Abstract
Primary hyperoxaluria type II is a recessive genetic disorder caused by mutations in the GRHPR gene. Although several dozen mutations have been described, all affect coding or transcript splicing. A man suspected of having primary hyperoxaluria type II was heterozygous for a novel single-nucleotide deletion (c.694delC) in GRHPR affecting Gln232, which introduced a premature termination (p.Gln232Argfs*3). Two 5′ UTR variants of unknown significance were also noted. We show that these two variants occur in cis, on the opposite allele, and introduce – immediately upstream of the canonical translation initiation site – a novel out-of-frame translational start site. In vitro studies using the GRHPR 5′ UTR fused to a luciferase reporter show that the variant start site pre-empted initiation at the canonical translational start site, and this was corroborated within the broader context of 1.3 kb of the GRHPR proximal promoter. This latter mechanism may be underappreciated in general; reports of clinically significant functional variation of this type are extremely rare.
Keywords: hyperoxaluria, mutation, translation, human
INTRODUCTION
Primary hyperoxaluria is a rare but potentially devastating autosomal recessive disorder characterized by excessive urinary oxalate excretion, refractory calcium oxalate nephrolithiasis, and often progressive kidney dysfunction (1). Primary hyperoxaluria type II (OMIM 604296, 260000) results from mutations in glyoxylate reductase/hydroxypyruvate reductase (GRHPR; (2, 3)). The phenotype is generally milder than type I, but is marked by severe nephrolithiasis frequently accompanied by progressive chronic kidney disease.
Essentially all mutations described for primary hyperoxaluria directly affect coding or splicing. The most common mutation in primary hyperoxaluria type II, affecting GRHPR, is the c.103delG in exon 2. This mutation, accounting for approximately 40% of cases of type II (4, 5), introduces a frameshift followed by a premature stop at codon 45 (2). An actively curated online database collates and annotates GRHPR mutations in the context of primary hyperoxaluria type II (Primary hyperoxaluria mutation database; http://www.uclh.nhs.uk/OURSERVICES/SERVICEA-Z/PATH/PATHBIOMED/CBIO/Pages/Phmdatabase.aspx). No functional variants have been reported to affect the 5′ UTR of GRHPR or to involve alternative translational initiation.
We report here a novel GRHPR deletion mutation in a case of primary hyperoxaluria type II, resulting in premature truncation of the protein product. More strikingly, the other affected allele in this compound heterozygote involves the introduction of a new and highly active translational start site in the 5′ UTR immediately upstream of – and out of frame with respect to – the canonical GRHPR translational initiation site. This is an exceedingly uncommon mechanism for generating a clinically important hypomorphic or amorphic allele.
CASE REPORT
A man presented in the sixth decade of life for evaluation for recurrent calcium oxalate nephrolithiasis and associated chronic kidney disease. He passed his first stone 40 years prior, and averaged one stone passage episode per month over the preceding two decades. There was no family history of kidney disease or nephrolithiasis and there was no known consanguinity. He underwent multiple urological interventions in the past including extracorporeal shock-wave lithotripsy (ESWL). Stone composition via prior crystallography was 100% calcium oxalate monohydrate. 24-h urinary oxalate on two recent determinations showed 116 and 152 mg/d (normal: < 40 mg/d) on a reduced oxalate diet. Physical examination was unremarkable. Serum Cr was 1.7 mg/dL. Non-contrast abdominal CT demonstrated six non-obstructing stones in the right kidney and four non-obstructing stones in the left kidney.
METHODS
Cloning and mutagenesis of the canonical human GRHPR 5′ UTR
Commercial genotyping was conducted by Mayo Medical Laboratories. Human GRHPR 5′ UTR inclusive of the translational (ATG) start site was cloned into the vector pGL3-Promoter in a two-stage process using site directed mutagenesis. This construct includes the entirety of the 41-bp human GRHPR 5′ UTR in accordance with the RefSeq entry for this gene and the canonical GRHPR cDNA, NM_012203.1. The vector includes an SV40 promoter upstream of the luciferase gene to drive transcription from the (promoterless) 5′ UTR in the context of GRHPR. Wild-type start site was mutated to mimic the patient variant, with the resultant variant start site context as follows (vector sequence, lower case; GRHPR-derived sequence, UPPER CASE; start site, underlined; variant nucleotides, bold): ggcctaggcttttgcaaaaagcttggCGCTTCTGTACTGCCAGGTCCGGGTCGGCGGCTGCACTATGGATGgaagacgccaaaaacataaagaaagg. We called these plasmids hGRHPR-41ATG-Pro-WT and hGRHPR-41ATG-Pro-Var, abbreviated -41ATG-WT and -41ATG-Var, respectively.
Cloning and mutagenesis of the canonical human GRHPR 5′ proximal promoter sequence and human GRHPR cDNA
1.3 kb of the human GRHPR promoter sequence (inclusive of the 5′ UTR and translational start site) was amplified from anonymized human genomic DNA using Bgl2- and Ncol-tailed primers and sub-cloned into Ncol- and Bgl2-cleaved and gel-purified pGL3-Enhancer (Promega Corporation). This vector was selected because it has an SV40 enhancer downstream of the luciferase reporter. Mismatches introduced through this cloning strategy were corrected via mutagenesis to reconstitute the wild-type human GRHPR start site context (sequence convention as above): TCCGGGTCGGCGGCTGCACTGCGGATGAGagacgccaaaaacataaagaaag. We call this plasmid hGRHPR-1321ATG-Enh-WT, abbreviated -1321ATG-WT, which was then mutagenized to the variant context (TCCGGGTCGGCGGCTGCACTATGGATGAGagacgccaaaaacataaagaaag) to create plasmid hGRHPR-1321ATG-Enh-Var (abbreviated -1321ATG-Var). hGRHPR cDNA was mutagenesized to introduce the patient p.Gln232Argfs*3 (c.694delC) mutation, sequence-confirmed, and subjected to functional testing.
GRHPR enzymatic activity and immunoblotting
Chinese Hamster Ovary cells (CHO cells) were transfected (Lipofectamine 3K; Invitrogen) with a mammalian expression vector pcDNA 3.1D/V5-His TOPO (Invitrogen) carrying the coding sequence for the mutant or wild-type GRHPR, and geneticin-selected for one week (6, 7). Glyoxylate reductase catalytic activity in cell lysates or mouse liver (positive control) was tested as described (8, 9). Lysates (10 ug) were immunoblotted with a rabbit polyclonal anti-human GRHPR antibody (6) and anti-actin loading control and imaged via chemiluminescence.
RESULTS
Genotyping
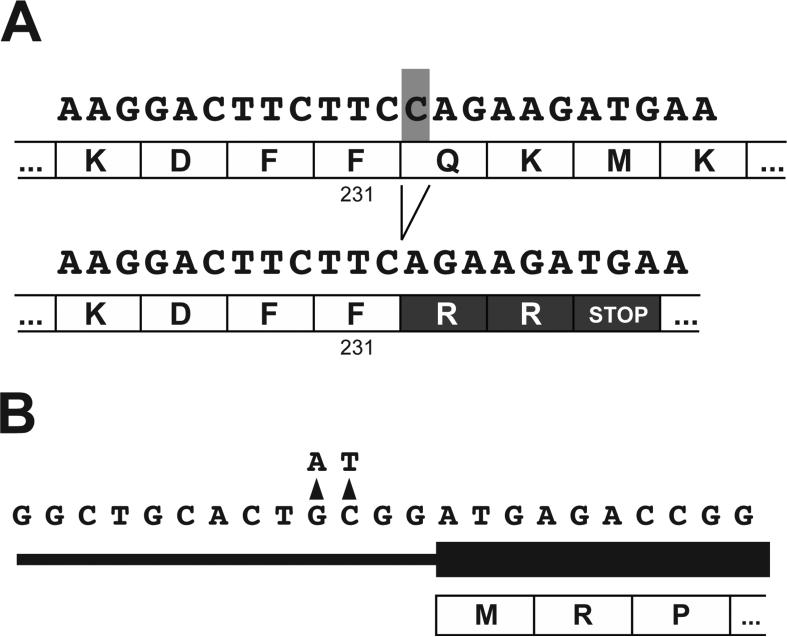
Blood was obtained and referred for commercial genotyping (see Methods). Initially, the AGXT gene was tested (for primary hyperoxaluria type I) and no mutations were detected. Next, GRHPR exonic sequencing was commercially performed, which revealed the c.694delC mutation affecting a single allele. This deletion of nucleotide 694 affected codon 232. The initial 231 amino acids were unaffected by the frameshift; however residue 232 was predicted to be mutated from Q -> R and residue 233 was mutated from K -> R, followed by a premature stop codon (p.Gln232Argfs*3; Figure 1A).
Figure 1. Mutations in the proband.
A. Deletion mutation c.694delC/p.Gln232Argfs*3, affecting the first nucleotide (shaded in gray) in the codon for amino acid residue 232 (Gln, Q) of GRHPR, and resulting in a change in reading frame and a premature stop codon. Upper sequence, wild-type; lower-sequence, mutant. B. Mutations affecting the GRHPR 5′ UTR. Coding sequence shown by the thicker black bar. Note the optimal Kozak consensus sequence of the novel translation initiation site (ANNATGG) but not the canonical site (CNNATGA).
Two additional variants were noted in the 5′ UTR (Figure 1B) and were described by the commercial lab as “variant of uncertain significance.” However, close inspection revealed that, if in cis, this dinucleotide change introduced an alternative translational start site immediately upstream of – and out of frame with – the canonical GRHPR translational start site (Figure 1B). Furthermore, this variant start site exhibited a more idealized Kozak consensus sequence (10) than did the canonical start site. There were no additional downstream variants correcting the reading frame. These variants (NM_012203.1:c.-4G>A , and NM_012203.1:c.-3C>T) match rs201826196 and rs200847843, respectively in dbSNP138; the source was given as 1000 Genomes (www.1000genomes.org/) and the allele frequency was estimated at 0.001 +/− 0.021 (n = 1 occurrence for each). There was no functional/clinical correlate or linkage data. The coding variant/mutation does not appear in online databases.
Confirmation of compound heterozygosity
Peripheral blood was obtained from an asymptomatic adult child of the proband for GRHPR sequencing by the same clinical laboratory. This individual was a carrier of only the p.Gln232Argfs*3 truncation mutant, confirming that the coding and 5′ UTR variants affected separate GRHPR alleles, and that the two 5′ UTR variants occurred in cis in the proband (i.e., c.-4_-3delGCinsAT). This allele is predicted to direct premature out-of-frame translational initiation and synthesis of a 20-amino acid peptide (MDETGATHEGVRHPQDTRRG) bearing no relation to GRHPR.
Functional testing of mutations
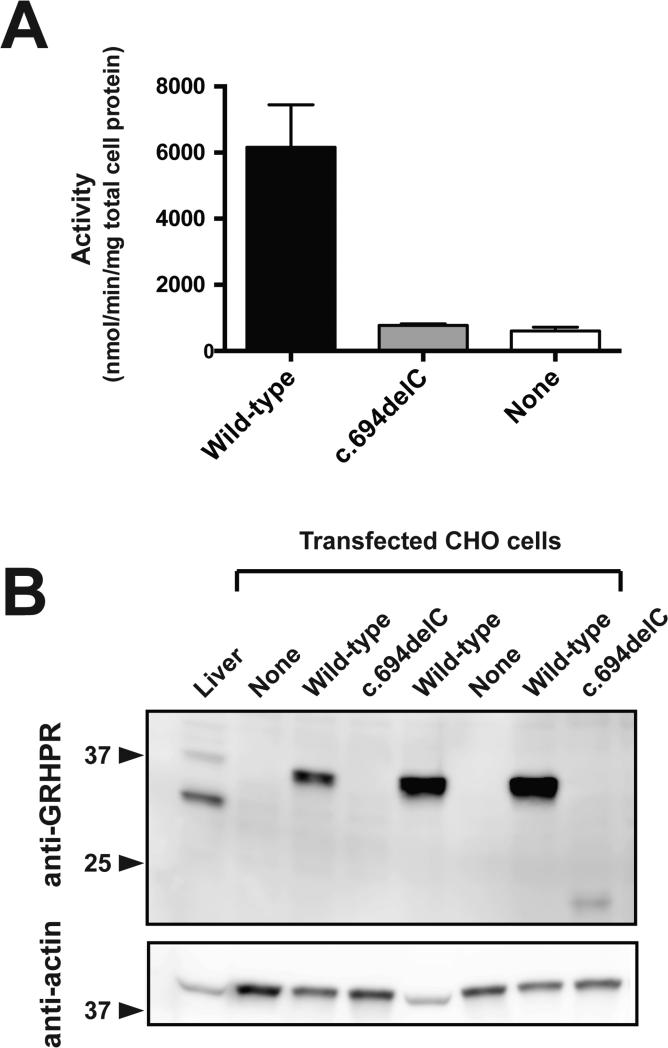
In transfected CHO cells, activity of the p.Gln232Argfs*3 mutant was indistinguishable from background in untransfected cells (Figure 2A). The variant protein also could not be detected by polyclonal anti-GRHPR antibody, consistent with rapid degradation of the severely truncated product (Figure 2B).
Figure 2. Effect of the p.Gln232Argfs*3 mutation upon GRHPR activity and expression in vitro.
Lysates from CHO cells transfected with wild-type (Wild-type) or mutant c.694delC/p.Gln232Argfs*3 (c.694delC) GRHPR or from untransfected CHO cells (None) were assayed for enzymatic activity (A) and immunodetectable GRHPR (B). In A, enzymatic activity of the mutant (expressed as nmol glycolate produced/min/mg total cell protein) was not different from background activity in untransfected cells (None), whereas activity was detected in cells expressing wild-type GR (Wild-type), and in mouse liver homogenate (positive control; 3620 ± 730; not shown). In B, GRHPR and actin (loading control) immunoreactivity is shown for lysates prepared from untransfected CHO cells (None) and CHO cells transfected with wild-type (Wild-type) or mutant (c.694delC) cDNA. Mouse liver serves as an additional control. Data are mean ± SEM.
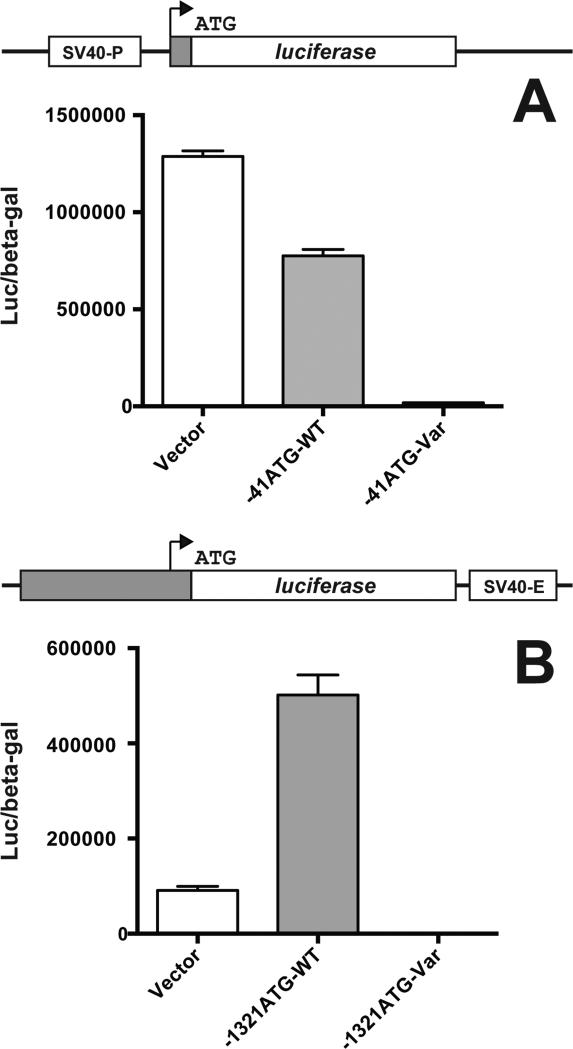
An SV40-driven luciferase reporter downstream of the variant 41-bp GRHPR 5′ UTR exhibited marked hypoactivity relative to the wild-type 5′ UTR (see Methods; Figure 3A). This confirmed that, in the presence of the upstream out-of-frame translational start site, negligible translation occurred from the more distal native initiation site, and likely giving rise to a profoundly hypomorphic allele in vivo.
Figure 3. Effect of the 5′ UTR GRHPR variants upon in-frame translation of a luciferase reporter gene in transient transfection.
For A, the entire 41-bp GRHPR 5′ UTR (cDNA NM_012203.1) was subcloned immediately upstream of the initiator Met of the luciferase gene in plasmid pGL3-Promoter. This vector has an SV40 promoter upstream of the reporter gene (SV40-P; see diagram). In this assay, only in-frame translation from the canonical luciferase translational start site gives rise to functional luciferase. CHO cells were transfected with empty vector alone (Vector), or with vector containing wild-type 5′ UTR-luciferase reporter gene chimera (-41ATG-WT) or the variant with out-of-frame upstream translational initiation site (-41ATG-Var). P-values: Vector vs. -41ATG-WT, 0.0003; -41ATG-WT vs -41ATGVar, 2 × 10-5. For B, 1.3 kb of the 5′ flanking region/proximal promoter sequence of human GRHPR inclusive of the 41-bp 5′ UTR was subcloned immediately upstream of the luciferase reporter gene in plasmid pGL3-Enhancer. This vector has an SV40 enhancer downstream of the reporter gene (SV40-E; see diagram). CHO cells were transfected with empty vector alone (Vector), or with vector containing wild-type (-1321ATG-WT) or mutant (-1321ATG-Var) GRHPR promoter and 5′ UTR. P-values: Vector vs. -1321ATG-WT, 0.0007; -1321ATG-WT vs -1321ATGVar, 0.0003. Diagrams of constructs are not to scale. Data are mean ± SEM.
When a full 1.3 kb of adjacent upstream GRHPR genomic sequence was included with the canonical or variant ATG start site (along with a downstream SV40 enhancer to ensure adequate expression level), a similar phenomenon was noted (Figure 3B). Therefore, the novel dinucleotide 5′ UTR variant, through introduction of a high-efficiency out-of-frame alternative translational start site, was a strongly hypomorphic allele – in both a minimal context and a more comprehensive promoter environment. Functionally, the patient was a compound heterozygote with two essentially null alleles for GRHPR.
DISCUSSION
The p.Gln232Argfs*3 GRHPR mutation reported here is novel and is predicted to result in premature truncation of the protein after 233 (of 328) amino acids. This truncation occurs immediately upstream of the predicted motif, D-isomer specific 2-hydroxyacid dehydrogenases signature sequence (ProSite motif PS00671; http://prosite.expasy.org), characteristic of this enzyme family and implying a loss of catalytic activity. Our in vitro data confirmed absence of GRHPR enzymatic activity; moreover, the predicted truncated protein product was not immunodetectable.
The mechanism through which the other allele is functionally inactivated is unique for this gene and phenotype, and is seldom encountered in any context. This pair of 5′UTR variants, uncovered through commercial genotyping, was recently included in the 1000 Genomes database (as SNPs rs201826196 and rs200847843); however, only here do we show that these two adjacent variants occur in cis and introduce a novel upstream out-of-frame aberrant translational initiation site that pre-empts the canonical start site. We did not exclude an ancillary effect on transcript abundance for either variant allele.
There have been few reports of alternative or aberrant translational initiation as a form of functional inter-individual genomic variation or disease-causing mutation. Such variants are exceedingly rare (11) and can be readily overlooked when emphasis is placed on coding exons (12). Calvo et al, in a comprehensive assessment (13), identified only two such instances with genotype—phenotype correlations (14, 15). Using an exhaustive bioinformatics-based approach, they detected five new instances of disease-causing variants/mutations for which they demonstrated aberrant initiation from the canonical translational start site in vitro (13). Five more instances were identified but not confirmed experimentally. Subsequently, a mutation in the IFITM5 gene was shown to cause osteogenesis imperfecta by introducing a novel upstream translational initiation site with a strong Kozak consensus sequence (11, 12).
This case and others (13) point out a limitation in the reporting and interpretation of single-gene commercial genotyping. Most is exon-centric, relying upon manual inspection and interpretation. In contrast, whole-genome, whole-exome, or transcriptome studies increasingly incorporate an algorithm-driven analytical approach to assign putative function to variants; extending this approach to even single-gene analyses may increase sensitivity for mutation detection.
Acknowledgement
This project was supported by the National Institutes of Health and by the Department of Veterans Affairs.
Footnotes
Conflict of Interest: none
REFERENCES
- 1.Hoppe B. An update on primary hyperoxaluria. Nat Rev Nephrol. 2012;8:467–475. doi: 10.1038/nrneph.2012.113. [DOI] [PubMed] [Google Scholar]
- 2.Cramer SD, Ferree PM, Lin K, et al. The gene encoding hydroxypyruvate reductase (GRHPR) is mutated in patients with primary hyperoxaluria type II. Hum Mol Genet. 1999;8:2063–2069. doi: 10.1093/hmg/8.11.2063. [DOI] [PubMed] [Google Scholar]
- 3.Webster KE, Ferree PM, Holmes RP, et al. Identification of missense, nonsense, and deletion mutations in the GRHPR gene in patients with primary hyperoxaluria type II (PH2). Hum Genet. 2000;107:176–185. doi: 10.1007/s004390000351. [DOI] [PubMed] [Google Scholar]
- 4.Cregeen DP, Williams EL, Hulton S, et al. Molecular analysis of the glyoxylate reductase (GRHPR) gene and description of mutations underlying primary hyperoxaluria type 2. Hum Mutat. 2003;22:497. doi: 10.1002/humu.9200. [DOI] [PubMed] [Google Scholar]
- 5.Webster KE, Cramer SD. Genetic basis of primary hyperoxaluria type II. Molecular urology. 2000;4:355–364. [PubMed] [Google Scholar]
- 6.Behnam JT, Williams EL, Brink S, et al. Reconstruction of human hepatocyte glyoxylate metabolic pathways in stably transformed Chinese-hamster ovary cells. Biochem J. 2006;394:409–416. doi: 10.1042/BJ20051397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fargue S, Lewin J, Rumsby G, et al. Four of the most common mutations in primary hyperoxaluria type 1 unmask the cryptic mitochondrial targeting sequence of alanine:glyoxylate aminotransferase encoded by the polymorphic minor allele. J Biol Chem. 2013;288:2475–2484. doi: 10.1074/jbc.M112.432617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giafi CF, Rumsby G. Kinetic analysis and tissue distribution of human D-glycerate dehydrogenase/glyoxylate reductase and its relevance to the diagnosis of primary hyperoxaluria type 2. Ann Clin Biochem. 1998;35(Pt 1):104–109. doi: 10.1177/000456329803500114. [DOI] [PubMed] [Google Scholar]
- 9.Baker PR, Cramer SD, Kennedy M, et al. Glycolate and glyoxylate metabolism in HepG2 cells. Am J Physiol Cell Physiol. 2004;287:C1359–1365. doi: 10.1152/ajpcell.00238.2004. [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semler O, Garbes L, Keupp K, et al. A mutation in the 5′-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012;91:349–357. doi: 10.1016/j.ajhg.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho TJ, Lee KE, Lee SK, et al. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012;91:343–348. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Dilworth D, Gao L, et al. Mutation of the CDKN2A 5' UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet. 1999;21:128–132. doi: 10.1038/5082. [DOI] [PubMed] [Google Scholar]
- 15.Wen Y, Liu Y, Xu Y, et al. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nat Genet. 2009;41:228–233. doi: 10.1038/ng.276. [DOI] [PubMed] [Google Scholar]