Abstract
Multidrug resistance protein 4 (MRP4), a member of the ATP binding cassette transporter family, functions as a plasma membrane exporter of cyclic nucleotides. Recently, we demonstrated that fibroblasts lacking the Mrp4 gene migrate faster and contain higher cyclic-nucleotide levels. Here, we show that cAMP accumulation and protein kinase A (PKA) activity are higher and polarized in Mrp4−/− fibroblasts, versus Mrp4+/+ cells. MRP4-containing macromolecular complexes isolated from these fibroblasts contained several proteins, including actin, which play important roles in cell migration. We found that actin interacts with MRP4, predominantly at the plasma membrane, and an intact actin cytoskeleton is required to restrict MRP4 to specific microdomains of the plasma membrane. Our data further indicated that the enhanced accumulation of cAMP in Mrp4−/− fibroblasts facilitates cortical actin polymerization in a PKA-dependent manner at the leading edge, which in turn increases the overall rate of cell migration to accelerate the process of wound healing. Disruption of actin polymerization or inhibition of PKA activity abolished the effect of MRP4 on cell migration. Together, our findings suggest a novel cAMP-dependent mechanism for MRP4-mediated regulation of fibroblast migration whereby PKA and actin play critical roles as downstream effectors.
Keywords: MRP4, migration, cyclic nucleotides, actin, PKA
1.1 INTRODUCTION
Cell migration is a carefully orchestrated process requiring cross talk between cell adhesion, cytoskeleton re-organization and membrane extensions [1, 2]. Conventionally, the process of cell migration consists of four distinct steps; 1) formation of leading-edge protrusions, 2) adhesion of the leading edge to the extracellular matrix, 3) forward translocations of the cell body and 4) retraction of the trailing edge by releasing attachment at the cell rear [1, 2]. The spatial segregation of the intracellular signaling can dynamically regulate diverse biochemical events that ensure cell polarization and efficient cell migration [3, 4]. The extracellular signal can be sensed and communicated inside the cell by specific receptors, which in turn directs the cell to extend membrane protrusions via actin polymerization [2]. Establishment of polarity is very important for directional cell migration and the spatial distribution of signaling molecules, and compartmentalized signaling plays a critical role in directing cell movement [5]. For example, adhesive biomolecules aggregate at the cell front to promote tethering of the cell protrusions to the substrum, whereas the removal of those molecules from the cell posterior facilitates detachment of the trailing edge [6]. In addition, the preferential contraction of actomyosin at the cell front pulls the cell body in a forward direction. However, several regulatory steps are involved in directional cell migration and dysregulation of any of those steps leads to catastrophic consequences [1].
Localized increases in cAMP concentration and cAMP-dependent protein kinase A (PKA) at the leading edge, both play pivotal roles in ensuring the polarity of migrating cells [3, 4, 7]. The diversity of PKA substrates permits the regulation of multiple signaling events, based on the subcellular localization of PKA [8]. At the leading edge, PKA activates small GTPses called Ras-related C3 botulinum toxin substrate (Rac) and cell division control protein 42 homolog or Cdc42 by phosphorylation, which subsequently activate localized actin-related protein 2/3 (Arp 2/3) in a Wiskott-Aldrich syndrome proteins (WASP)/WASP-family verprolin-homologous protein or WAVE-dependent manner [9, 10]. Activated Arp 2/3 binds to the sides or tips of the pre-existing actin filaments and induces the formation of new actin filaments from the pointed end at a 70° angle. The formation of branched dendritic networks of actin filament promotes a flat protrusion called lamellipodia, [2, 11, 12]. Another PKA substrate, vasodilator-stimulated phosphoprotein (VASP) regulates actin polymerization as an anti-capping agent, and phosphorylation of VASP can regulate the oscillatory cycles of membrane extension and retraction [9, 13, 14].
Multidrug resistance protein 4 (MRP4) is a member of the ATP binding cassette or ABC transporter family that localizes to the plasma membrane and functions as an endogenous transporter of cyclic nucleotides. Therefore MRP4 can regulate various cyclic nucleotide dependent signaling events by modulating intracellular cAMP and cGMP levels (Cheepala et al 2013). Recently, we demonstrated that Mrp4−/− fibroblasts migrated faster compared to Mrp4+/+ fibroblasts and this was associated with moderately higher levels of total intracellular cyclic nucleotides. We also found that inhibition of MRP4 enhances the compartmentalized cAMP levels at or near the leading edge of a migrating cell [15]. Therefore, we hypothesized that this polarized elevation of cAMP is responsible for localized PKA activation at the cell front, which is the early hallmark step in directional cell migration [3, 7]. In this study, we identified actin as an integral part of the MRP4 interactome. In previous studies, cAMP/PKA has been found to regulate actin polymerization and therefore the overall cell migration [16–18]. For this study, we investigated the role of MRP4 in regulating actin dynamics at the leading edge of migrating cells and the relationship between MRP4 and PKA in this process of regulation. Together, our data suggest a novel cAMP-dependent mechanism for MRP4-mediated regulation of fibroblast migration where PKA and actin play critical roles as downstream targets.
1.2 MATERIALS AND METHODS
1.2.1 Reagents
MRP4 inhibitor MK571 was obtained from Cayman Chemical (Ann Arbor, MI). Forskolin was obtained from Tocris (Ellisville, MO). IBMX, Lat B, cpt-cAMP, and cpt-cGMP were purchased from Sigma-Aldrich (St. Louis, MO). Zaprinast and PKA inhibitor H-89, 2HCl were purchased from Enzo Life Sciences (Farmingdale, NY).
1.2.2 Cell Culture and Transfections
MEF cells and NIH 3T3 cells were cultured in DMEM media (MRP4 overexpressed) and HEK293 cells were grown in DMEM F-12 media (Invitrogen; Carlsbad, CA), both containing 10% FBS and 1% penicillin/streptomycin, and cell cultures were maintained in a 5% CO2 incubator at 37°C. Mrp4+/+ and Mrp4−/− primary MEFs were transfected with 1 µg of total SV40 genomic DNA using Lipofectamine LTX (Invitrogen) according to the manufacturer’s instructions. On the next day, the transfected cells were serially diluted (1000 to 10 cells per well) and plated in 96-well plates. SV40 immortalized clones were selected from the lower-dilutions wells [15]. Lipofectamine 2000 (Invitrogen) was used for all transient transfections according to the manufacturer’s instructions, and MRP4-overexpressing cell lines were generated using lentivirus vector and selected using puromycin (2 µg/ml).
1.2.3 Wound-Healing Assay
Cell migration was measured according to the previously described method [15]. Briefly, confluent monolayers of fibroblast cells were wounded by scraping with a pipette tip across the monolayer. Cells were washed with PBS and incubated with appropriate media. Images of 100× magnification were taken at the initial time of wounding and then at 6 h or 10 h post-wounding using a cooled electron microscope (EM)-CCD camera (Hamamatsu; Bridgewater; Denver, CO). Cell migration was analyzed by Image J software and represented as a percentage of initial wound length [19].
1.2.4 High-Content Microscopy
Mrp4+/+ and Mrp4−/− Cells were grown in a fibronectin-coated 96-well microplates (Image-Lock micropklates, Essen BioScience; Ann Arbor, MI) in a standard CO2 incubator for 24 hours. Precise wounds were made using the 96-pin Wound-Maker provided with the IncuCyte™ live-cell imaging system (Essen BioScience). Cells were washed thoroughly with PBS to remove the detached cells and then placed in the incubator imaging system with appropriate media. The wound images were acquired from the incubated cells at 1-h intervals and the kinetics of the RWD was analyzed using IncuCyte™ software [15].
1.2.5 Immunoblotting
For Western blot, cells were lysed in lysis buffer (1× PBS, containing 0.2% Triton X-100 and protease inhibitors: 1 mM phenylmethylsulfonyl fluoride or PMSF, 1 µg/ml pepstatin A, 1 µg/ml leupeptin, and 1 µg/ml aprotinin). The lysate was centrifuged at 16,000 × g for 10 min at 4°C, and the clear supernatant was mixed with 5× Laemmli sample buffer (containing 2.5% β-mercaptoethanol), denatured, subjected to SDS-PAGE on a 4–15% gradient gel (Bio-Rad; Hercules, CA), transferred to a PVDF membrane, and the membranes were then incubated with specific antibodies. We used anti-MRP4 antibody (rabbit polyclonal antibody) for MRP4 detection. Anti-beta actin antibody (mouse) was purchased from Sigma-Aldrich Co. (St. Louis, MO). VASP and phosphorylated VASP (Ser157) were purchased from Cell Signaling Technology (Danvers, MA), and PKA RIIα antibody was obtained from Transduction Laboratories (Lexington, KY). The protein bands were detected by chemiluminescence using ECL™ Western blotting detection reagents from GE Healthcare (Buckinghamshire, UK). We loaded 50µg of total protein per lane [20, 21].
1.2.6 Fluorescence Resonance Energy Transfer Microscopy
Cells were transiently transfected with a CFP-regulatory and YFP-catalytic subunit of PKA (a generous gift from Dr. Manuela Zaccolo, Department of Physiology, Anatomy & Genetics, University of Oxford, UK) and were grown in 35-mm glass-bottomed dishes (MatTek Corporation; Ashland, MA) for 24–48 h (117,118); after washing with HBSS, cells were mounted on an Olympus microscopy system for FRET imaging. Using the cooled CCD camera Hamamatsu ORCA285 (Hamamatsu, Japan) mounted on the Olympus microscope IX51 (U-Plan Fluorite 60 × 1.25 NA oil-immersion objective) images were recorded. The system was controlled by SlideBook 4.1 software (Intelligent Imaging Innovations; Denver, CO) and ratio modules were used to obtain and analyze the images. A 300W Xenon lamp attenuated with a ND filter with 50% light transmission provided the excitation light. Images were taken using a JP4 CFP/YFP filter set (Chroma; Brattleboro, VT), including a 430/25-nm excitation filter, a double dichroic beam splitter, and two emission filters (470/30-nm for CFP and 535/30-nm for FRET) alternated with a filter-changer Lambda 10–3 (Sutter Instruments; Novato, CA). With 100 ms exposure time, time-lapse images were taken at 1-min intervals. Multiple regions of interest were selected on the cell after background subtraction for quantitative data analysis (4–6 cells per condition). At different time points, the emission ratio images (CFP/FRET) were obtained as described previously [22]. The representative pseudo color images at 60× magnification indicate the changes in the ratio of CFP to FRET intensity.
Membrane-targeted pmAKAR3 and pmAKAR3-TA sensors were generous gifts from Dr. Jin Zhang, (Department of Pharmacology and Molecular Sciences, The Johns Hopkins University School of Medicine, Baltimore, MD, USA). These sensors were used for monitoring PKA activity. PKA-mediated phosphorylation increases the FRET between the CFP and YFP tags. Cells transfected with pmAKAR3 sensor were subjected to FRET measurement after background subtraction. FRET was normalized with donor CFP intensity (FRET/CFP) to obtain the normalized corrected FRET or N-FRET, which was represented as an index of PKA activity [22, 23].
1.2.7 Purification of HA-MRP4
HEK293 cells, stably overexpressing HA-MRP4 were seeded on ten 100 mm dishes and grown until completely confluent. After lysis, the cell lysate was mixed with anti-HA beads overnight at 4°C for immunoprecipitating HA-MRP4. The beads were washed and the bead-bound MRP4 was eluted with 100 mM glycine (pH 2.2) and neutralized with 150 mM tris (pH 8.8). The eluted MRP4-containing macromolecular complex was run by SDS-PAGE on 4–15% gradient gels (Bio-Rad, Hercules, CA) and visualized with GelCode Blue (manufacturer’s information or add to reagents section). The MRP4-containing macromolecular complex was subjected to mass-spectrometric analysis for identifying the MRP4-interacting partners [22].
1.2.8 Co-Immunoprecipitation
5–10µg of anti-MRP4 rabbit polyclonal antibody was added to protein A/G and cross-linked with DSS for 60 min at room temperature. After crosslinking, the beads were washed, resuspended in PBS-0.2% TX100 and used for immunoprecipitation of endogenous MRP4 from MEFs. MRP4-containing macromolecular complex was eluted with 100 mM glycine (pH 2.2) and neutralized with 150 mM Tris (pH 8.8). The eluted proteins were mixed with sample buffer (5×; containing 1% β-mercaptoethanol), denatured, subjected to SDS-PAGE, transferred to PVDF membrane, and immunoblotted using goat polyclonal α-MRP4 antibody and anti-beta-actin antibody (mouse) [22].
1.2.9 Immunofluorescence
Cells were washed with PBS and fixed with 4% formaldehyde. Fixed cells were permeabilized with 0.2% Triton X-100 and blocked with 2.5% normal horse serum overnight at 4°C. For immunostaining, samples were incubated overnight with respective primary antibodies and then incubated with secondary antibody, Alexa Fluor® 488 phalloidin or anti-rabbit Alexa Fluor® 568 (Molecular Probes, Invitrogen) for 1 h and mounted in DAPI-containing Vectashield (Vector Labs; Burlingame, CA). Fluorescence images were taken using a NikonA1R-A1 confocal microscope system [20].
1.2.10 Mass-Spectrometric Identification of MRP4-Interacting Proteins
Purified HA-MRP4-containing macromolecular complexes was subjected to 1D SDS-PAGE, stained with GelCode Blue, and visualized bands were excised and subjected to in-gel protein digestion by sequence-grade pig trypsin (20 µg/ml 100 mM HCO3-) as previously described [24]. Following digestion, eluted tryptic peptides in 1% acetic acid were directly analyzed for protein identification by electrospray ionization tandem mass spectrometry, as previously described [24–27]. Briefly, samples were loaded in a Dionex Ultimate 3000 HPLC system autosampler (manufacturer’s information) and eluted by reverse-phase chromatography into a Thermo Scientific LTQ velos pro mass spectrometer fitted with a nanospray ion source for analysis. The mass analyzer was set up for data-dependent mode using dynamic exclusion settings of: repeat count = 1; repeat duration = 0.5 min; exclusion list size = 50; exclusion duration = 1.5 min; and exclusion mass width = 1.5 amu. Collision-induced dissociation was used to fragment peptides and CID spectra were searched against a human fasta database using the Proteome Discoverer™ software. A decoy database was used to control for false discovery. A threshold filter of > 2.0 for peptide XCorr score was used for sequence identification. This produced a range of coverage for identified proteins of 2.05 – 49.91% and an average of 10.03 % for all proteins identified. PD scores shown in the supplementary data reflect the sum of the XCorr scores for all of the peptides identified for each protein.
1.2.11 Ingenuity Pathway Analysis
Using Ingenuity Pathway Analysis, we assigned MRP4 as seed and the MRP4-interacting proteins as high-confidence interactome. We then checked the interconnectivity between those proteins in the context of cell migration using the “shortest path” explorer, which also included the intermediate proteins and molecules involved in the pathway based on literature and experimental evidence. Subsequently, we identified specific migration-related signaling cascades that are regulated by certain nodes in the interactome network.
1.2.12 PKA Activity Assay
Cells were lysed in Tris-based lysis buffer with 1 mM PMSF and 10 mM activated orthovanadate (phosphatase inhibitor) and used for PKA activity assay using the DetectX, PKA (Protein Kinase A) Activity kit (Arbor Assays; Ann Arbor, MI). Lysate containing PKA phosphorylated the immobilized PKA substrate on the microtiter plate in the presence of ATP. Antibody specific for phospho-PKA substrate recognized the immobilized phosphorylated substrate and was detected by peroxidase-conjugated anti-rabbit IgG. The Fluostar Omega microplate reader (BMG Labtech) was used to measure the optical density at 450 nm. Standards, supplied with the kit were run in parallel for quantitation of total PKA activity [28].
1.2.13 Single-Particle Tracking
NIH 3T3 cells stably expressing HA-MRP4 were grown on 35-mm glass-bottom dishes (MatTek). Cells were washed two times with PBS containing 6 mM glucose and 1 mM sodium pyruvate (PBS/Glu/NaPyr). PBS/Glu/NaPyr containing 4% BSA was used to block the washed cells for 10 min. After blocking, cells were incubated with biotin-conjugated anti-HA antibody (1 µg/ml, Sigma) for 15 min, followed by an incubation with streptavidin-conjugated Qdot-655 (0.1 nM, Invitrogen) for 2 min in the absence of light. Cells were then washed extensively and mounted immediately on the Olympus inverted microscope (IX51). The images were captured using a Hamamatsu EM-CCD camera at 1–3 frames per second for 1–3 min with 50-ms exposure time, 100× oil-immersion objective (NA 1.40), xenon (300-W lamp) light source, and SlideBook 4.2 software. Qdot 655-A BrightLine high brightness and a contrast single-band filter set (Semrock, Rochester, NY) were used for collecting data. SPT was monitored by the particle-tracking module of SlideBook 4.2 software. The software generated trajectories for single Qdot and calculated the mean squared displacement or MSD and the diffusion coefficient D. Data obtained from 5–10 cells were used for plotting histograms of the diffusion coefficient. To monitor changes in lateral diffusion of MRP4 in fibroblasts with cytoskeletal disruption, cells were pretreated with Lat B (1 µM, 30 min) or DMSO (as control), which were also added to the buffer during the course of the experiment [22].
1.2.14 Statistical Analysis
The mean values of different groups were compared using a two-tailed Student’s t-test; P values less than 0.05 were considered as significant. All the data were presented as mean ± SEM where n indicates the number of experiments.
1.3 RESULTS
1.3.1 Actin is an integral part of the MRP4 interactome
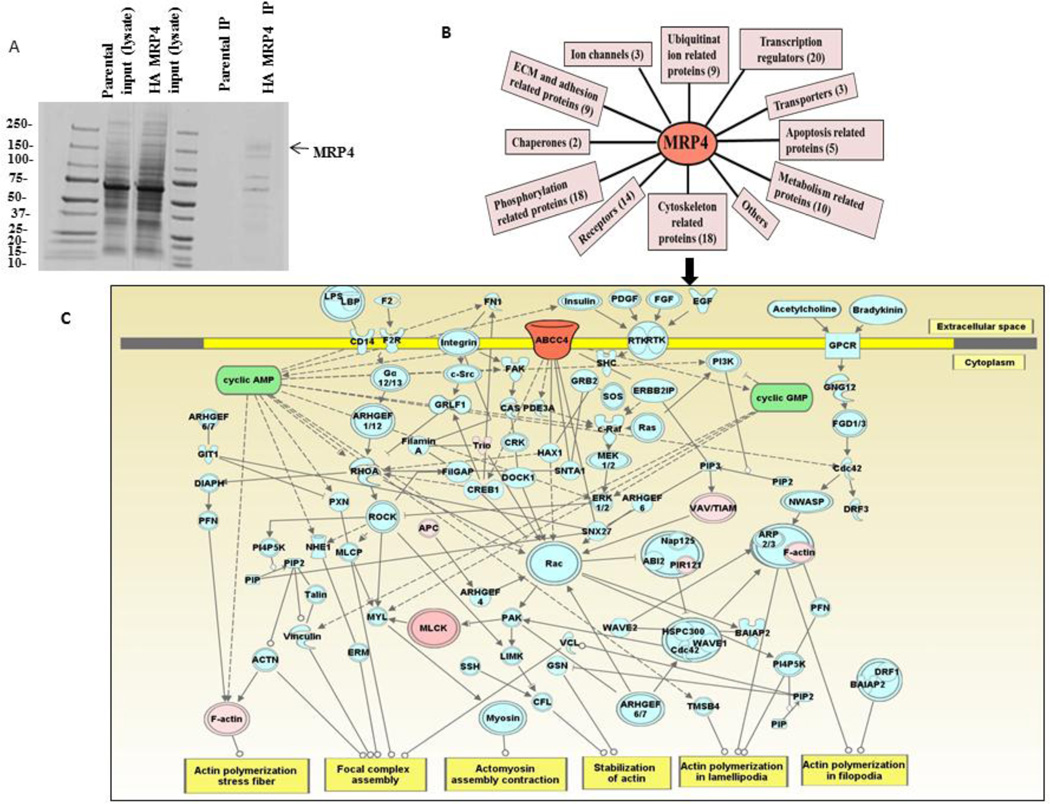
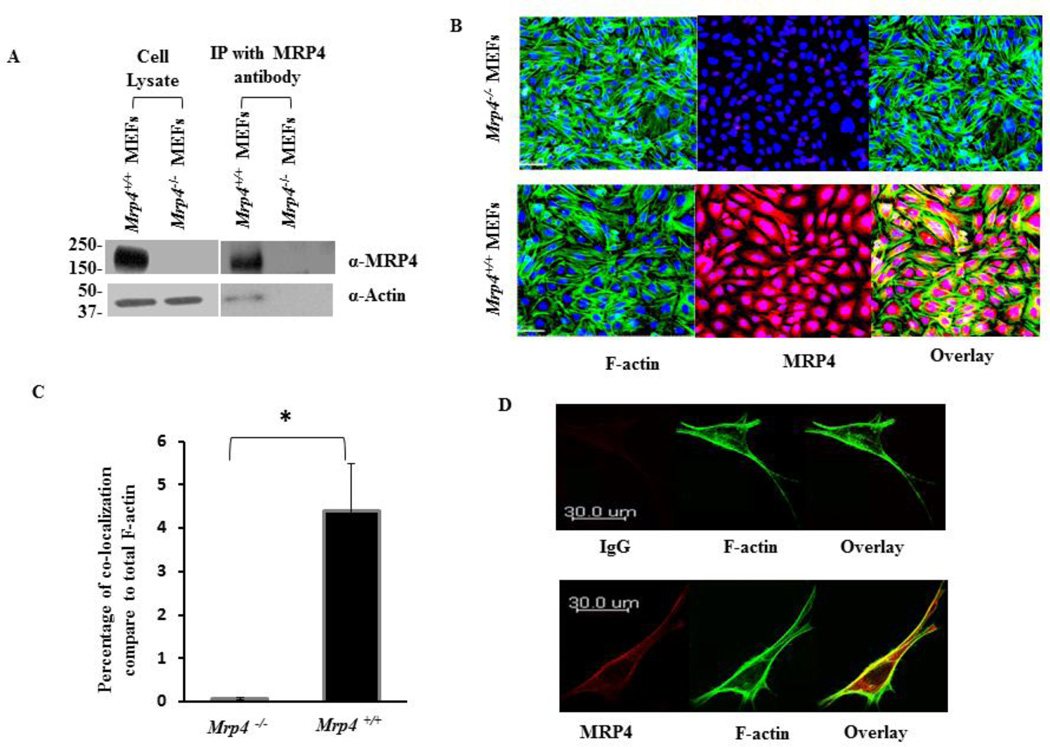
We recently demonstrated that MRP4 plays an important role in the regulation of fibroblast migration via alteration of cyclic nucleotide levels [15]. In order to understand migratory phenomenon and its relationship with MRP4, we wanted to identify proteins that interact with MRP4 to allow us to identify the downstream effectors of MRP4. To investigate the MRP4 interactome, we stably overexpressed human influenza hemagglutinin (HA)-tagged MRP4 (HA tag between residues 751 and 752) in HEK293 cells using a lentivirus vector and purified MRP4 using anti-HA beads. Fig. 1A shows a Coomassie-stained gel of immunoprecipitated full-length HA-MRP4 and its interacting partners. After mass-spectrometric analysis, we identified several MRP4-interacting proteins and calculated the plasma desorption or PD scores. We removed the randomized interactions and categorized the MRP4-interacting proteins based on their molecular function (Fig 1B). The complete list of the MRP4 interactome is provided in the supplementary material (Tables 1–11). A large number of the interacting proteins, including actin, are cytoskeleton-related proteins, suggesting possible interplay between MRP4 and the cytoskeleton (Fig 1C). Using Ingenuity Pathway Analysis or IPA, we demonstrated that a significant number of the MRP4-interacting proteins, including actin, are involved in various signaling cascades associated with the process of cell migration. We used the “shortest path” explorer module that also includes the intermediate proteins and molecules involved in those pathway based on literature and experimental evidences. Since both cAMP and cGMP were present as key nodal points and F-actin served as a major target, we suggest that MRP4 might play a role in actin regulation in a cyclic nucleotide-dependent manner. Although no previous report to our knowledge has proposed a direct interaction between MRP4 and actin, it has been established that the actin cytoskeleton plays an important role during the course of cell migration and dynamic rearrangement of actin at the leading edge (referred to as cortical actin) promoting cellular protrusion formation and efficient cell migration [9, 11, 12, 18, 29–32]. To verify the association of actin with endogenous MRP4 in fibroblasts, we immunoprecipitated MRP4 from Mrp4+/+ mouse embryo fibroblasts (MEFs) using anti-MRP4 rabbit polyclonal antibody and found that actin co-immunoprecipitated with MRP4 (Fig. 2A) compared to Mrp4−/− MEFs used as negative control. Using immunofluorescence staining, we demonstrated that a significant proportion of actin co-localized with MRP4 in Mrp4+/+ MEFs (Fig. 2B and 2C). Additionally, actin interacted with MRP4 predominantly at the plasma membrane in NIH 3T3 cells (Fig. 2D). Taken ttogether, our immunofluorescence and co-immunoprecipitation data support the mass-spectrometry-based detection of MRP4 and actin interaction from which we hypothesized the possible role actin as a key downstream effector protein for MRP4-mediated regulation of fibroblast migration.
Figure 1. Actin is an integral part of the MRP4 interactome.
(A) MRP4 was affinity purified using anti-HA beads from HA-MRP4-overexpressing HEK293 cells. The Coomassie-stained gel shows the immunoprecipitated full-length HA-MRP4 and its interacting partners. (B) MRP4-interacting partners were identified by mass-spectrometric analysis and categorized based on their molecular function. (C) Ingenuity Pathway Analysis showed the interconnectivity between the MRP4 interactome components in the context of cell migration. We assigned MRP4 as seed (orange) and the MRP4-interacting proteins as high-confidence interactome (pink) and indicated the intermediate proteins involved in the pathways based on the literature and experimental evidence (blue).
Figure 2. Actin interacts with MRP4 in fibroblast cells.
(A) Endogenous MRP4 was immunoprecipitated from Mrp4+/+ and Mrp4−/− MEFs. Immunoblot data showing that actin is co-immunoprecipitated with MRP4. (B) Co-localization of MRP4 and actin in Mrp4+/+ and Mrp4−/− MEFs by confocal fluorescence microscopy (20× objective) using anti-MRP4 antibody (red) and phalloidin antibody (green). (C) Bar graph showing the percentage co-localization of MRP4 and actin compare to total F-actin in Mrp4+/+ and Mrp4−/− MEFs. Error bars indicate SEM. (D) Co-localization of MRP4 and actin in NIH 3T3 cells by confocal fluorescence microscopy (40× objective) using anti-MRP4 antibody (red) and phalloidin antibody (green); normal rabbit IgG was used as a control. All data represent at least three independent experiments. *, P < 0.05.
1.3.2 Disruption of the actin cytoskeleton increases MRP4 mobility on the cell membrane
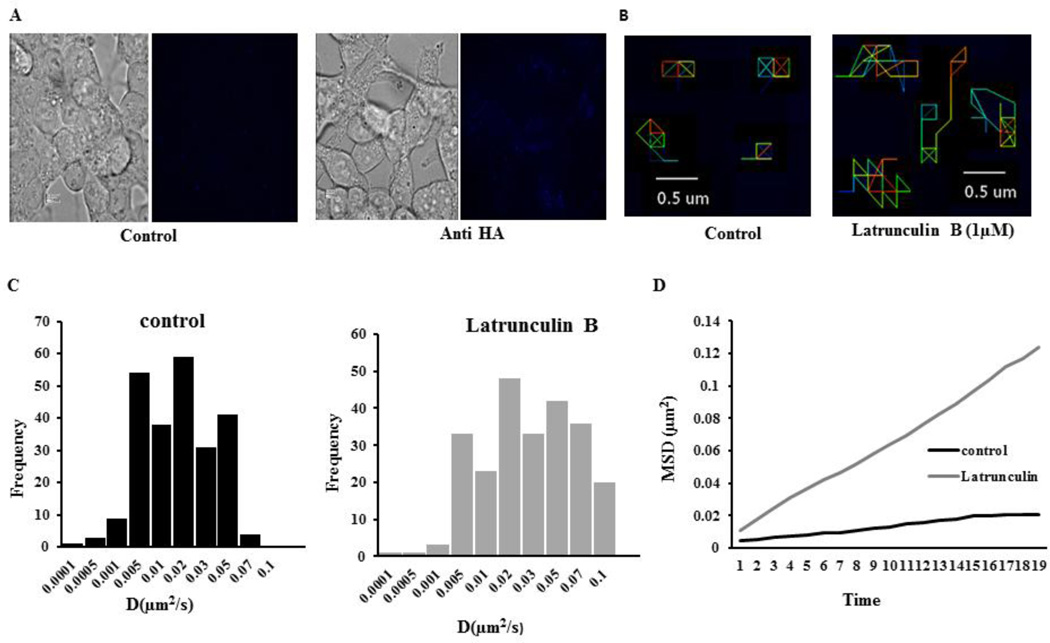
After revealing that MRP4 can form a macromolecular complex with actin at the plasma membrane, we wanted to further confirm the association of MRP4 with the actin cytoskeleton. Since the actin cytoskeleton is known to play important roles in maintaining several membrane proteins at particular domains of the plasma membrane [22, 33], we hypothesized that interaction of MRP4 with actin would restrict the surface mobility of MRP4. To determine whether actin affects MRP4 mobility, we disrupted the actin cytoskeleton with Latrunculin B (Lat B), which induces de-polymerization of actin filaments. The dynamics of individual protein molecules in the plasma membrane of a live cell can be successfully monitored by single-particle tracking (SPT) [34], therefore we used the SPT method to investigate the effects of actin cytoskeleton disruption on HA-MRP4 dynamics in live NIH 3T3 cells. HA-MRP4 was labeled with biotin-conjugated anti-HA antibody and streptavidin-conjugated Qdot-655 was added for monitoring lateral mobility of the protein on the plasma membrane (Fig. 3A). The mean diffusion coefficient of MRP4 (0.016 µm2/s) denoted a confined lateral diffusion of MRP4 on the plasma membrane, which is comparable with the diffusion coefficient of other membrane proteins [22]. After pretreatment with Lat B (1 µM), the diffusion coefficient of MRP4 increased by nearly 2 fold (mean diffusion coefficient 0.03µm2/s) and the MRP4 exhitibed longer trajectory paths (Fig 3B) and had greater overall mobility (Fig. 3C and 3D). Our SPT data suggested that MRP4 is highly associated with the actin cytoskeleton and this interaction is pivotal in restricting MRP4 to certain micro-domains of plasma membrane.
Figure 3. Disruption of the actin cytoskeleton increases MRP4 mobility on the cell membrane.
(A) Representative bright-field and quantum-dot-labeled images of NIH 3T3 cells expressing HA-MRP4 treated with or without biotin anti-HA antibody. (B) SPT of HA-MRP4 in NIH 3T3 cells represented by pseudocolor images showing trajectories for individual HA-MRP4 molecules with either DMSO (control) or Lat B treatment. (C) Histograms representing the diffusion coefficients of a population of HA-MRP4 in the plasma membrane of NIH 3T3 cells in DMSO- or Lat B-treated cells. (D) Representative mean squared displacement curve showed the overall mobility kinetics of HA-MRP4 in DMSO- or Lat B-treated cells (n = 5–6 cells, 150–250 trajectories).
1.3.3 MRP4-deficient fibroblasts exhibit altered β-actin dynamics
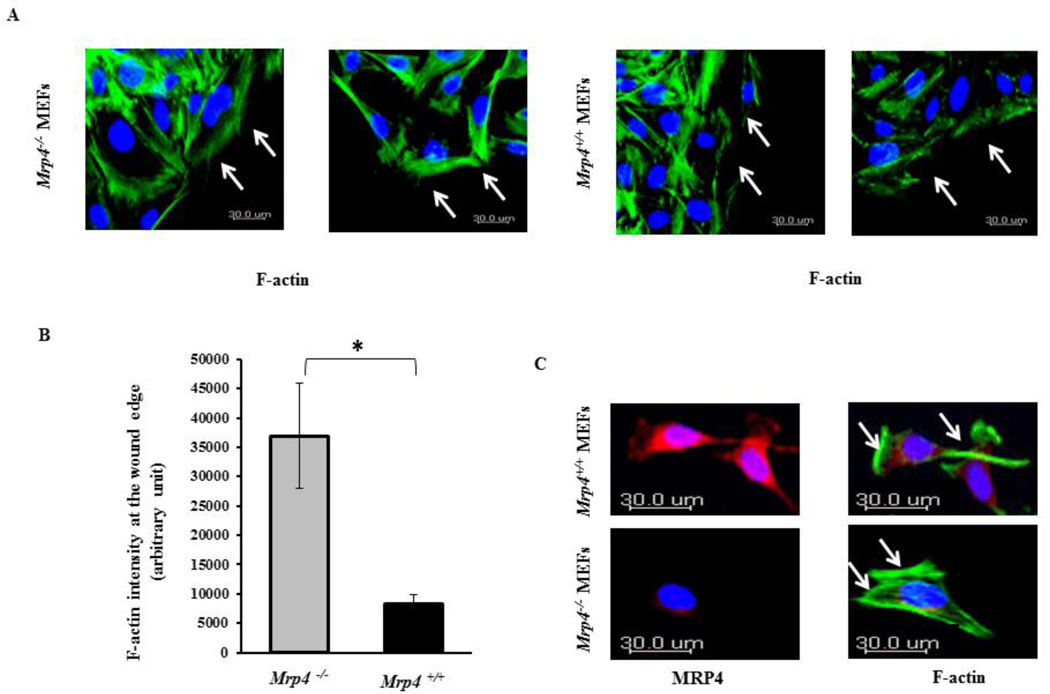
Our SPT data confirmed that the actin cytoskeleton is responsible for restraining MRP4 in a specific membrane locale. Hence, we investigated whether MRP4 itself can exert an effect on cytoskeleton rearrangement during the course of cell migration. To study the effect of MRP4 on actin dynamics, we fixed wounded monolayers of Mrp4+/+ and Mrp4−/− MEFs and stained them with Alexa Fluor® 488-conjugated phalloidin antibody. During cell migration, the actin cytoskeleton polymerizes at the cortex, which is a narrow zone called cortical actin just beneath the plasma membrane at the leading edge [2, 12]. We observed that a clear cortical actin band formed at the wound edge for both of the cell types. However, the F actin intensity at the wound edge was significantly higher for Mrp4−/− MEFs compared to wild-type MEFs (Fig. 4A and 3B). Similarly, migrating Mrp4−/− MEFs exhibit greater actin polymerization compared to Mrp4+/+ MEFs (Fig. 4C). In wild-type cells, distinct MRP4 expression was observed in the lamellipodia. These observations suggest that at the leading edge, MRP4 exerts a negative impact on cortical actin organization and therefore, MRP4-deficient fibroblasts exhibit more intense cortical actin and a faster migration rate.
Figure 4. MRP4-deficient fibroblasts exhibit altered β-actin dynamics.
(A) Cortical actin dynamics at the wound edges by confocal fluorescence microscopy (20× objective) are shown in Mrp4+/+ and Mrp4−/− MEFs using phalloidin antibody (green). (B) Bar graph representing the total actin intensity at the wound edge in Mrp4+/+ and Mrp4−/− MEFs. Error bars indicate SEM. (C) Actin dynamics at the leading edges of migrating Mrp4+/+ and Mrp4−/− MEFs by confocal fluorescence microscopy (20× objective) are shown using phalloidin antibody (green) and anti-MRP4 antibody (red). All data represent at least three independent experiments. *, P < 0.05.
1.3.4 MRP4-deficient fibroblasts exhibit higher polarized cAMP and PKA activity
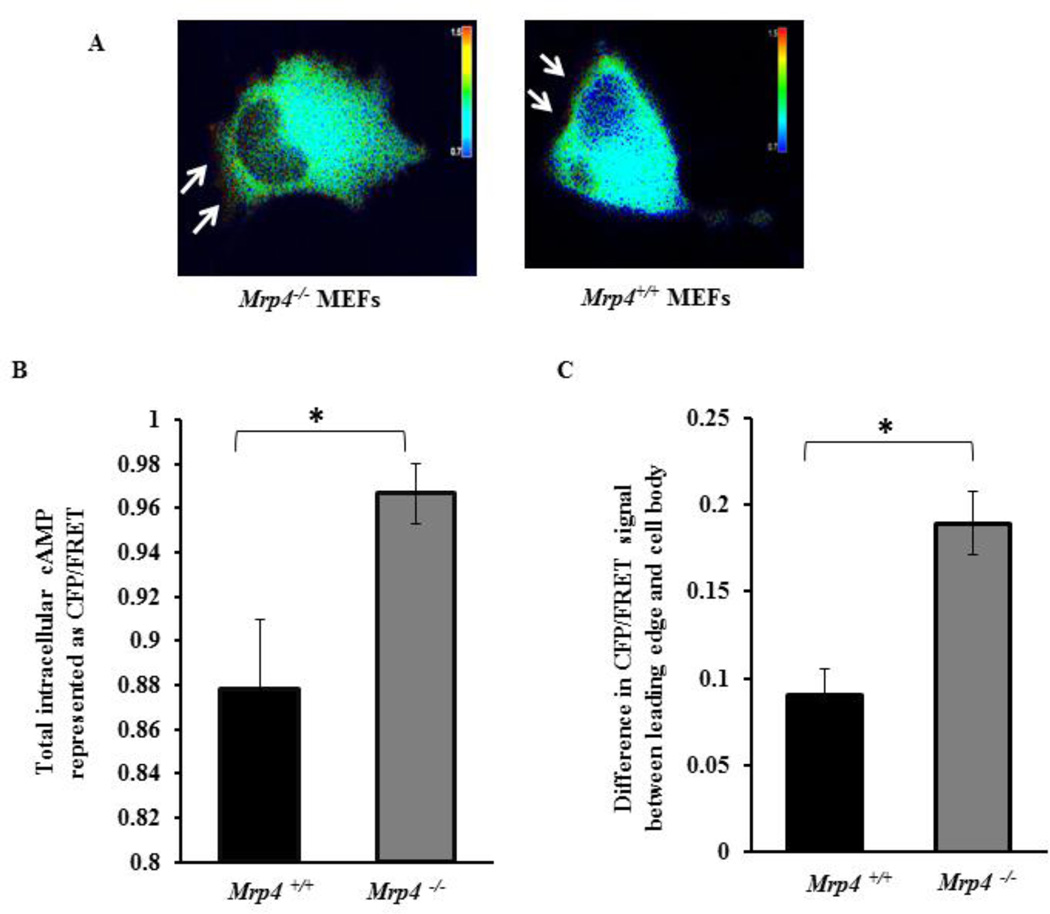
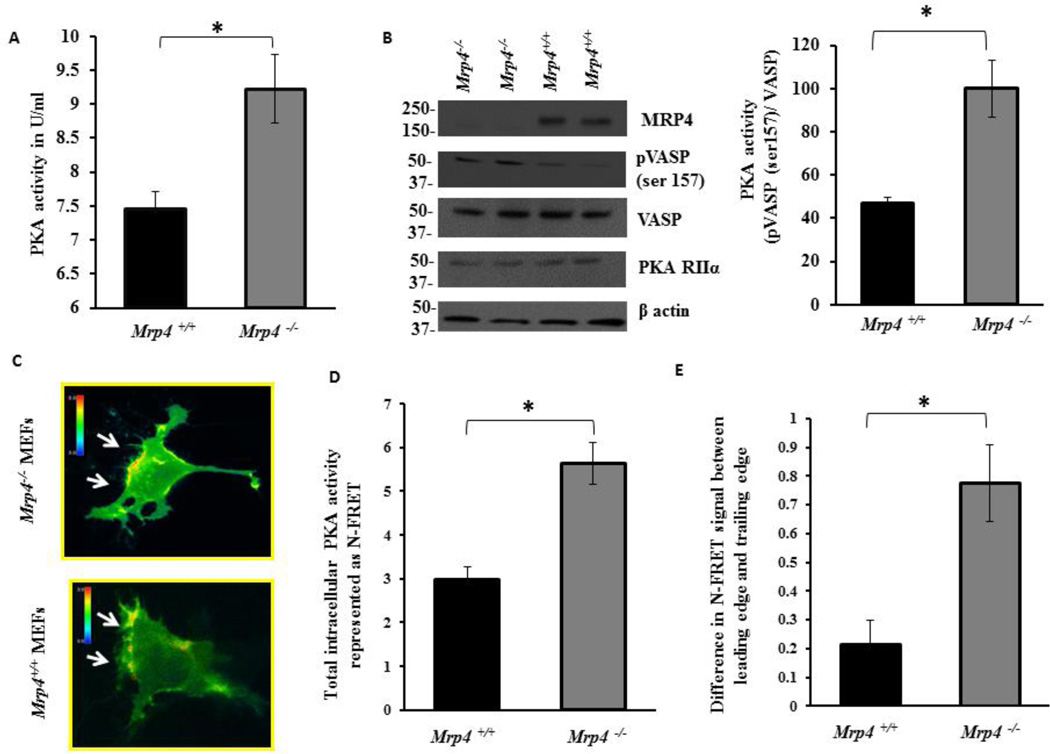
The polarized activation of the cAMP-dependent kinase PKA is an essential early step for directional cell migration that is also involved in actin polymerization and the regulation of cytoskeleton dynamics [3, 16, 18]. We previously have demonstrated that in migrating fibroblasts, inhibition of MRP4 enhances the polarized cAMP level at or near the leading edge [15]. Here, we used CFP- and YFP-tagged PKA regulatory and catalytic subunits as a FRET-based cAMP sensor to visualize the spatiotemporal dynamics of cAMP at the wound edge of Mrp4+/+ and Mrp4−/− MEFs. After making a precise wound in the confluent monolayer of fluorescent cAMP sensor-transfected MEFs, we analyzed cells expressing the sensor particularly at the wound edge. In both cell types, we observed polarized cAMP at the leading edge (Fig. 5A). However, the total intracellular cAMP concentration and the cAMP gradient reflecting the difference in cAMP amount between leading edge and trailing edge was significantly higher in Mrp4−/− MEFs compared to Mrp4+/+ MEFs (Fig. 5B and 5C). Using a competitive immunoassay, we discovered that total PKA activity was significantly higher in cells lacking MRP4 (Fig. 6A). These data were supported by the phosphorylation status of the PKA substrate VASP in the absence of MRP4. Mrp4−/− MEFs exhibited higher levels of Ser157 phosphorylated VASP [a site specifically phosphorylated by PKA [14]] compared to wild-type cells (Fig. 6B). To monitor the PKA dynamics in real time, we made precise wounds on the confluent monolayers of MEF cells transfected with a FRET-based sensor of PKA activity, pmAKAR3 [3, 35]. For analysis, we selected cells expressing pmAKAR3 at the wound edge. By monitoring the FRET signal, we observed polarized PKA activity at the wound edge in both Mrp4+/+ and Mrp4−/− MEFs (Fig. 6C). We also found that the total intracellular PKA activity was higher in Mrp4−/− compared to Mrp4+/+ MEFs (Fig. 6D). Additionally, the difference in PKA activity between the leading edge and trailing edge was also significantly higher for Mrp4−/− MEFs than for Mrp4+/+ MEFs (Fig. 6E). These findings indicated that, in absence of MRP4, elevated levels of cAMP induce polarized PKA activation, which in turn, augments cell migration by facilitating the cortical actin network formation at the leading edge.
Figure 5. MRP4-deficient fibroblasts exhibit increased polarized cAMP.
Mrp4+/+ and Mrp4−/− MEFs were transfected with a FRET-based cAMP sensor. (A) The pseudocolor images of CFP/FRET with 60× magnification for Mrp4+/+ and Mrp4−/− MEFs expressing cAMP sensor are shown. Images in each panel were captured from the same field of view. Color bar shows the magnitude of the CFP/FRET. (B) Bar graph representing the CFP/FRET ratio indicating total intracellular cAMP levels in Mrp4+/+ and Mrp4−/− MEFs. (C) Bar graph representing the difference in CFP/FRET signal between the leading and trailing edges of Mrp4+/+ and Mrp4−/− MEFs. Error bars indicate SEM. All data represent at least three independent experiments. *, P < 0.05.
Figure 6. MRP4 deficient fibroblasts exhibit higher PKA activity at the wound edge.
(A) Bar graph representing total intracellular PKA activity in Mrp4+/+ and Mrp4−/− MEFs measured by competitive immunoassay. (B) Immunoblot data represented the phosphorylation status of VASP and bar graph represented the ratio of phosphorylated VASP to the total VASP. Mrp4+/+ and Mrp4−/− MEFs were transfected with a FRET-based PKA sensor pmAKAR. (C) The pseudocolor images of N-FRET with 60× magnification for Mrp4+/+ and Mrp4−/− MEFs expressing pmAKAR sensor were shown. Images in each panel were captured from the same field of view. Color bar shows the magnitude of the N-FRET. (D) Bar graph representing the N-FRET signal indicating total intracellular PKA activity in Mrp4+/+ and Mrp4−/− MEFs. (E) Bar graph representing the difference in N-FRET signal between the leading and trailing edges of Mrp4+/+ and Mrp4−/− MEFs. Error bars indicate SEM. All data represent at least three independent experiments. *, P < 0.05.
1.3.5 Disruption of the actin cytoskeleton abrogates the effect of MRP4 on cell migration
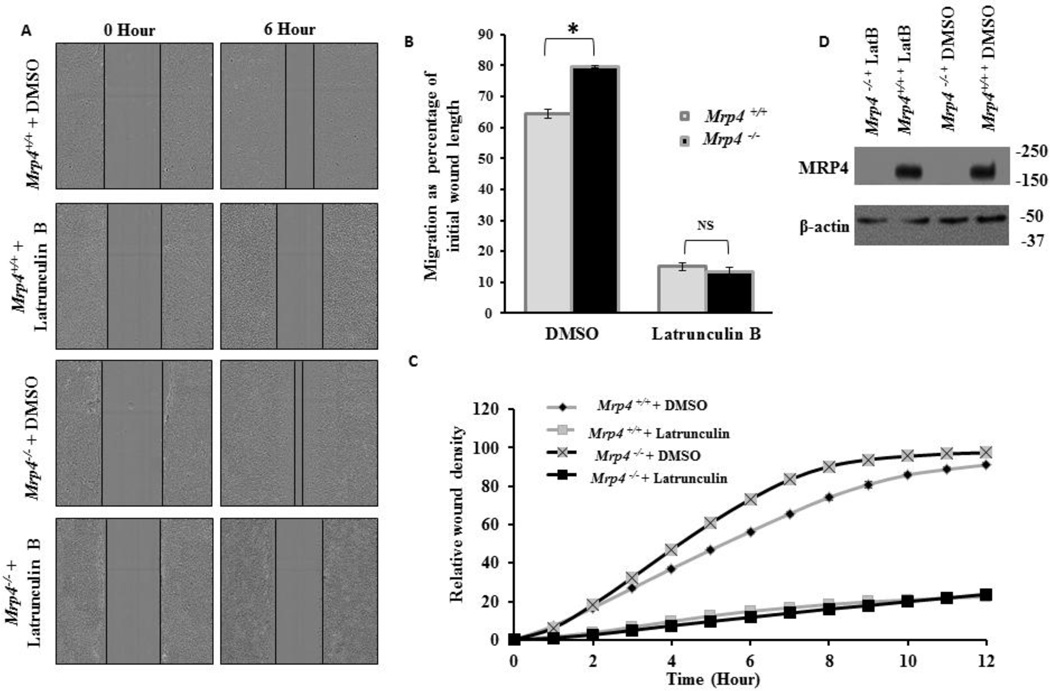
The altered actin dynamics in MRP4-deficient cells suggested that the effect of MRP4 on cell migration is actin dependent. Therefore, we investigated the effect of actin disruption using Lat B on MRP4-dependent cell migration. In agreement with our previous study, the in vitro wound-healing assay revealed that Mrp4−/− MEFs move significantly faster compare to Mrp4+/+ MEFs. However, after Lat B treatment, both cell types migrated at a similar rate that was much slower than the basal migration rate for wild-type fibroblasts (Fig. 7A and 7B). In this assay, cell migration was monitored for 6 h and was represented as a percentage of initial wound length. High-content microscopy was used to confirm the effect of Lat B on fibroblast migration in vitro. After making precise wounds in the confluent monolayers of MEFs, cells were treated with DMSO or Lat B and images were taken at 1-h intervals for 12 h. We did not continue the assay for a longer time as the effect of Lat B was not sustained after 12 h. At each time point, we measured a self-normalized parameter called relative wound density (RWD) that indicates the spatial cell density inside the wound area relative to the spatial cell density outside the wound area. Although the basal RWD kinetics were higher for Mrp4−/− MEFs, Lat B treatment reduced the overall migration rate such that both types of MEFs exhibited similar RWD kinetics during the entire period of time (Fig. 7C). However, our immunoblot data suggested that Lat B has no effect on MRP4 expression in MEFs (Fig. 7D). Altogether, our data indicate that actin is the downstream target of MRP4 and therefore, disruption of the actin cytoskeleton abrogates the effect of MRP4-mediated cell migration.
Figure 7. Disruption of the actin cytoskeleton abrogates the effect of MRP4 on cell migration.
(A) A conventional wound-healing assay was performed on Mrp4+/+ and Mrp4−/− MEFs treated with DMSO (control) or Lat B, and images were taken (10× magnification) at 0 and 6 hours. (B) The initial and final wound lengths were calculated. Bar graph representing cell migrations as a percentage of initial wound length. Error bars indicate SEM. (C) Mrp4+/+ and Mrp4−/− MEFs were grown on fibronectin-coated dishes, and precise wounds were made. The kinetics of the relative wound density were analyzed after treatment with DMSO or Lat B. (D) Immunoblots showing the expression of MRP4 in Mrp4+/+ and Mrp4−/− MEFs treated with DMSO or Lat B. All data represent at least three independent experiments. *, indicates P < 0.05.
1.3.6. Inhibition of PKA attenuates the effect of MRP4 on cell migration by alteration of cortical actin dynamics
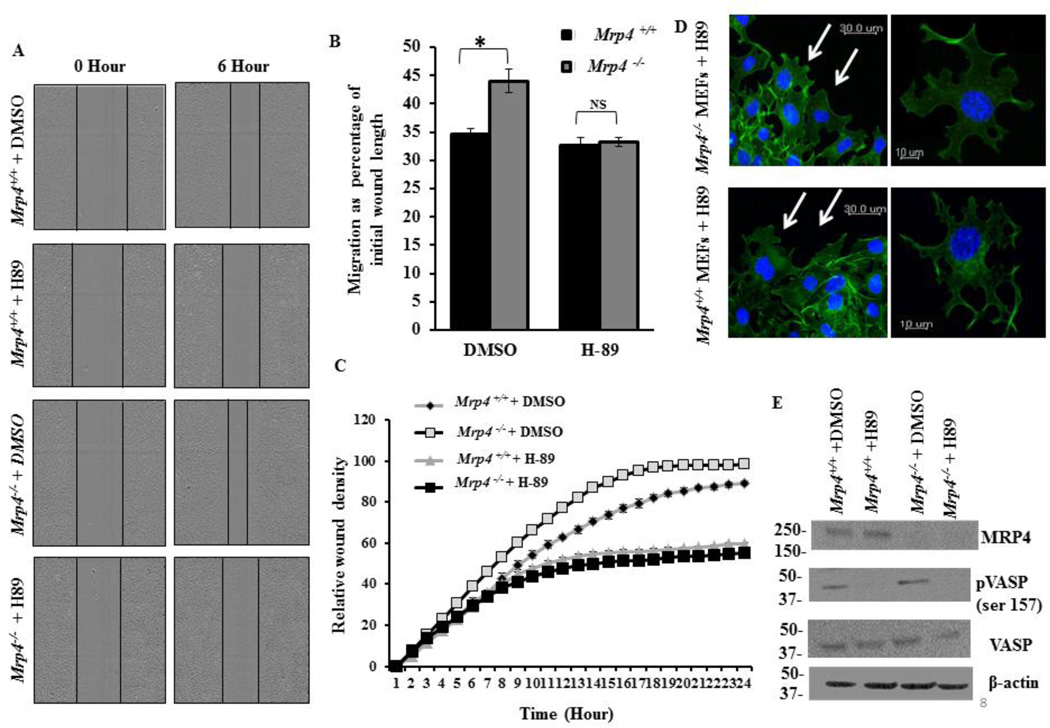
When we observed that MRP4 affects cell migration and modulates actin polymerization at a migrating cell’s leading edge, we hypothesized that PKA may act as an intermediate effector protein for transmitting the effect of MRP4 on cortical actin dynamics. Using an in vitro wound-healing assay, we found that inhibition of PKA by 50 µM of N-[2-p-bromocinnamylamino-ethyl]-5-isoquinolinesulphonamide (H-89) [36] attenuated the effect of MRP4 on cell migration (Fig. 8A), and after 6 h of H-89 treatment, both Mrp4+/+ and Mrp4−/− MEFs migrated at a similar rate (Fig. 8B). Using high-content microscopy to monitor the RWD kinetics for 24 hours, we found that H-89-treated cells exhibited significantly slower RWD kinetics compared to DMSO-treated controls. However the RWD kinetics for H-89-treated Mrp4+/+ and Mrp4−/− MEFs were identical for the entire 24 h period of time (Fig. 8C). To understand the inhibitory effect of H-89 on cell migration, we treated wounded monolayers of Mrp4+/+ and Mrp4−/− MEFs with H-89 and after 4 h, cells were fixed and stained with Alexa Fluor® 488-conjugated phalloidin antibody. The resulting images revealed that inhibition of PKA diminished cortical actin formation at the wound edge, and both Mrp4+/+ and Mrp4−/− MEFs showed similar, but reduced actin polymerization after H-89 treatment. Importantly, inhibition of PKA induced the formation of multiple pseudopods for both of the cell types, denoting that PKA activity is essential for promoting localized protrusion formation particularly in the direction of cell migration (Fig. 8D). Our immunoblot data confirmed that H-89 inhibits the PKA activity in MEFs, since diminished levels of phosphorylated VASP were detected in H-89-treated Mrp4+/+ and Mrp4−/− fibroblasts (Fig. 8E). Together, these results confirmed that PKA acts as a downstream target for MRP4 for executing its effect on actin polymerization and therefore on overall cell migration.
Figure 8. Inhibition of PKA attenuates the effect of MRP4 on cell migration by alteration of cortical actin dynamics.
(A) A conventional wound-healing assay was performed on Mrp4+/+ and Mrp4−/− MEFs treated with DMSO (control) or PKA inhibitor H-89, and images were taken (10× magnification) at 0 and 6 hours. (B) The initial and final wound lengths were calculated. Bar graph representing cell migrations as a percentage of initial wound length. Error bars indicate SEM. (C) Mrp4+/+ and Mrp4−/− MEFs MEFs were grown on fibronectin-coated, and precise wounds were made. The kinetics of the relative wound density were analyzed after treatment with DMSO or H-89. (D) Actin dynamics at the wound edges of migrating Mrp4+/+ and Mrp4−/− MEFs by confocal fluorescence microscopy (20× objective) are shown using phalloidin antibody (green). (E) Immuneoblots showing the expression of MRP4, phosphorylated VASP and total VASP in Mrp4+/+ and Mrp4−/− MEFs treated with DMSO or H-89. All data represent at least three independent experiments. *, indicates P < 0.05.
1.4 DISCUSSION
MRP4 is a membrane protein that was first identified in T-lymphoid cell lines as the efflux transporter of nucleoside-based antiviral drugs [37, 38]. However, MRP4 since has been reported to transport various endogenous substrates, particularly cAMP, with a high affinity (Km = 45 µM) and therefore, plays a pivotal role in regulating some cAMP-dependent signaling processes in normal physiological and pathological conditions [14, 39]. We recently demonstrated that MRP4-deficient fibroblasts contain higher levels of cyclic nucleotides and can migrate significantly faster than can wild-type fibroblasts [15]. Cell migration is important for embryonic development, wound healing, and the immune response, and a defective cell migration may lead to various pathologic conditions such as vascular diseases, tumor formation and cancer metastasis [1, 30, 40]. Establishment of cell polarity is indispensable for directional cell migration, and restricted activation of PKA at the leading edge of the cell is a key initiator ensuring efficient directional cell migration [3, 4]. We also found that the absence of MRP4 elevates the cAMP concentration at the leading edge, inducing polarized PKA in the migrating fibroblast, which augments the overall cell migration rate. Studies have shown that at the leading edge, PKA activates small GTPases such as Rac and Cdc42, which are important for lamellipodia and filopodia formation, respectively, during cell migration [2]. Phosphorylation-activated Rac induces the WASP/WAVE-mediated activation of Apr 2/3 and promotes the formation of dendritic-actin-network-containing lamellipodia [2, 11, 18]. Additionally PKA-dependent phosphorylation of VASP can also regulate actin polymerization and hence, can control protrusion formation during cell migration. In the current study, we purified a MRP4 protein complex from HEK293 cells and using a network- and systems biology-based approach, we developed a comprehensive understanding of the MRP4 interactome and related this to migratory signaling events. In this migration-related network, we observed an interaction between actin and MRP4 and showed that actin restricted MRP4 to specific plasma-membrane microdomains and disruption of the actin cytoskeleton caused a nearly two-fold increase in lateral diffusion of MRP4 at the cell surface. While investigating the effect of MRP4 on actin dynamics, we observed that migrating Mrp4−/− MEFs exhibit a higher degree of actin polymerization and therefore a more intense cortical actin band formation at or near the leading edge. The simultaneous localization of MRP4 at the lamellipodia suggested a novel role for MRP4 in regulating cytoskeleton dynamics and actin rearrangement at the cell protrusions.
Questions remain, such as how does MRP4 regulate actin cytoskeleton? Because inhibition of MRP4 leads to a polarized elevation of cAMP in migrating cells [15] and cAMP/PKA regulates actin-based cell migration [18], we assessed the difference in cAMP dynamics between Mrp4+/+ and Mrp4−/− fibroblasts. We used a PKA-based FRET sensor to monitor the spatiotemporal dynamics of cAMP in the migrating fibroblasts at the wound’s edge. In agreement with previous studies, we observed elevated cAMP levels at the wound edges for both types of migrating fibroblasts [3, 15]. In addition, not only was the total intracellular cAMP greater in Mrp4−/− MEFs, but the polarized gradient of cAMP was also significantly higher compared to the Mrp4+/+ fibroblasts. Using a FRET-based membrane-bound PKA sensor, pmAKAR3, we found that the PKA activity was also polarized to a greater extent and significantly higher in Mrp4−/− MEFs. These data indicated that, in absence of MRP4, the increased accumulation of cAMP at the leading edge activates more PKA, which in turn enhances formation of a cell’s structured extension by inducing actin polymerization and improving the overall rate of cell migration. We confirmed this role for MRP4 by studying the contribution of MRP4 and actin in cell migration using a wound-healing assay. We observed that disruption of the actin cytoskeleton, while not affecting MRP4 expression, abrogated the influence of MRP4 on cell migration. To establish the role of PKA as a possible intermediate effector protein in this process of regulation, we also checked for the effect of PKA inhibition on fibroblast migration. In Mrp4−/− fibroblasts, the higher levels of cAMP led to a high PKA activity, and thereby elevated VASP phosphorylation at the Ser157 residue. However, the PKA inhibitor H-89 treatment eliminated phosphorylated VASP expression in Mrp4+/+ and Mrp4−/− MEFs without affecting total VASP levels. We found that after inhibition of PKA, both Mrp4+/+ and Mrp4−/− MEFs migrated with a rate similar to the untreated wild-type MEFs after 6 h. Although the migration rate further reduced after that, both Mrp4+/+ and Mrp4−/− MEFs maintained similar RWD kinetics. When we assessed the actin dynamics in cells treated with H-89, we observed a reduced cortical actin formation at the wound edges for both of the cell types. Importantly, the inhibition of PKA induced multiple pseudopod formations and impaired directionality in both Mrp4+/+ and Mrp4−/− MEFs, informing a pivotal role for PKA in promoting polarization and localized protrusion formation during the course of directional migration.
Together, our data suggest that inhibition of MRP4 elevates cAMP and therefore activates more PKA at or near the leading edge of a migrating fibroblast cell (Fig. 9). This segregated and confined PKA activation enhanced cell polarization and is the early hallmark step in directional cell migration. However, in the absence of MRP4-mediated cAMP efflux, the augmented PKA activity stimulated actin polymerization probably via VASP phosphorylation or small-GTPase activation and facilitated the formation of restricted and localized structured extensions specifically in the direction of cell migration.
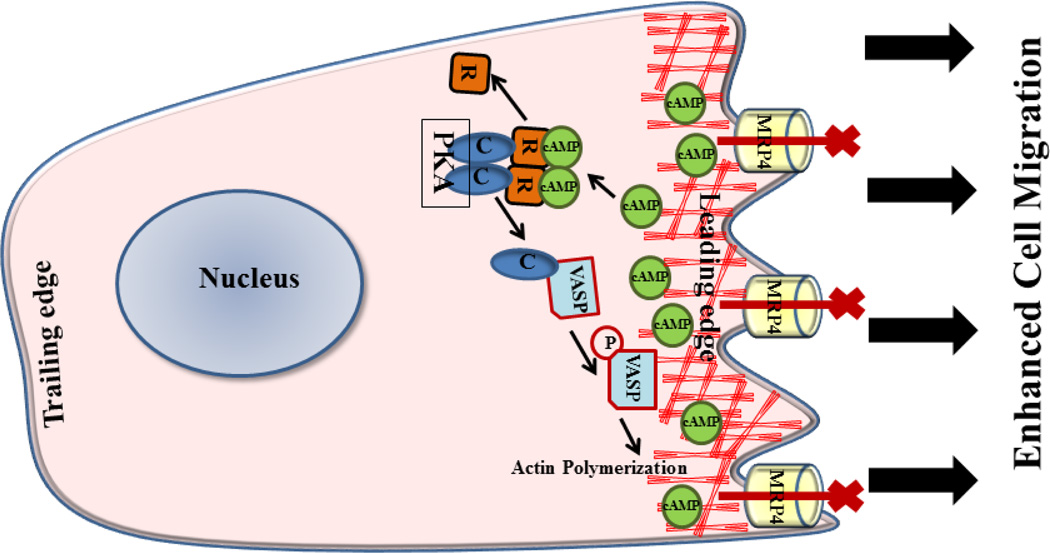
Figure 9. Schematic illustration of mechanisms of MRP4-dependent fibroblast migration.
Inhibition of MRP4 increases intracellular cAMP levels primarily near the leading edge of a migrating fibroblast, which in turn activates PKA. Activated PKA induces phosphorylation of VASP, which enhances polymerization of the cortical actin at the leading edge and facilitates cell migration.
1.5 CONCLUSIONS
In conclusion our study demonstrated a novel cAMP-dependent mechanism for MRP4-mediated regulation of fibroblast migration and indicated that PKA and actin play important roles as downstream effectors. Further study is still required to identify the intermediate molecules that carry on the effect of PKA on actin dynamics and unravel the entire molecular mechanism behind MRP4-dependent cell migration.
Supplementary Material
HIGHLIGHTS.
-
➢
MRP4 deficient MEFs migrate faster than the wild type fibroblasts.
-
➢
Actin interacts with MRP4 and restricts MRP4 to specific subcellular domains.
-
➢
Inhibition of MRP4 induces cAMP-dependent polarized PKA activity.
-
➢
Mrp4−/− MEFs contain higher level of cortical actin at the leading edge.
-
➢
PKA and actin are involved in MRP4-dependent regulation of fibroblast migration.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grants R01-DK080834 and R01-DK093045 (APN) and R01-GM60904, P30-CA21745, and CA21865 (JDS) and by the American Lebanese Syrian Associated Charities (JDS). The authors thank J. Denise Wetzel, CCHMC Medical Writer, for review and editing of the manuscript.
Abbreviations
- MEFs
mouse embryo fibroblasts
- MRP4
multidrug resistance protein 4
- PKA
protein kinase A
- Rac
Ras-related C3 botulinum toxin substrate
- SPT
single-particle tracking
- VASP
vasodilator-stimulated phosphoprotein
- WASP
Wiskott-Aldrich syndrome proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Lim CJ, Kain KH, Tkachenko E, Goldfinger LE, Gutierrez E, Allen MD, Groisman A, Zhang J, Ginsberg MH. Integrin-mediated protein kinase A activation at the leading edge of migrating cells. Mol Biol Cell. 2008;19:4930–4941. doi: 10.1091/mbc.E08-06-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe AK, Baldor LC, Hogan BP. Spatial regulation of the cAMP-dependent protein kinase during chemotactic cell migration. Proc Natl Acad Sci U S A. 2005;102:14320–14325. doi: 10.1073/pnas.0507072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha C, Arora K, Moon CS, Yarlagadda S, Woodrooffe K, Naren AP. Forster Resonance Energy Transfer - An approach to visualize the spatiotemporal regulation of macromolecular complex formation and compartmentalized cell signaling. Biochim Biophys Acta. 2014;1840:3067–3072. doi: 10.1016/j.bbagen.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newell-Litwa KA, Horwitz AR. Cell migration: PKA and RhoA set the pace. Curr Biol. 2011;21:R596–R598. doi: 10.1016/j.cub.2011.06.032. [DOI] [PubMed] [Google Scholar]
- 7.Paulucci-Holthauzen AA, Vergara LA, Bellot LJ, Canton D, Scott JD, O'Connor KL. Spatial distribution of protein kinase A activity during cell migration is mediated by A-kinase anchoring protein AKAP Lbc. J Biol Chem. 2009;284:5956–5967. doi: 10.1074/jbc.M805606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora K, Sinha C, Zhang W, Ren A, Moon CS, Yarlagadda S, Naren AP. Compartmentalization of cyclic nucleotide signaling: a question of when, where, and why? Pflugers Arch. 2013;465:1397–1407. doi: 10.1007/s00424-013-1280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 10.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Weaver AM, Young ME, Lee WL, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23–30. doi: 10.1016/s0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- 12.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 13.Guo D, Tan YC, Wang D, Madhusoodanan KS, Zheng Y, Maack T, Zhang JJ, Huang XY. A Rac-cGMP signaling pathway. Cell. 2007;128:341–355. doi: 10.1016/j.cell.2006.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara Y, Sassi Y, Guibert C, Gambaryan N, Dorfmuller P, Eddahibi S, Lompre AM, Humbert M, Hulot JS. Inhibition of MRP4 prevents and reverses pulmonary hypertension in mice. J Clin Invest. 2011;121:2888–2897. doi: 10.1172/JCI45023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha C, Ren A, Arora K, Moon CS, Yarlagadda S, Zhang W, Cheepala SB, Schuetz JD, Naren AP. Multi-drug resistance protein 4 (MRP4)-mediated regulation of fibroblast cell migration reflects a dichotomous role of intracellular cyclic nucleotides. J Biol Chem. 2013;288:3786–3794. doi: 10.1074/jbc.M112.435925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostenko S, Johannessen M, Moens U. PKA-induced F-actin rearrangement requires phosphorylation of Hsp27 by the MAPKAP kinase MK5. Cell Signal. 2009;21:712–718. doi: 10.1016/j.cellsig.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Spurzem JR, Gupta J, Veys T, Kneifl KR, Rennard SI, Wyatt TA. Activation of protein kinase A accelerates bovine bronchial epithelial cell migration. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1108–L1116. doi: 10.1152/ajplung.00148.2001. [DOI] [PubMed] [Google Scholar]
- 18.Howe AK. Regulation of actin-based cell migration by cAMP/PKA. Biochim Biophys Acta. 2004;1692:159–174. doi: 10.1016/j.bbamcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh MC, Makena PS, Gorantla V, Sinclair SE, Waters CM. CXCR4 regulates migration of lung alveolar epithelial cells through activation of Rac1 and matrix metalloproteinase-2. Am J Physiol Lung Cell Mol Physiol. 2012;302:L846–L856. doi: 10.1152/ajplung.00321.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren A, Zhang W, Yarlagadda S, Sinha C, Arora K, Moon CS, Naren AP. MAST205 competes with cystic fibrosis transmembrane conductance regulator (CFTR)-associated ligand for binding to CFTR to regulate CFTR-mediated fluid transport. J Biol Chem. 2013;288:12325–12334. doi: 10.1074/jbc.M112.432724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Dandridge KS, Di A, Marrs KL, Harris EL, Roy K, Jackson JS, Makarova NV, Fujiwara Y, Farrar PL, Nelson DJ, Tigyi GJ, Naren AP. Lysophosphatidic acid inhibits cholera toxin-induced secretory diarrhea through CFTR-dependent protein interactions. J Exp Med. 2005;202:975–986. doi: 10.1084/jem.20050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penmatsa H, Zhang W, Yarlagadda S, Li C, Conoley VG, Yue J, Bahouth SW, Buddington RK, Zhang G, Nelson DJ, Sonecha MD, Manganiello V, Wine JJ, Naren AP. Compartmentalized cyclic adenosine 3',5'-monophosphate at the plasma membrane clusters PDE3A and cystic fibrosis transmembrane conductance regulator into microdomains. Mol Biol Cell. 2010;21:1097–1110. doi: 10.1091/mbc.E09-08-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galperin E, Sorkin A. Visualization of Rab5 activity in living cells by FRET microscopy and influence of plasma-membrane-targeted Rab5 on clathrin-dependent endocytosis. J Cell Sci. 2003;116:4799–4810. doi: 10.1242/jcs.00801. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Kinter M, Shank S, Cotton C, Kelley TJ, Ziady AG. Dysfunction of Nrf-2 in CF epithelia leads to excess intracellular H2O2 and inflammatory cytokine production. PLoS One. 2008;3:e3367. doi: 10.1371/journal.pone.0003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Shank S, Davis PB, Ziady AG. Nucleolin-mediated cellular trafficking of DNA nanoparticle is lipid raft and microtubule dependent and can be modulated by glucocorticoid. Mol Ther. 2011;19:93–102. doi: 10.1038/mt.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziady AG, Kinter M. Protein sequencing with tandem mass spectrometry. Methods Mol Biol. 2009;544:325–341. doi: 10.1007/978-1-59745-483-4_21. [DOI] [PubMed] [Google Scholar]
- 27.Ziady AG, Sokolow A, Shank S, Corey D, Myers R, Plafker S, Kelley TJ. Interaction with CREB binding protein modulates the activities of Nrf2 and NF-kappaB in cystic fibrosis airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2012;302:L1221–L1231. doi: 10.1152/ajplung.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purohit JS, Hu P, Chen G, Whelan J, Moustaid-Moussa N, Zhao L. Activation of nucleotide oligomerization domain containing protein 1 induces lipolysis through NF-kappaB and the lipolytic PKA activation in 3T3-L1 adipocytes. Biochem Cell Biol. 2013;91:428–434. doi: 10.1139/bcb-2013-0049. [DOI] [PubMed] [Google Scholar]
- 29.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 30.Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J Cell Sci. 2005;118:4917–4919. doi: 10.1242/jcs.02662. [DOI] [PubMed] [Google Scholar]
- 31.Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 32.Zigmond SH. Beginning and ending an actin filament: control at the barbed end. Curr Top Dev Biol. 2004;63:145–188. doi: 10.1016/S0070-2153(04)63005-5. [DOI] [PubMed] [Google Scholar]
- 33.Jin S, Haggie PM, Verkman AS. Single-particle tracking of membrane protein diffusion in a potential: simulation, detection, and application to confined diffusion of CFTR Cl- channels. Biophys J. 2007;93:1079–1088. doi: 10.1529/biophysj.106.102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Lagerholm BC, Yang B, Jacobson K. Methods to measure the lateral diffusion of membrane lipids and proteins. Methods. 2006;39:147–153. doi: 10.1016/j.ymeth.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Sachs BD, Baillie GS, McCall JR, Passino MA, Schachtrup C, Wallace DA, Dunlop AJ, MacKenzie KF, Klussmann E, Lynch MJ, Sikorski SL, Nuriel T, Tsigelny I, Zhang J, Houslay MD, Chao MV, Akassoglou K. p75 neurotrophin receptor regulates tissue fibrosis through inhibition of plasminogen activation via a PDE4/cAMP/PKA pathway. J Cell Biol. 2007;177:1119–1132. doi: 10.1083/jcb.200701040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Candas M, Griko NB, Taussig R, Bulla LA., Jr A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc Natl Acad Sci U S A. 2006;103:9897–9902. doi: 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuetz JD, Connelly MC, Sun D, Paibir SG, Flynn PM, Srinivas RV, Kumar A, Fridland A. MRP4: A previously unidentified factor in resistance to nucleoside-based antiviral drugs. Nat Med. 1999;5:1048–1051. doi: 10.1038/12487. [DOI] [PubMed] [Google Scholar]
- 38.Russel FG, Koenderink JB, Masereeuw R. Multidrug resistance protein 4 (MRP4/ABCC4): a versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol Sci. 2008;29:200–207. doi: 10.1016/j.tips.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Cheepala S, Hulot JS, Morgan JA, Sassi Y, Zhang W, Naren AP, Schuetz JD. Cyclic nucleotide compartmentalization: contributions of phosphodiesterases and ATP-binding cassette transporters. Annu Rev Pharmacol Toxicol. 2013;53:231–253. doi: 10.1146/annurev-pharmtox-010611-134609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackay CR. Moving targets: cell migration inhibitors as new anti-inflammatory therapies. Nat Immunol. 2008;9:988–998. doi: 10.1038/ni.f.210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.