Abstract
HOXC6 is a homeobox-containing gene associated with mammary gland development and is overexpressed in variety of cancers including breast and prostate cancers. Here, we have examined the expression of HOXC6 in breast cancer tissue, investigated its transcriptional regulation via estradiol (E2) and bisphenol-A (BPA, an estrogenic endocrine disruptor) in vitro and in vivo. We observed that HOXC6 is differentially over-expressed in breast cancer tissue. E2 induces HOXC6 expression in cultured breast cancer cells and in mammary glands of Sprague Dawley rats. HOXC6 expression is also induced upon exposure to BPA both in vitro and in vivo. Estrogen-receptor-alpha (ERα) and ER-coregulators such as MLL-histone methylases are bound to the HOXC6 promoter upon exposure to E2 or BPA and that resulted in increased histone H3K4-trimethylation, histone acetylation, and recruitment of RNA polymerase II at the HOXC6 promoter. HOXC6 overexpression induces expression of tumor growth factors and facilitates growth 3D-colony formation, indicating its potential roles in tumor growth. Our studies demonstrate that HOXC6, which is a critical player in mammary gland development, is upregulated in multiple cases of breast cancer, and is transcriptionally regulated by E2 and BPA, in vitro and in vivo.
Introduction
Homeobox containing genes (HOX genes) are evolutionarily conserved family of genes that are critical players in embryogenesis and fetal development (1, 2). Each HOX protein contains a highly conserved homeodomain through which it binds DNA, recognizes gene promoters and regulates transcription of a variety of target genes (3). Though HOX genes were primarily considered as developmental genes associated with embryogenesis, recent studies demonstrate that they are also involved in postnatal development and are expressed in a variety of adult tissues (4, 5). The human genome contains 39 HOX genes that are arranged in four different clusters (and 13 paralogs): HOXA, HOXB, HOXC, and HOXD loci, which are located in chromosomes 7p15.3, 17q21.3, 12q13.3, and 2q31, respectively (1, 6). HOX gene expression specifies the identity of the body segments along the anterior-posterior axis and secondary axis in animals (7). Recent studies demonstrate that HOX gene expressions are disrupted in various cancers (8–17). Many HOX genes appear to play critical roles in tumor cell proliferation, metastasis, and angiogenesis and the level of expression of these genes is correlated with patient outcome (17–20). Thus, HOX genes are emerging as key players in variety of cancer and are potential targets for disease diagnosis and novel therapy.
HOXC6 is one of the 39 HOX genes present in human and is associated with mammary gland development and milk production (21, 22). HOXC6 null female mice show complete absence of mammary epithelium in thoracic glands and dilated ducts in inguinal glands (23). HOXC6 is crucial in the development and proliferation of epithelial cells in response to hormonal signals (24). HOXC6 overexpression has been detected in variety of cancers including breast (15, 21), lung (25) prostate (26), leukemia (27), osteosarcomas (28), and medulloblastomas (29). HOXC6 regulates proteins such as bone morphogenic protein 7 (BMP7), fibroblast growth factor receptor 2 (FGFR2), and platelet-derived growth factor receptor α (PDGFRA) that are crucial players in tumor cell proliferation, growth, and metastasis (30, 31). It also regulates the PI3K/Akt, Notch, and Wnt signaling pathways (30–35). Though mechanism is not clear, HOXC6 has been shown to be associated with both transcription repression (36) as well as activation of various genes (24, 37, 38). In a recent study, we demonstrated that HOXC6 expression is transcriptionally induced by estradiol (E2) in estrogen-receptor (ER) positive placenta choriocarcinoma cells (JAR) (15). Herein, we have investigated the potential association of HOXC6 with breast cancer and studied its regulatory mechanisms.
As HOXC6 is transcriptionally induced by E2 in JAR cells and is potentially associated with breast cancer, we further examined E2-mediated transcriptional regulation of HOXC6 in breast cancer cells in vitro and in vivo (using animal model). Furthermore, we also investigated if HOXC6 expression could be affected upon exposure to estrogenic endocrine disrupting chemicals such as bisphenol-A (BPA). BPA is a well-known environmental contaminant that is routinely used as a plasticizer in plastics. Similar to estradiol, BPA contains multiple phenolic groups and hence behaves as a synthetic estrogen (39–42). Once BPA enters in the body, it has potential to bind estrogen-receptors (ERs) and interfere with normal estrogen-signaling processes contributing towards various human diseases including reproductive and developmental defects and metabolic disorders (41, 43). BPA is a well-known estrogenic endocrine disruptor. Here, we show that HOXC6 is differentially overexpressed in breast cancer tissue and is transcriptionally induced by E2 and BPA both in vitro and in vivo.
Experimental section
Cell culture and treatment with E2 and BPA
MCF7 (ER-positive, mammary adenocarcinoma), T47D (ER-positive, ductal carcinoma mammary), and MDA-MB-231 (ER-negative, adenocarcinoma mammary) cells were obtained from ATCC and grown in DMEM (Dulbecco’s Modified Eagle’s Medium) containing 10 % FBS (fetal bovine serum), 2 mM L-glutamine and 1 % penicillin/streptomycin (100 unit and 0.1 mg/mL respectively) in presence of 5 % CO2 in a humidified incubator at 37°C (16, 41, 44). For the E2 and BPA-treatments, MCF7 cells were grown for two generations in phenol-red-free DMEM-F-12 supplemented with 10 % charcoal stripped FBS, 2 mM L-glutamine and penicillin/streptomycin (100 units and 0.1 mg/mL, respectively). Cells were grown to 60 % confluency and then treated with E2 or BPA. RNA and proteins were isolated and analyzed by real-time PCR (qPCR) and western blotting, respectively (14, 16, 41, 45–47).
Flag-HOXC6 stable cell line
Human full-length HOXC6 gene was cloned in a human expression construct pflag-CMV4. The Flag-HOXC6-CMV4 construct was transfected into HEK293 cells using Lipofectamine-2000 (Invitrogen) and stably transfected cells were isolated using G418 selection procedure as described by us earlier (16, 17). In brief, 5 µg of the Flag-HOXC6 construct was incubated with 8 µL of lipofectamine transfection reagent for 30 min in 500 µL DMEM at room temperature and the mixture was added to HEK293 cells (at 80% confluency). After 48 h, cells were washed with PBS, trypsinized and replated in medium supplemented with 1 mg/mL of G418 (Sigma). After about 3 weeks, stably transfected cells expressing Flag-HOXC6 were isolated, cultured separately grown and maintained in G418 containing media. Flag-HOXC6 overexpression was confirmed by Western blot using Flag-antibody.
RNA extraction, reverse transcription, and qPCR
For RNA extraction, cells were harvested by centrifugation at 700 rpm, the cell pellets were resuspended in diethyl pyrocarbonate (DEPC) treated buffer A (20 mM Tris-HCl, pH 7.9; 1.5 mM MgCl2; 10 mM KCl and 0.5 mM DTT; 0.2 mM PMSF), incubated on ice for 10 min, and centrifuged at 3500 rpm for 5 min. The supernatant containing the RNA was subjected to phenol-chloroform extraction followed by ethanol precipitation of RNA by incubating 1h at −80 °C (17). Reverse transcription (RT) reactions were performed in a total volume of 25 µL of cDNA containing 500 ng of RNA, 2.4 µM of oligo dT (Promega), 100 units of MMLV reverse transcriptase, 1X first strand buffer (Promega), 100 µM each of dATP, dGTP, dCTP and dTTP (Invitrogen), 1 mM DTT, and 20 units of RNaseOut (Invitrogen). The cDNA was diluted to 100 µL. PCR was performed in a 10 µL reaction volume containing 5 µL diluted cDNA and gene specific primer pairs (Table 1). For qPCR analyses, the cDNA was amplified using SsoFast EvaGreen supermix (Bio-Rad) using CFX96 real-time PCR detection system. The qPCR results were analyzed using the CFX manager software. The experiments were repeated at least thrice with three replicates each time (16, 46–48).
Table 1.
Nucleotide sequence of primers
| Gene | Forward Primer (5'-3') | Reverse Primer (5'-3') |
|---|---|---|
| PCR primers | ||
| GAPDH | CAATGACCCCTTCATTGACC | GACAAGCTTCCCGTTCTCAG |
| hHOXC6 | CAGACCCTGGAACTGGAGAA | CTTCCCGCTTTTCCTCTTTT |
| rHOXC6 | GTTTTGTGCCCGGATCTCTA | CCGAGTTAGGTAGCGGTTGA |
| bFGF | TTCTTCCTGCGCATCCAC | CGGTTAGCACACACTCCTTT |
| VEGF | TCCACCATGCCAAGTGGTCC | TGGATGGCAGTAGCTGCGCT |
| PDGFRA | GAAGCTGTCAACCTGCATGA | CTTCCTTAGCACGGATCAGC |
| Antisense | ||
| ERα | CATGGTCATGGTCAGb | |
| Scramble | CGTTTGTCCCTCCAGCATCTb | |
| Cloning primers | ||
| hHOXC6 ERE1 | AGCCTCATAGCTCAGGTCCAa | CTCCTTCTCAGGACCCCTCTa |
| HOXC6-Flag-CMV4 | AAGCTTCTCTCAAACTGCAGACAAAACAa | GGATCCTCACTCTTTCTGCTTCTCCTCTTCTa |
cloning primers flanked by appropriate restriction sites
Phosphodiester linkages replaced by phosphorothioate linkages
Immunohistological analysis of breast cancer tissue microarray
The breast cancer tissue microarray slide containing six different cases (duplicates of each) of breast cancer and their corresponding adjacent normal tissues were purchased from US Biomax Inc. and subjected to immunohistological staining. Prior to staining, the paraffin embedded tissue microarray slides were immersed twice in xylene (for 10 min) and then sequentially immersed in 100%, 95% and 70 % ethanol (5 min each) to deparafinize the tissue. Antigen retrieval was carried out by incubating the slides in 0.01 M sodium citrate buffer at 95 °C for 15 min according to the supplier's instructions. For immunohistological staining, the tissue microarray slide was incubated with 3% H2O2 for 15 min, washed with PBS thrice, and then blocked with blocking buffer containing donkey serum. The slides were then incubated with HOXC6 antibody overnight, washed thrice in PBS, and then incubated with biotinylated donkey secondary antibody for 1.5 h. The slide was washed thrice with PBS, incubated with avidin-biotin complex (ABC, Vector laboratories) for 1.5 h, washed twice with PBS and then twice with 0.1 M Tris-HCl (pH 7.4). Next, the slides were incubated with diaminobenzidine (DAB) substrate (Vector laboratories) for peroxidase labeling. The tissue microarray slides were dehydrated with sequential immersion in 70%, 95%, and 100% ethanol and then cleaned by sequential incubation (1, 5 and 10 min) in CitriSolv clearing agent (Fisherband). Tissue sections were finally mounted with DPX mounting solution (Sigma), photographed, and examined under microscope (Nikon Eclipse TE2000-U, Japan) (14).
Dual luciferase reporter assay
The HOXC6 ERE located at −125 nt upstream to the transcription start site along with its flanking region (total 396 nt) was cloned and inserted upstream of the promoter of a fire fly luciferase gene in pGL3-promoter vector (Promega) (primers in Table 1). MCF7 cells were co-transfected with the ERE-pGL3 construct along with reporter plasmid containing, renilla luciferase (pRLTk, Promega) as an internal transfection control using lipofectamine transfection reagent (Invitrogen). Control transfections were done using empty pGL3 promoter vector without any ERE insertion. At 30 h post-transfection, cells were treated with 1 nM E2 or 10 nM BPA and incubated for an additional 8 h. Total protein was extracted and then subjected to luciferase assay using Dual-Glo Luciferase Assay System (Promega) as instructed and detected using a microplate reader (Flowstar-Omega) (14, 15, 41, 44, 48–50). Each treatment was done in four replicates and the experiment was repeated at least twice.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using EZ Chip kit (Upstate) as described by us previously (51). For ChIP assay, cells were fixed in 4% formaldehyde for 15 min at room temperature, excess formaldehyde was quenched with 1.25 M glycine, washed twice with cold PBS containing protease inhibitor cocktail (5 µL.mL−1) and PMSF (0.2 mM). The cells were then lysed in SDS lysis buffer and sonicated to shear the chromatin to ~150–450 nt in length. The sonicated chromatin was pre-cleaned using protein G agarose (Millipore) beads and subjected to immuno-precipitation using antibodies specific to ERα (Santa cruz; 2Q418), H3K4-trimethyl (EMD-Millipore; 07-473), RNAPII (Abcam; ab5408), MLL1 (Abgent; AO1146a), MLL2 (Bethyl laboratories; A300-113A), MLL3 (Abgent; AP6184a), MLL4 (Sigma; AV33704), CBP (Santa Cruz, sc-369), p300 (Santa Cruz; sc-585), N-CoR (Santa Cruz; sc-1609), and histone-acetylation (EMD-Millipore; 06-599). The ChIP chromatin was deproteinized by incubating at 65°C in presence of NaCl, followed by incubation with proteinase K (Sigma) to obtain purified DNA fragments. The immuno-precipitated DNA was PCR-amplified using HOXC6 promoter specific primers (shown in Table 1) to analyze the level of recruitment of different proteins (45).
3D-colony formation assay (soft agar assay)
Colony forming ability of Flag-HOXC6 overexpressed cells were assessed using soft agar method using a similar protocol described by us earlier (16, 17). In brief, 1.2 % agar was prepared in PBS (pH 7.4), autoclaved, cooled down (to ~40°C) and added to an equal volume of 2X DMEM with 20 % FBS, 4 mM L-glutamine and 2% Penicillin/Streptomycin (100 unit and 0.1 mg/mL, respectively) to form the base agar layer. The base agar layer mixture was plated onto 6 well culture plates (16, 17), cooled to room temperature and the base agar layer was solidified. Then ~5000 HEK293 (control) or Flag-HOXC6 cells were added to a mixture of 0.5 mL 2X DMEM and 0.5 ml of 0.7 % agar (in PBS, pH 7.4) at room temperature and then plated on top of the base agar layer. The dishes were kept at room temperature for 15 min (to allow the top agar to solidify) and then incubated in the cell culture incubator under normal growth conditions. The cells were fed with 0.5 mL of normal growth media at 2 days interval for 21 days, until the colonies were visible. Prior to counting the colonies, the media was removed from the plates and the plates were rinsed with PBS, stained with 0.005 % crystal violet (2 h), then washed with PBS and colonies were examined under light microscope (16, 17).
Animal experiments
Subjects
Adult, 90 day old, experimentally naïve, female Sprague Dawley rats (n=12) were triple housed and maintained in a temperature and humidity-controlled environment under a 12 h reversed light/dark cycle with lights on at 7 p.m. and off at 7 a.m. All the animals were maintained and cared for in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. The University of Texas at Arlington’s Animal Care and Use Committee approved all experimental procedures and protocols. Notably, all the animals (controls and treatments) were maintained under same caging, bedding, food, and water bottles conditions to normalize effects of any exogenous source of estrogens in our results.
Ovariectomy
Sprague-Dawley rats were anesthetized with 2–3% isoflurane-oxygen vapor mixture and ovariectomized (OVX) as previously described by us (52, 53). The animals were allowed to recover for 4–5 days post-surgery, daily vaginal lavage testing was performed for eight consecutive days to confirm cessation of estrous cycling, thus verifying the completion of the OVX procedure. All OVXs performed were confirmed complete and thus, no animals were eliminated on the basis of an incomplete procedure (54). Treatments with E2 and BPA: BPA (Bisphenol-A): 10 mg of BPA was dissolved in 1 mL of ethanol to create a stock solution that was stored at −4 °C. Estradiol (E2; 17β-estradiol) was dissolved in peanut oil to yield final concentrations of 5 µg/mL. BPA was dissolved in ethanol and brought up to a final concentration of 50 µg/mL with saline. Rats were given subcutaneous injections of either BPA (25 µg/kg) or E2 (5 µg/kg), (n=4) 24 and 4 hours prior to sacrifice. Animals were sacrificed via rapid decapitation and mammary gland tissue was collected from each rat and flash frozen on dry ice and then stored at −80 °C) until RNA extraction (52, 53, 55). RNA and protein extraction: For RNA extraction, the flash frozen mammary glands were homogenized. RNA extraction was carried out using a ZyGEM kit (Hamilton, New Zealand) according to the manufacturer’s protocol. The RNA was reverse transcribed to cDNA and subjected to qPCR using rat-specific primers (Table 1) (46). Protein extraction of the excised mammary glands was performed using ZyGEM kit (Hamilton, New Zealand). The protein extracts were analyzed using western blots (46, 47).
Statistical analysis
Each experiment was performed in two to three replicates and then cells were pooled together and treated as one sample, subjected to RNA extraction, RT-PCR and ChIP analyses and each experiment was repeated at least three times (n = 3). The qPCR analyses were done in three parallel replicate reactions and repeated thrice (n = 3). For luciferase assay, each treatment was carried out in four parallel replicates and the experiment was repeated at least twice (n = 2). Normally distributed data was analyzed by ANOVA and non-normally distributed data were analyzed using student-t tests (SPSS) to determine the level of significance between individual treatments. The treatments were considered significantly different at p≤ 0.05 (42).
Results
HOXC6 is overexpressed in breast carcinomas and in ER-positive breast cancer cells
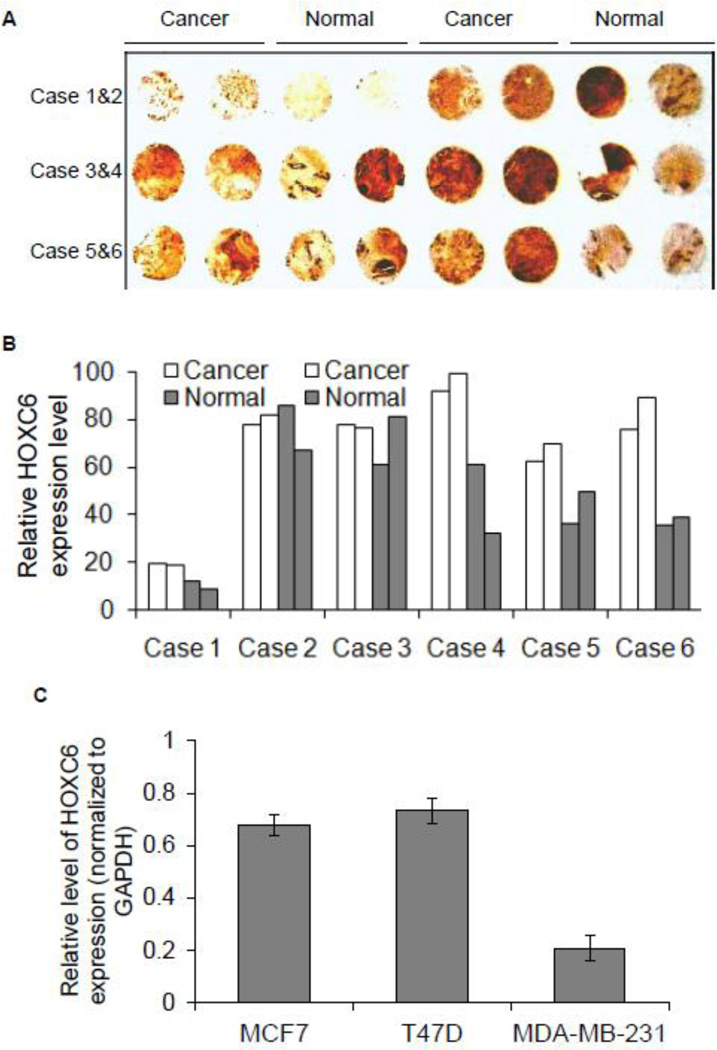
HOXC6 is known to be associated with mammary gland development and our previous studies demontrated that HOXC6 is transcriptionally regulated by estrogen in placenta choricarcinoma cells (15). To further understand the expression and transcriptional regulation of HOXC6 in breast carcinomas, initially we examined its expression in different breast cancer tissue samples (Figures 1A–B). In brief, we performed immunohistochemical staining (DAB staining) of breast cancer tissue microarray that contains six cases of breast cancer (in duplicates) along with corresponding adjacent normal tissue, using HOXC6 antibody. HOXC6 appeared to be overexpressed in some of the breast cancer tissue samples in comparison to its corresponding adjacent normal breast tissues (such as cases 4, 5 and 6, Figure 1A, quantification is shown in 1B). To further understand its mechanism of gene expression, we examined its expression levels in different types of breast cancer cells such as estrogen-receptor (ER)-positive (MCF7 and T47D) and ER-negative (MDA-MB-231) breast cancer cells (Figure 1C). Briefly, RNA was isolated from different cell lines, reverse-transcribed and subjected to qPCR analyses for expression of HOXC6 (relative to GAPDH, as control). These analyses demonstrate that HOXC6 expression is relatively higher in ER-positive breast cancer cells (MCF7 and T47D) in comparison to the ER-negative MDA-MB-231 cells (Figure 1C). These analyses indicated that HOXC6 expression is potentially upregulated in breast cancer and regulated by ERs in breast cancer cells.
Figure 1.
HOXC6 expression in breast cancer cells and tissues. (Panel A-C) Immunohistological analyses of HOXC6 expression in breast cancer tissues: Human breast cancer tissue microarray (6 cases of breast cancer along with their matched adjacent normal breast tissues) were immunostained (DAB staining) with HOXC6 antibody (panel A) and the relative quantification of HOXC6 expression is shown in panel B. (C) RNA from different breast cancer cells (MCF7, T47D, and MDA-MB-231) was reverse transcribed and analyzed by qPCR using primers specific to HOXC6. HOXC6 expression relative to GAPDH is plotted. Each experiment was repeated at least thrice with three parallel replicates (n=3). Bars indicate standard errors.
HOXC6 expression is induced by E2 and BPA in breast cancer cells
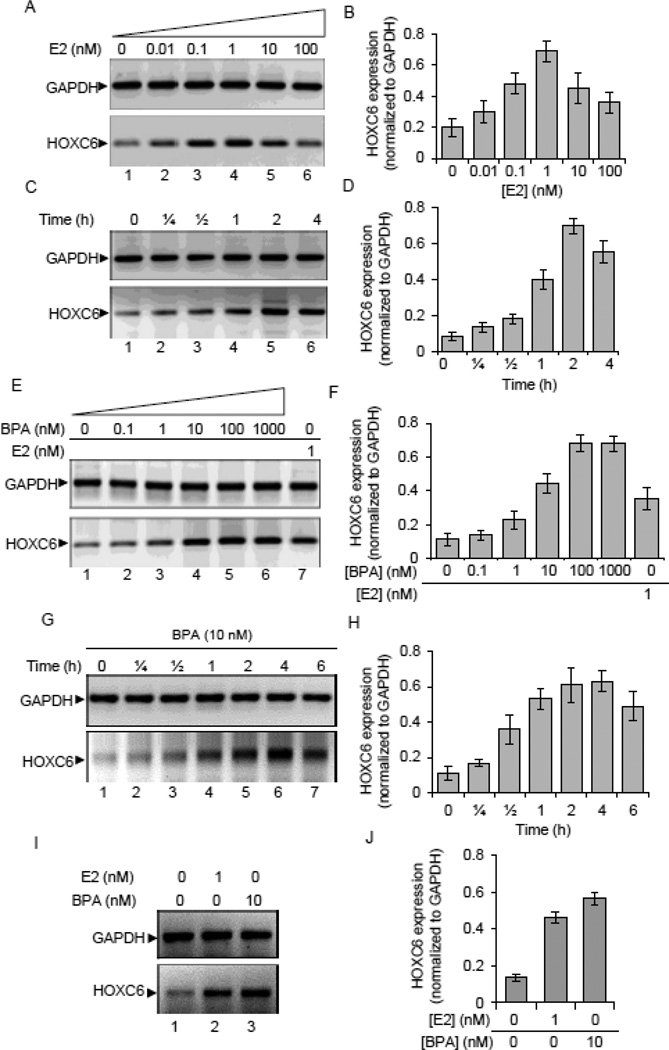
As HOXC6 is overexpressed in multiple cases of breast cancer tissues and in cultured ER-positive breast cancer cells, we examined its transcriptional regulation by E2 in ER-positive breast cancer cells MCF7. Furthermore, to investigate the potential endocrine disruption of HOXC6, we also examined the expression of HOXC6 upon exposure to a well-known estrogenic endocrine disruptor, BPA. Briefly, MCF7 cells that were grown in phenol-red-free media, treated with varying concentrations of E2 and BPA. The RNA from the control, E2 and BPA treated samples were reverse-transcribed and analyzed by qPCR and regular RT-PCR. This analysis showed that HOXC6 transcription is induced upon treatment with E2 (Figure 2A–B) and BPA in a concentration dependent manner in MCF7 cells (Figure 2E–F). The highest levels of HOXC6 induction was observed at 1 nM of E2 (6.9 fold increase) and 10 nM of BPA (4.16 fold increase) (Figures 2A–B and E–F). The time-dependent analysis showed that HOXC6 induction in MCF7 cells was optimum at 4 h post-treatment with 1 nM E2 or 10 nM BPA (Figures 2C–D and G–H). These studies demonstrate that HOXC6 is an E2-regulated gene and exposure to estrogenic endocrine disrupting chemicals (such as BPA) may induce its expression even in the absence of E2. Notably, exposure to E2 and BPA also induced the expression of HOXC6 in other ER-positive breast cancer cells such as T47D (Figure 2I–J), but not in ER-negative cell line such as MDA-MB-231(data not shown). The induction in HOXC6 expression by E2 and BPA in ER-positive breast cancer cells (MCF7 and T47D) indicate its potential regulation by E2 and BPA mediated via ER.
Figure 2.
Effects of E2 and BPA on HOXC6 expression in MCF7 cells. MCF7 cells grown in phenol red free DMEM-F-12 were treated with varying concentrations (or time) of E2 or BPA. RNA from the control and E2 or BPA-treated cells were analyzed by RT-PCR and qPCR using HOXC6 specific primers. GAPDH was used as the loading control. Panel A-B: RT-PCR and qPCR analysis of HOXC6 expression upon exposure varying concentrations of E2, respectively. Panel C-D: RT-PCR and qPCR analysis of HOXC6 expression upon exposure to 1 nM E2 for varying time periods. Panel E-F: RT-PCR and qPCR analysis of HOXC6 expression upon exposure varying concentrations of BPA, respectively. Panel G-H: RT-PCR and qPCR analysis of HOXC6 expression upon exposure to 10 nM BPA for varying time periods. Panel I-J; RT-PCR and qPCR analyses of HOXC6 expression in T47D cells upon exposure to 1nM E2 and 10 nM BPA. Each qPCR experiment was repeated for thrice times with three parallel replicates (n = 3). Bars indicate standard errors (p < 0.05).
BPA induced the expression of HOXC6, in vivo; rat mammary glands
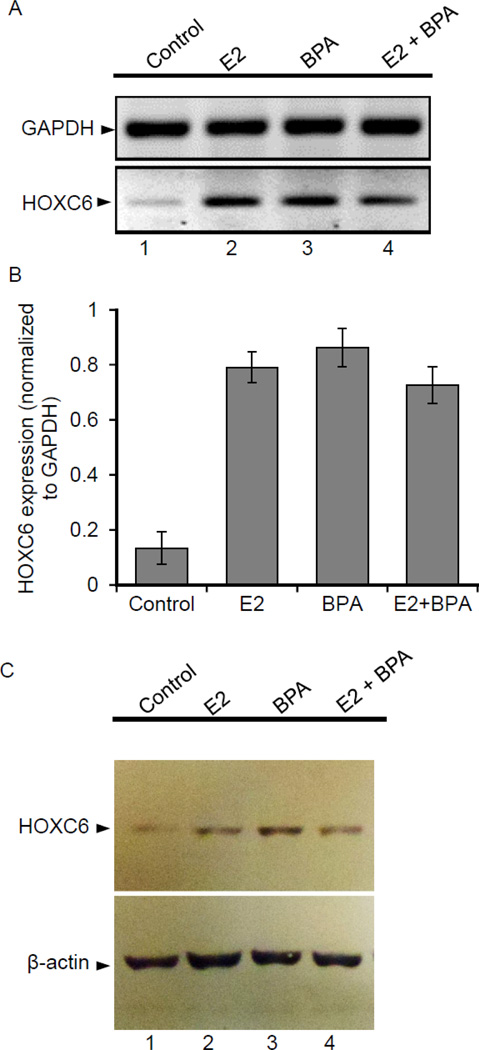
As HOXC6 is found to be overexpressed in breast cancer tissues and it is transcriptionally induced by E2 and BPA in breast cancer cells (in vitro), we examined its regulation by E2 and BPA in vivo. We exposed overiectomized (OVX) Sprague Dawley rats with E2 and BPA, either seperately or in combination, and examined its impacts on HOXC6 expression in mammary tissue. Briefly, OVX rats were treated with acute doses of E2 (5 µg/kg) and BPA (25 µg/kg) individually or in combination for up to 24 hrs. Notably, these doses were selected on the basis of the studies previously performed by other laboratories and shown to be effective in in vivo analyses of gene expression and to influence behaviors and other neural functions (14, 46, 47, 56–58). OVX rats were used to minimize the effects of endogenous estrogen levels in female rats. RNA and proteins were isolated from the mammary gland tissues and analyzed by qPCR and western blots for the expression of HOXC6. These analyses demonstrated that HOXC6 expression was significanlty increased (both at RNA and protein levels) in the rat’s mammary tissues upon treatment with E2 (Figures 3A–C, compare lane 1 and 2). BPA treatments also resulted in upregulation of HOXC6 in the mammary tissue, either independently or in combination with E2 (Figures 3A–C, compare lane 1, 3 and 4). These observations demonstrate that HOXC6 is an estrogen-regulated gene in vivo, in the mammary tissue of the Sprague Dawley rats and importantly, its expression is also upregulated upon BPA exposure, even in the absence of E2.
Figure 3.
Effects of E2 and BPA on HOXC6 gene expression, in ovariectomized (OVX) rats, in vivo. (Panel A-C): OVX female rats were administered with acute doses of E2 (5 µg) and BPA (25 µg/kg), for 24 h, either separately or in combination. Control animals were administered with the vehicle (peanut oil/saline). RNA and protein were isolated from the mammary glands of the control, E2 and BPA treated animals. RNA was analyzed by regular RT-PCR (Panel A); GAPDH was used as a loading control and qPCR (Panel B) using rat specific HOXC6 primers (relative to GAPDH). Each experiment was repeated thrice with three parallel replicates. Bars indicate standard errors, (p < 0.05)). Western blot analysis of the HOXC6 protein levels from the protein samples obtained from control, E2 and BPA treated mammary gland tissues of animals are shown in panel C. β-actin was used as the loading control. Each experiment was repeated at least thrice (n = 3).
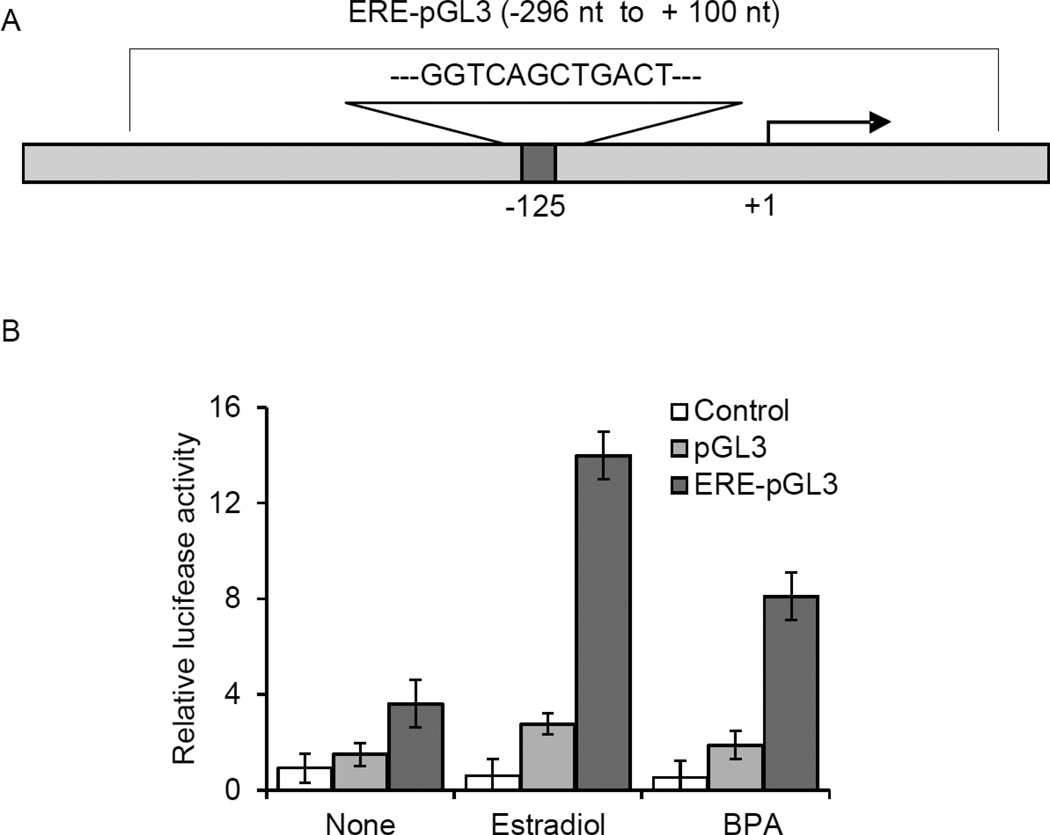
HOXC6 promoter ERE is responsive to BPA
HOXC6 promoter contains an imperfect (GGTCAnnTGACT) estrogen-response element (ERE) with one base pair mismatch and one base-pair less spacing between two half-sites, compared to a typical full EREs (GGTCAnnnTGACC), located at the 125 bp upstream of the transcription start site (Figure 4A) (15). Herein, we examined if this ERE also participates in E2 and BPA-mediated HOXC6 activation in MCF7 cells, using luciferase-based reporter assay. The promoter of HOXC6 containing the ERE region (from +100 to −296 nt) was cloned into luciferase construct pGL3 (15). The ERE-pGL3 construct along with renilla vector (transfection control) was tranfected into MCF7 cells followed by expsoure to E2 (1 nM) and BPA (10 nM) and the induction of luciferase was determined by using dual luciferase detection kit (15). Empty pGL3 transfection was used as negative control. As seen in Figure 4B, E2-treatment induced (5 fold) the luciferase expression in ERE-pGL3 transfected MCF7 cells. Interestingly, exposure to BPA also induced luciferase activity by 4.3 fold in comparison to untreated samples (Figure 4B). These analyses demonstrate that HOXC6 promoter ERE is functional towards E2 and BPA-induced HOXC6 expression in breast cancer cells. Notably, our recent study also showed the presence of another ERE1/2-site located at −1143 bp upstream of the transcription start site at the HOXC6 promoter (15), however, this ERE1/2-site (at −1143 bp) was found to be much less E2-responsive that ERE located at −125 bp region (in Figure 4A) (15). Therefore, in the current study, we studied the contributions of the ERE located at −125 bp region in E2 and BPA-induced HOXC6 expression. Our luciferase data indeeded demonstrate that this HOXC6 promoter is responsive towards E2 and BPA-treatments (Figure 4).
Figure 4.
HOXC6 promoter ERE is responsive to E2 and BPA treatment (luciferase assay). Panel A-B: (A) HOXC6 promoter showing the ERE that was cloned into luciferase construct, pGL3. (B) Luciferase assay: ERE-pGL3 construct was transfected into MCF7 cells for 30 h. Control cells were treated with empty pGL3 vector and no transfection controls were done in parallel. Cells were also co-transfected with renilla luciferase expression construct as an internal transfection control. Cells were then treated with 1 nM E2 or 10 nM BPA and then subjected to luciferase assay by using Dual-Glo Luciferase Assay System (Promega). The luciferase activities in presence of E2 and BPA (over untreated controls and normalized against renilla luciferase expression) were plotted. The experiment with four replicate treatments was repeated at least thrice (n = 3). Bars indicate standard errors (p < 0.05).
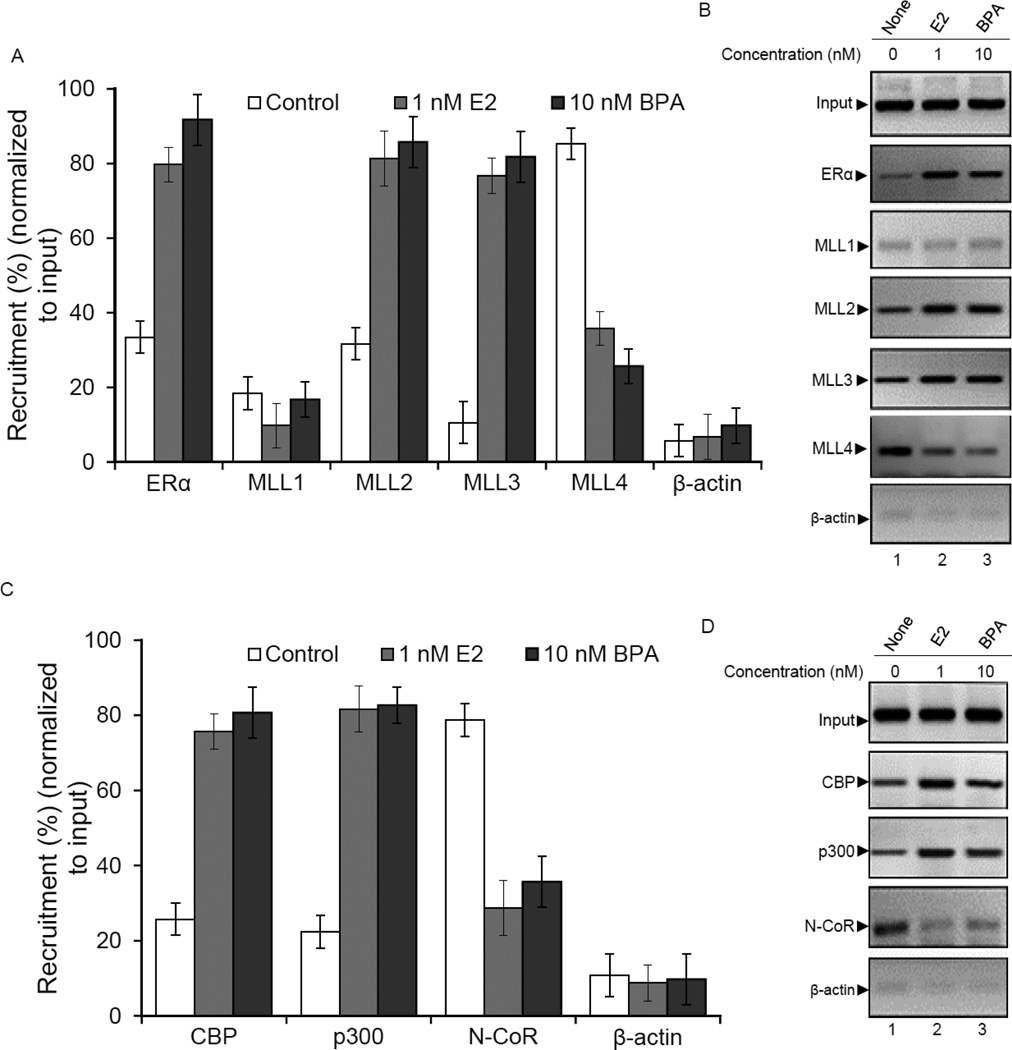
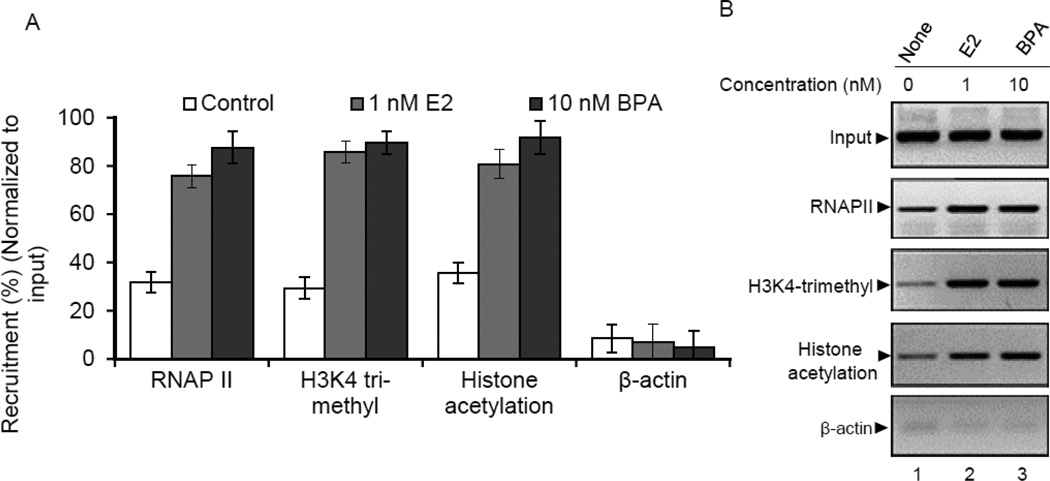
E2 and BPA modulates the recruitment of ERα and ER-coactivators and modify histones at the HOXC6 promoter
ER-coregulators are integral components of estrogen-mediated gene activation (59, 60). ER-coregulators interact with ERs, bind to the promoter of ER-target genes, modify and remodel chromatins and interact with general transcription machineries and all these contributes to gene activation (61–63). Examples of ER-coregulators include SRC1-family of coactivators, CBP/p300, etc. (60, 64, 65). Studies demonstrate that mixed lineage leukemia (MLL) family of histone methylases act as ER-coregulators during E2-mediated gene activation (14, 15, 44, 48, 49, 64–74). Herein, we examined, if ER and MLLs are involved in the E2 and BPA induced HOXC6 expression in MCF7 cells. We examined the binding of ERα, MLLs (MLL1–4), CBP/p300 and N-CoR at the HOXC6 promoter in the presence and absence of E2 and BPA separately (Figures 5A–D), using chromatin immuno-precipitation (ChIP) assay. We also analyzed the levels of histone modifications (H3K4-methylation and acetylation) at the HOXC6 promoter upon treatments with E2 or BPA (Figure 6A–B). Notably, H3K4-trimethyl and histone acetylation are key to transcriptional activation. MLLs and CBP/p300 are well known H3K4-trimethylases and histone acetylases, respectively, that aid in gene activation (14, 15, 44, 48, 49, 51, 75). We treated MCF7 cells with E2 and BPA separately, fixed with formaldehyde and subjected to ChIP assay using antibodies against ERα, MLL1, MLL2, MLL3, MLL4, CBP, p300, N-CoR, H3K4-trimethyl, histone acetyl, RNAPII and β-actin (negative control). N-CoR is a nuclear-receptor co-repressor recently shown to be involved in HOXC6 gene repression under basal conditions (76–78). ChIP DNA were analyzed by qPCR using HOXC6 promoter ERE primers. The analyses showed that ERα, histone methylases MLL2 and MLL3, histone acetylases CBP/p300 were enriched at the HOXC6 promoter ERE region upon treatment with either E2 or BPA (Figures 5A–D). On the contrary, the occupancy levels of N-CoR and histone methylase MLL4 at the HOXC6 promoter are significantly reduced upon treatment with E2 or BPA (Figure 5A – D). Levels of histone H3K4-trimethylation, histone acetylation and RNA polymerase II (RNAP II) recruitment were increased at the HOXC6 promoter upon E2 and BPA treatments (Figures 6A–B). These observations demsontrated that ERα and histone methylases MLL2 and MLL3, histone acetylases CBP/p300 are associated not only with E2 but also with BPA-dependent HOXC6 activation in MCF7 cells. In contrast to MLL2 and MLL3, the level of recruitment of MLL4 as well as the N-CoR was decreased upon treatment with either E2 or BPA, indicating potential involvment of MLL4 and N-CoR in the maintenace of basal transcription of HOXC6. This may indicate that there is an exchange of negative regulatory transcription factors with positive regulatory factors upon treatment with E2 or BPA, at the HOXC6 promoter and this type of exchange in transcription factors has been previously observed HOXC6 and other genes (14, 15, 41, 42, 44, 45, 48, 49, 73). Similar to E2, BPA exposure induced the epigenetic changes (histone modifications) at the HOXC6 promoter affecting its gene expression. Upon exposure to E2 or BPA, the levels of H3K4-trimethylation, histone acetylation, recruitment of histone methylase MLL2, MLL3, and histone acetylase CBP, p300 and RNAP II were enriched at the HOXC6 promoter.
Figure 5.
Enrichment of ERα, MLLs (MLL1–4), CBP/p300 and N-CoR at the HOXC6 promoter in presence of E2 and BPA. (Panel A-D): MCF7 cells were treated with 1nM E2 or 10 nM BPA for 4 h and analyzed by ChIP assay using ERα, MLL1, MLL2, MLL3, MLL4, CBP, p300, N-CoR and β-actin (negative control) antibodies. The ChIP DNA was PCR-amplified using primers specific to ERE region of HOXC6 promoter. The recruitment of ERα, MLL1, MLL2, MLL3, MLL4 and β-actin in the HOXC6 promoter are shown in the panel A (qPCR) and panel B (RT-PCR). Panel C-D shows the recruitment of CBP, p300, N-CoR and β-actin upon treatment with E2 or BPA. Bars indicate standard errors (p < 0.05).
Figure 6.
Enrichment of RNA polymerase II (RNAPII), histone acetylation and H3K4-trimethylation at the HOXC6 promoter in presence of E2 or BPA. (Panel A-B): MCF7 cells were treated with 1 nM E2 or 10 nM BPA for 4 h and analyzed by ChIP assay using RNAPII, H3K4-trimethyl, histone acetylation and β-actin (negative control) antibodies. The ChIP DNA was PCR amplified using primers specific to ERE region of HOXC6 promoter. Panel A-B (qPCR and regular RT-PCR data respectively) shows the levels of RNAPII, H3K4-trimethyl and histone acetylation at the HOXC6 promoter. Bars indicate standard errors (p < 0.05).
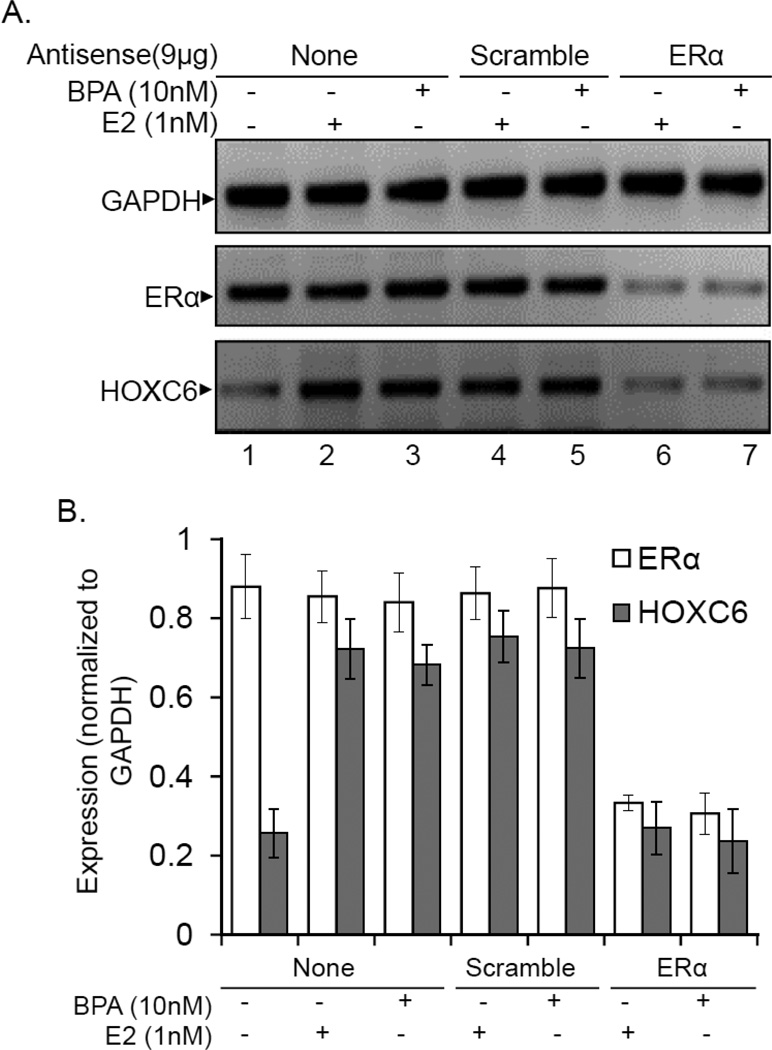
To examine further about the involvement of ERα, we knocked it down in MCF7 cells and examined its impact on E2 and BPA-induced HOXC6 expression. Briefly, we transfected MCF7 cells with ERα and scramble antisenses (no homology to ERα) and then exposed to E2 or BPA followed by analysis of ERα and HOXC6 expression using RT-PCR and qPCR. Our results demonstrate that upon transfection with ERα-antisense, the level of ERα was decreased and that resulted in decreased E2 and BPA induced HOXC6 expression (Figure 7A–B). These results further support that ERα plays critical roles in E2/BPA-induced HOXC6 gene expression.
Figure 7.
ERα knockdown downregulates E2 and BPA induced expression of HOXC6. (Panels A-B): MCF7 cells were transfected with antisense specific for ERα or scramble antisense for 48 h and then treated with 1 nM E2 and 10 nM BPA separately for additional 4 h. RNA was subjected to RT-PCR (Panel A) and qPCR analyses (Panel B) using primers specific to ERα, HOXC6 and GAPDH. Bars indicate standard error (n = 3; P < 0.05).
HOXC6 over expression induces tumor growth factors and facilitates 3D-colony formation
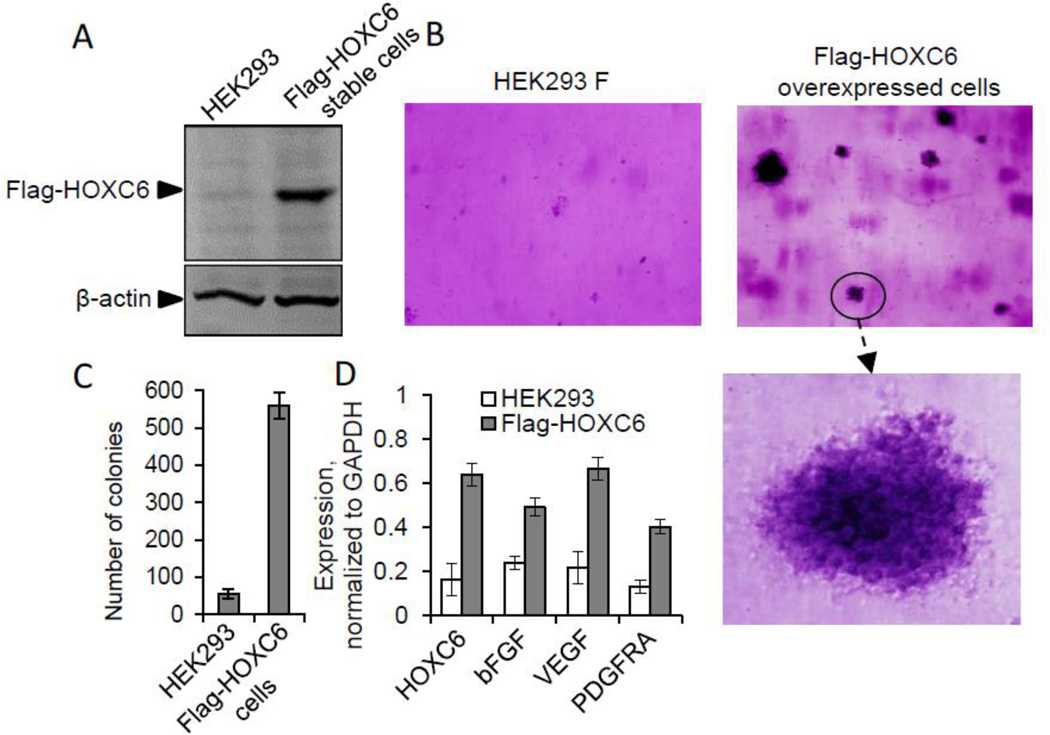
To understand potential roles of HOXC6 in tumorigenesis, we examined the impact of HOXC6 overexpression on regulation of tumor growth factors and also evaluated its ability towards 3-dimensional (3D) colony formation using a soft agar assay (16, 17). In brief, we generated a stable transfected cell line overexpressing HOXC6 (Flag tagged HOXC6) in HEK293 cells (16, 17). Human HOXC6 gene was cloned in pFlag-CMV4 human expression construct, transfected into HEK293 cells and stably transfected cells expressing Flag-HOXC6 were selected using G418 antibiotic selection procedure as described by us earlier (16, 17). The overexpression of Flag-HOXC6 was confirmed by western blot (using Flag-antibody) analysis (Figure 8A).
Figure 8.
HOXC6 over expression induces carcinogenesis. (A) Western blots of HOXC6 overexpression analysis. Proteins were isolated from HEK293 cells and stable cells expressing Flag-HOXC6 and subjected to Western blotting using β-actin (control) and Flag antibodies. (B) 3D-colony formation ability of HOXC6 overexpressed cells. HEK293 and Flag-HOXC6 stable cells were incubated in soft agar for 4–5 weeks stained with 0.005% crystal violet and analyzed under microscope. Magnified view of one HOXC6 overexpressed colony (C) Number of colonies grown in soft agar assays were counted using light microscope and plotted. (D) Analysis of expression of growth factors. The RNA from HEK293 and Flag-HOXC6 overexpressed cells were subjected to RT-qPCR with primers specific to HOXC6, bFGF, VEGF, PDGFRA, and GAPDH (control). Bars indicate standard error ((n = 3, p<0.05)
In order to study the impact of HOXC6 overexpression in tumorigenesis, we examine the 3D-colony formation ability of Flag-HOXC6 overexpressed stable cell line using a soft-agar assay (16, 17). Briefly, HEK293 (control) and Flag-HOXC6 stable cells were plated in soft agar separately, grown for about 21 days (until distinct colonies were visible), stained with crystal violet, and then the visible colonies were examined under a light microscope. Our analysis showed that while very few small colonies were observed in the control HEK293 cells, HOXC6-overexpressing cells (Flag-HOXC6 stable cell line) formed many more distinctly visible large three dimensional (3D) colonies embedded in different layers of soft agar (Figure 8B, an enlarged colony is below). The number of colonies were about 6–7 fold more in the HOXC6 overexpressed cells than in the control HEK293 cells (Figure 8C). Colonies in the HOXC6-overexpressed cells appeared three dimensional (3D) and dense in cell population. These observations indicated that HOXC6 overexpression enhanced the cell proliferation facilitating the formation of large 3D-growth of cell colonies. Furthermore, we isolated the RNA from the HEK293 and Flag-HOXC6 stable cells and analyzed by RT-qPCR analysis for the expression of various tumor growth factors. These analysis showed that overexpression of HOXC6 (Flag-HOXC6) resulted in upregulation of VEGF (vascular epithelial growth factor), bFGF (basic fribroblast growth factor), and PDGFRA (platelate derived growth factor receptor-alpha) (Figure 8D). VEGF, bFGF and PDGFRA are well known tumor grwoth factors what are over expressed in variety tumors and contribute towards tumor growth, angiogenesis, and metastasis (23, 24, 27, 31, 36–38, 79, 80). Theorefore, the upregulation of these tumor growth factor upon overexpression of HOXC6 indicate potential roles of HOXC6 in tumor cell proliferation and growth.
Discussion
HOXC6 is a key player in mammary gland development and milk production (15). HOXC6 expression is associated with several human carcinomas such as cancers of gastrointestinal, breast, lung, prostate, head and neck squamous cell carcinomas, osteosarcomas, medulloblastomas and leukemias (15, 19, 21, 24, 25, 28, 29, 81). HOXC6 is also transcriptionally upregulated in lymph node metastases (31). HOXC6 regulates the expression of various tumor growth factors such as connective tissue growth factor (CTGF), T-cell receptor alternate reading frame protein (TARP), insulin-like growth factor binding protein 3 (IGFBP3), and neutral endopeptidase/membrane metallo-endopeptidase (NEP/MME) and therefore contribute to tumorigenesis (24, 30, 80). Since, HOXC6 expression is associated with the development of mammary glands and breast cancer, its expression is likely regulated by E2. Indeed, our recent findings showed that HOXC6 is transcriptionally induced by E2 in placenta choriocarcinoma (JAR) cells (15). Here, we further investigated the epigenetic mechanisms of E2-mediated transcriptional regulation of HOXC6 in breast cancer cells, in vitro and in animal models, in vivo. We also examined its expression levels in primary breast cancer tissues.
Our studies demonstrated that HOXC6 expression is upregulated in some of the breast cancer tissues, (50–60% cases of the tissue we examined) in comparison to the surrounding normal breast tissue. We also observed that HOXC6 expression is higher in ER-positive breast cancer cells such as MCF7 and T47D, than in ER-negative breast cancer cells (MDA-MB-231). The overexpression of HOXC6 in multiple cases of breast cancer tissues and it upregulation in ER-positive breast cancer cells indicate its potential association with breast cancer and its transcriptional regulation via involvement of estrogen-receptors (ERs). To understand further the potential regulation of HOXC6 by estrogen, we exposed ovariectomized (OVX) Sprague Dawley rats with acute levels of E2 and analyzed the HOXC6 expression levels in rat mammary gland tissue. These analyses demonstrated that HOXC6 expression (both mRNA and protein levels) is indeed upregulated in mammary gland upon exposure to E2. These observations further demonstrate that HOXC6 expression is transcriptionally regulated by E2 both in vitro and in vivo. Notably, in agreement with our observations, HOXC6 expression was previously shown to be upregulated in cultured breast cancer cells (40, 82, 83). In particular, affymetrix GeneChip microarray analyses revealed that HOXC6 expression was upregulated in MCF7 and T47D cells upon treatment with E2 (40, 82, 83). Futherthermore, RNA-Seq analysis by Frasor et al demonstrated that HOXC6 expression is upregulated in MCF7 upon exposure to E2 (82–84). Thus, our results showing the E2-induced up regulation of HOXC6 in vitro and in vivo, are in agreement with previous observations.
Since HOXC6 is an estrogen-regulated gene and is overexpressed in breast cancer, we examined its potential misregulation by hazardous endocrine disrupting chemicals using both in vitro and in vivo models. BPA is well recognized as a estrogenic endocrine disrupting chemical that is commonly found in plastics, metallic storage containers and various other routinely used consumables, and due to its presence in plastic objects, humans are ubiquitously exposed to varying levels of BPA (39, 41, 85). Exposure to BPA has been associated with adverse and harmful perinatal, childhood, and adult health outcomes, including reproductive and developmental defects, metabolic disease, and other health defects (41, 43). BPA exposure has been shown to alter the expression of developmental genes, such as HOX genes, in the reproductive tracts of animals (41, 86). Studies of perinatal and in utero BPA exposure (mice and rats) show abnormalities in mammary gland development, alterations in the overall morphology of the mammary tissue, increased sensitivity to E2, increased cell proliferation, decreased apoptosis, and altered timing of physical development (87, 88). BPA also enhances the incidence of carcinoma in the mammary glands and pre-neoplastic lesions (87). Our recent studies also demonstrated that BPA induces the expression of long non-coding RNA HOTAIR, a crucial player in breast and other types of cancer (6, 41, 42, 44). Thus, BPA exposure poses a major health risk (41, 76). Herein, we observed that, similar to E2, exposure to BPA induced the expression of HOXC6 in the rat mammary glands as well as in cultured MCF7 and T47D cells, even in the absence of E2. Notably, the upregulation of HOXC6 by E2 and BPA in breast cancer cells has been previously observed by other researchers based on genome wide RNA-Seq analyses (40, 82–84). BPA has been shown to regulate a subset of genes that are regulated by E2 (84). Furthermore, we also observed that treatment with BPA in the presence of E2, showed no significant additional impact on HOXC6 expression in comparison to E2 treatment alone and these observations were also supported by previous studies (40). We predict that being relatively weaker than E2, BPA may not be able to effectively compete with E2 in binding to ER for target gene regulation (89, 90) and hence there is a limited impact of BPA in presence of E2. Our data support that HOXC6 is an estrogen-regulated gene in breast cancer cells and its expression may be induced upon exposure to estrogenic endocrine disrupting chemicals such as BPA both in vitro and in vivo.
Promoter analysis revelaed that HOXC6 promoter contains two putative EREs (half-site, GGTCA) that are located at −125 bp and −1143 bp upstream of the transcription start site (15). Recently, we reported that out of these two EREs, the ERE located at −125 bp is much more responsive to E2-treatment (in JAR cells) than the other ERE1/2-site located at −1143 nt. ERs and ER-coregulators were found to be enriched the this ERE located at −125 bp, in an E2-dependent manner, indicating its functionality in the E2-induced HOXC6 expression (15). Therefore, in this study, we primiarily foussed on analyzing the involvment of this more responsive ERE (at −125 bp site, termed as HOXC6 ERE here, Figure 4A) during BPA/E2-induced HOXC6 expression in breast cancer cells. Here, luciferase based reporter analysis indeed demonstrated that the HOXC6-ERE is indcued upon exposure to E2 and BPA, in MCF7 cells (see Figure 4). ChIP analysis demosntrated that ERs and ER-coregulators are recruited to ERE region in an E2/BPA-dependent manner (Figures 5 an 6). Analysis of the neighbouring sequences indicated that the HOXC6 ERE (shown in Figure 4A) is a putative imperfect full ERE (GGTCAnnTGACT) with one base pair mismatch and one base-pair less between two half-sites, compared to a typical consensus full ERE (GGTCAnnnTGACC) (15). Notably, numerous ChIP-Seq analysis demonstrated that ERs are enriched at both consensus full ERE-sites as well as in ERE-half sites (GGTCA) in an E2-dependent manner (91–97), indicating pontential functional signifcance of ERE-half sites along with typical full EREs, during E2-mediated gene regulation. In this stydy, we also observed the E2-dependent enrichment of ERs and ER-coregulators at the HOXC6 ERE. Thus, irrespective of being a ERE1/2-site or an imperfect full ERE, the observed the E2/BPA-dependent enrichment of ERs at the HOXC6 promoter ERE-region, indicate its potential involvement in E2/BPA-induced HOXC6 gene regulation.
Mechanistic analyses showed that, similar to E2, BPA-treatment also induced the recrutiment of ERs and ER-coactivators such as histone methylases MLL2 and MLL3 and histone acetylases such as CBP and p300 to the HOXC6 promoter promiximal ERE region. Notably, MLLs are well-known human histone H3K4-specific methyl-transferases that play key roles in gene activation (51, 71). There are several MLLs: MLL1, MLL2, MLL3, MLL4 and MLL5 (51, 71). Recent studies from our (and others) laboratory demonstrated that MLLs (MLL1–4) acts as ER-coregulators and associated with ER-mediated gene activation (14, 15, 45, 49, 63, 69). MLLs (MLL1–4) contains one or more LXXLL (nuclear receptor box or NR-box) domains through which they interact with nuclear receptors (NRs), get recruited to the NR-regulated gene promoters, introduces H3K4-trimethylation leading to gene activation (51, 71). Here, our results demonstrated that along with ERs, histone methylases MLL2 and MLL3, are enriched at the HOXC6 promoter upon treatment with E2 and BPA. Exposure to E2 or BPA altered the epigenetic states at the HOXC6 promoter, such as increased histone H3K4-trimethylation and histone acetylation levels at the HOXC6 promoter that ultimately resulted in HOXC6 gene activation. We also found that histone methylase MLL4 and nuclear receptor co-repressor N-CoR are pre-occupied at the HOXC6 promoter in the absence of E2 or BPA-treatment and get delocalized from the HOXC6 promoter upon exposure to either E2 or BPA. These observation indicated that MLL4 and N-CoR are likely involved in maintence of basal expression levesl of HOXC6 (14, 98). A schematic model showing the epigenetic mechanism of BPA induced endocrine disruption of HOXC6 is shown in Figure 9.
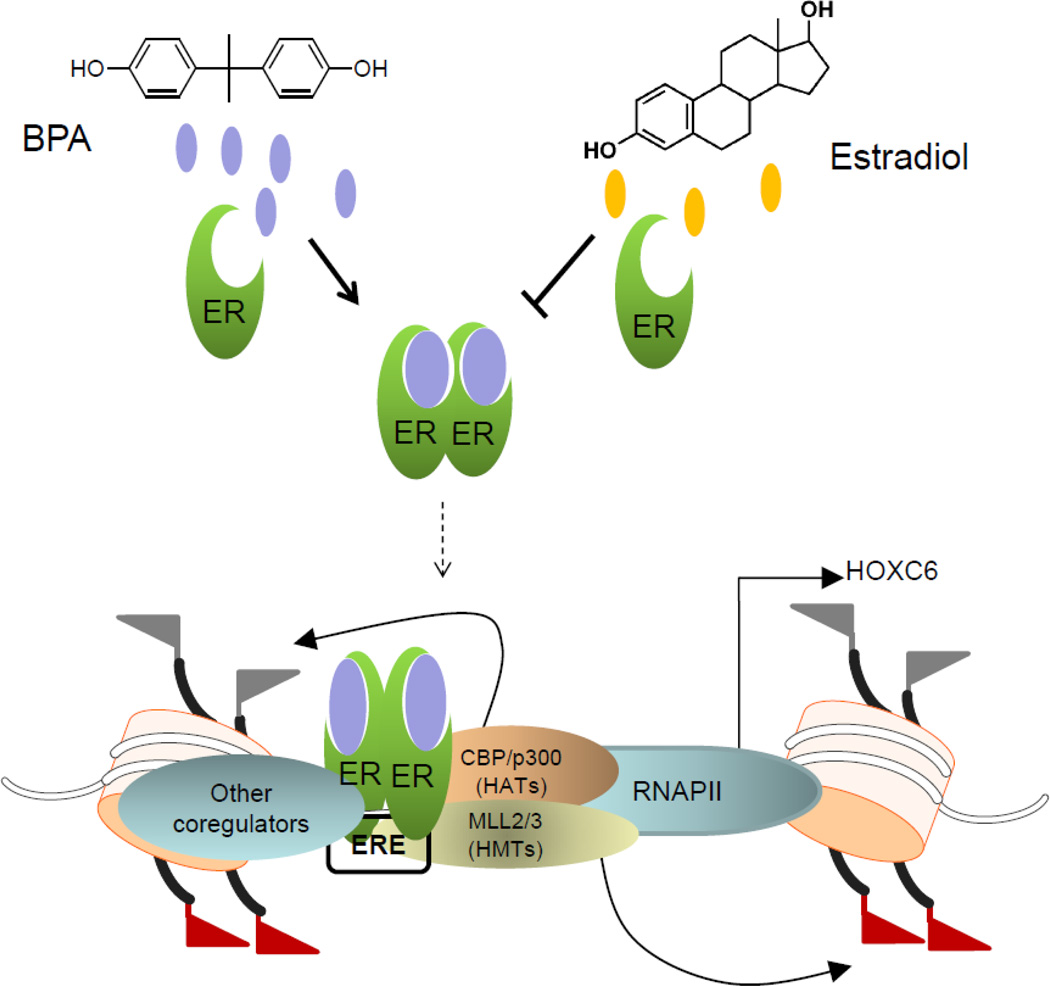
Figure 9.
Models showing the roles of ERs, MLLs and other ER-coregulators during E2 and BPA mediated upregulation of HOXC6. During a typical estradiol mediated HOXC6 gene activation, ERs upon bindinig to E2 dimerize and then dimerized (activated) ERs bind to the EREs of the HOXC6 promoter. ER-coregulators such as MLL2, MLL3, CBP, p300, and other ER-coregulators are also recruited to the HOXC6 promoter. Promoter histones are methylated via MLL-methylases and acetylated via p300 activity, allowing access to RNA polymerase II (RNAP II) to the promoter, ultimately resulting in HOXC6 expression activation. Steroidogenic endocrine disrupting chemicals (EDCs) like BPA when enters into the cells, competes with esrtradiol and binds to ERs leading to activation of ERs and associated cell signaling and target gene activation, in a fashion very similar to E2. Thus, even in the absence of E2, BPA can induce ER-target genes such as HOXC6 leading to their misregulation.
Notably, though we demonstrated that ERs and MLLs are enriched at the ERE-region of the HOXC6 promoter in a E2/BPA dependent manner, the mode of their recruitments remain unclear. Multiple studies have demonstrated that ERs can directly bind to the EREs and that allows recruitment of other factors at the promoters ultimately resulting in gene activation (91, 93, 94). Addtionally, studies also showed that ERs may not be directly interacting with ERE-sequences for their recruitment, rather they can be enriched at the gene promoter via interactions with other factors and/or via chromatin looping (99–104). Here, based on ChIP analyses, we observed that the ERs and ER-coregulators are enriched at the HOXC6 promoters in an E2/BPA-dependent manner. As this observed enrichement of ERs/MLLs at the HOXC6 promoter ERE-region is based on ChIP analyses, the mode of their recruitment could follow either direct or indirect mode of interactions including chromatin looping.
The potential impacts of HOXC6 overexpression on cell proliferation and tumor growth was assesed using a soft-agar based 3D-colony formation assay. Our studies demonstrated that HOXC6 overexpression facilitated 3D-colony formation. HOXC6 overexpression also resulted in upregulation of multiple tumor growth factors including VEGF, bFGF and PDGFRA. The HOXC6-induced overexpression of the factors, that are critical players in tumor cell proliferation and growth, indicated potential roles of HOXC6 in cell proliferation and tumor growth. In summary, our studies demonstrated that HOXC6, which is critical in mammary gland development and is potentially upregulated in some of the breast cancer and is transcriptionally regulated by estrogen in vitro and in vivo. Furthermore, our studies showing the upregulation of HOXC6 expression in presence of BPA, may indicate its potential endocrine disruption upon exposure to estrogenic endocrine disrupting chemicals and that may contribute towards the increased risk of cancer and other associated developmental disorders.
Highlights.
HOXC6 is a homeobox containing gene associated with breast cancer
HOXC6 expression is induced by estradiol (E2) in vitro and in vivo
HOXC6 expression is also induced by endocrine disrupting chemicals BPA
ERs coordinate with MLLs during E2/BPA-induced HOXC6 expression
Results provide mechanistic insight of HOXC6 gene regulation and its endocrine disruption
Acknowledgement
We thank all the Mandal and Perrotti lab members for helpful discussions. Research in Mandal laboratory is supported by grant from NIH (1R15 ES019129-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lappin TR, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. Ulster Med J. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- 2.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 3.Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi Y, Hamada J-i, Murakawa K, Takada M, Tada M, Nogami I, Hayashi N, Nakamori S, Monden M, Miyamoto M, Katoh H, Moriuchi T. Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Experimental Cell Research. 2004;293:144–153. doi: 10.1016/j.yexcr.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Mallo M, Alonso CR. The regulation of Hox gene expression during animal development. Development. 2013;140:3951–3963. doi: 10.1242/dev.068346. [DOI] [PubMed] [Google Scholar]
- 6.Bhan A, Mandal SS. Long Noncoding RNAs: Emerging Stars in Gene Regulation, Epigenetics and Human Disease. ChemMedChem. 2014;9:1932–1956. doi: 10.1002/cmdc.201300534. [DOI] [PubMed] [Google Scholar]
- 7.Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30:965–979. doi: 10.1002/bies.20823. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Sukumar S. HOX genes: emerging stars in cancer. Cancer Biol Ther. 2003;2:524–525. doi: 10.4161/cbt.2.5.525. [DOI] [PubMed] [Google Scholar]
- 9.Friedmann Y, Daniel CA, Strickland P, Daniel CW. Hox genes in normal and neoplastic mouse mammary gland. Cancer Res. 1994;54:5981–5985. [PubMed] [Google Scholar]
- 10.Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, Tamoto E, Shindo G, Matsunaga A, Teramoto K, Komuro K, Kondo S, Katoh H, Koike T, Moriuchi T. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep. 2005;13:673–679. [PubMed] [Google Scholar]
- 11.Maroulakou IG, Spyropoulos DD. The study of HOX gene function in hematopoietic, breast and lung carcinogenesis. Anticancer Res. 2003;23:2101–2110. [PubMed] [Google Scholar]
- 12.Nunes FD, Almeida FCSd, Tucci R, Sousa SCOMd. Homeobox genes: a molecular link between development and cancer. Pesquisa Odontológica Brasileira. 2003;17:94–98. doi: 10.1590/s1517-74912003000100018. [DOI] [PubMed] [Google Scholar]
- 13.Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005;3:240–252. doi: 10.1089/lrb.2005.3.240. [DOI] [PubMed] [Google Scholar]
- 14.Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. J Mol Endocrinol. 2012;48:61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- 15.Ansari KI, Hussain I, Shrestha B, Kasiri S, Mandal SS. HOXC6 Is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. Journal of Molecular Biology. 2011;411:334–349. doi: 10.1016/j.jmb.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrestha B, Ansari KI, Bhan A, Kasiri S, Hussain I, Mandal SS. Homeodomain-containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth in vitro and is overexpressed in breast cancer tissue. FEBS J. 2012;279:3715–3726. doi: 10.1111/j.1742-4658.2012.08733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasiri S, Ansari KI, Hussain I, Bhan A, Mandal SS. Antisense oligonucleotide mediated knockdown of HOXC13 affects cell growth and induces apoptosis in tumor cells and over expression of HOXC13 induces 3D-colony formation. RSC Adv. 2013;3:3260–3269. doi: 10.1039/C2RA22006G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Sukumar S. Role of Homeobox Genes in Normal Mammary Gland Development and Breast Tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:159–175. doi: 10.1023/a:1025996707117. [DOI] [PubMed] [Google Scholar]
- 19.Kelly ZL, Michael A, Butler-Manuel S, Pandha HS, Morgan RG. HOX genes in ovarian cancer. J Ovarian Res. 2011;4:16. doi: 10.1186/1757-2215-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita T, Tazawa S, Yawei Z, Katayama H, Kato Y, Nishiwaki K, Yokohama Y, Ishikawa M. Suppression of invasive characteristics by antisense introduction of overexpressed HOX genes in ovarian cancer cells. Int J Oncol. 2006;28:931–938. [PubMed] [Google Scholar]
- 21.Bodey B, Bodey B, Jr, Siegel SE, Kaiser HE. Immunocytochemical detection of the homeobox B3, B4, and C6 gene products in breast carcinomas. Anticancer Res. 2000;20:3281–3286. [PubMed] [Google Scholar]
- 22.Castronovo V, Kusaka M, Chariot A, Gielen J, Sobel M. Homeobox genes: potential candidates for the transcriptional control of the transformed and invasive phenotype. Biochem Pharmacol. 1994;47:137–143. doi: 10.1016/0006-2952(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Gasca A, Spyropoulos DD. Differential mammary morphogenesis along the anteroposterior axis in Hoxc6 gene targeted mice. Dev Dyn. 2000;219:261–276. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1048>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran S, Liu PB, Young AN, Yin-Goen QQ, Lim SD, Laycock N, Amin M, Carney JK, Marshall FF, Petros JA, Moreno CS. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- 25.Bodey B, Bodey B, Jr, Groger AM, Siegel SE, Kaiser HE. Immunocytochemical detection of homeobox B3, B4, and C6 gene product expression in lung carcinomas. Anticancer Res. 2000;20:2711–2716. [PubMed] [Google Scholar]
- 26.Ramachandran S, Liu P, Young AN, Yin-Goen Q, Lim SD, Laycock N, Amin MB, Carney JK, Marshall FF, Petros JA, Moreno CS. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- 27.Bijl J, van Oostveen JW, Kreike M, Rieger E, van der Raaij-Helmer LM, Walboomers JM, Corte G, Boncinelli E, van den Brule AJ, Meijer CJ. Expression of HOXC4, HOXC5, and HOXC6 in human lymphoid cell lines, leukemias, and benign and malignant lymphoid tissue. Blood. 1996;87:1737–1745. [PubMed] [Google Scholar]
- 28.Bodey B, Bodey B, Jr, Siegel SE, Luck JV, Kaiser HE. Homeobox B3, B4, and C6 gene product expression in osteosarcomas as detected by immunocytochemistry. Anticancer Res. 2000;20:2717–2721. [PubMed] [Google Scholar]
- 29.Bodey B, Bodey B, Jr, Siegel SE, Kaiser HE. Immunocytochemical detection of the homeobox B3, B4, and C6 gene products in childhood medulloblastomas/primitive neuroectodermal tumors. Anticancer Res. 2000;20:1769–1780. [PubMed] [Google Scholar]
- 30.McCabe CD, Spyropoulos DD, Martin D, Moreno CS. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68:1988–1996. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon S-M, Kim S-A, Yoon J-H, Ahn S-G. HOXC6 Is Deregulated in Human Head and Neck Squamous Cell Carcinoma and Modulates Bcl-2 Expression. Journal of Biological Chemistry. 2012;287:35678–35688. doi: 10.1074/jbc.M112.361675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grishina IB, Kim SY, Ferrara C, Makarenkova HP, Walden PD. BMP7 inhibits branching morphogenesis in the prostate gland and interferes with Notch signaling. Dev Biol. 2005;288:334–347. doi: 10.1016/j.ydbio.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricort JM, Binoux M. Insulin-like growth factor-binding protein-3 activates a phosphotyrosine phosphatase. Effects on the insulin-like growth factor signaling pathway. J Biol Chem. 2002;277:19448–19454. doi: 10.1074/jbc.M200439200. [DOI] [PubMed] [Google Scholar]
- 34.Geer PV, Hunter T, Lindberg RA. Receptor Protein-Tyrosine Kinases and their Signal Transduction Pathways. Annual Review of Cell Biology. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y, Liu G, Zhang Y, Hu Y-P, Yu K, Lin C, McKeehan K, Xuan JW, Ornitz DM, Shen MM, Greenberg N, McKeehan WL, Wang F. Fibroblast growth factor receptor 2 tyrosine kinase is required for prostatic morphogenesis and the acquisition of strict androgen dependency for adult tissue homeostasis. Development. 2007;134:723–734. doi: 10.1242/dev.02765. [DOI] [PubMed] [Google Scholar]
- 36.Chariot A, Castronovo V, Le P, Gillet C, Sobel ME, Gielen J. Cloning and expression of a new HOXC6 transcript encoding a repressing protein. Biochem J. 1996;319(Pt 1):91–97. doi: 10.1042/bj3190091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Hamada J, Nishimoto A, Takahashi Y, Murai T, Tada M, Moriuchi T. HOXC6 and HOXC11 increase transcription of S100beta gene in BrdU-induced in vitro differentiation of GOTO neuroblastoma cells into Schwannian cells. J Cell Mol Med. 2007;11:299–306. doi: 10.1111/j.1582-4934.2007.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones FS, Holst BD, Minowa O, De Robertis EM, Edelman GM. Binding and transcriptional activation of the promoter for the neural cell adhesion molecule by HoxC6 (Hox- 3.3) Proc Natl Acad Sci U S A. 1993;90:6557–6561. doi: 10.1073/pnas.90.14.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA) Reprod Toxicol. 2007;24:139–177. doi: 10.1016/j.reprotox.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Gertz J, Reddy TE, Varley KE, Garabedian MJ, Myers RM. Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome Res. 2012;22:2153–2162. doi: 10.1101/gr.135681.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–170. doi: 10.1016/j.jsbmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Histone Methyltransferase EZH2 Is Transcriptionally Induced by Estradiol as Well as Estrogenic Endocrine Disruptors Bisphenol-A and Diethylstilbestrol. Journal of Molecular Biology. 2014 doi: 10.1016/j.jmb.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 43.Schonfelder G, Wittfoht W, Hopp H, Talsness CE, Paul M, Chahoud I. Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ Health Perspect. 2002;110:A703–A707. doi: 10.1289/ehp.110-1241091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. Journal of Molecular Biology. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ansari KI, Kasiri S, Hussain I, Bobzean SA, Perrotti LI, Mandal SS. MLL histone methylases regulate expression of HDLR-SR-B1 in presence of estrogen and control plasma cholesterol in vivo. Mol Endocrinol. 2013;27:92–105. doi: 10.1210/me.2012-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ansari KI, Kasiri S, Mandal SS. Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene. 2013;32:3359–3370. doi: 10.1038/onc.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansari KI, Kasiri S, Mishra BP, Mandal SS. Mixed lineage leukaemia-4 regulates cell-cycle progression and cell viability and its depletion suppresses growth of xenografted tumour in vivo. Br J Cancer. 2012;107:315–324. doi: 10.1038/bjc.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ansari KI, Shrestha B, Hussain I, Kasiri S, Mandal SS. Histone methylases MLL1 and MLL3 coordinate with estrogen receptors in estrogen-mediated HOXB9 expression. Biochemistry. 2011;50:3517–3527. doi: 10.1021/bi102037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansari KI, Kasiri S, Hussain I, Mandal SS. Mixed lineage leukemia histone methylases play critical roles in estrogen-mediated regulation of HOXC13. FEBS J. 2009;276:7400–7411. doi: 10.1111/j.1742-4658.2009.07453.x. [DOI] [PubMed] [Google Scholar]
- 50.Kasiri S, Ansari KI, Hussain I, Bhan A, Mandal SS. Antisense oligonucleotide mediated knockdown of HOXC13 affects cell growth and induces apoptosis in tumor cells and over expression of HOXC13 induces 3D-colony formation. Rsc Advances. 2013;3:3260–3269. doi: 10.1039/C2RA22006G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ansari KI, Mishra BP, Mandal SS. Human CpG binding protein interacts with MLL1, MLL2 and hSet1 and regulates Hox gene expression. Biochim Biophys Acta. 2008;1779:66–73. doi: 10.1016/j.bbagrm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Betancourt AM EI, Desmont RA, Russo J, Lamartiniere CA. In-utero exposure to bisphenol A shifts the window of susceptibility for mammary carcinogenesis in the rat. Environ Health Perspect. 2010;118:1614–1619. doi: 10.1289/ehp.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maclusky H, Leranth The environmental estrogen bisphenol A inhibits estradiol-induced hippocampal synaptogenesis. 2005. Environ Health Perspect. 2005;11:675–679. doi: 10.1289/ehp.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inagaki T, Frankfurt M, Luine V. Estrogen-induced memory enhancements are blocked by acute bisphenol A in adult female rats: role of dendritic spines. Endocrinology. 2012;153:3357–3367. doi: 10.1210/en.2012-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eilam-Stock T, Serrano P, Frankfurt M, Luine V. Bisphenol-A impairs memory and reduces dendritic spine density in adult male rats. Behav Neurosci. 2012;126:175–185. doi: 10.1037/a0025959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Betancourt AM, Eltoum IA, Desmond RA, Russo J, Lamartiniere CA. In utero exposure to bisphenol A shifts the window of susceptibility for mammary carcinogenesis in the rat. Environ Health Perspect. 2010;118:1614–1619. doi: 10.1289/ehp.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casals-Casas C, Desvergne B. Endocrine disruptors: from endocrine to metabolic disruption. Annu Rev Physiol. 2011;73:135–162. doi: 10.1146/annurev-physiol-012110-142200. [DOI] [PubMed] [Google Scholar]
- 58.MacLusky NJ, Hajszan T, Leranth C. The environmental estrogen bisphenol a inhibits estradiol-induced hippocampal synaptogenesis. Environ Health Perspect. 2005;113:675–679. doi: 10.1289/ehp.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nilsson S, Gustafsson JA. Estrogen receptor transcription and transactivation: Basic aspects of estrogen action. Breast Cancer Res. 2000;2:360–366. doi: 10.1186/bcr81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nilsson S, Gustafsson JA. Estrogen receptor action. Crit Rev Eukaryot Gene Expr. 2002;12:237–257. doi: 10.1615/critreveukaryotgeneexpr.v12.i4.10. [DOI] [PubMed] [Google Scholar]
- 61.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. Journal of Biological Chemistry. 2006;281:15714–15720. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 62.Dreijerink KMA, Mulder KW, Winkler GS, Hoppener JWM, Lips CJM, Timmers HTM. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Research. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 63.Lee S, Lee DK, Dou YL, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. P Natl Acad Sci USA. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barkhem T, Nilsson S, Gustafsson JA. Molecular mechanisms, physiological consequences and pharmacological implications of estrogen receptor action. Am J Pharmacogenomics. 2004;4:19–28. doi: 10.2165/00129785-200404010-00003. [DOI] [PubMed] [Google Scholar]
- 65.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 66.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 68.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 69.Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW. Crucial Roles for Interactions between MLL3/4 and INI1 in Nuclear Receptor Transactivation. Molecular Endocrinology. 2009;23:610–619. doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mandal SS, Ansari KI, Hussain I, Kasiri S, Shrestha B. MLL histone methylases in estrogen-mediated regulation of HOX genes involved in hair follicle development and leukemia. Faseb J. 2010;24 [Google Scholar]
- 71.Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Front Biosci. 2009;14:3483–3495. doi: 10.2741/3466. [DOI] [PubMed] [Google Scholar]
- 72.Mandal SS. Mixed lineage leukemia: versatile player in epigenetics and human disease. Febs Journal. 2010;277:1789–1789. doi: 10.1111/j.1742-4658.2010.07605.x. [DOI] [PubMed] [Google Scholar]
- 73.Ansari KI, Mandal SS. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS J. 2010;277:1790–1804. doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- 74.Ansari KI, Hussain I, Das HK, Mandal SS. Overexpression of human histone methylase MLL1 upon exposure to a food contaminant mycotoxin, deoxynivalenol. Febs Journal. 2009;276:3299–3307. doi: 10.1111/j.1742-4658.2009.07055.x. [DOI] [PubMed] [Google Scholar]
- 75.Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. FEBS J. 2009;276:1629–1640. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- 76.Jiang S, Meyer R, Kang K, Osborne CK, Wong J, Oesterreich S. Scaffold attachment factor SAFB1 suppresses estrogen receptor alpha-mediated transcription in part via interaction with nuclear receptor corepressor. Mol Endocrinol. 2006;20:311–320. doi: 10.1210/me.2005-0100. [DOI] [PubMed] [Google Scholar]
- 77.Oesterreich S, Zhang Q, Hopp T, Fuqua SA, Michaelis M, Zhao HH, Davie JR, Osborne CK, Lee AV. Tamoxifen-bound estrogen receptor (ER) strongly interacts with the nuclear matrix protein HET/SAF-B, a novel inhibitor of ER-mediated transactivation. Mol Endocrinol. 2000;14:369–381. doi: 10.1210/mend.14.3.0432. [DOI] [PubMed] [Google Scholar]
- 78.Shibata H, Spencer TE, Onate SA, Jenster G, Tsai SY, Tsai MJ, O'Malley BW. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. discussion 164-145. [PubMed] [Google Scholar]
- 79.Velghe AI, Van Cauwenberghe S, Polyansky AA, Chand D, Montano-Almendras CP, Charni S, Hallberg B, Essaghir A, Demoulin JB. PDGFRA alterations in cancer: characterization of a gain-of-function V536E transmembrane mutant as well as loss-of-function and passenger mutations. Oncogene. 2014;33:2568–2576. doi: 10.1038/onc.2013.218. [DOI] [PubMed] [Google Scholar]
- 80.Fujiki K, Duerr EM, Kikuchi H, Ng A, Xavier RJ, Mizukami Y, Imamura T, Kulke MH, Chung D. HOXC6 is overexpressed in gastrointestinal carcinoids and interacts with JunD to regulate tumor growth. Gastroenterology. 2008;134:A297–A297. doi: 10.1053/j.gastro.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, Lucia MS, Nordeen SK. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 82.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 83.Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol. 2007;21:2112–2123. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- 84.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FCGJ, Span PN, Stunnenberg HG. ChIP-Seq of ER alpha and RNA polymerase II defines genes differentially responding to ligands. Embo J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krishnan AV, Stathis P, Permuth SF, Tokes L, Feldman D. Bisphenol-A: an estrogenic substance is released from polycarbonate flasks during autoclaving. Endocrinology. 1993;132:2279–2286. doi: 10.1210/endo.132.6.8504731. [DOI] [PubMed] [Google Scholar]
- 86.Smith CC, Taylor HS. Xenoestrogen exposure imprints expression of genes (Hoxa10) required for normal uterine development. Faseb J. 2007;21:239–246. doi: 10.1096/fj.06-6635com. [DOI] [PubMed] [Google Scholar]
- 87.Vandenberg LN, Maffini MV, Wadia PR, Sonnenschein C, Rubin BS, Soto AM. Exposure to environmentally relevant doses of the xenoestrogen bisphenol-A alters development of the fetal mouse mammary gland. Endocrinology. 2007;148:116–127. doi: 10.1210/en.2006-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Munoz-de-Toro M, Markey CM, Wadia PR, Luque EH, Rubin BS, Sonnenschein C, Soto AM. Perinatal exposure to bisphenol-A alters peripubertal mammary gland development in mice. Endocrinology. 2005;146:4138–4147. doi: 10.1210/en.2005-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Washington W, Hubert L, Jones D, Gray WG. Bisphenol a binds to the low-affinity estrogen binding site. In Vitr Mol Toxicol. 2001;14:43–51. doi: 10.1089/109793301316882531. [DOI] [PubMed] [Google Scholar]
- 90.Blair RM, Fang H, Branham WS, Hass BS, Dial SL, Moland CL, Tong W, Shi L, Perkins R, Sheehan DM. The estrogen receptor relative binding affinities of 188 natural and xenochemicals: structural diversity of ligands. Toxicol Sci. 2000;54:138–153. doi: 10.1093/toxsci/54.1.138. [DOI] [PubMed] [Google Scholar]
- 91.Akbas GE, Song J, Taylor HS. A HOXA10 estrogen response element (ERE) is differentially regulated by 17 beta-estradiol and diethylstilbestrol (DES) Journal of Molecular Biology. 2004;340:1013–1023. doi: 10.1016/j.jmb.2004.05.052. [DOI] [PubMed] [Google Scholar]
- 92.Anderson I, Gorski J. Estrogen receptor alpha interaction with estrogen response element half-sites from the rat prolactin gene. Biochemistry. 2000;39:3842–3847. doi: 10.1021/bi9924516. [DOI] [PubMed] [Google Scholar]
- 93.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klinge CM, Studinski-Jones AL, Kulakosky PC, Bambara RA, Hilf R. Comparison of tamoxifen ligands on estrogen receptor interaction with estrogen response elements. Mol Cell Endocrinol. 1998;143:79–90. doi: 10.1016/s0303-7207(98)00130-0. [DOI] [PubMed] [Google Scholar]
- 95.Lopez D, Sanchez MD, Shea-Eaton W, McLean MP. Estrogen activates the high-density lipoprotein receptor gene via binding to estrogen response elements and interaction with sterol regulatory element binding protein-1A. Endocrinology. 2002;143:2155–2168. doi: 10.1210/endo.143.6.8855. [DOI] [PubMed] [Google Scholar]
- 96.Shu FJ, Sidell N, Yang DZ, Kallen CB. The tri-nucleotide spacer sequence between estrogen response element half-sites is conserved and modulates ER alpha-mediated transcriptional responses. J Steroid Biochem. 2010;120:172–179. doi: 10.1016/j.jsbmb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sidell N, Mathad RI, Shu FJ, Zhang ZJ, Kallen CB, Yang DZ. Intercalation of XR5944 with the estrogen response element is modulated by the tri-nucleotide spacer sequence between half-sites. J Steroid Biochem. 2011;124:121–127. doi: 10.1016/j.jsbmb.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ansari AZ, Peterson-Kaufman KJ. A partner evokes latent differences between Hox proteins. Cell. 2011;147:1220–1221. doi: 10.1016/j.cell.2011.11.046. [DOI] [PubMed] [Google Scholar]
- 99.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fullwood MJ, Wei CL, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Holwerda S, de Laat W. Chromatin loops, gene positioning, and gene expression. Front Genet. 2012;3:217. doi: 10.3389/fgene.2012.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Theodorou V, Carroll JS. Estrogen receptor action in three dimensions - looping the loop. Breast Cancer Res. 2010;12:303. doi: 10.1186/bcr2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu MH, Cheung E. Estrogen receptor-mediated long-range chromatin interactions and transcription in breast cancer. Mol Cell Endocrinol. 2014;382:624–632. doi: 10.1016/j.mce.2013.09.019. [DOI] [PubMed] [Google Scholar]