Abstract
MicroRNAs (miRNAs) induce messenger RNA (mRNA) degradation and repress mRNA translation. Several miRNAs control the expression of the brain-derived neurotrophic factor (BDNF) in the prefrontal cortex (PFC). The BDNF signaling pathway is activated by moderate intake of alcohol to prevent escalation to excessive drinking. Here, we present data to suggest that the transition from moderate to uncontrolled alcohol intake occurs, in part, upon a breakdown of this endogenous protective pathway via a miRNA-dependent mechanism. Specifically, a mouse paradigm that mimics binge alcohol drinking in humans produced a robust reduction in BDNF mRNA levels in the medial PFC (mPFC), which was associated with increased expression of several miRNAs including miR-30a-5p. We show that miR-30a-5p binds the 3′ untranslated region of BDNF, and that overexpression of miR-30a-5p in the mPFC decreased BDNF expression. Importantly, overexpression of miR-30a-5p in the mPFC produced an escalation of alcohol intake and a preference over water. Conversely, inhibition of miR-30a-5p in the mPFC using a Locked Nucleic Acid sequence that targets miR-30a-5p restored BDNF levels and decreased excessive alcohol intake. Together, our results indicate that miR-30a-5p plays a key role in the transition from moderate to excessive alcohol intake.
Introduction
MicroRNAs (miRNAs) are small non coding RNAs of ∼20 nucleotides derived from longer precursor molecules that induce messenger RNA (mRNA) degradation and repression of mRNA to protein translation.1 In the brain, miRNAs contribute to number of functions such as dendritic spine development2 as well as plasticity,3 learning and memory,3 and malfunction of miRNAs have been reported to contribute to numerous psychiatric disorders4 including addiction.5 Several studies suggested a link between several miRNAs and alcohol's actions in the central nervous system. For example, experiments conducted in striatal neuronal cultures revealed that the expression of miR-9 was increased in response to alcohol exposure, which was associated with a decreased mRNA expression of the alpha subunit of the big potassium (BK) channel, and with the development of tolerance to alcohol,6 and systemic administration of alcohol (1 g kg-1, intraperitoneal) was shown to reduce the expression of miR-382 in the nucleus accumbens of rats.7 Furthermore, alcohol was shown to induce changes in the expression of miRNAs during brain development,8 and the expression of 35 miRNAs were found to be increased in the prefrontal cortex (PFC) of postmortem human alcoholics.9
One of the genes whose expression is controlled by miRNAs is the brain-derived neurotrophic factor (BDNF) in cultured cells10,11 and in the brain.12–14 BDNF is an essential growth factor that promotes neuronal proliferation, differentiation and survival,15 as well as synaptic plasticity, and learning and memory.16 Previously, we identified BDNF as an endogenous factor that reduces the development of adverse behaviors associated with alcohol exposure including consumption.17–20 Specifically, we found that BDNF expression is increased in the dorsal striatum of rodents that consume moderate levels of alcohol,17,19 which is comparable to humans that consume alcohol socially. Furthermore, we showed that increasing BDNF levels in the dorsal striatum of mice17 or rats,18 or the activation of the BDNF receptor TrkB in the dorsolateral striatum of rats,19,20 attenuated self-administration of moderate levels of alcohol. Conversely, a global reduction of the BDNF gene17 or small-interfering RNA-mediated knockdown of BDNF expression in the dorsolateral striatum19 produced an increase in mice and rats self-administration of moderate levels of alcohol. In addition, we observed that prolonged exposure of mice to alcohol led to a dysregulation of BDNF expression in cortico–striatal regions including the PFC.21 PFC hypofunction contributes to the development of compulsive and excessive drug intake,22 and more specifically, the medial PFC (mPFC) was shown to have a major role in the transition from a moderate to excessive alcohol intake.23 As miRNAs in the PFC control BDNF levels,12,13 we tested the possibility that miRNA-dependent reduction of BDNF expression in the mPFC has a role in the development of excessive and uncontrolled alcohol intake.
Materials and Methods
Reagents
Reverse Transcription System and PCR master mix were purchased from Promega Corporation (Madison, WI, USA). DNase and the mouse anti-GFAP antibodies were obtained from Sigma-Aldrich (St Louis, MO, USA). All miRNA reagents, including miRCURY LNA Universal RT miRNA PCR, SybrGreen master mix and the miRCURY LNA miRNA inhibitors were obtained from Exiqon (Vedbaek, Denmark). The Q5 Site-Directed Mutagenesis Kit and rabbit anti-maltose binding protein (MBP) antibodies were purchased from New England Biolabs (Ipswich, MA, USA). pRNAT-H1.1/Shuttle was purchased from GenScript (Piscataway, NJ, USA). The adenoviral vector Adeno-X, the Adeno-X Virus Purification Kit and the Adeno-X Rapid Titer Kit were purchased from Clontech (Mountain View, CA, USA). pUSEamp(+) vector and mouse anti-NeuN antibodies were purchased from Millipore (Billerica, MA, USA). All real-time PCR reagents (including TaqMan Gene Expression Assays), secondary antibodies Alexa Fluor 488-labeled donkey anti-rabbit and Alexa Fluor 594-labeled donkey anti-mouse, the Lipofectamine 2000 and ProLong Gold medium were obtained from Life Technologies (Carlsbad, CA, USA). Rabbit anti-green fluorescent protein (anti-GFP) antibodies for immunohistochemical detection were purchased from Abcam (Cambridge, MA, USA). Mouse anti-Flag antibodies were obtained from Cell Signaling Technology (Beverly, MA, USA) and goat anti-Actin and mouse anti-GFP antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The mirVana miRNA Isolation Kit was purchased from Ambion (Austin, TX, USA). Horseradish peroxidase-conjugated secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA, USA). Enhanced Chemiluminescence was acquired from GE Healthcare Life Sciences (Piscataway, NJ, USA).
Animals
Male C57BL/6J mice were obtained at 8 weeks of age from the Jackson Laboratory (Bar Harbor, ME, USA). Animals were housed under a reverse 12 h light/dark cycle, with lights off at 10:00 hours and lights on at 22:00 hours, and were provided with continuous ad libitum access to food and water. Because of the reversal of normal light/dark cycle, animals were given 2 weeks to adjust to the new housing conditions prior to the beginning of the procedures. All animal procedures were approved by the Gallo Center and the University of California San Francisco (UCSF) Institutional Animal Care and Use Committee (IACUC) and were conducted in agreement with the Guide for the Care and Use of Laboratory Animals (Gallo Center), National Research Council (1996) and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC, UCSF).
mRNA and miRNAs extraction and purification
Mice were killed immediately or 24 h after the final alcohol access session; brain regions were isolated by microdissection and snap frozen to minimize RNA degradation. Areas designated for microdissection were defined according to coordinates from ‘the mouse brain in stereotaxic coordinates’.24 The mPFC and hippocampus were removed, and total RNA fraction and a fraction enriched in miRNAs were extracted using the mirVana miRNA Isolation kit according to the manufacturer's instructions. RNA yield and purity were evaluated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies/Thermo Scientific, Waltham, MA, USA).
Reverse transcriptase-PCR
Total RNAs were reverse transcribed using a Reverse Transcription System kit at 42 °C for 30 min. BDNF, nerve growth factor (NGF) and GFP expressions were analyzed by PCR as previously described17 and with temperature cycling parameters consisting of initial denaturation at 94 °C for 2 min followed by 32 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min, and a final incubation at 72 °C for 7 min. Glyceraldehyde-3-phosphatedehydrogenase (GAPDH) and β-actin expression were analyzed using the same cycling parameters for 27 cycles and postsynaptic density protein 95 for 30 cycles. The following primers were used: mouse BDNF, upstream: 5′-TTGAGCACGTGATCGAAGAGC-3′ and downstream: 5′-GTTCGGCATTGCGAGTTCCAG-3′; mouse GAPDH, upstream: 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′ and downstream: 5′-CATGTAGGCCATGAGGTCCACCAC-3′; GFP, upstream: 5′-CACATGAAGCA GCACGACTT-3′and downstream: 5′-CATTGTGGGCGTTGTAGTTG-3′; mouse postsynaptic density protein 95, upstream: 5′-GACTGCGGTTTCTTGAGC-3′ and downstream: 5′-GTTGGCACGGTCTTTGGT-3′; mouse β-actin, upstream: 5′-CACTGTGCCCATCTACGA-3′ and downstream: 5′-CAGGATTCCATACCCAA G-3′; mouse NGF, up stream: 5′-ACACTCTGGATCTAGACTTCCAGG-3′ and downstream: 5′-AGGCAAGTCAGCCTCTTCTTGTAG-3′. PCR products were separated as we described17 and photographed by ChemiDoc XRS+ (BioRad, CA, USA). The signals of the PCR products were quantified by densitometry using the NIH ImageJ 1.61 program (National Institutes of Health, Bethesda, MD, USA). The intensities of signals were normalized to GAPDH.
Real-time quantitative reverse transcriptase PCR (qRT-PCR)
qRT-PCR for miRNA detection was performed using the miRCURY LNA Universal RT miRNA PCR system according to the manufacturer's instructions. An RNA spike-in (internal control) was added to the RNA just before the complementary DNA synthesis to control the quality of the RT reaction for each sample. Each data point, derived from qRT-PCR assays, represents an average of three replicates and data were normalized using small nuclear U6 RNA. Relative miRNA expression was determined by calculating the mean difference between the cycle threshold (CT) of the miRNA of interest and the U6 small nuclear RNA normalization control for each sample (ΔCT). Data (ΔCT) were averaged over independently replicated experiments and expressed as the mean fold change over the mean of the control group (2−ΔΔCT) ±s.e.m. Experiments were run on an Applied Biosystems 7900HT real-time PCR instrument using the SYBR green-based real-time PCR reaction kit with specific miRNA primer sets (mmu-miR-1: MIMAT0000123, targeted sequence: 5′-UGGAAUGUAAAGAA GUAUGUAU-3′; mmu-miR-30a-5p: MIMAT0000128, targeted sequence: 5′-UGUAAACAUCCUCGACUGGAAG-3′; mmu-miR-195: MIMAT0000225, targeted sequence: 5′-UAGCAGCACAGAAAUAUUGGC-3′; mmu-miR-124: MIMAT0004527, targeted sequence: 5′-CGUGUUCACAGCGGACCUUG AU-3′). Primer sets for 5s ribosomal RNA (rRNA) and an internal control were used as controls.
qRT-PCR for BDNF RNA samples were treated with DNase prior to RT using the Reverse Transcription System. The resulting complementary DNA samples were amplified by TaqMan quantitative PCR using commercially available primer/probe kits from Applied Biosystems for BDNF (Gene Expression Assay Mm00432069_m1) and GAPDH Gene Expression Assay (Mm99999915_g1) was used as an internal control as described in ref. 21.
Generation of wild type untranslated region (Wt-3′UTR)(BDNF) and mutant (Mut)-3′UTR(BDNF) plasmids
Wt-3′UTR(BDNF) RNA was isolated and reverse transcribed to complementary DNA. The PCR product of 976 bp contained a putative miR-30a-5p binding site (1824-GTTTACA-1832).
Site directed mutagenesis
Site-Directed mutagenesis of 1824-GTTTACA-1832 (Wt-3′UTR(BDNF)) to 1824-ACATTTC-1832 (Mut-3′UTR(BDNF) was conducted using Q5 Site-Directed Mutagenesis Kit. The following primers were used for the mutagenesis: 5′-ACATTTCTTTTAGACACTAAGTATCTTC-3′ and 5′-GGA ATG TTT TGG TTC AAA T-3′. The generation of Wt-3′UTR(BDNF) and Mut-3′UTR(BDNF) was confirmed by sequencing.
Preparation of reporter plasmids
MBP-Flag-GFP sequence used as reporter detection method was inserted into NheI and KpnI sites of pUSEamp(+) vector, downstream of the cytomegalovirus promoter of the vector. Wt-3′UTR(BDNF) and Mut-3′UTR(BDNF) were then PCR cloned into pUSE-MBP-Flag-GFP at the KpnI sites to generate pUSE-MBP-Flag-GFP-Wt-3′UTR(BDNF) and pUSE-MBP-Flag-GFP-Mut-3′UTR(BDNF).
Generation of plasmids for miR-30a-5p and scrambled (SCR) sequence expression
miR-30a-5p MIMAT0000128, (sense, 5′-GATCCCGCGACTGTAAACATCCTCG ACTGGAAGCTGTGAAGCCACAGATGGGCTTTCAGTCGGATGTTTGCAGCTGCT TTTTTCCAAA-3′; antisense 5′-AGCTTTTGGAAAAAAGCAGCTGCAAACAT CCGACTGAAAGCCCATCTGTGGCTTCACAGCTTCCAGTCGAGGATGTTTACAGT CGCGG-3′), and a SCR sequence (sense, 5′-GATCCCGCGACGAAGGTCA GCTCCTACAAATGTCTGTGAAGCCACAGATGGGCGACGTTTGTAGGCTGACTTT CTGCTTTTTTCCAAA-3′; antisense, 5′-AGCTTTTGGAAAAAAGCAGAAAGTCA GCCTACAAACGTCGCCCATCTGTGGCTTCACAGACATTTGTAGGAGCTGACCTT CGTCGCGG-3′) with overhangs of BamHI and HindIII restriction sites were synthesized and cloned into a small RNA expression vector, pRNAT-H1.1/Shuttle, downstream of an H1 promoter. The plasmid cassette containing the miR-30a-5p (pRNAT-H1.1-miR-30a-5p) or SCR sequence (pRNAT-H1.1-SCR) also includes GFP for detection.
Construction of the adenovirus (Adv)
The miR-30a-5p or SCR sequences were subcloned into the Adv backbone vector pAdeno-X using the I-Ceu I and PI-Sce I restriction sites. AdV-miR-30a-5p and Adv-SCR were prepared and purified using the Adeno-X Virus Purification Kit and titered using the Adeno-X Rapid Titer Kit.
Cell culture
HEK 293FT cells (Life Technologies) were plated at a density of 2×105 cells per well (six-well plates) in Dulbecco's Modified Eagle Medium supplemented with fetal bovine serum (10%), penicillin/streptomycin, and non-essential amino acids. Cells were co-transfected with either pRNAT-H1.1-miR-30a-5p or pRNAT-H1.1-SCR (4 μg per well) and pUSE-MBP-Flag-GFP-Wt-3′UTR(BDNF) or pUSE-MBP-Flag-GFP-Mut-3′UTR(BDNF) (2 μg per well). Cells were harvested 24 h after transfection and the levels of Wt-3′UTR(BDNF) and Mut-3′UTR(BDNF) were analyzed by western blot.
Western blot analysis
HEK 293FT cells were harvested in radio-immunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 5 mM EDTA, 120 mM NaCl, 1% NP-40, 0.1% deoxycolate and 0.5% SDS) at 4 °C. Thirty μg of HEK 293FT homogenates were resolved on a NuPAGE 10% Bis-Tris gel and transferred onto nitrocellulose membranes. Blots were blocked with 5% milk-phosphate buffered saline with tween 20, and then incubated overnight at 4 °C in the blocking solution including mouse anti-Flag (1:2000), rabbit anti-MPB (1:1000), mouse anti-GFP (1:2000) or goat anti-Actin (1:2000) antibodies. Membranes were then washed and probed with the appropriate Horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Membranes were visualized using Enhanced Chemiluminescence. Band intensities were quantified by ImageJ program (National Institutes of Health).
Immunochemistry
Five days after viral infusion, mice were deeply anesthetized with pentobarbital and perfused with 0.9% NaCl, followed by 4% paraformaldehyde in phosphate buffer, pH 7.4. Brains were removed, fixed in the same fixative for 2 h, and transferred to PBS at 4 °C. The following day, brains were transferred into 30% sucrose and stored at 4 °C until the brain sank to the bottom of the tube. Frozen 50 μm thick coronal sections were cut on a cryostat (Microm; Thermo Scientific, Wilmington, DE, USA) and collected into 24-well dishes. Free-floating sections containing the injection site in the mPFC were selected. Coronal sections were blocked with 5% normal donkey serum in PBS for 1 h and then incubated for 24 h at 4 °C on an orbital shaker with antibodies for either a neuronal marker (anti-NeuN antibody, 1:500) or a glial marker (anti-GFAP antibody, 1:1000) in combination with the anti-GFP antibody (1:5,000) and diluted in PBS plus 3% BSA and 0.05% Triton X-100. The sections were then washed three times for 5 min each in PBS followed by incubation for 4 h with the following secondary antibodies: Alexa Fluor 488-labeled donkey anti-rabbit and Alexa Fluor 594-labeled donkey anti-mouse (both at 1:500). After staining, the sections were washed three times for 5 min each in PBS, and mounted in ProLong Gold mounting medium. Images were acquired using Zeiss LSM 510 META laser confocal microscope (Zeiss, Jena, Germany).
Adenoviral infection of the mPFC
Mice were anesthetized using a mixture of ketamine (120 mg kg-1) and xylazine (8 mg kg-1). Bilateral microinfusions were made using stainless-steel injectors (33 gauge, Small Parts) into the mice mPFC (the stereotaxic coordinates were anteroposterior +2.4 mm from bregma; mediolateral ± 0.35 mm from bregma and dorsoventral −2.25 mm from the skull surface). Animals were infused with AdVs miR-30a-5p and AdV-SCR (1.0 μl per injection) at a concentration of 1 × 108 infectious units (ifu) per ml and at an injection rate of 0.1 μl min-1. After each infusion, the injectors were left in place for an additional 10 min to allow the virus to diffuse. Mice recovered for 5 days before experiments were initiated. For each subject, the infected area was verified in 50 μm coronal sections using Zeiss LSM 510 META laser confocal microscope (Zeiss). Animals showing localized infections in the mPFC were included in the studies.
Locked Nucleic Acid (LNA)-miRNA administration
Fluorescently-labeled in vivo LNA targeting miRNA-30a-5p (MIMAT0000128, LNA-miR-30a-5p, sequence: 5′-GTC GAG GAT GTT TAC-3′) and SCR (LNA-SCR, sequence: 5′-ACG TCT ATA CGC CCA-3′) were synthesized. LNA-miR-30a-5p and LNA-SCR were dissolved in PBS at a concentration of 25 mM.25 Guide cannulae were placed in the mPFC and cemented into place (anteroposterior +2.4 mm from bregma; mediolateral±0.4 mm from bregma and dorsoventral −2.25 mm from the skull surface) and 7 days after recovery the in vivo LNA miRNA inhibitor and SCR were infused 2 h before access to alcohol. LNA sequences (1ul) were infused over 2 min and the injectors remained in position for an additional 2 min. At the end of the procedure, the site of cannulae implantation was verified by histology (Supplementary Figure 6). Animals showing correct cannulae placements in the mPFC were included in the studies.
Preparation of solutions
Alcohol solution was prepared from ethyl alcohol absolute anhydrous (190 proof) diluted to 10 or 20% (v/v) in tap water. Saccharin (Sigma-Aldrich) was dissolved in tap water.
Drinking paradigms
Intermittent access (IA) to 20% alcohol, two bottle choice paradigm (2-BC)
Mice had access to one bottle of 20% alcohol (v/v) and one bottle of water for 24 h, starting at 1200 hours (that is, 2 h after the lights turn off). Bottles were available on Mondays, Wednesdays and Fridays for 21 sessions over a total of 49 days.26 Alcohol and water intake were recorded immediately after the first 4 h of access (to assess binge-like drinking) and at the end of the 24 h of access to alcohol.
Continuous access of 10% alcohol
Mice had continuous access to one bottle of 10% alcohol (v/v) and one bottle of water in their home cage for 21 consecutive days.17 Alcohol and water consumption were recorded daily.
Saccharin consumption
Mice were tested for saccharin intake using a 2-BC paradigm. The procedure was similar to the 20% alcohol IA described above, except that saccharin (0.01%) solution was used instead of alcohol.
Histology
Mice with implanted cannulae received an intraperitoneal injection of pentobarbital followed by transcardial perfusion with 4% paraformaldehyde. Cannula placement was verified using a classic Bright Field microscope.
Statistical analyses
Statistical analyses were performed using SIGMASTAT statistical software (Systat, San Jose, CA, USA). Gene expression and miRNA data were analyzed by unpaired two-tailed t-test. Correlations were analyzed by linear regression, and the effect size (R2 value) was calculated. Western blot analysis data were analyzed by two-way analysis of variance. Behavioral experiments were analyzed by two-way analysis of variance with repeated measures. All post-hoc analyses were performed using the Student– Newman–Keuls test.
Results
Excessive consumption of alcohol decreases BDNF mRNA and increases miRNA levels in the mPFC
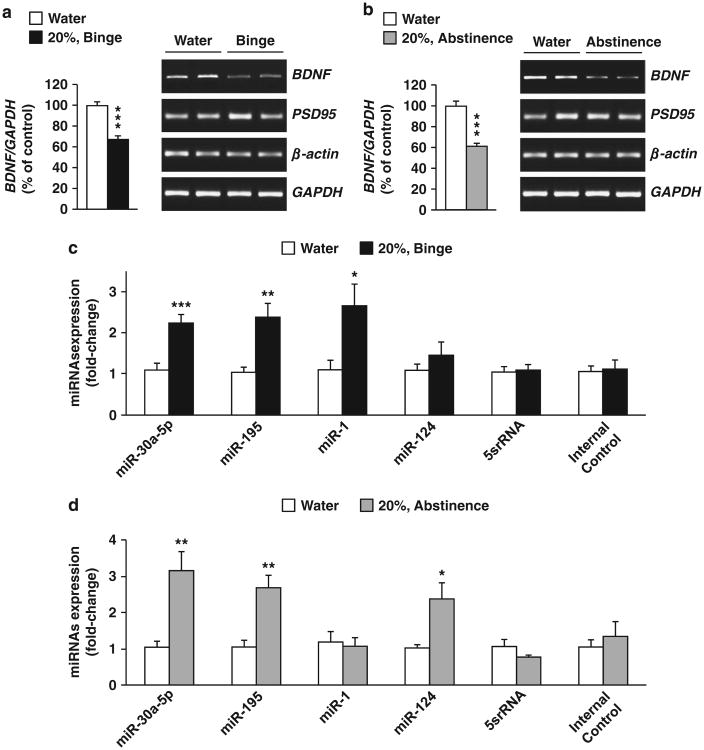
As the mPFC has a major role in the transition from a moderate to an excessive alcohol intake,23 we focused our studies on this cortical region and first tested whether repeated cycles of binge drinking and abstinence alter BDNF expression in the mPFC. Mice underwent IA to 20% alcohol for 7 weeks, a total of 21 sessions of 24 h access (Supplementary Figure 1A). This procedure generates an escalation in voluntary alcohol consumption that models episodic heavy drinking in humans.27 Specifically, repeated cycles of alcohol intake and abstinence produced over time a high level of intake in a 24-h session, and a binge-like alcohol drinking pattern during the first 4 h of alcohol access (Supplementary Table 1). Binge-like alcohol drinking in mice produces blood alcohol concentration of ∼100 mg%,28 that corresponds to binge drinking in humans.29 As shown in Figures 1a and b and Supplementary Figure 2A, repeated cycles of excessive alcohol intake and abstinence produced a robust reduction in BDNF expression in the mPFC, which was tightly correlated with levels of alcohol intake (Supplementary Figure 2B). Alcohol-mediated reduction of BDNF levels in the mPFC was specific as the expression of Actin and postsynaptic density protein 95 were unaltered (Figures 1a and b).
Figure 1.
Excessive alcohol drinking decreases brain-derived neurotrophic factor (BDNF) expression and increases microRNA (miRNA) expression in the mPFC. (a–d) Mice underwent IA to 20% alcohol using a 2-BC procedure for a total of 21 drinking sessions while a second group consumed water only. The medial PFC (mPFC) was collected 4 h after the beginning of the last alcohol drinking session (a and c, Binge) or 24 h after the end of the last alcohol drinking session (b and d, Abstinence). (a and b) BDNF, PSD95, β-actin and GAPDH messenger RNA (mRNA) were measured by reverse transcriptase PCR (RT-PCR). Left, BDNF mRNA data are expressed as a mean ration of BDNF/ GAPDH and plotted as percentage of water control ±s.e.m. Right, representative images of the RT-PCR results. (c and d) miRNAs expression were measured by qRT-PCR. Data are expressed as mean ± s.e.m. fold change (2–ΔΔCt), normalized to control U6 small nuclear RNA (snRNA) levels. ***P<0.001, **P<0.01 and *P<0.05, two-tailed unpaired t-test. (a) BDNF (t(6) = 7.32, P<0.001), (b) BDNF (t(6) = 6.65, P<0.001), (c) miR-30a-5p (t(15) = 4.27, P<0.001), miR-195 (t(15) = 3.37, P= 0.004), miR-1 (t(15) = 2.46, P = 0.03), miR-124 (t(15)= 0.94, P=0.37), 5srRNA, (t(15) = 0.05, P = 0.79), Internal Control (t(15) = 0.06, P = 0.82), (d) miR-30a-5p (t(10) = 2.14, P = 0.007), miR-195 (t(10) = 3.80, P = 0.003), miR-1 (t(10) = 0.13, P = 0.90), miR-124 (t(10) = 2.60, P =0.03), 5srRNA (t(10) = 1.72, P = 0.12) and Internal Control (t(10) = 0.54, P = 0.60). (a, b) n = 4 (c) n =7–10 and (d) n=5–7.
Next, we tested whether the levels of miRNAs were altered in the mPFC in response to excessive alcohol intake using the same samples used to generate the data shown in Figures 1a and b. We focused our analysis on miR-30a-5p and miR-195, that were shown to negatively regulate BDNF levels in the cortex,12,13 and miR-124 and miR-1 that were reported to reduce the expression of BDNF in NG108-15 cells and HEK293 cells, respectively.10,11 As shown in Figures 1c and d, binge-like alcohol drinking and abstinence increased the levels of miR-30a-5p, miR-195 whereas miR-1 levels were increased only in response to a binge drinking session and miR-124 only during abstinence. In contrast, the expression of BDNF (Supplementary Figures 3A and B) and the miRNAs (Supplementary Figures 3C and D) were unaltered in the hippocampus of mice consuming high levels of alcohol. Furthermore, the expression of the endogenous control (5srRNA) and the internal control (an RNA spike-in that was added to the sample just before the complementary DNA synthesis to control the quality of the reverse transcription) were unaltered in response to alcohol intake in both regions (Figures 1c and d and Supplementary Figures 3C and D). Together, these results show that binge drinking and abstinence reduced BDNF expression in the mPFC, which was associated with elevated levels of miR-30a-5p and miR-195.
Moderate consumption of alcohol does not alter BDNF or miRNAs levels in the mPFC
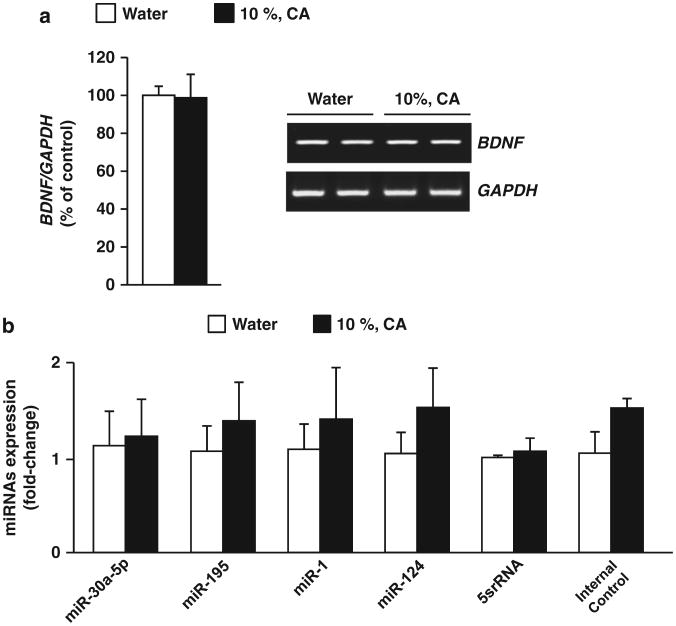
Next, we used a model in which mice had continuous access to 10% alcohol and water for 21 days (Supplementary Figure 1A). This paradigm leads to a moderate level of alcohol intake (Supplementary Table 1). We measured BDNF mRNA and miRNAs targeting BDNF in the mPFC at the end of the final alcohol drinking session and found that the expression of BDNF (Figure 2a) or the tested miRNAs (Figure 2b) were unaltered in response to consumption of moderate levels of alcohol. Similarly, BDNF mRNA expression (Supplementary Figure 3E) and the levels of miRNAs (Supplementary Figure 3F) were also unchanged in the hippocampus. Together, these results suggest that miR-30a-5p, miR-195, miR-1 and miR-124 in the mPFC are elevated only in response to excessive but not moderate alcohol drinking, and are tightly correlated with BDNF expression.
Figure 2.
Moderate alcohol intake does not alter brain-derived neurotrophic factor (BDNF) and MicroRNA (miRNA) expression in the medial PFC (mPFC). Mice underwent a procedure of continuous access to alcohol 10% or water only for 21 days and mPFC was collected 6 h after the beginning of the dark cycle of the last drinking session (a) BDNF mRNA in the mPFC was measured by reverse transcriptase PCR (RT-PCR). Data are expressed as a mean ratio of BDNF/GAPDH and plotted as percentage of water control ±s.e.m. Right, representative image of the RT-PCR results. (b) miRNAs expression were measured by quantitative RT-PCR. Data are expressed as mean ±s.e.m. fold change (2–ΔΔCt), normalized to control U6 small nuclear RNA (snRNA) levels. (a) BDNF (t(5) = 0.78, P = 0.47) (b) miR-30a-5p (t(6) = -0.17, P = 0.87), miR-195 (t(6) = -0.56, P = 0.60), miR-1 (t(6) = -0.42, P = 0.69), miR-124 (t(6) = - 0.828, P = 0.44), 5srRNA (t(6) = -0.33, P = 0.75) and Internal Control (t(6) = -1.615, P = 0.20), (a) n = 3-4 and (b) n = 4.
miR-30a-5p targets the 3′UTR of BDNF and downregulates BDNF expression in the mPFC
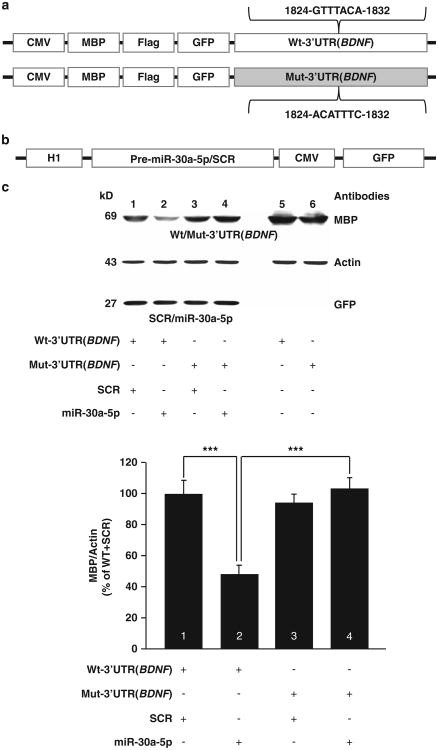
We next sought to determine whether the increases of miRNA levels, and the consequent decreases in BDNF expression in the mPFC have a role in the development of excessive alcohol drinking. We focused our analysis on miR-30a-5p, a miRNA is enriched in layer III pyramidal neurons, that inversely associated with BDNF protein levels in human PFC12,13 and in mouse cortex.13 Furthermore, this miRNA was increased in response to both binge drinking of alcohol (Figure 1c) and after a period of 24 h of abstinence (Figure 1d). First, we verified that miR-30a-5p is indeed targeting BDNF by conducting an in silico analysis to identify a putative miR-30a-5p binding site within the mouse BDNF gene (NM_007540). Using microRNA.org30 we discovered a potential mi-R30a-5p binding site (1824-GTTTACA-1832) within the 3′UTR region of BDNF. Next, we designed plasmids that express the Wt form of 3′UTR(BDNF) (Wt-3′UTR(BDNF) or a mutated form of the 3′UTR(BDNF) in which the putative mi-R30a-5p binding site was mutated (Mut-3′UTR(BDNF)) (Figure 3a). We then co-transfected HEK293 cells with plasmids expressing miR-30a-5p or a scramble sequence of miR-30a-5p (SCR) (Figure 3b), and plasmids expressing Wt-3′UTR(BDNF) or Mut-3′ UTR(BDNF). 3′UTR(BDNF) expression was tested by the use of MBP (Figure 3c) and Flag (Supplementary Figure 4) as surrogate detection tags. As shown in Figure 3c and Supplementary Figure 4, overexpression of miR-30a-5p but not SCR produced a reduction in Wt-3′UTR(BDNF) expression (lanes 1 versus 2), which was abolished upon co-transfection of miR-30a-5p with Mut-3′ UTR(BDNF) (lanes 2 versus 4). Together, these results show the miR-30a-5p targets BDNF for degradation by binding to its 3′-UTR sequence.
Figure 3.
miR-30a-5p downregulates brain-derived neurotrophic factor (BDNF) expression by binding to the 3′UTR of BDNF (a) Structure of plasmids for expression of wild-type (Wt) or mutated (Mut) form 3′ UTR sequence of the mouse BDNF gene. (b) Structure of plasmids for expression of miR-30a-5p or a scramble (SCR) sequences. (c) miRNA-30a-5p directly targets the 3′UTR(BDNF) for messenger RNA degradation. HEK 293FT cells were transfected with pUSE-MBP-Flag-GFP-Wt-3′UTR(BDNF) and pRNAT-H1.1-SCR (1), pUSE-MBP-Flag-GFP-Wt-3′UTR(BDNF) and pRNAT-H1.1-miR-30a-5p (2), pUSE-MBP-Flag-GFP-Mut-3′UTR(BDNF) and pRNAT-H1.1-SCR (3), pUSE-MBP-Flag-GFP-Mut-3′UTR(BDNF) and pRNAT-H1.1-miR-30a-5p (4). Confirmation of equal transfection levels of pUSE-MBP-Flag-GFP-Wt-3′UTR(BDNF) (5) and pUSE-MBP-Flag-GFP-Mut-3′UTR(BDNF) (6). Cells were harvested 24 h after transfection and western blot analysis was used for detection. Anti-MBP antibodies were used to detect the levels of Wt-3′UTR(BDNF) and Mut-3′UTR(BDNF), anti-actin antibodies were used as a loading control and anti-GFP antibodies were used to confirm equal levels of transfection of miR-30a-5p and SRC. Data are expressed as a mean ratio of MBP/Actin and plotted as percentage of control (Wt-3′UTR(BDNF) and SCR)±s.e.m. Two-way RM-ANOVA revealed a significant interaction between Wt-3′UTR (BDNF) or Mut-3′UTR(BDNF) and SCR or miR-30a-5p (F(1,20) = 15.5, P<0.001). SNK post-hoc test: ***P<0.001. n =6.
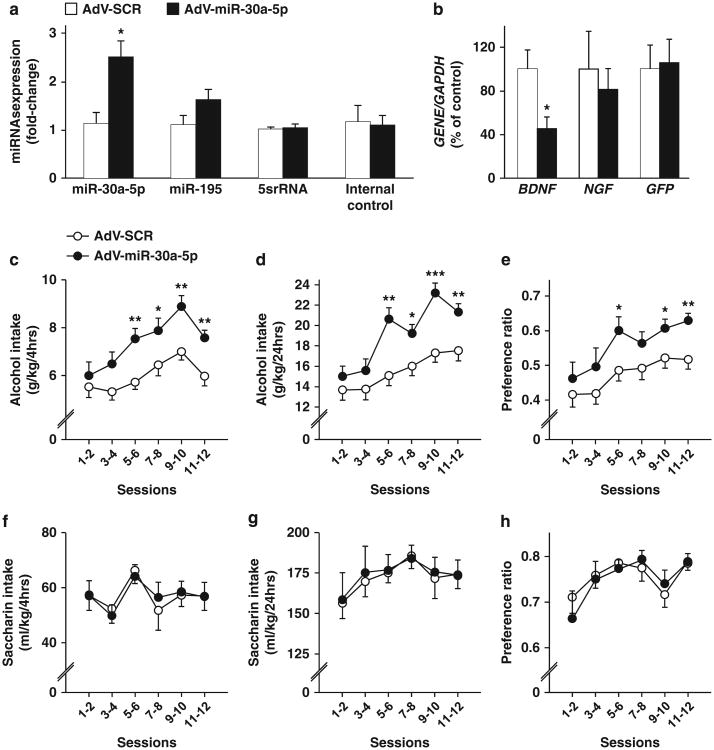
Next, we generated an adenovirus expressing miR-30a-5p (AdV-miR-30a-5p) or the SCR sequence of miR-30a-5p (AdV-SCR). AdV-miR-30a-5p or AdV-SCR was stereotaxically infused into the mPFC and molecular measurements were performed 5 days later (Supplementary Figure 1B). AdV-miR-30a-5p selectively infected mPFC neurons but not glia (Supplementary Figure 5A) and produced a significant and specific increase in the level of miR-30a-5p as the levels of miR-195, 5srRNA as well as the internal control were unaltered (Figure 4a). As shown in Figure 4b and Supplementary Figure 5B, overexpression of AdV-miR-30a-5p but not AdV-SCR led to a significant decrease in BDNF expression in naïve mice. To test the specificity of miR-30a-5p in targeting BDNF, we examined the mRNA levels of the NGF, a member of the BDNF neurotrophin family15 whose expression is not predicted to be targeted by miR-30a-5p (miRWalk online database31). As shown in Figure 4b and Supplementary Figure 5B, NGF expression was unaltered in response to the overexpression of miR-30a-5p.
Figure 4.
Overexpression of miR-30a-5p in the medial PFC (mPFC) decreases brain-derived neurotrophic factor messenger RNA (BDNF mRNA) and enhances the development of excessive alcohol drinking without altering saccharin intake. Five days after the infusion of AdV-SCR or AdV-miR-30a-5p, the mPFCs were collected and analyzed by quantitative reverse transcriptase PCR (qRT-PCR) for micro RNA (miRNA) analysis (a) and RT-PCR for mRNA analysis (b). (a) Data are expressed as mean± s.e.m. fold change (2–ΔΔCt), normalized to control U6 small nuclear RNA (snRNA) levels (miR-30a-5p (t(8) = - 3.29, P = 0.01), miR-195(t(8) = - 1.63, P = 0.14), 5srRNA (t(8) = - 0.74, P = 0.47) and Internal Control (t(8) = -0.14, P = 0.90), *P<0.05, two-tailed t-test). (b) Data are expressed as mean a mean ratio of GENE/GAPDH and plotted as percentage of Adv-SCR (control) ± s.e.m. (BDNF (t(8) = 2.85, P = 0.02), NGF (t(8) = 0.48, P = 0.64) and GFP (t(8) = 0.18, P =0.86)), *P<0.05, two-tailed t-test. (c–h) After 5 days of recovery from the surgery, mice were tested in the IA to 20% alcohol procedure (c–e) or consumption of 0.01% saccharin (f–h). Amount of alcohol (g kg-1) (c) or saccharin (ml kg-1) (f) consumed during the first 4 h of 20% alcohol access. Amount of alcohol (g kg-1) (d) or saccharin (ml kg-1) (g) consumed during the 24 h of 20% alcohol access. Alcohol (e) or saccharin (h) preference was calculated as the percentage of alcohol or saccharin solution consumed relative to total fluid intake (alcohol or saccharin+water). Results are expressed as mean±s.e.m. Two-way analysis of variance with repeated measures (RM-ANOVA) with Student–Newman–Keuls post-hoc tests. (c) Two-way RM-ANOVA, revealed a main effect of the overexpression of miR-30a-5p in the mPFC (F(1,24) = 13.9, P=0.001) and a main effect of the session (F(5,120) = 8.7, P<0.001), but no interaction between infusion and session (F(5,120) = 0.90, P=0.48). (d) Two-way RM-ANOVA revealed a main effect of the miR-30a-5p overexpression (F(1,24) = 9.44, P<0.01), a main effect of the session (F(5,120) = 31.21, P<0.001), and an interaction between miR-30a-5p overexpression and session (F(5,120) =4.64, P<0.001). (e) Two-way RM-ANOVA revealed a main effect of the overexpression of miR-30a-5p (F(1,24) = 4.13, P= 0.05) and a main effect of the session (F(5,120) = 12.06, P<0.001, but no significant interaction between miR-30a-5p overexpression and session (F(5,120) = 0.64, P = 0.67), (f) (F(1,14) = 0.001, P = 0.97), (g) (F(1,14) = 0.04, P = 0.84) and (h) (F(1,14) = 0.06, P =0.81), *P<0.05, **P<0.01, ***P<0.001 compared with SCR. (a, b) n = 5. (c–e) n = 12–14, (f–h) n = 8.
Overexpression of miR-30a-5p in the mPFC increases alcohol consumption
To test the effect of overexpression of miR-30a-5p in mPFC on alcohol drinking, mice received a stereotaxic infusion of AdV-miR-30a-5p or AdV-SCR into the mPFC. After 5 days of recovery from surgery, mice underwent an IA to 20% alcohol 2-BC procedure (Supplementary Figure 1B). As shown in Figure 4c, overexpression of miR-30a-5p in the mPFC increased alcohol intake during the first 4 h of alcohol access, which was also detected during the whole session (Figure 4d). Alcohol preference was also increased as a consequence of the overexpression of miR-30a-5p in the mPFC (Figure 4e), however, total fluid intake was unaltered (data not shown, two-way RM-ANOVA (F(1,24) = 0.57, P= 0.46)). We then assessed whether miR-30a-5p alters the propensity to consume another rewarding solution, saccharin (Supplementary Figure 1B), and as shown in Figures 4f–h, saccharin drinking and preference or total body fluid (data not shown, two-way RM-ANOVA, (F(1,24) = 0.07, P=0.80) were unaltered in response to overexpression of miR-30a-5p in the mPFC. Together, these results suggest that an increase of miR-30a-5p, a miRNA that decreases BDNF mRNA expression in mPFC, leads to an escalation from moderate to excessive alcohol consumption.
Inhibition of miR-30a-5p levels in the mPFC adjusts the level of alcohol drinking
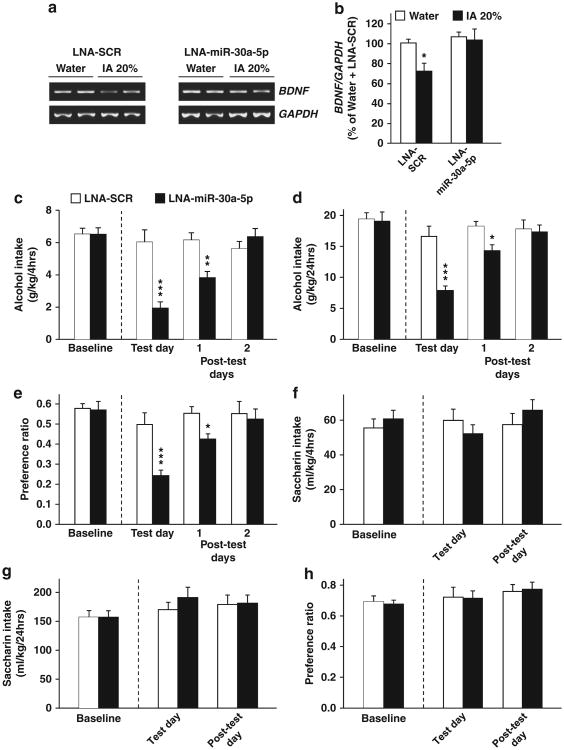
Since overexpression of miR-30a-5p in the mPFC led to a decrease in BDNF levels and increased alcohol drinking, inhibiting the action of miR-30a-5p should result in the opposite effects. To address this prediction, we used the LNA approach, in which a complementary sequence is designed to bind a specific miRNA with high specificity and inhibit its function.3,25 After 21 sessions of IA to 20% alcohol, mice were implanted with bilateral guide cannulae in the mPFC, and after 7 days of recovery, LNA-miR-30a-5p or its control LNA-SCR (1 μl per injection site, 25 mM of LNA in PBS) was infused into the mPFC 22 h after the last drinking session, and BDNF expression was measured at the end of the 24 h abstinence session (Supplementary Figures 1C and 6). As shown in Figure 5a left panel and Figure 5b, repeated cycles of excessive alcohol drinking produced a reduction in BDNF expression in the mPFC of mice infused with LNA-SCR. A similar reduction in BDNF levels was observed in naïve mice that underwent the same drinking paradigm (Figure 1b). Importantly, alcohol-mediated reduction in BDNF levels was abolished following intra-mPFC infusion of LNA-miR-30a-5p (Figures 5a and b).
Figure 5.
Inhibition of miR-30a-5p in the medial PFC (mPFC) restores brain-derived neurotrophic factor (BDNF) expression and reduces alcohol intake and preference. Mice were trained to consume alcohol using the Intermittent access (IA) to 20% alcohol procedure for 21 sessions before they were implanted with a cannula guide in the mPFC. Seven days later, mice were microinjected with Locked Nucleic Acid (LNA)-miR-30a-5p or LNA-SCR in the mPFC. (a, b) Inhibition of miR-30a-5p in the mPFC restores BDNF mRNA expression in response to excessive alcohol intake. LNA-SCR or LNA miR-30a-5p (25 mM) was infused into the mPFC after 22 h of abstinence, and the mPFC was removed 2 h after the infusion. Data are expressed as a mean ratio of BDNF/GAPDH and plotted as percentage of mice consuming water and infused with LNA-SCR± s.e.m. (b) Two-way analysis of variance (ANOVA) revealed a significant main effect of alcohol (F(1,11) = 4.43, P<0.05), and a significant main effect of the LNA-miR-30a-5p (F(1,11) = 5.40, P<0.05); n=3–4. (c–e) Mice undergoing intermittent IA to 20% alcohol were infused with LNA-SCR or LNA-miR-30a-5p (25 mM) into the mPFC after 22 h of abstinence. (c) Amount of alcohol (g kg-1) consumed during the first 4 h of alcohol access. (d) Amount of alcohol (g kg-1) consumed during 24 h of alcohol access. (e) Preference for alcohol was calculated as the ratio of the volume of alcohol solution intake/volume of total fluid intake during a 24 h session. (f–h) After 1 week of abstinence from alcohol, mice underwent intermittent access to 0.01% saccharin 2-bottle choice for 24 h and were then infused with LNA-SCR or LNA miR-30a-5p (25 mM) into the mPFC 2 h before the beginning of the drinking session. (f) Amount of saccharin (ml kg-1) consumed during the first 4 h of access. (g) Amount of saccharin (ml kg-1) consumed during the 24 h of access. (h) Preference for saccharin solution was calculated as described in e. (c–h) Results are expressed as mean±s.e.m., Two-way ANOVA with repeated measures (RM-ANOVA) with Student–Newman–Keuls post-hoc tests, (c) Two-way RM-ANOVA showed a main effect of LNA-miR-30a-5p treatment (F(1,14) = 19.1, P<0.001), and a main effect of session (F(2,28) = 81.7, P<0.001) and a significant interaction between LNA-miR-30a-5p treatment and session (F(2,28) = 89.8, P<0.001). (d) Two-way RM-ANOVA revealed a main effect of LNA-miR-30a-5p treatment (F(1,14) = 13.1, P<0.01), a main effect of the session (F(2,28) = 16.3, P<0.001), and a significant interaction between LNA-miR-30a-5p treatment and session (F(2,28) = 9.2, P<0.001), (e) Two-way RM-ANOVA showed a main effect of LNA-miR-30a-5p treatment (F(1,14) = 9.3, P<0.01), a significant main effect of the session (F(2,28) = 10.3, P<0.001) and a significant interaction LNA-miR-30a-5p treatment and session (F(2,28) = 4.5, P<0.05). (f) (F(1,14) = 0.001, P = 0.97). (g) (F(1,14) = 0.39, P = 0.54) and (h) (F(1,14) = 0.003, P=0.96). *P<0.05, *P<0.01 and ***P<0.001 compared with mice infused with LNA-SCR the same day. n=8.
We then examined the possibility that reducing miR-30a-5p function and the corresponding restoration of BDNF levels would then result in a decrease in alcohol drinking. To do so, mice underwent 21 sessions of IA to 20% alcohol, LNA-miR-30a-5p or LNA-SCR was then infused into the mPFC 2 h before the beginning of the drinking session (Supplementary Figure 1C). As shown in Figures 5c and d, intra-mPFC infusion of LNA-miR-30a-5p but not LNA-SCR reduced alcohol drinking during the first 4 h of alcohol access and over the whole 24-h drinking session, which corresponded with a decrease in preference for alcohol (Figure 5e). Interestingly, LNA-miR-30a-5p-mediated decrease in alcohol drinking and preference was still observed 48 h after LNA-miR-30a-5p administration (Figures 5c and e). Saccharin intake was unaltered in response to intra-mPFC infusion of LNA-miR-30a-5p (Figures 5f and h), and similar levels of total fluid intake were obtained for alcohol and water (two-way RM-ANOVA, (F(1,14) = 0.27, P= 0.61)) or saccharin and water (two-way RM-ANOVA, (F(1,14) = 0.40, P = 0.53)). Together, these results suggest that inhibiting miR-30a-5p function in the mPFC and restoring BDNF levels decreases the development of excessive alcohol intake.
Discussion
In this study we provide evidence to suggest that excessive but not moderate intake of alcohol produces a reduction in BDNF expression in the mPFC of mice, which correlates with increased levels of several miRNAs including miR-30a-5p. We further suggest miR-30a-5p has an important role in the development of excessive alcohol drinking. Specifically, we show that the expression of miR-30a-5p was increased, whereas BDNF expression was decreased in the mPFC of mice that consumed large but not moderate amounts of alcohol. We show that miR-30a-5p binds a sequence within the 3′-UTR of BDNF targeting the gene for degradation. Importantly, we report that overexpression of miR-30a-5p in the mPFC increased, whereas the inhibition of the miR-30a-5p attenuated excessive alcohol drinking without altering the consumption of saccharin solution. The mPFC has been implicated in the transition from a moderate to an excessive alcohol intake in rats that consumed 20% alcohol in an IA 2-BC paradigm,23 and more recently, in vivo optogenetic excitation of the mPFC was shown to reduce compulsive cocaine seeking in rats.32 Thus, our results suggest that cortical miR-30a-5p represents an example of a pro-alcohol addiction response element that facilitates the emergence of an alcohol abuse phenotype by inhibiting cortical BDNF mRNA expression.
We show that miR-30a-5p binds to the 3′UTR of BDNF. The 3′UTR(BDNF) is found in two forms, a long and a short form,33 and our results suggest that the miR-30a-5p binding site is located within the long form of 3′UTR(BDNF), and that removing this binding site is sufficient to abolish the reduction of BDNF expression induced by miR-30a-5p. The majority of the long form of 3′UTR(BDNF) is localized in dendrites,33,34 whereas the short form is localized in soma.34 Dendritic BDNF modulates neuronal morphology,33,34 whereas BDNF localized in the soma is important for neuronal survival.34 Thus, it is plausible that miR-30a-5p targets dendritic BDNF mRNA, which in turn, may lead to alterations in synaptic structures after excessive alcohol drinking. This intriguing possibility merits further investigation.
We also report that repeated cycles of excessive alcohol intake and abstinence produce an increase in the levels of other miRNAs (miR-195, miR-124 and miR-1) which have been reported to, or predicted to, target BDNF mRNA, suggesting that these miRNAs may also have a role in regulating alcohol intake. Interestingly, high levels of miR-195 coupled with low levels of BDNF have been found in the PFC of human with schizophrenia,13,35 suggesting that alterations in miR-195-dependent regulation of BDNF expression may have a role in psychiatric disorders including addiction. We found that miR-124 expression is increased in mPFC; however, the increase was detected only after 24 h of abstinence. Interestingly, Bahi and Dreyer36 showed that 2 weeks of continuous access to a low concentration of alcohol (5%) led to a decrease in miR-124 in dorsolateral striatum of rats. In addition, overexpression or silencing of miR-124 in the nucleus accumbens of rats was shown to reduce or promote, respectively, place preference and reinstatement to cocaine.37 These results indicate that miR-124 and its targets, such as BDNF, may have a role in the regulation of drug-related behaviors. We also detected an increase in miR-1 levels but only after a binge drinking session and interestingly this miRNA was shown to be increased in the PFC of human alcoholics.9 Further studies are needed to determine if these miRNAs also contribute to mechanisms underlying uncontrolled alcohol drinking. Interestingly, Tapocik et al.14 recently reported that another miRNA which targets BDNF, miR-206, is increased in the mPFC of rats that were physically dependent on alcohol. The authors further report that overexpression of miR-206 in the mPFC of rats increased operant lever presses for alcohol in non-dependent animals.14 It is of interest to compare and contrast the interaction between miRNA levels in the mPFC and BDNF expression in paradigms that model ‘problem drinkers’ used herein and in models of physical dependence.
The mechanism by which alcohol modulates miRNAs levels will require further investigation. Interestingly, miRNAs expression has been associated with increased neuronal excitability.38 Alcohol withdrawal results in increased neuronal activity.39–41 Thus, it is plausible that microRNAs including miR-30a-5p are increased in response to enhanced neuronal excitability that occurs during periods of abstinence. Another possibility is the potential contribution of chromatin remodeling in response to alcohol to changes in the expression of miRNAs. We, and others, previously showed that voluntary alcohol drinking induces posttranslational modifications on DNA and histones, resulting in alterations in chromatin structure.26,42–45 A recent study suggested that the expression of some miRNAs is associated with Histone methylation,46 thus it is possible that changes in chromatin structure by alcohol is upstream of the alterations in miRNAs expression. These possibilities should be further explored.
Drug addiction is a psychiatric disorder characterized by chronic episodes of relapse that results from alterations in signaling, synaptic plasticity and neuronal connectivity.47,48 These adaptations are orchestrated in part by persistent changes in gene expression,49 which in turn contribute to the maintenance, development and relapse of drug-taking behaviors.48 Our results suggest that alcohol's actions in the brain may be controlled by miRNAs. Each miRNA can potentially target numerous genes,1,4 and therefore it is reasonable to assume that the alterations in miRNAs levels in the mPFC affect the expression of genes other than BDNF. The possibility that miRNAs are the master keys of alterations in gene expression in response to alcohol is intriguing and should also be further explored.
The drinking paradigm presented here models ‘problem drinkers’, people who engage in harmful excessive alcohol use but do not meet all criteria for alcohol use disorders,50 and our study is the first to implicate altered levels of specific miRNA in mechanisms underlying the transition from moderate to uncontrolled excessive alcohol drinking. Excessive use of alcohol results in health, economical and societal burden.51–53 Thus, the identification of biomarkers and new therapeutic targets to treat this disorder is of great importance. Recently, a miRNA expression profile was suggested as a peripheral biomarker for cancer and central nervous system disorders such as schizophrenia.54,55 Therefore, our findings may lead to the identification of potential new biomarkers that could be useful for the identification of people with increased vulnerability to develop alcohol abuse disorders. Furthermore, antisense inhibition of human miRNAs is in early stages of development for cancer therapy.56 Thus, it is plausible that miR-30a-5p represents a new and promising target for the development of therapeutic strategies for the treatment of alcohol abuse disorders.
Supplementary Material
Acknowledgments
We thank Dr Dao Yao He, Ms Melissa Lin, Ms Wenheng Zhu for technical assistance, and Dr Jacob Beckley and Dr Virginia Long for careful review of the manuscript. This research was supported by NIAAA R01 AA016848 (DR), NIAAA R37 AA016848 (DR), NIAAA P50 AA017072 (DR), and the State of California for medical research on alcohol and substance abuse through UCSF (DR).
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 3.Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li MD, van der Vaart AD. MicroRNAs in addiction: adaptation's middlemen? Mol Psychiatry. 2011;16:1159–1168. doi: 10.1038/mp.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, et al. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Liu X, Qin S, Guan Y, Liu Y, Cheng Y, et al. MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction. EMBO Mol Med. 2013;5:1402–1414. doi: 10.1002/emmm.201201900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miranda RC. MicroRNAs and fetal brain development: implications for ethanol teratology during the second trimester period of neurogenesis. Front Genet. 2012;3:77. doi: 10.3389/fgene.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res. 2011;35:1928–1937. doi: 10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrasekar V, Dreyer JL. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci. 2009;42:350–362. doi: 10.1016/j.mcn.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Varend K, Kumar A, Harma MA, Andressoo JO. miR-1, miR-10b, miR-155, and miR-191 are novel regulators of BDNF. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1628-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2009;65:1006–1014. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A, et al. microRNA -206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci. 2014;34:4581–4588. doi: 10.1523/JNEUROSCI.0445-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- 16.Minichiello L. TrkB signalling pathways in LTP and learning. Nat Rev Neurosci. 2009;10:850–860. doi: 10.1038/nrn2738. [DOI] [PubMed] [Google Scholar]
- 17.McGough NN, He DY, Logrip ML, Jeanblanc J, Phamluong K, Luong K, et al. RACK1 and brain-derived neurotrophic factor: a homeostatic pathway that regulates alcohol addiction. J Neurosci. 2004;24:10542–10552. doi: 10.1523/JNEUROSCI.3714-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeanblanc J, He DY, McGough NN, Logrip ML, Phamluong K, Janak PH, et al. The dopamine D3 receptor is part of a homeostatic pathway regulating ethanol consumption. J Neurosci. 2006;26:1457–1464. doi: 10.1523/JNEUROSCI.3786-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeanblanc J, He DY, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. J Neurosci. 2009;29:13494–13502. doi: 10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeanblanc J, Logrip ML, Janak PH, Ron D. BDNF-mediated regulation of ethanol consumption requires the activation of the MAP kinase pathway and protein synthesis. Eur J Neurosci. 2013;37:607–612. doi: 10.1111/ejn.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logrip ML, Janak PH, Ron D. Escalating ethanol intake is associated with altered corticostriatal BDNF expression. J Neurochem. 2009;109:1459–1468. doi: 10.1111/j.1471-4159.2009.06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalivas PW, Volkow ND. The neural basis of addiction: a pathology of motivation and choice. Am J Psychiatry. 2005;162:1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- 23.George O, Sanders C, Freiling J, Grigoryan E, Vu S, Allen CD, et al. Recruitment of medial prefrontal cortex neurons during alcohol withdrawal predicts cognitive impairment and excessive alcohol drinking. Proc Natl Acad Sci USA. 2012;109:18156–18161. doi: 10.1073/pnas.1116523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. Academic press; New York: 2001. [Google Scholar]
- 25.Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling—a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koob GF. Addiction is a reward deficit and stress surfeit disorder. Front Psychiatry. 2013;4:72. doi: 10.3389/fpsyt.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwa LS, Chu A, Levinson SA, Kayyali TM, DeBold JF, Miczek KA. Persistent escalation of alcohol drinking in C57BL/6J mice with intermittent access to 20% ethanol. Alcohol Clin Exp Res. 2011;35:1938–1947. doi: 10.1111/j.1530-0277.2011.01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.NIAAA. Council appoves definition of binge drinking. NIAAA News Lett. 2004;3:3. [Google Scholar]
- 30.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Chen BT, Yau HJ, Hatch C, Kusumoto-Yoshida I, Cho SL, Hopf FW, et al. Rescuing cocaine-induced prefrontal cortex hypoactivity prevents compulsive cocaine seeking. Nature. 2013;496:359–362. doi: 10.1038/nature12024. [DOI] [PubMed] [Google Scholar]
- 33.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orefice LL, Waterhouse EG, Partridge JG, Lalchandani RR, Vicini S, Xu B. Distinct roles for somatically and dendritically synthesized brain-derived neurotrophic factor in morphogenesis of dendritic spines. J Neurosci. 2013;33:11618–11632. doi: 10.1523/JNEUROSCI.0012-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahi A, Dreyer JL. Striatal modulation of BDNF expression using microRNA124a-expressing lentiviral vectors impairs ethanol-induced conditioned-place preference and voluntary alcohol consumption. Eur J Neurosci. 2013;38:2328–2337. doi: 10.1111/ejn.12228. [DOI] [PubMed] [Google Scholar]
- 37.Chandrasekar V, Dreyer JL. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology. 2011;36:1149–1164. doi: 10.1038/npp.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eacker SM, Dawson TM, Dawson VL. The interplay of microRNA and neuronal activity in health and disease. Front Cell Neurosci. 2013;7:136. doi: 10.3389/fncel.2013.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hopf FW, Martin M, Chen BT, Bowers MS, Mohamedi MM, Bonci A. Withdrawal from intermittent ethanol exposure increases probability of burst firing in VTA neurons in vitro. J Neurophysiol. 2007;98:2297–2310. doi: 10.1152/jn.00824.2007. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Lanfranco MF, Gibb SL, Yowell QV, Carnicella S, Ron D. Long-lasting adaptations of the NR2B-containing NMDA receptors in the dorsomedial striatum play a crucial role in alcohol consumption and relapse. J Neurosci. 2010;30:10187–10198. doi: 10.1523/JNEUROSCI.2268-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovinger DM, Roberto M. Synaptic effects induced by alcohol. Curr Top Behav Neurosci. 2013;13:31–86. doi: 10.1007/7854_2011_143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolstenholme JT, Warner JA, Capparuccini MI, Archer KJ, Shelton KL, Miles MF. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PloS One. 2011;6:e21100. doi: 10.1371/journal.pone.0021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stragier E, Massart R, Salery M, Hamon M, Geny D, Martin V, et al. Ethanol-induced epigenetic regulations at the Bdnf gene in C57BL/6J mice. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.38. in press. [DOI] [PubMed] [Google Scholar]
- 46.Mei S, Liu Y, Bao Y, Zhang Y, Min S, Liu Y, et al. Dendritic cell-associated miRNAs are modulated via chromatin remodeling in response to different environments. PloS One. 2014;9:e90231. doi: 10.1371/journal.pone.0090231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCool BA. Ethanol modulation of synaptic plasticity. Neuropharmacology. 2011;61:1097–1108. doi: 10.1016/j.neuropharm.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luscher C, Malenka RC. Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron. 2011;69:650–663. doi: 10.1016/j.neuron.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enoch MA, Goldman D. Problem drinking and alcoholism: diagnosis and treatment. Am Fam Physician. 2002;65:441–448. [PubMed] [Google Scholar]
- 51.Nutt DJ, King LA, Phillips LD. Drug harms in the UK: a multicriteria decision analysis. Lancet. 2010;376:1558–1565. doi: 10.1016/S0140-6736(10)61462-6. [DOI] [PubMed] [Google Scholar]
- 52.World Health Organization (WHO) Global Status Report on Alcohol and Health. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 53.Collins PY, Patel V, Joestl SS, March D, Insel TR, Daar AS, et al. Grand challenges in global mental health. Nature. 2011;475:27–30. doi: 10.1038/475027a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 55.Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PloS One. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.