tumors display expression of at least one of the hormone receptors, and these patients display reduced mortality compared to those who express neither13, in part due to the efficacy of endocrine therapies. Triple-negative breast cancers, which account for approximately 12–17% of all cases14, are associated with poor prognosis.
The relation between imprinted genes and hormone receptor status in breast cancer has not been well elucidated. Correlations between the methylation of four tumor-suppressor genes and the expression of the hormone receptors has been observed3, but similar studies have not been performed for imprinted genes. Elucidating such a relationship may provide insight into tumorigenesis and the determination of prognostic factors.
In this study, we investigated the aberrant methylation of nine imprinted regions in samples of breast tissue taken from healthy individuals and patients with benign breast disease and invasive breast cancer. The interrogated imprinted regions were: DIRAS3 DMR; GRB10 ICR; H19 ICR; IGF2 DMR0 and DMR2; KvDMR; MEST ICR; PEG3 DMR; and SNPRN/SNURF ICR. The intra-individual tissue-specificity of the aberrant methylation events was determined by comparison of methylation in DNA from breast tissue and peripheral blood within individuals. Variation of DNA methylation in imprinted genes and LINE-1 global methylation were measured, and possible associations between aberrant methylation and the status of the estrogen, progesterone and HER2 receptors in breast cancer patients were identified. Finally, we analyzed the allele-specific expression of the genes in order to establish the relative impact of the aberrant methylation events.
Materials and methods
Study populations
The Clinical Breast Care Project (CBCP) is a clinical and research program that began enrolling patients in 2001. The primary clinical arm of the CBCP was the Clinical Breast Care Center at Walter Reed Army Medical Center (Washington DC, USA). Additional recruitment centers include the Joyce Murtha Breast Care Center (Windber, PA, USA), and the Anne Arundel Medical Center (Annapolis, MD, USA). Enrolled patients were required to be 18-years-old or older, mentally competent and willing to provide informed consent, and presenting with evidence of possible breast disease, attending for routine screening mammograms, or undergoing elective reductive mammoplasty. Patients were provided with layered consent forms that included permission to obtain samples of blood, breast and metastatic tissues, and a description of the primary research uses. Once informed consent was granted, the core questionnaire, with over 500 fields of information, was completed with the help of a nurse case manager, and an extensive pathology checklist was completed by the dedicated breast pathologist. Ethical approval for the collection of blood and tissue samples and their use in this study was provided by the Walter Reed Army Medical Center Human Use Committee and Institutional Review Board.
The Susan G. Komen for the Cure Tissue Bank (Indianapolis, IN, USA) is a charitable organization that enrolls healthy volunteers to donate blood and up to four breast biopsies. Eligible individuals must be at least 18 years of age and able to provide informed consent. Volunteers are asked to complete a questionnaire regarding health and lifestyle factors. Approval for the collection of blood and tissue, and for their use in this study, was provided by the Indiana University Institutional Review Board.
Blood and tissue samples
DNA and RNA from benign and tumor tissue and blood were obtained from the CBCP for 38 patients with invasive breast cancer and 30 with benign breast disease (13 fibrocystic changes, 8 fibroadenoma, 3 post-surgical changes, 2 stromal fibrosis, and 4 other) (Table 1, Supplementary Table 1). Genomic DNA was extracted from frozen tumor samples following laser microdissection (Leica Microsystems, Wetzlar, Germany) and homogenized benign tissue by incubation with proteinase K at 37°C overnight and passage through purification columns (Millipore, Billerica, MA, USA). DNA extractions from blood were performed using Clotspin and Puregene DNA purification kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions.
Table 1. Characteristics of the participants.
The characteristics of the individuals involved in the study from whom samples of breast tissue and blood were taken. Healthy individuals were matched to the cancer patients by age, ethnicity, BMI, menopausal status and familial history of cancer. The ages and BMIs are the means of each category. Post-menopausal status includes patients who were surgically post-menopausal, such as following hysterectomy. Familial history of cancer refers to primary and secondary history. For one healthy individual, the familial history was not known. BMI: body mass index.
| Healthy | Benign breast disease | Invasive cancer | ||
|---|---|---|---|---|
| Number of participants | 36 | 30 | 38 | |
| Age at diagnosis | 50.3 | 47.5 | 51.5 | |
| Ethnicity (%) | White | 29 (80.6) | 21 (70.0) | 28 (73.7) |
| African-American | 7 (19.4) | 5 (16.7) | 7 (18.4) | |
| Other | 0 (0.0) | 4 (13.3) | 3 (7.9) | |
| BMI | 28.0 | 25.2 | 28.5 | |
| Menopausal status (%) | Pre- | 21 (58.3) | 20 (66.7) | 19 (50.0) |
| Post- | 15 (41.7) | 10 (33.3) | 19 (50.0) | |
| Familial history of cancer (%) | 17 (48.6) | 11 (36.7) | 18 (47.4) | |
Samples from 36 healthy individuals were obtained from the Susan G. Komen for the Cure Foundation Tissue Bank (Table 1). DNA was extracted from 25mg tissue using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s protocol, following proteinase K digestion at 56°C for 3–6 hours. RNA extractions were performed using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol, following mechanical disruption of 25mg tissue in a bead mill. Between 2 and 41µg of DNA isolated from blood was made available for this study.
Bisulfite conversion of DNA
250–500ng of DNA was converted using the EZ DNA Methylation Gold kit (Zymo Research, Irvine, CA, USA), according to the manufacturer’s Alternative 2 protocol, with elution in 40µl of elution buffer.
DNA methylation analysis by pyrosequencing
Pyrosequencing was performed on the Pyromark Q24 Pyrosequencer (Qiagen). Assays were designed using the PyroMark Assay Design and PyroMark Q24 software programs (Qiagen), with the exceptions of those for H19 ICR, IGF2 DMR0 and IGF2 DMR2, which have previously been reported15–17. Nine DMRs/ICRs were analyzed: DIRAS3 DMR (5 CpG dinucleotides); GRB10 ICR (6 CpGs); H19 ICR (8 CpGs); IGF2 DMR0 (6 CpGs); IGF2 DMR2 (7 CpGs); KvDMR (9 CpGs); MEST ICR (9 CpGs) PEG3 DMR (6 CpGs); and SNRPN/SNURF ICR (8 CpGs) (Supplementary Table 2).
Regions of interest were amplified by polymerase chain reaction (PCR), using 3µl of bisulfite-treated DNA and 0.2µM of each primer with HotStar Taq Plus Master Mix (Qiagen) in a final volume of 20µl. Pyrosequencing was performed according to the manufacturer’s instructions. Duplicate bisulfite-conversions were run for each sample and mean methylation levels were calculated across all CpG sites per replicate. Studies in healthy human tissues have reported methylation levels of between 30 and 70% at DMRs and ICRs16,17, and we therefore defined hypomethylation as values below 30% and hypermethylation as values above 70%. It was not possible to ascertain methylation values for all samples.
Identification of allelic origins of mRNA
Allele-specific expression was performed by pyrosequencing, using single nucleotide polymorphisms (SNPs) to determine the allelic origins of mRNA transcripts in heterozygous patients, identified by pyrosequencing using 10ng of DNA from blood. For heterozygous individuals, 20ng of RNA was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, USA) according to the manufacturer’s instructions, and 2µl of cDNA used for PCR-based amplification prior to allele quantification by pyrosequencing. Primer sequences and SNPs are provided in Supplementary Table 2.
Gene expression microarray data
Expression data for ten genes (DIRAS3, DNMT1, DNMT3A, DNMT3B, GRB10, IGF2, KCNQ1, MEST, PEG3, and SNRPN) from 302 breast tumors were made available by the Walter Reed Army Medical Center (Washington DC, USA). Of the tumors, 199 were ER-positive and 103 were ER-negative, and 153 were PR-positive and 149 PR-negative.
Statistical analysis
Correlations between DNA methylation and age at diagnosis were calculated by Pearson correlation for all genes except GRB10, for which Spearman’s rank correlation was used as the data was not normally distributed. Associations with tumor stage, receptor status and familial history of cancer were calculated by ANOVA. For the FAM group, associations were calculated by Fisher’s exact test, and associations with age at diagnosis calculated by t-test or Wilcoxon rank sum test, according to the distribution of the data. P values below 0.05 were deemed statistically significant.
For the modeling of variation, deviation was calculated as the absolute difference between the median methylation level in blood or breast tissue from healthy individuals and the methylation level for the individual. This was conducted for each gene, for comparisons of samples from patients with benign breast disease and cancer to the healthy controls, in both blood and breast tissue. A general linearized model was used to model deviation with a gamma distribution, and a log link. Model coefficients and 95% confidence intervals were exponentiated to give the relative change in deviation between tissues. The adjusted model controlled for family history (binary), menopausal status (binary), BMI category (normal, <25; overweight, 25 – 30; obese, >30), and age (continuous). Results were corrected for multiple hypothesis testing using the Bonferroni correction, and p values below 0.05 deemed statistically significant.
Results
Aberrant DNA methylation of imprinted genes is a frequent event in breast tumors
We investigated the DNA methylation at nine imprinted regions in breast tissue and peripheral blood from 36 healthy individuals, 30 patients with benign breast disease and 38 with invasive cancer. We have previously reported methylation levels at six of the imprinted regions (GRB10 ICR, H19 ICR, IGF2 DMR0, IGF2 DMR2, KvDMR, and SNRPN/SNURF ICR) in patients with benign breast disease and breast cancer18, and we integrated these results with methylation levels measured in healthy individuals to identify the relative frequency of aberrant methylation (hypo- and hypermethylation) in the disease states.
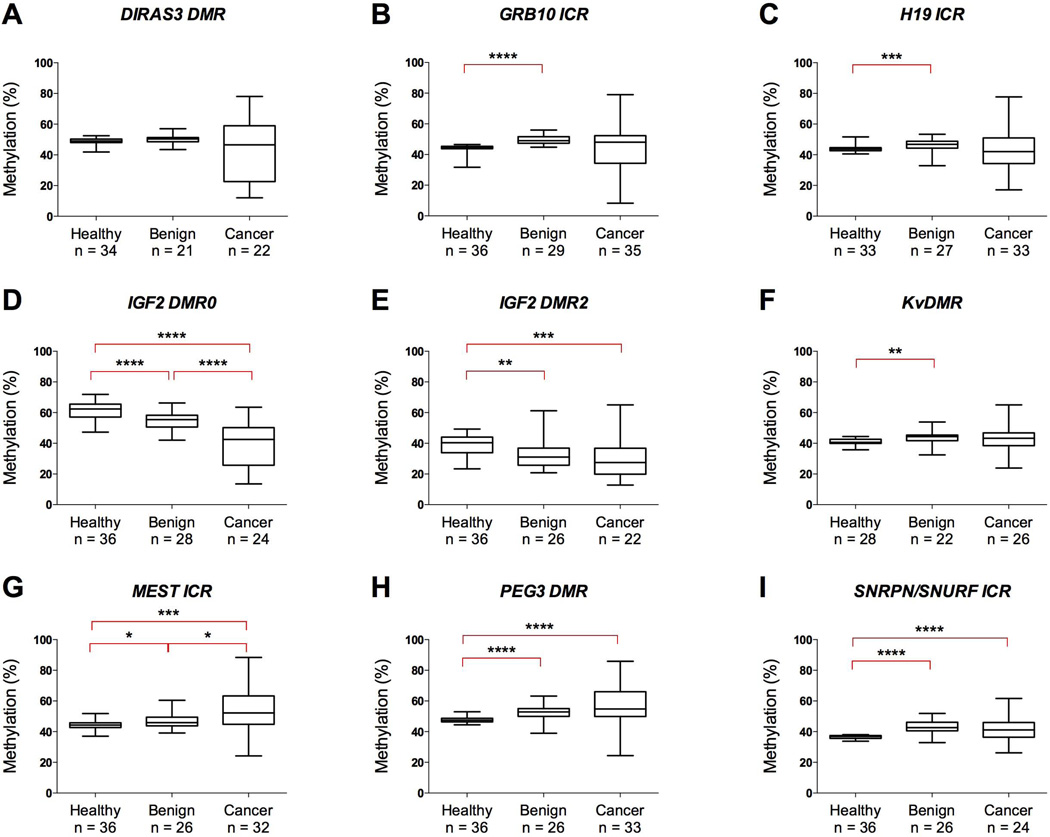
In breast tissue, median methylation levels were close to the expected 50% with the exception of the IGF2 sites, which also displayed greater disparities in the median levels between normal, benign and tumor tissue (Figure 1). Aberrant methylation was frequently observed in invasive breast cancer, observed at IGF2 DMR2 in 59% of patients, IGF2 DMR0 in 38%, DIRAS3 DMR in 36%, GRB10 ICR in 23%, PEG3 DMR in 21%, MEST ICR in 19%, H19 ICR in 18%, KvDMR in 8%, and SNRPN/SNURF ICR in 4% (Supplementary Table 3). Hypomethylation was more common than hypermethylation at all sites except MEST ICR and PEG3 DMR, and was not associated with whether the genes are maternally or paternally expressed. Among patients with benign breast disease, aberrant methylation was only observed at IGF2 DMR2, which was hypomethylated in 12 of the 26 successfully-analyzed samples. No aberrant methylation was observed in healthy individuals at seven of the imprinted regions, with IGF2 DMR0 hypermethylated in one sample and IGF2 DMR2 hypomethylated in three.
Figure 1. DNA Methylation at nine imprinted regions in breast tissue from patients and healthy individuals.
Measured methylation values in breast tissue for DIRAS3 DMR (A), GRB10 ICR (B), H19 ICR (C), IGF2 DMR0 (D), IGF2 DMR2 (E), KvDMR (F), MEST ICR (G), PEG3 DMR (H) and SNRPN/SNURF ICR (I) in healthy individuals and patients with benign breast disease and invasive cancer. Boxes correspond to the median and interquartile range, and the whiskers to the full range of measured values. Statistically significant differences are indicated (Mann-Whitney U Test; * = p < 0.05; ** = p <0.01; *** = p < 0.0005; **** = p < 0.0001).
To determine the intra-individual tissue specificity of the methylation changes observed in breast tumors, we analyzed peripheral blood samples taken from the same individuals. Median values for all nine regions were between 39.1 and 53.6% across the three groups (Supplementary Table 3). No samples met the criteria of being hypo- or hypermethylated, indicating that the aberrant methylation events were unique to breast tissue.
Variation of DNA methylation is significantly greater in breast tissue and blood from patients with invasive breast cancer and benign breast disease
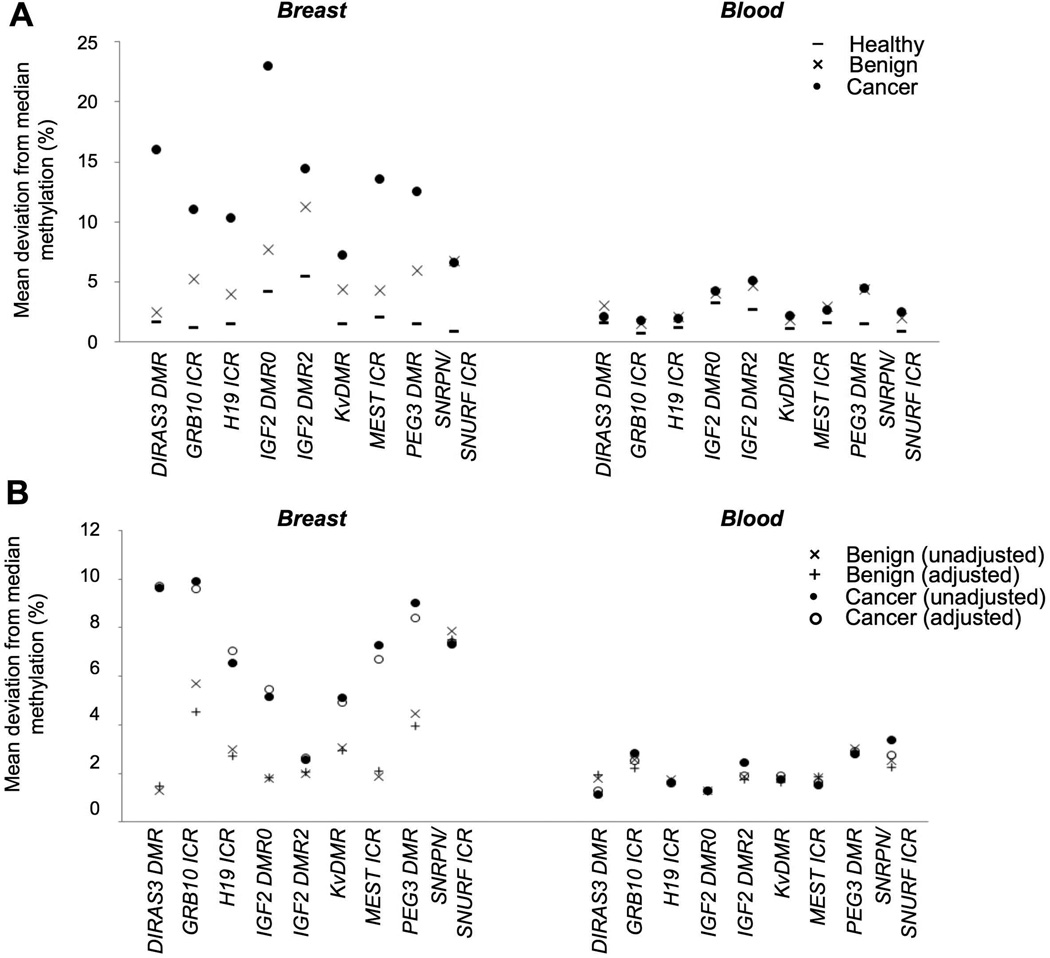
Variation of DNA methylation in the healthy and disease states was estimated by the absolute difference between the methylation level in an individual and the median level measured in breast tissue or peripheral blood from healthy individuals, with the latter representing the normal value. The mean deviation from the median methylation levels observed in normal breast tissue was greatest in invasive tissue, ranging from 6.6% for SNRPN/SNURF ICR to 23.0% for IGF2 DMR0 (Figure 2A, Supplementary Table 4A). The mean deviations were lowest in normal breast tissue, where they were under 2.0%, with the exceptions of IGF2 DMR0 (4.2%) and IGF2 DMR2 (5.5%). Benign breast tissue displayed intermediate values, with median deviation ranging from 2.5% for DIRAS3 DMR to 11.2% for IGF2 DMR2.
Figure 2. Variation in DNA methylation in breast tissue and blood.
(A) Mean deviation from the measured median value in normal breast tissue or blood from healthy individuals for each of the nine imprinted regions. Results are from patients with invasive cancer (•), benign breast disease (×) and healthy individuals (−). (B) Change in mean deviation from median methylation values measured in normal tissue and blood, relative to values observed in normal breast tissue and blood from healthy individuals. Results for cancer patients from the unadjusted (•) and adjusted (○) models, and from patients with benign breast disease in the unadjusted (×) and adjusted (+) models.
The mean deviations in healthy individuals were lower in blood than in breast tissue, with the exceptions of PEG3 DMR in healthy individuals and DIRAS3 DMR in patients with benign breast disease (Figure 2A, Supplementary Table 4A). The deviations were greatest in blood from invasive breast cancer patients, where median deviations ranged between 1.7% and 6.6%. The values ranged between 1.5 and 6.7% among benign breast disease patients, and between 0.7 and 3.2% in healthy individuals.
The relative deviations from the median methylation levels were significantly greater at all interrogated regions in tumor tissue than in normal breast tissue, ranging from 2.6-fold (IGF2 DMR2) to 9.7-fold (DIRAS3 DMR) greater (Figure 2B, Supplementary Table 4B). When the model was adjusted for the age, BMI, menopausal status and familial history of cancer of the individuals, statistical significance was retained at all sites. Variation was also significantly greater in benign breast disease tissue than normal breast tissue for all DMRs except DIRAS3 DMR, with the relative deviations between 1.5-fold (DIRAS3 DMR) and 7.5-fold (SNRPN/SNURF ICR) greater. Significance was retained for all sites except IGF2 DMR0 and MEST ICR with the adjusted model.
Significantly greater variation of methylation was similarly observed in peripheral blood. Mean deviations in blood from patients with invasive breast cancer were between 1.3-fold (DIRAS3 DMR) and 7.4-fold (SNRPN/SNURF ICR) greater than in healthy individuals, and were significant for GRB10 ICR, KvDMR, PEG3 DMR and SNPRN/SNURF ICR. In the adjusted model, the relative changes were significant for GRB10 ICR, IGF2 DMR2, PEG3 DMR and SNRPN/SNURF ICR (Supplementary Table 4B). Mean deviations were also between 1.3-fold (IGF2 DMR0) and 7.5-fold (SNRPN/SNURF ICR) greater in blood taken from patients with benign breast disease than in healthy individuals (Figure 2B, Supplementary Table 4B). The differences were significant for GRB10 ICR, MEST ICR, PEG3 DMR and SNRPN/SNURF ICR in both the adjusted and unadjusted models (Supplementary Table 4B).
Aberrant DNA methylation is Associated with Hormone Receptor Status
We investigated possible associations between the measured methylation values at each imprinted loci and expression of the estrogen (ER), progesterone (PR) and HER2 receptors (details in Supplementary Table 1). Hypermethylation of PEG3 DMR (ANOVA, p<0.01) and IGF2 DMR0 (p=0.04) were associated with negative ER status, while hypermethylation of PEG3 DMR (p=0.02) and MEST ICR (p=0.03) were associated with negative PR status (Table 2). No associations were observed between the methylation of imprinted genes and the expression of HER2 or tumor stage.
Table 2. Correlations between DNA methylation of imprinted genes and tumor pathology.
Correlations between aberrant methylation of nine imprinted genes and the tumor and patient characteristics were identified using the Pearson correlation (age at diagnosis) and ANOVA (tumor stage, receptor status and familial history of cancer). For the frequently altered methylation group (FAM, tumors with more than three aberrantly methylated imprinted regions), Fisher’s exact test was used to identify associations with tumor stage, receptor status and familial history, while a t-test or Wilcoxon rank sum test was used to identify associations with age at diagnosis. P values are provided with rho values below where appropriate. Statistically significant (p < 0.05) associations are highlighted in bold.
| Imprinting regions |
Age at diagnosis (rho values) |
Tumor stage |
ER | PR | ER/PR | HER2 | Familial history |
|---|---|---|---|---|---|---|---|
| DIRAS3 DMR | 0.88 (−0.04) | 0.85 | 0.20 | 0.36 | 0.44 | 0.53 | 0.55 |
| GRB10 ICR | 0.96 (<0.01) | 0.06 | 0.09 | 0.45 | 0.21 | 0.16 | 0.20 |
| H19 ICR | 0.39 (−0.16) | 0.29 | 0.67 | 0.52 | 0.45 | 0.57 | 0.89 |
| IGF2 DMR0 | 0.18 (−0.28) | 0.15 | 0.04 ↓ | 0.98 | 0.02 ↓ | 0.22 | 0.23 |
| IGF2 DMR2 | 0.49 (−0.15) | 0.08 | 0.96 | 0.65 | 0.82 | 0.70 | 0.32 |
| KvDMR | 0.45 (−0.15) | 0.24 | 0.70 | 0.31 | 0.59 | 0.33 | 0.38 |
| MEST ICR | 0.94 (−0.01) | 0.58 | 0.23 | 0.03 ↓ | 0.10 | 0.40 | 0.42 |
| PEG3 DMR | 0.33 (−0.18) | 0.71 | <0.01 ↓ | 0.02 ↓ | 0.01 ↓ | 0.99 | 0.66 |
| SNRPN ICR | 0.58 (0.12) | 0.89 | 0.89 | 0.46 | 0.56 | 0.61 | 0.66 |
| FAM | 0.91 | 0.15 | 0.02 - | 0.02- | 0.02- | 0.32 | 0.28 |
Down-arrows indicate reduced levels of methylation with expression of the estrogen or progesterone receptor. For the FAM grouping,
minus symbols indicate that aberrant methylation is associated with negative status of the receptor.
Frequently altered methylation (FAM) and hormone receptor status
Twelve of the invasive tumors analyzed displayed aberrant methylation of at least three of the imprinted regions that were interrogated. We categorized these together as “frequently altered methylation” (FAM). FAM was associated with negative ER (Fisher’s exact test, p=0.02) and PR status (p=0.02), but not HER2 (p=0.32) or tumor stage (p=0.15) (Table 2). All five triple-negative tumors displayed aberrant methylation at three or more sites.
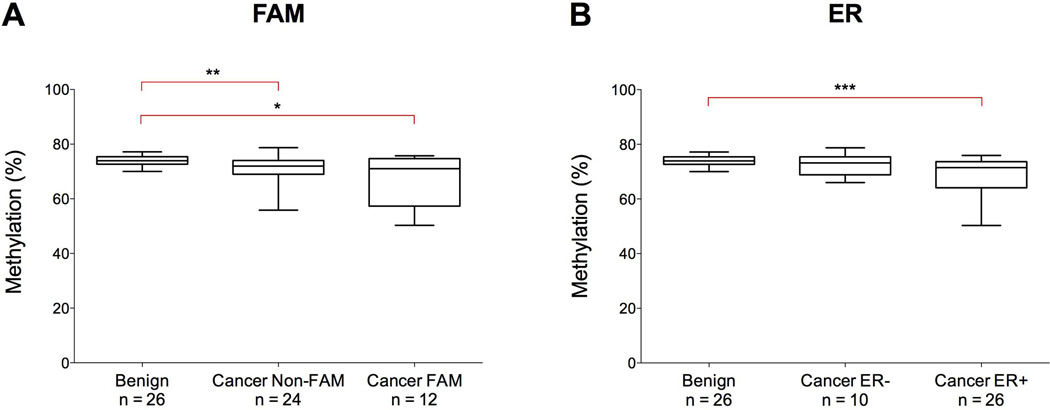
Methylation at LINE-1 repetitive elements was measured in breast tissue from patients with benign breast disease and invasive cancer, and analyzed according to the FAM grouping of cancers and the expression of the estrogen receptor (Figure 3A–B). Greater variation was observed in breast tumors, with values ranging between 50 and 79% compared to 69–77% in benign breast disease tissue (Figure 3A). Within the tumor samples, median values were similar in the FAM (71.1%) and non-FAM (71.8%) groups, but slightly higher in ER-negative tumors (73.2%) than in ER-positive ones (71.5%). Tumors expressing the estrogen receptor also displayed a greater range of values (50 to 76%) in comparison to those that do not (66 to 79%) (Figure 3B).
Figure 3. DNA methylation at LINE-1 elements in benign breast disease tissue and breast tumors.
Results are displayed in relation to the FAM status of the tumors (A) and by expression of the estrogen receptor (ER) (B). Boxes correspond to the median and interquartile range, and the whiskers to the full range of measured values. Statistically significant differences are indicated (Mann-Whitney U Test; * = p < 0.05; ** = p <0.01; *** = p < 0.0005; **** = p < 0.0001).
Monoallelic expression of imprinted genes is maintained in breast tumors
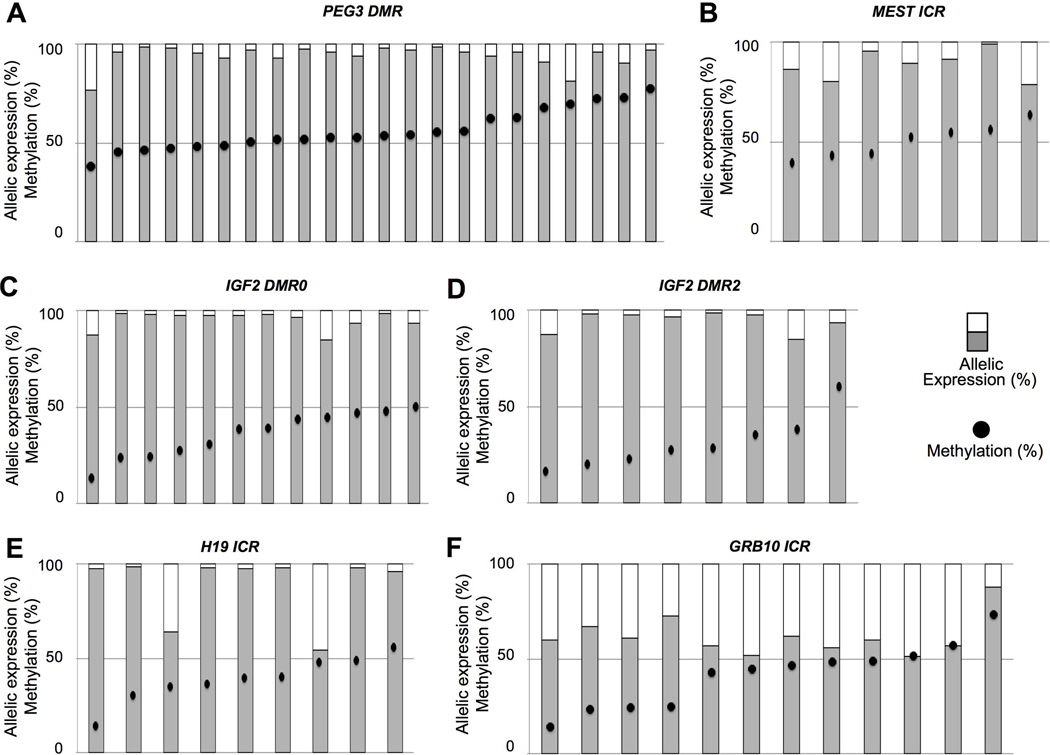
To determine the impact of the observed methylation changes, we examined the allele-specific expression of the genes. Firstly, we genotyped the cancer patients for SNPs in the DIRAS3, GRB10, H19, IGF2, MEST and PEG3 genes. KCNQ1 and SNRPN were not analyzed due to the infrequency of aberrant methylation in the tumor samples. We identified 24 patients who were heterozygous for SNPs in the PEG3 gene, 18 for IGF2, 14 for GRB10, 10 for H19, 7 for MEST and 4 for DIRAS3.
Monoallelic expression, defined here as >85% of transcripts from a single allele, was almost exclusively retained (Figure 4). PEG3 was monoallelically expressed in 17 of 18 patients with normal methylation levels and 5 of 6 patients displaying hypermethylation of the gene (Figure 4A). In the other two patients, 71 and 81% of transcripts originated from a single allele. Monoallelic expression was observed in five of the seven patients informative for MEST, with 79% and 80% of transcripts expressed from a single allele in the other two patients (Figure 4B). All 18 patients informative for IGF2 monoallelically expressed the gene, including the 4 patients with hypomethylation of DMR0 and 4 displaying hypomethylation of DMR2 (Figures 4C–D). Monoallelic expression of H19 was observed in 8 of the 10 informative patients, including the one displaying hypomethylation of the gene (Figure 4E). Two individuals displayed biallelic expression despite normal methylation profiles, with the relative expression of the two alleles 64/36% and 55/45%. GRB10 displayed a markedly different pattern of expression, with the proportional expression of the two alleles between 51/49% and 60/40% in 7 of the 14 informative patients, and only three patients expressed >70% of transcripts from a single allele (Figure 4F). DIRAS3 results are not shown due to the lack of informative patients.
Figure 4. Expression of imprinted genes relative to DNA methylation status.
Allelic-expression of PEG3 (A), MEST (B) IGF2 relative to DMR0 (C) and DMR2 (D), H19 (E) and GRB10 (F). For each patient, DNA methylation values (•) and allelic expression (bar graph) are provided. Expression values correspond to the percentage of transcripts originating from the more transcribed allele.
GRB10 and IGF2 are differentially expressed in ER-positive tumors
To further examine the relation between hormone receptor status and the expression of imprinted genes, we utilized gene expression microarray data from 302 breast tumor samples, made available through the Walter Reed Army Medical Center. Expression of seven of the imprinted genes was analyzed between ER-positive (n=199) and ER-negative (n=103) tumors, and between PR-positive (n=153) and PR-negative (n=149) tumors. H19 expression was not covered by the array. GRB10 expression was 1.4-fold lower (p=1.7×10−10) and IGF2 expression was 1.9-fold higher (p=6.0×10−7) in ER-positive tumors (Supplementary Table 5). No significant differences were observed by PR status.
Expression of DNA methyltransferases are not associated with hormone receptor status
To investigate how DNA methylation at imprinted regions may be related to estrogen and progesterone receptor status, we utilized gene expression microarray data from 302 breast tumor samples (Supplementary Table 5). We observed no significant differences in the expression of the DNMT1 (−1.1 fold-change), DNMT3A (−1.3 fold-change) and DNMT3B (−1.6 fold-change) genes in ER-positive tumors in comparison to ER-negative ones. Similarly, no significant difference was observed for any of the three genes (−1.1, −1.1 and −1.3 fold-changes respectively) in PR-positive tumors versus PR-negative tumors.
Discussion
We found the imprinted regions DIRAS3 DMR, GRB10 ICR, H19 ICR, IGF2 DMR0, IGF2 DMR2, MEST DMR and PEG3 DMR to be frequently aberrantly methylated in the patients with invasive breast cancer included in this study. These alterations are highly tissue- and tumor-specific, with no such changes observed in blood and only highly infrequently in normal breast tissue and benign breast disease tissue. This result confirmed our previous work in which we observed no correlation between the methylation at six imprinted regions in breast tumors and matched peripheral blood18. Variation of DNA methylation was significantly greater at all nine imprinted regions in breast tumors comparison to normal breast tissue, and at eight (unadjusted model) and six (adjusted) of the regions in benign breast disease tissue. Aberrant methylation of more than three of the imprinted regions was significantly associated with negative status of the estrogen and progesterone receptors. Despite this disruption of DNA methylation, monoallelic expression of the imprinted genes was frequently maintained.
This is the first study to identify a correlation between the methylation of imprinted genes and expression of the estrogen and progesterone receptors in breast cancer. ER-positive and ER-negative tumors have distinct global DNA methylation profiles19,20, and work in animal models and cell lines has demonstrated that hormone receptor signaling, including that by agonists such as bisphenol A, can affect the expression of DNA methyltransferases21–23 and directly lead to changes in the methylation and expression of imprinted genes such as IGF2 and PEG321,24,25. Furthermore, in vitro studies have shown that expression of the imprinted gene CDKN1C is repressed via epigenetic mechanisms induced by estrogen signaling in breast cancer cell lines26. Taken together with our observations in primary human breast tumors, this may suggest that normal hormone receptor signaling is important for the maintenance of methylation for imprinted genes, and absence of such signaling may result in aberrant regulation in malignant cells. Interestingly, in vitro work has demonstrated that mutations in the DNMT1 gene lead to loss of IGF2 and PEG3 imprinting, with KCNQ1 less susceptible to such changes27, and we similarly observed that aberrant methylation of the IGF2 and PEG3 DMRs was substantially more frequent than at the KvDMR (Figure 2A), although we did not observe an impact upon allele-specific expression. However, we did not observe significantly different expression of DNMT1, DNMT3A or DNMT3B between ER-positive and ER-negative tumors (Supplementary Table 5). Our findings may either suggest that the relation between DNA methylation at imprinted loci and ER signaling may be confined to a subset of ER-negative tumors, or that the association is independent of the regulation of DNA methyltransferase expression reported in vitro.
Similar associations between DNA methylation and hormone receptor status have been identified for non-imprinted genes3,28. However, the differential methylation of alleles and roles in regulating proliferation and differentiation make imprinted genes particularly susceptible to driving tumorigenesis. Indeed, changes in the methylation of imprinted genes have been reported in esophageal dysplasia29 and localized ovarian tumors30. As the ER is frequently silenced by promoter methylation in breast tumors31, it is not clear whether hormone receptor status is the cause or product of wider epigenetic dysregulation. Further work is required to establish whether a subset of breast tumors may display a characteristic epigenetic profile. The identification of such a group may open new therapeutic options to the patients, including the combined use of 5-Azacytidine and S-adenosylmethionine, which inhibit the growth of breast cancer cells32.
We observed increasing variation in DNA methylation from normal breast tissue to benign breast disease to cancer. This finding is consistent with observations made by Teschendorff and colleagues, who suggested increased variability as a marker for early detection of cervical cancer, even being present in cytologically-normal cells of individuals who later developed12. Thus, this variability may be a key part of the neoplastic transformation process. It may arise through the shifting of methylation boundaries, rather than gene-specific changes11. As we have employed a candidate-gene approach, with comparatively short reads obtained by pyrosequencing, it remains uncertain whether this variability is confined to the interrogated DMRs or whether it may be the result of wider boundary-shifts. Furthermore, the degree of variation of methylation within the tumors remains unknown. Mosaicism is a well-known phenomenon in cancer, and may provide some explanation for our observations.”.
Increased variability in DNA methylation has been proposed as a blood-based marker for ovarian cancer33, and we similarly observed significantly increased variation in both the adjusted and unadjusted models at GRB10 ICR, PEG3 DMR and SNRPN/SNURF ICR in peripheral blood from cancer patients. However, the statistical significance may be the product of the tight clustering of values observed in healthy individuals, with mean deviations from median values below 3.5% at all sites. While group differences may be significant, the value for risk prediction in individuals remains uncertain.
The sites displaying the most frequent aberrant methylation and greatest variation were the two IGF2 DMRs. IGF2 is a widely studied oncogene that stimulates cellular proliferation, and hypomethylation of DMR0 is associated with poor prognosis with colorectal cancer34. We observed that hypomethylation of DMR0 was associated with negative ER status, which is itself associated with poor prognosis in breast cancer13. The observed frequency of hypomethylation was similar to that reported elsewhere16. While loss of IGF2 imprinting in peripheral blood has been suggested as a potential diagnostic marker for colorectal cancer8, changes in methylation at DMR0 were specific to the tumors, and we have previously reported a lack of correlation between blood and tumor tissues18. This may suggest that peripheral blood cannot serve as a surrogate tissue, although correlations may be gene-specific, as there is evidence that methylation of ATM35 and targets of ER signaling36 could be used as blood-based markers of breast cancer risk.
Interestingly, the disruption of methylation did not impact upon allele-specific expression in the tumors, with monoallelic expression almost exclusively retained. Conversely, our results suggest that GRB10, which is monoallelically expressed in fetal brain tissues and placenta but not in other fetal tissues37–39, may not be imprinted in breast tissue in adult humans. Loss of IGF2 imprinting has been reported in colorectal and ovarian tumors8,40 and PEG3 in gynecological cell lines41, but we observed >77% of transcripts originating from a single allele in all the tumor samples informative for these genes. Furthermore, our observation of monoallelically-expressed MEST in all seven informative patients is in direct contrast to findings elsewhere of biallelic expression in breast cancer patients42. While H19 was biallelically expressed in two patients, methylation of the H19 ICR was not disrupted in these tumors, suggesting an alternative cause of LOI. Hypomethylation of H19 ICR has been correlated with LOI in lung cancer43, but there is evidence that the IGF2/H19 competition model does not hold true in colorectal44,45 and ovarian40 tumors. Similarly, a lack of correlation has been observed between hypomethylation of IGF2 DMR0 and LOI in colon and breast tumors16, while hypomethylation of IGF2 DMR0 in ovarian serous tumors increases overall expression but does not result in biallelic expression40. While the classical model of imprinting suggests that monoallelic expression is the product of differential methylation of the two alleles, this does not appear to hold as strongly in humans as it does in mice. However, it is not clear why the frequency of LOI is lower here than reported elsewhere, as this phenomenon has been reported in primary breast tumors42,46. An alternative explanation for our observations could be that the changes in DNA methylation are confined to a subset of cells in which expression is silenced. As pyrosequencing enables the measurement of the relative abundance of the transcripts from the two alleles, but not their overall quantity, our observations of monoallelic expression being retained may be based upon an inability to detect silencing of the expressed allele in the affected cells.
There have been few studies investigating epigenetic dysregulation in benign breast disease. Although proliferative benign disease is associated with a greater relative risk of developing cancer47, we have previously reported no significant difference in methylation at six of the imprinted loci between proliferative and non-proliferative conditions18. There is limited evidence of loss of IGF2 imprinting in benign breast disease48, and we observed that hypomethylation of IGF2 DMR2 was highly frequent. Although aberrant methylation was not observed at the other regions, there was significantly greater variability in methylation at six (adjusted model) and eight (unadjusted) sites, and increased variability in cytologically-normal epithelial cells is associated with increased risk of developing neoplasia12. Further work is required to establish the possible role of epigenetic dysregulation in the etiology of benign breast disease.
A limitation of our study is the potential difference in the proportions of cell types between the normal, benign and tumor tissues. Laser-microdissection of the tumor tissue was performed to isolate malignant cells, but was not conducted with normal tissue to enrich the epithelial cells. While median DNA methylation values in normal tissue were consistently close to the expected 50% value, we cannot rule out that the variation modeling may have been affected. We also cannot exclude the possibility of copy number changes influencing measured methylation values in the samples, as insufficient quantities of DNA remained following epigenetic analyses to also perform genetic analyses. Furthermore, the number of samples in this study may also have precluded us from identifying other significant correlations between DNA methylation and tumor subtype, such as with triple-negative tumors. A particular strength of this study is the access to breast tissue from healthy individuals. There is evidence that histologically-normal tissue adjacent to breast tumors can possess genomic alterations seen in the cancer49, and therefore it is important to use breast tissue from healthy women as controls in order to accurately identify changes associated with malignant transformation. A further strength was the availability of both DNA and RNA from blood and breast tissue that has enabled us to investigate the specificity of changes in DNA methylation and the effect upon allele-specific expression.
This is the first study, to our knowledge, to identify an association between the aberrant methylation of imprinted genes and hormone receptor status in breast cancer. We have established that 1) aberrant events in the DNA methylation of these imprinted regions are frequent in breast tumors, 2) variation is greater in both breast tissue and in blood from patients with invasive breast cancer than in healthy individuals, and 3) aberrant changes in DNA methylation are associated with hormone receptor status in the tumors. Further work is required to establish whether such methylation changes are the direct result of loss of hormone receptor signaling or are the product of more widespread changes in global DNA methylation.
Supplementary Material
Supplementary Table 1. Pathology and receptor status for the cancer patients.
The AJCC stage and status of the estrogen (ER), progesterone (PR) and HER2 receptors for each of the 38 cancer patients from whom blood and tumor tissue was taken. The pathology and HER2 status were not known for two patients. Mammary tissue was collected from patients undergoing medical procedures and immediately stored on ice for analysis to be performed by a licensed pathologist. Excess tissue was frozen for research purposes.
Supplementary Table 2. Primers used for methylation and expression analysis.
For each DMR analyzed, the sequences for the forward and reverse primers (PCR) and the sequencing primer (pyrosequencing) are provided. For H19 ICR, the forward primer was also used as the sequencing primer, while for IGF2 DMR0 two sequencing primers were used with the same PCR product. DNA methylation at LINE-1 elements was analyzed using the PyroMark Q24 CpG LINE-1 assay (Qiagen). Assays were validated by running a scale of 0%, 20%, 40%, 60%, 80% and 100% methylated DNA. Stocks of 0% and 100% methylated DNA were created by whole-genome amplification using the RepliG kit (Qiagen) and treatment with MssI (New England Biolabs, Ipswich, USA) respectively, with the two mixed proportionally to create the others. Standards were bisulfite-converted in quadruplicate and the correlation in results calculated (r2>0.97). The SNPs used to identify informative patients for analysis of allele-specific expression are provided along with the gene name.
Supplementary Table 3. DNA Methylation at nine imprinted regions in blood and breast tissue from healthy individuals and patients.
For each individual, values were calculated from two separate bisulfite-converted DNA replicates. The median and the range in mean values for each subgrouping is provided for each of the nine imprinted regions analyzed, along with the number of aberrantly methylated samples (hypomethylated / normally methylated / hypermethylated). We have previously reported raw methylation values for GRB10 ICR, H19 ICR, IGF2 DMR0 and DMR2, KvDMR and SNPRN/SNURF ICR in benign and cancer samples18 and we have reanalyzed this data using values from healthy individuals to determine the frequency of aberrant methylation in the disease states.
Supplementary Table 4. Modeling of variation in DNA methylation in breast tissue and blood.
(A) The mean deviations from the median methylation levels measured in breast tissue or blood from healthy individuals were calculated for healthy individuals, patients with benign breast disease, and patients with invasive cancer. (B) The relative fold change in mean deviation was calculated for the benign and invasive cancer patients relative to the values measured in healthy individuals. 95% confidence intervals are given in brackets. Asterisks (*) denote statistical significance, defined as p<0.05 following Bonferroni correction; †The adjusted model was adjusted for the age, BMI category (‘normal’, ‘overweight’, and ‘obese’), menopausal status and familial history of cancer of the individuals.
Impact: This is the first study, to our knowledge, to identify an association between the aberrant DNA methylation of imprinted genes and negative status of the estrogen and progesterone receptors in breast cancer. Furthermore, we describe how variation in methylation of these genes increases from normal breast tissue to benign breast disease to cancer. Our findings may imply an important role for epigenetic disruption of imprinted genes in the development of different breast cancer subtypes.
Acknowledgements
This work was supported by grants from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services (grant number R03CA143967) and the Breast Cancer Research Foundation (both to Karin Michels). We would like to thank Jennifer Kane for the identification and histological analysis of samples from the Walter Reed National Military Medical Center, and Jill Henry and Julia McCarty from the Susan G. Komen for the Cure Foundation for the identification and supply of matched samples from healthy volunteers. We would also like to thank Dr Rebecca Rancourt for the pyrosequencing assays she designed, and Dr Amy Non, Dr Benedetta Izzi, Dr Jessica LaRocca, Dr Sabrina Böhm and Dr Aggeliki Tserga in the Michels lab for their general comments and advice. The views expressed in this article are those of the authors and do not reflect the official policy of the Department of Army/Navy/Air Force, Department of Defense, or U.S. Government.
Abbreviations
- CBCP
Clinical Breast Care Project
- CpG
cytosine-phosphate-guanine
- DMR
differentially methylated region
- ER
estrogen receptor
- FAM
frequently altered methylation
- FISH
fluorescence in situ hybridization
- HER2
human epidermal growth factor receptor 2
- ICR
imprinting control region
- LOI
loss of imprinting
- PR
progesterone receptor
Footnotes
Supplementary Table 5: Gene expression by hormone receptor status Gene expression microarray data from 302 breast tumor samples. Fold-change in expression (positive versus negative) and statistical significance are provided for ten genes (n/s = not significant). H19 was not present on the microarray. Of the 302 tumors, 199 were ER-positive and 103 were ER-negative, while 153 were PR-positive and 149 PR-negative.
References
- 1.Cerrato F, Sparago A, Verde G, De Crescenzo A, Citro V, Cubellis MV, Rinaldi MM, Boccuto L, Neri G, Magnani C, D'Angelo P, Collini P, Perotti D, Sebastio G, Maher ER, Riccio A. Different mechanisms cause imprinting defects at the IGF2/H19 locus in Beckwith-Wiedemann syndrome and Wilms' tumour. Hum Mol Genet. 2008;17(10):1427–1435. doi: 10.1093/hmg/ddn031. [DOI] [PubMed] [Google Scholar]
- 2.Rabinovitz S, Kaufman Y, Ludwig G, Razin A, Shemer R. Mechanisms of activation of the paternally expressed genes by the Prader-Willi imprinting center in the Prader-Willi/Angelman syndromes domains. Proc Natl Acad Sci U S A. 2012;109(19):7403–7408. doi: 10.1073/pnas.1116661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng W, Shen L, Wen S, Rosen DG, Jelinek J, Hu X, Huan S, Huang M, Liu J, Sahin AA, Hunt KK, Bast RC, Shen Y, Issa JP, Yu Y. Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res. 2007;9(4):R57. doi: 10.1186/bcr1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, Fishman DM, Yu Y, Bast RC. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112(7):1489–1502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- 5.Dalai I, Missiaglia E, Barbi S, Butturini G, Doglioni C, Falconi M, Scarpa A. Low Expression of ARHI Is Associated with Shorter Progression-Free Survival in Pancreatic Endocrine Tumors. Neoplasia. 2007;9(3) doi: 10.1593/neo.06838. 181-IN2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin D, Cui F, Bu Q, Yan C. The Expression and Clinical Significance of GTP-Binding RAS-Like 3 (ARHI) and MicroRNA 221 and 222 in Prostate Cancer. J Int Med Res. 2011;39(5):1870–1875. doi: 10.1177/147323001103900530. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Meng G, Guo QN. Changes in genomic imprinting and gene expression associated with transformation in a model of human osteosarcoma. Exp Mol Pathol. 2008;84(3):234–239. doi: 10.1016/j.yexmp.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299(5613):1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, Craig JM, Robinson WP, Saffery R. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry RA, Feinberg AP. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teschendorff AE, Jones A, Fiegl H, Sargent A, Zhuang JJ, Kitchener HC, Widschwendter M. Epigenetic variability in cells of normal cytology is associated with the risk of future morphological transformation. Genome Med. 2012;4(3):24. doi: 10.1186/gm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentzon N, Düring M, Rasmussen BB, Mouridsen H, Kroman N. Prognostic effect of estrogen receptor status across age in primary breast cancer. Int J Cancer. 2008;122(5):1089–1094. doi: 10.1002/ijc.22892. [DOI] [PubMed] [Google Scholar]
- 14.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 15.Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, Uxa R, Keating S, Kingdom J, Weksberg R. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol. 2008;320(1):79–91. doi: 10.1016/j.ydbio.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 16.Ito Y, Koessler T, Ibrahim AE, Rai S, Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE, Uribe-Lewis S, Woodfine K, Jagodic M, Nativio R, Dunning A, Moore G, Klenova E, Bingham S, Pharoah PD, Brenton JD, Beck S, Sandhu MS, Murrell A. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17(17):2633–2643. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodfine K, Huddleston JE, Murrell A. Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin. 2011;4(1):1. doi: 10.1186/1756-8935-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barault L, Ellsworth RE, Harris HR, Valente AL, Shriver CD, Michels KB. Leukocyte DNA as surrogate for the evaluation of imprinted Loci methylation in mammary tissue DNA. PLoS One. 2013;8(2):e55896. doi: 10.1371/journal.pone.0055896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bediaga NG, Acha-Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J, Montaner D, Renobales M, Fernández AF, Field JK, Fraga MF, Liloglou T, de Pancorbo MM. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010;12(5):R77. doi: 10.1186/bcr2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fackler MJ, Umbricht CB, Williams D, Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P, Visvananthan K, Marks J, Ethier S, Gray JW, Wolff AC, Cope LM, Sukumar S. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71(19):6195–6207. doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao HH, Zhang XF, Chen B, Pan B, Zhang LJ, Li L, Sun XF, Shi QH, Shen W. Bisphenol A exposure modifies methylation of imprinted genes in mouse oocytes via the estrogen receptor signaling pathway. Histochem Cell Biol. 2012;137(2):249–259. doi: 10.1007/s00418-011-0894-z. [DOI] [PubMed] [Google Scholar]
- 22.Cui M, Wen Z, Yang Z, Chen J, Wang F. Estrogen regulates DNA methyltransferase 3B expression in Ishikawa endometrial adenocarcinoma cells. Mol Biol Rep. 2009;36(8):2201–2207. doi: 10.1007/s11033-008-9435-9. [DOI] [PubMed] [Google Scholar]
- 23.Shi JF, Li XJ, Si XX, Li AD, Ding HJ, Han X, Sun YJ. ERα positively regulated DNMT1 expression by binding to the gene promoter region in human breast cancer MCF-7 cells. Biochem Biophys Res Commun. 2012;427(1):47–53. doi: 10.1016/j.bbrc.2012.08.144. [DOI] [PubMed] [Google Scholar]
- 24.Takeo C, Ikeda K, Horie-Inoue K, Inoue S. Identification of Igf2, Igfbp2 and Enpp2 as estrogen-responsive genes in rat hippocampus. Endocr J. 2009;56(1):113–120. doi: 10.1507/endocrj.k08e-220. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XF, Zhang LJ, Feng YN, Chen B, Feng YM, Liang GJ, Li L, Shen W. Bisphenol A exposure modifies DNA methylation of imprint genes in mouse fetal germ cells. Mol Biol Rep. 2012;39(9):8621–8628. doi: 10.1007/s11033-012-1716-7. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez BA, Weng YI, Liu TM, Zuo T, Hsu PY, Lin CH, Cheng AL, Cui H, Yan PS, Huang TH. Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1C in breast cancer cells. Carcinogenesis. 2011;32(6):812–821. doi: 10.1093/carcin/bgr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver JR, Sarkisian G, Krapp C, Mager J, Mann MR, Bartolomei MS. Domain-specific response of imprinted genes to reduced DNMT1. Mol Cell Biol. 2010;30(16):3916–3928. doi: 10.1128/MCB.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Lee KM, Han W, Choi JY, Lee JY, Kang GH, Park SK, Noh DY, Yoo KY, Kang D. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet. 2010;19(21):4273–4277. doi: 10.1093/hmg/ddq351. [DOI] [PubMed] [Google Scholar]
- 29.Alvi MA, Liu X, O'Donovan M, Newton R, Wernisch L, Shannon NB, Shariff K, di Pietro M, Bergman JJ, Ragunath K, Fitzgerald RC. DNA methylation as an adjunct to histopathology to detect prevalent, inconspicuous dysplasia and early-stage neoplasia in Barrett's esophagus. Clin Cancer Res. 2013;19(4):878–888. doi: 10.1158/1078-0432.CCR-12-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, Nagata Y, Nakao M, Wake N. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005;115(5):690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- 31.Prabhu JS, Wahi K, Korlimarla A, Correa M, Manjunath S, Raman N, Srinath BS, Sridhar TS. The epigenetic silencing of the estrogen receptor (ER) by hypermethylation of the ESR1 promoter is seen predominantly in triple-negative breast cancers in Indian women. Tumour Biol. 2012;33(2):315–323. doi: 10.1007/s13277-012-0343-1. [DOI] [PubMed] [Google Scholar]
- 32.Chik F, Machnes Z, Szyf M. Synergistic anti-breast cancer effect of a combined treatment with the methyl donor S-adenosyl methionine and the DNA methylation inhibitor 5-aza-2'-deoxycytidine. Carcinogenesis. 2014;35(1):138–144. doi: 10.1093/carcin/bgt284. [DOI] [PubMed] [Google Scholar]
- 33.Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Gayther SA, Apostolidou S, Jones A, Lechner M, Beck S, Jacobs IJ, Widschwendter M. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4(12):e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baba Y, Nosho K, Shima K, Huttenhower C, Tanaka N, Hazra A, Giovannucci EL, Fuchs CS, Ogino S. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139(6):1855–1864. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennan K, Garcia-Closas M, Orr N, Fletcher O, Jones M, Ashworth A, Swerdlow A, Thorne H, Riboli E, Vineis P, Dorronsoro M, Clavel-Chapelon F, Panico S, Onland-Moret NC, Trichopoulos D, Kaaks R, Khaw KT, Brown R, Flanagan JM. Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Res. 2012;72(9):2304–2313. doi: 10.1158/0008-5472.CAN-11-3157. [DOI] [PubMed] [Google Scholar]
- 36.Widschwendter M, Apostolidou S, Raum E, Rothenbacher D, Fiegl H, Menon U, Stegmaier C, Jacobs IJ, Brenner H. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS One. 2008;3(7):e2656. doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbaux S, Gascoin-Lachambre G, Buffat C, Monnier P, Mondon F, Tonanny MB, Pinard A, Auer J, Bessières B, Barlier A, Jacques S, Simeoni U, Dandolo L, Letourneur F, Jammes H, Vaiman D. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics. 2012;7(9):1079–1090. doi: 10.4161/epi.21495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blagitko N, Mergenthaler S, Schulz U, Wollmann HA, Craigen W, Eggermann T, Ropers HH, Kalscheuer VM. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum Mol Genet. 2000;9(11):1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- 39.Yoshihashi H, Maeyama K, Kosaki R, Ogata T, Tsukahara M, Goto Y, Hata J, Matsuo N, Smith RJ, Kosaki K. Imprinting of human GRB10 and its mutations in two patients with Russell-Silver syndrome. Am J Hum Genet. 2000;67(2):476–482. doi: 10.1086/302997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy SK, Huang Z, Wen Y, Spillman MA, Whitaker RS, Simel LR, Nichols TD, Marks JR, Berchuck A. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol Cancer Res. 2006;4(4):283–292. doi: 10.1158/1541-7786.MCR-05-0138. [DOI] [PubMed] [Google Scholar]
- 41.Dowdy SC, Gostout BS, Shridhar V, Wu X, Smith DI, Podratz KC, Jiang SW. Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. Gynecol Oncol. 2005;99(1):126–134. doi: 10.1016/j.ygyno.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen IS, Dervan PA, Broderick D, Harrison M, Miller N, Delany E, O'Shea D, Costello P, McGoldrick A, Keating G, Tobin B, Gorey T, McCann A. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 1999;59(21):5449–5451. [PubMed] [Google Scholar]
- 43.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10(6):1193–1198. [PubMed] [Google Scholar]
- 44.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62(22):6442–6446. [PubMed] [Google Scholar]
- 45.Tian F, Tang Z, Song G, Pan Y, He B, Bao Q, Wang S. Loss of imprinting of IGF2 correlates with hypomethylation of the H19 differentially methylated region in the tumor tissue of colorectal cancer patients. Mol Med Rep. 2012;5(6):1536–1540. doi: 10.3892/mmr.2012.833. [DOI] [PubMed] [Google Scholar]
- 46.Shetty PJ, Movva S, Pasupuleti N, Vedicherlla B, Vattam KK, Venkatasubramanian S, Ahuja YR, Hasan Q. Regulation of IGF2 transcript and protein expression by altered methylation in breast cancer. J Cancer Res Clin Oncol. 2011;137(2):339–345. doi: 10.1007/s00432-010-0890-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartmann LC, Sellers TA, Frost MH, Lingle WL, Degnim AC, Ghosh K, Vierkant RA, Maloney SD, Pankratz VS, Hillman DW, Suman VJ, Johnson J, Blake C, Tlsty T, Vachon CM, Melton LJ, Visscher DW. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353(3):229–237. doi: 10.1056/NEJMoa044383. [DOI] [PubMed] [Google Scholar]
- 48.McCann AH, Miller N, O'Meara A, Pedersen I, Keogh K, Gorey T, Dervan PA. Biallelic expression of the IGF2 gene in human breast disease. Hum Mol Genet. 1996;5(8):1123–1127. doi: 10.1093/hmg/5.8.1123. [DOI] [PubMed] [Google Scholar]
- 49.Heaphy CM, Bisoffi M, Fordyce CA, Haaland CM, Hines WC, Joste NE, Griffith JK. Telomere DNA content and allelic imbalance demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer. 2006;119(1):108–116. doi: 10.1002/ijc.21815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Pathology and receptor status for the cancer patients.
The AJCC stage and status of the estrogen (ER), progesterone (PR) and HER2 receptors for each of the 38 cancer patients from whom blood and tumor tissue was taken. The pathology and HER2 status were not known for two patients. Mammary tissue was collected from patients undergoing medical procedures and immediately stored on ice for analysis to be performed by a licensed pathologist. Excess tissue was frozen for research purposes.
Supplementary Table 2. Primers used for methylation and expression analysis.
For each DMR analyzed, the sequences for the forward and reverse primers (PCR) and the sequencing primer (pyrosequencing) are provided. For H19 ICR, the forward primer was also used as the sequencing primer, while for IGF2 DMR0 two sequencing primers were used with the same PCR product. DNA methylation at LINE-1 elements was analyzed using the PyroMark Q24 CpG LINE-1 assay (Qiagen). Assays were validated by running a scale of 0%, 20%, 40%, 60%, 80% and 100% methylated DNA. Stocks of 0% and 100% methylated DNA were created by whole-genome amplification using the RepliG kit (Qiagen) and treatment with MssI (New England Biolabs, Ipswich, USA) respectively, with the two mixed proportionally to create the others. Standards were bisulfite-converted in quadruplicate and the correlation in results calculated (r2>0.97). The SNPs used to identify informative patients for analysis of allele-specific expression are provided along with the gene name.
Supplementary Table 3. DNA Methylation at nine imprinted regions in blood and breast tissue from healthy individuals and patients.
For each individual, values were calculated from two separate bisulfite-converted DNA replicates. The median and the range in mean values for each subgrouping is provided for each of the nine imprinted regions analyzed, along with the number of aberrantly methylated samples (hypomethylated / normally methylated / hypermethylated). We have previously reported raw methylation values for GRB10 ICR, H19 ICR, IGF2 DMR0 and DMR2, KvDMR and SNPRN/SNURF ICR in benign and cancer samples18 and we have reanalyzed this data using values from healthy individuals to determine the frequency of aberrant methylation in the disease states.
Supplementary Table 4. Modeling of variation in DNA methylation in breast tissue and blood.
(A) The mean deviations from the median methylation levels measured in breast tissue or blood from healthy individuals were calculated for healthy individuals, patients with benign breast disease, and patients with invasive cancer. (B) The relative fold change in mean deviation was calculated for the benign and invasive cancer patients relative to the values measured in healthy individuals. 95% confidence intervals are given in brackets. Asterisks (*) denote statistical significance, defined as p<0.05 following Bonferroni correction; †The adjusted model was adjusted for the age, BMI category (‘normal’, ‘overweight’, and ‘obese’), menopausal status and familial history of cancer of the individuals.