Abstract
An imbalance in molecular signaling cascades and transcriptional regulation in nucleus accumbens (NAc) medium spiny neuron (MSN) subtypes, those enriched in dopamine D1 versus D2 receptors, is implicated in the behavioral responses to psychostimulants. To provide further insight into the molecular mechanisms occurring in MSN subtypes by cocaine, we examined the transcription factor early growth response 3 (Egr3). We evaluated Egr3 because it is a target of critical cocaine-mediated signaling pathways and because Egr3-binding sites are found on promoters of key cocaine-associated molecules. We first used a RiboTag approach to obtain ribosome-associated transcriptomes from each MSN subtype and found that repeated cocaine administration induced Egr3 ribosome-associated mRNA in NAc D1-MSNs while reducing Egr3 in D2-MSNs. Using Cre-inducible adeno-associated viruses combined with D1-Cre and D2-Cre mouse lines, we observed that Egr3 overexpression in D1-MSNs enhances rewarding and locomotor responses to cocaine, whereas overexpression in D2-MSNs blunts these behaviors. miRNA knock-down of Egr3 in MSN subtypes produced opposite behavioral responses from those observed with overexpression. Finally, we found that repeated cocaine administration altered Egr3 binding to promoters of genes that are important for cocaine-mediated cellular and behavioral plasticity. Genes with increased Egr3 binding to promoters, Camk2α, CREB, FosB, Nr4a2, and Sirt1, displayed increased mRNA in D1-MSNs and, in some cases, a reduction in D2-MSNs. Histone and the DNA methylation enzymes G9a and Dnmt3a displayed reduced Egr3 binding to their promoters and reduced mRNA in D1-MSNs. Our study provides novel insight into an opposing role of Egr3 in select NAc MSN subtypes in cocaine action.
Keywords: cocaine, Egr3, medium spiny neurons, nucleus accumbens, RiboTag, transcription
Introduction
The nucleus accumbens (NAc) of the ventral striatum, a major brain reward region, is critical for reward seeking and reinforcement behaviors. The main projection neurons of the NAc and the dorsal striatum are medium spiny neurons (MSNs), which can be divided into two major subpopulations on the basis of their enrichment of dopamine receptor D1 versus D2 receptors and their differential outputs through the brain (Gerfen et al., 1990; Smith et al., 2013). Previous studies using optogenetic, chemogenetic, and neurotransmission blocking approaches demonstrates opposing roles for these two MSN subtypes in behavioral responses to psychostimulants (Hikida et al., 2010; Lobo et al., 2010; Ferguson et al., 2011; Lobo and Nestler, 2011; Bock et al., 2013; Chandra et al., 2013; Lenz and Lobo, 2013). The opposing role of activity in MSN subtypes in mediating psychostimulant-induced behavioral responses potentially occurs through an imbalance in molecular signaling cascades, transcription factors, and chromatin-modifying enzymes in MSNs, because recent studies have demonstrated that these processes in each MSN subtype lead to differential synaptic plasticity events and behavioral responses to psychostimulants (Bateup et al., 2010; Lobo et al., 2010; Lobo and Nestler, 2011; Arango-Lievano et al., 2014; Maze et al., 2014).
Dopamine (DA) and brain-derived neurotrophic (BDNF) signaling in the NAc are critical for psychostimulant synaptic and behavioral plasticity (Hyman et al., 2006; Russo et al., 2009; Volkow et al., 2009; McGinty et al., 2010). Both DA and BDNF signaling can alter behavioral responses to cocaine differentially depending on which MSN subtype it is acting through (Self, 2004; Lobo et al., 2010; Lobo and Nestler, 2011). Here, we investigated a downstream molecular target of both DA and BDNF signaling, the transcription factor early growth response 3 (Egr3) (Patwardhan et al., 1991; Yamagata et al., 1994; O'Donovan and Baraban, 1999; Jouvert et al., 2002, Roberts et, al., 2006), to gain further insight into the transcriptional mechanisms that mediate psychostimulant-induced behaviors. Egr3 is induced in total striatum with acute cocaine use through the activation of D1 receptors (Yamagata et al., 1994; Jouvert et al., 2002). However, it is unknown which MSN subtypes display altered Egr3 expression after repeated cocaine exposure. Here, we examined Egr3 levels in MSN subtypes using Cre-inducible RiboTag (RT) mice (Sanz et al., 2009; Sanz et al., 2013), which have been crossed to D1-Cre and D2-Cre bacterial artificial chromosome (BAC) transgenic lines (Gong et al., 2007; Gerfen et al., 2013), to create an epitope-tagged ribosomal protein in each MSN subtype. This allowed the isolation of ribosome-associated mRNA from each MSN subtype for gene expression profiling in MSNs, similar to previous studies using the epitope-tagged ribosomal approach, FACS, or single-cell isolation (Lobo et al., 2006; Heiman et al., 2008; Ena et al., 2013; Rothwell et al., 2014). We then investigated the role of Egr3 in each MSN subtype in mediating cocaine conditioned place preference (CPP) and cocaine-induced locomotor activity using D1-Cre and D2-Cre mouse lines combined with Cre-inducible adeno-associated viruses (AAVs) to overexpress Egr3 or miRNA knock-down of Egr3 levels in each NAc MSN subtype. Finally, we assessed the function of Egr3 as a transcription factor by examining Egr3 binding to promoters of genes critical for psychostimulant-induced cellular and behavioral plasticity. Our studies provide novel insight into a role for Egr3 in each MSN subtype in cocaine-induced behavioral responses and its role in transcriptional regulation of cocaine-associated genes.
Materials and Methods
Animals.
D1-Cre hemizygote (line FK150) or D2-Cre hemizygote (line ER44) BAC transgenic mice from GENSAT (Gong et al., 2007; Gerfen et al., 2013; www.gensat.org) on a C57BL/6J background were used for behavioral experiments. Homozygous RT mice on a C57BL/6J background expressing a Cre-inducible HA-Rpl22 were purchased from The Jackson Laboratory (Sanz et al., 2009). These mice were crossed to D1-Cre or D2-Cre mouse lines to generate D1-Cre-RT and D2-Cre-RT mice and used for cell-type-specific ribosome-associated mRNA isolation. C57BL/6J mice obtained from The Jackson Laboratory were used for ChIP, RT-PCR, and Western blot experiments. Male mice were used for the studies and all mice were maintained on a 12 h light/dark cycle with ad libitum food and water. Male Sprague-Dawley rats, used for self-administration tissue, were maintained on a 12 h reverse light/dark cycle with ad libitum food and water. All studies were conducted in accordance with the guidelines set up by the Institutional Animal Care and Use Committee at The University of Maryland School of Medicine and The University at Buffalo, The State University of New York.
Cell culture.
Neuro2a cells (Invitrogen) were maintained at 37°C, 5% CO2 in DMEM with GlutaMAXI, 4500 mg/L glucose and 110 mg/L sodium pyruvate (Invitrogen) supplemented with 10% v/v FCS (Invitrogen).
Repeated cocaine treatment.
D1-Cre-RT, D2-Cre-RT, D1-Cre, D2-Cre, and C57BL/6J mice received 7 daily intraperitoneal injections of cocaine (20 mg/kg) or 0.9% saline in the home cage. NAc tissue was collected 24 h after the last injection for molecular biology experiments and was perfused for immunostaining and imaging. Cocaine hydrochloride (Sigma) was dissolved in sterile saline. The dose of cocaine and the 24 h withdrawal time point was selected based on previous studies showing important cocaine-mediated transcriptional and plasticity changes at this time point (Maze et al., 2010; Russo et al., 2010, Kim et al., 2011, Feng et al., 2014).
AAVs.
Recombinant Cre-dependent AAVs expressing EYFP, Egr3-EYFP, Egr3miR-mCitrine, and Scramble-mCitrine were used in this study. Egr3 sequences were PCR amplified (Phusion DNA polymerase; New England Biolabs) and cloned into the NheI and NcoI restriction site of the EF1a-DIO-EYFP vector. Egr3miR and Scramble-miR were generated from Egr3shRNA (see “Egr3miRNA engineering” section below) and cloned into NheI and StuI restriction sites of AAV-hSyn-DIO-mCitrine vector backbone gifted from Dr. B.L. Roth (University of North Carolina). The DIO-Egr3-EYFP was packaged into AAV (serotype 2) at University of North Carolina (UNC) Vector Core Facility. AAV-DIO-EYFP was also purchased from UNC Vector Core Facility. Virus packaging for AAV-DIO-Egr3miR-mCitrine and AAV-DIO-Scramble-mCitrine were performed as described previously (Ried et al., 2002; Prasad et al., 2011). In brief, AAV vectors were packaged into AAV2 capsids by the triple transfection method using helper plasmids pAdDF6 (providing the three adenoviral helper genes) and plasmid pAAV2/2 (kindly provided by Dr. James M Wilson, University of Pennsylvania). The AAV-generating plasmids and helper plasmid(s) were transfected into HEK 293 cells by the calcium phosphate coprecipitation method (Jordan et al., 1996), 40 μg of plasmid DNA (mixed in an equimolar ratio) was used per 15 cm cell culture plate. Three days after transfection, AAV vectors were purified by ammonium sulfate fractionation and iodixanol gradient centrifugation. Titers for the AAV vectors (in viral genome particles per milliter) were determined by quantitative RT-PCR (qRT-PCR) as described previously (Prasad et al., 2011).
Egr3miRNA engineering.
Egr3 shRNA expression vectors were purchased from Origene with four possible sets of Egr3 shRNA and a scrambled sequence cloned into HuSH pGFP-V-RS plasmid vector driven under human U6 promoter (see Fig. 4A). To check the in vitro efficiency of shRNA, plasmids were transfected into Neuro2a cells and processed as described in the Effectene transfection kit (QIAGEN). The knock-down efficiency of Egr3 was measured by qRT-PCR 24 h after transfection. The most efficient shRNA was chosen for miRNA engineering. We generated a vector-based miRNA by using the BLOCK-iT Kits (Invitrogen). This allowed us to express the Egr3 miRNA in mammalian cells under the control of a variety of promoters, including tissue-specific promoters. The engineered pre-miRNA sequence structure is based on the murine miR-155 sequence (Lagos-Quintana et al., 2002). We designed two single-stranded DNA oligonucleotides, one encoding the target pre-miRNA top strand oligonucleotide and the other the bottom strand oligonucleotide compliment. Top and bottom strand oligonucleotides were annealed to generate a double-stranded oligonucleotide (ds oligo) and cloned into the pcDNA-EmGFP-miR vector provided in kit. We used the five-step design to engineer the top single strand (from 5′ to 3′ end) by using the first 21 bp of the shRNA target sequence of Egr3 that resulted in the greatest knock-down (44%) in Neuro2a cells (see Fig. 4B). A five-nucleotide TGCTG overhang was added at the 5′ of the reverse compliment of the first 21 bp of the target sequence, 19 nucleotides were added to form a terminal loop (GTTTTGGCCACTGACTGAC), and finally the Egr3 target sequence was added with the ninth and tenth nucleotide removed to form a short internal loop. To design the bottom strand, four nucleotides (TGCT) were removed from the top strand, followed by reverse compliment, and then CCTG was added to the 5′ end. The miRNA top and bottom strand sequences were synthesized at the Biopolymer/Genomics Core Facility at the University of Maryland, Baltimore. The annealing of top and bottom strands was performed in 20 μl of reaction mixture by incubating 200 μm of each strand with the Oligo Annealing Buffer at 95°C for 4 min. Finally, the ds oligo was cloned into the linearized of pcDNA6.2-GW/± EmGFPmiR vector available in the kit. Egr3miR amplified by PCR and the amplicons were gel purified using a QIAquick gel extraction kit (QIAGEN) and finally cloned into destination Cre inducible vector AAV-DIO-mCitrine driven hySn promoter.
Figure 4.
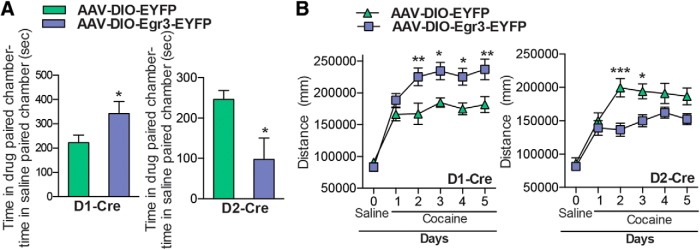
Egr3 overexpression in MSN subtypes alters cocaine CPP and cocaine-induced locomotor activity. A, Cocaine (7.5 mg/kg) CPP is enhanced with D1-Cre mice that received AAV-DIO-Egr3-EYFP into NAc, whereas cocaine CPP is decreased in D2-Cre mice that received AAV-DIO-Egr3-EYFP into NAc compared with AAV-DIO-EYFP controls. B, Repeated cocaine (10 mg/kg)-induced locomotor activity is increased in D1-Cre mice (days 2–5), whereas it is reduced in D2-Cre mice (day 2–3) expressing AAV-DIO-Egr3-EYFP in NAc compared with controls expressing AAV-DIO-EYFP. Error bars indicate SEM.
Mouse stereotaxic surgery.
D1-Cre or D2-Cre mice were anesthetized using 3% isoflurane in a small induction chamber. After the initial induction, isoflurane was maintained at 1% for the remainder of the surgery. Animals were placed in a stereotaxic instrument and their skull was exposed. Then, 33 gauge Hamilton syringe needles were used to inject 0.8 μl of AAV-DIO-EYFP, AAV-DIO-Egr3-EYFP, or AAV-DIO-Egr3miR-mCitrine or AAV-DIO-Scramble-mCitrine bilaterally into the NAc (anterior/posterior, AP +1.6; medial/lateral, ML ±1.5; dorsal/ventral, DV −4.4, 10° angle). Animals were then returned to the vivarium for 2 weeks to allow for recovery and maximal virus expression.
Cocaine self-administration.
Cocaine self-administration tissue used in this study was obtained from behavioral experiments described in Chandra et al. (2013). Briefly, 1 week after jugular catheter surgery, the rats were assigned to self-administer saline or 1.0 mg/kg/infusion cocaine. Two-hour daily testing occurred during the animals dark cycle in Med-Associates experimental test chambers fitted with two snout poke ports, using a fixed ratio 1 (FR1) schedule for 10 test sessions. Twenty-four hours after the last self-administration session, brains were harvested and NAc tissue punches were quickly collected and stored at −80°C.
CPP.
The CPP procedure was conducted as described previously (Lobo et al., 2010). Briefly, 14 d after intra-NAc infusions of AAV-DIO-Egr3-EYFP, AAV-DIO-EYFP, AAV-DIO-Egr3miR-mCitrine, and AAV-DIO-Scramble-mCitrine, D1-Cre or D2-Cre mice were placed into the conditioning boxes. The boxes consist of three distinct environments including gray and striped conditioning chambers with different mesh grating on the floor and a middle neutral chamber. Mice were allowed to explore the three chambers for 20 min and those that showed significant preference for one of the two conditioning chambers were excluded from the study (<10% of all animals). Groups were then balanced and adjusted for any chamber bias that may still exist. On subsequent days, animals were injected with saline and confined to one chamber before noon for 30 min and then injected with cocaine (7.5 mg/kg, i.p.) and further confined for 30 min to the other chamber in the afternoon for 2 d. On the test day, mice were placed back into the apparatus for 20 min without any treatment. Total time spent in the drug-paired chamber minus time spent in the saline-paired chamber was measured using Topscan tracking software (Clever Sys).
Cocaine-induced locomotor activity.
D1-Cre or D2-Cre mice injected with AAV-DIO-Egr3-EYFP, AAV-DIO-EYFP, AAV-DIO-Egr3miR-mCitrine, or AAV-DIO-Scramble-mCitrine underwent 1 d of 30 min habituation in a 42 × 42 × 42 cm open-field chamber. The next day, mice were habituated in the chambers to 0.9% saline intraperitoneal injections for 30 min. The animals then underwent five daily 30 min test sessions in which they were given 10 mg/kg intraperitoneal cocaine injections. Locomotion was video tracked with Topscan tracking software (Clever Sys).
ChIP.
Fresh NAc punches were prepared for ChIP as described previously with minor modifications (Lee et al., 2006b; Maze et al., 2010; Covington et al., 2011). Briefly, four 14 gauge NAc punches per animal (5 animals pooled per sample) were collected, cross-linked with 1% formaldehyde, and quenched with 2 m glycine before freezing at −80°C. Before sample sonication, IgG magnetic beads (Invitrogen; sheep anti-rabbit, catalog #11202D) were incubated with an anti-Egr3 antibody (rabbit polyclonal, Santa Cruz Biotechnology, catalog #SC-191) 15 μg per reaction, overnight at 4°C under constant rotation in blocking solution (0.5% w/v BSA in 1× PBS). NAc tissue was homogenized in 1 ml lysis buffer 1 (50 mm HEPES-KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and protease inhibitors) by douncing followed by constant rotation at 4°C for 10 min. Samples were centrifuged at 1350 × g for 5 min at 4°C and the pellet was resuspended in 1 ml of lysis buffer 2 (10 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, and protease inhibitors) and incubated gently on shaker at room temperature for 10 min. Pelleted cell nuclei by centrifugation at 1350 × g for 5 min at 4°C and resuspended in 300 μl of lysis buffer 3 (10 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, and protease inhibitors). Chromatin was sheared to an average length of 500–700 bp by the Diagenode Bioruptor Pico using eight cycles of 30 s on and off. Then, 1/10 volume of 10% Triton X-100 was added to the sonicated lysate to dissolve the nuclear membrane. Samples were centrifuged at 20,000 × g for 10 min at 4°C to pellet the debris. After washing and resuspension of the antibody–bead conjugates, the mixtures were added to each chromatin sample (600 μl) and incubated for ∼16 h under constant rotation at 4°C. Samples were then washed and reverse cross-linked at 65°C overnight and DNA was purified using a PCR purification kit (QIAGEN). After DNA purification, samples were used for qRT-PCR analysis, normalized to their appropriate input controls, and compared with IgG chipped samples as described previously (Renthal et al., 2009; Maze et al., 2010; Covington et al., 2011). The list of primers used for ChIP PCR is given below. Camk2α forward: CGTCCCCACAGCATCTTCT.
Camk2α reverse: CCTTGCTCCTCTTGTCCCC; CREB forward: CCGGGAAGTAGCCGAAGG, CREB reverse: GCCACTCACGGAAACAGC; Dnmt3a forward: AGGCAGAATACTATGGTGGCAG, Dnmt3a reverse: GGATATCCCTTGCCTCTCTCTG; FosB forward: GGATGGGACTCAGGTTGTCA, FosB reverse: ATCGGTGGCATGTGTTTAGC; Nr4a2 forward: TTAATGGGACCCTCCTATTTTG, Nr4a2 reverse: ACAAGCCTCTGCCTTCAATTC; G9a forward: GGGGCGGAGGAGGGGACCAGG, G9a reverse: CGCGCCCCTCGCGCGGGCT, mGluR2 forward: CTGGATGCTTTGAGCTGCC, mGluR2 reverse: TAGGCAGGCGATGGTGAC; NMDAR1forward: AGCATTTACGCCAACGCA, NMDAR1 reverse: GGACCTACGCGTTCTACCC; Sirt1 forward: TTAAATCTCCCGCAGCCGA, Sirt1 reverse: GCCCTCGCCTCCTCTG.
Immunoprecipitation of polyribosomes and RNA isolation from MSN subtypes.
Immunoprecipitation of polyribosome was prepared as described previously (Sanz et al., 2009) with minor modifications. We pooled NAc tissue from 4 D1-Cre-RT or D2-Cre-RT mice per sample and homogenized it by douncing in homogenization buffer containing 50 mm Tris, pH 7.4, 100 mm KCl, 12 mm MgCl2, and 1% NP-40 supplemented with 1 mm DTT, 100 μg/ml cyclohexamide, 1 mg/ml heparin, RNAase, and protein inhibitors (Promega). Samples were then centrifuged at 10,000 × g for 10 min to collect the clear supernatant. Then, 80 μl of supernatant was transferred to new tubes to serve as an input for sample validation and 800 μl of the supernatant was added directly to the HA-coupled beads (Invitrogen: 100.03D; Covance: MMS-101R) for constant rotation overnight at 4°C. The following day, magnetic beads were washed 3 times in a magnetic rack for 5 min in high-salt buffer (50 mm Tris, pH 7.4, 300 mm KCl, 12 mm MgCl2, 1% NP-40, 1 mm DTT, and 100 μg/ml cyclohexamide). Finally, RNA was extracted by adding TRK lysis buffer to the pellet provided in MicroElute Total RNA Kit (Omega) according to the manufacturer's instructions. RNA was quantified with a NanoDrop (Thermo Scientific). RNA integrity was performed using the Agilent 2100 Bioanalyzer at The Biopolymer/Genomics Core Facility at the University of Maryland, Baltimore. For cDNA synthesis and qRT-PCR, see below.
RNA extraction and qRT-PCR.
Mouse and rat NAc tissue punches were collected 24 h after the last cocaine administration and stored at −80°C. RNA was extracted using TRIzol (Invitrogen) and the MicroElute Total RNA Kit (Omega) with a DNase step (QIAGEN). Then, 300–400 ng of cDNA was synthesized using the reverse transcriptase iScript cDNA synthesis kit (Bio-Rad). mRNA expression changes were measured using qRT-PCR with PerfeCTa SYBR Green FastMix (Quanta). Quantification of mRNA changes was performed using the −ΔΔCT method described previously (Lobo et al., 2010; Chandra et al., 2013) using GAPDH as a housekeeping gene. The list of primers used in this study is given below. Mouse Egr3 forward: ACTCGGTAGCCCATTACAATC, Mouse Egr3 reverse: TGGCTGGAAAGAGCTCGAAT; Rat Egr3 forward: TTCCAGTGTCGGATCTGCAT, Rat Egr3 reverse: CAGAACTCACAGGCAAAGGG; Mouse GAPDH forward: AGGTCGGTGTGAACGGATTTG, Mouse GAPDH reverse: TGTAGACCATGTAGTTGAGGTCA; Rat GAPDH forward: AACGACCCCTTCATTGAC, Rat GAPDH reverse: TCCACGACATACTCAGCA; Mouse Drd1a forward: GAGCGTGGTCTCCCAGAT, Mouse Drd1a reverse: GGATGCTGCCTCTTCTTCTG; Mouse Pdyn forward: ATCCAGAAGCGGTAGCTGAA, Mouse Pdyn reverse: AGCGAATCCACTTGATCCT; Mouse Chrm4 forward: ATCGGCTACTGGCTCTGCTA, Mouse Chrm4 reverse: TACTGGCACAGCAAAAGGTG; Mouse Tac1 forward: TGTTGGACTAATGGGCAAAA, Mouse Tac1 reverse: GATAGTGCGTTCAGGGGTTT; Mouse Drd2 forward: TCAGATGCTTGCCATTGTTC, Mouse Drd2 reverse: GTGAAGGCGCTGTAGAGGAC; Mouse Gpr6 forward: GAGGATAGCCAGGCACACAG, Mouse Gpr6 reverse: ACCACTTGGGACTCGTTGAG; Mouse Penk forward: CCGCTTTACACTTGCCTTCT, Mouse Penk reverse: AGGTGCAAAGCCTCAGGAAC; Mouse Adora2a forward: CACGCAGAGTTCCATCTTCA, Mouse Adora2a reverse: AATGACAGCACCCAGCAAAT; Mouse Camk2α forward: TGCTTGTTCACAGAAGTCGC, Mouse Camk2α reverse: CCTGGGTACTTAAGCCGGAA; Mouse CREB forward: TTAGGGACAGTTACCCAGCC, Mouse CREB reverse: TTGGCAGTCAACACTTCTTCA; Mouse Dnmt3a forward: CTTGGCCTTGCAAAAGGGTT, Mouse Dnmt3a reverse: AGAAGGGGAGGGGCCTATTT; Mouse FosB forward: GTGCGAGCTTCCTTGTTTTC, Mouse FosB reverse: GTCTCCAACAGCCAGAGGAG; Mouse G9a forward: TGCCTATGTGGTCAGCTCAG, Mouse G9a reverse: GGTTCTTGCAGCTTCTCCAG; Mouse Nr4a2 forward: GAATTGAAGGCAGAGGCTTG, Mouse Nr4a2 reverse: CACATGCTTGTCCCCTTTTT; and Mouse Sirt1 forward: ACTTGGTGCTCAGTGTGTGA, Mouse Sirt1 reverse: AGGGGCCTGTTTGGACATTA.
Western blots.
All NAc tissue punches were collected 24 h after the last cocaine administration and stored at −80°C. NAc punches were homogenized in 30 μl of lysis buffer containing 320 mm sucrose, 5 nm HEPES buffer, 1% SDS, phosphatase inhibitor cocktails I and II (Sigma) and protease inhibitors (Roche) using a ultrasonic processor (Cole Parmer). Protein concentrations were determined using the DC protein assay (Bio-Rad) and then 15–20 μg samples of total protein were loaded onto Tris-HCl polyacrylamide gel (Bio-Rad). The samples were transferred to a nitrocellulose membrane and blocked for 1 h in blocking buffer, 5% nonfat dry milk in Tris-buffered saline, pH 7.6, with 0.1% Tween. Blocked membranes were incubated overnight at 4°C in blocking buffer with primary antibodies using either 1:3000 rabbit-anti-EGR3 (Santa Cruz Biotechnology, catalog #SC-191) or 1:8000 GAPDH (Cell Signaling Technology, catalog #14C10). Membranes were then incubated with goat anti-rabbit peroxidase-labeled secondary antibodies (Vector Laboratories, catalog #PI-1000, 1:20,000 or 1:40,000 depending on the primary antibody used) in blocking buffer. The bands were visualized using SuperSignal West Dura Extended Duration substrate (Pierce, catalog #34075). Bands were quantified with Image Lab Software (Bio-Rad) and normalized to GAPDH to control for equal loading.
Immunohistochemistry.
D1-Cre or D2-Cre mice were perfused with 0.1 m PBS followed by 4% paraformaldehyde (PFA). Brains were immersed in PFA overnight and then cryopreserved in 30% sucrose. Brains were cryosectioned (Leica) at 35 μm into 0.1 m PBS. Brain sections were blocked in 3% normal donkey serum with 0.3% Triton X-100 for 30 min at room temperature. Sections were then incubated overnight at room temperature in primary antibodies, 1:8000 chicken anti-GFP (Aves) diluted in the above blocking solution. On the second day, tissue sections were rinsed in 0.1 m PBS followed by a 1 h incubation at room temperature in secondary antibodies, 1:1000 donkey anti-chicken Alexa Fluor 488 (Jackson ImmunoResearch). Sections were rinsed in PBS, mounted onto slides, and coverslipped. Immunofluorescence was imaged on an Olympus Bx61 confocal microscope.
Results
Egr3 is oppositely induced in NAc D1-MSNs versus D2-MSNs after repeated exposure to cocaine
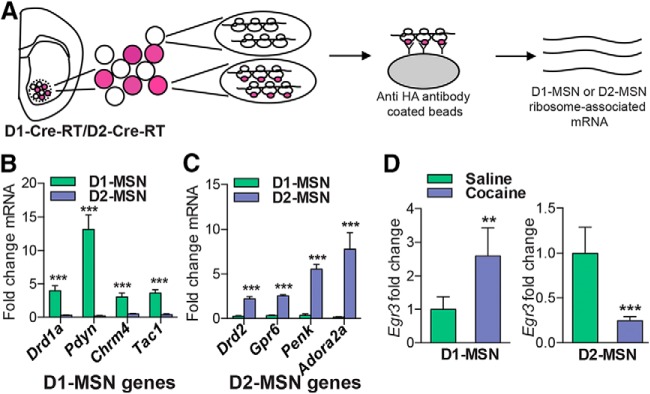
We first investigated Egr3 mRNA and protein expression in total NAc tissue after repeated cocaine administration (20 mg/kg) given as seven daily injections followed by 24 h withdrawal, a time point at which critical transcriptional and plasticity changes occur in the NAc (Maze et al., 2010; Russo et al., 2010; Kim et al., 2011; Feng et al., 2014), using qRT-PCR and Western blots. We found that repeated cocaine decreases Egr3 mRNA and protein levels in NAc tissue (Student's t test, *p < 0.05; mRNA n = 5–6 per group, t(9) = 1.86, p < 0.05; protein n = 7–9 per group, t(14) = 2.23, p < 0.05; Fig. 1A). We observed a similar decrease in Egr3 mRNA and protein levels in NAc after 10 d of rat cocaine self-administration in NAc tissue (1 mg/kg/infusion, FR1 schedule; Student's t test, *p < 0.05; mRNA n = 6 per group, t(10) = 3.37, p < 0.05; protein n = 4–7 per group, t(9) = 2.73, p < 0.05; Fig. 1B). We then examined Egr3 levels in each MSN subtype. We used Cre-inducible RT mice (Sanz et al., 2009) that were crossed to D1-Cre and D2-Cre mouse lines (Gong et al., 2007; Gerfen et al., 2013) to generate D1-Cre-RT and D2-Cre-RT mice. In these mice, a Cre-dependent expression of the ribosomal subunit, Rpl22 labeled with hemagglutinin (HA) protein allows immunoprecipitation with an HA antibody of polyribosomes from D1-MSNs and D2-MSNs. This results in isolation of ribosome-associated mRNA from each MSN subtype (Fig. 2A). Ribosome-associated mRNA isolated from D1-Cre-RT and D2-Cre-RT NAc displays enrichment of the D1-MSN genes Drd1a, Pdyn, Chrm4, and Tac1 and the D2-MSN genes Drd2, Gpr6, Penk, and Adora2a in their respective cell subtypes (Student's t test, ***p < 0.001; Drd1a: n = 10 per group, t(18) = 4.98, p < 0.001; Pdyn: n = 11 per group, t(20) = 6.02, p < 0.001: Chrm4: n = 11–12 per group, t(21) = 4.29, p < 0.001; Tac1: n = 6 per group, t(10) = 6.20, p < 0.001; Drd2: n = 6–9 per group, t(13) = 5.84, p < 0.001; Gpr6: n = 7–8 per group, t(13) = 13.38, p < 0.001; Penk: n = 4–5 per group, t(7) = 8.66, p < 0.001; Adora2a: n = 6 per group, t(10) = 4.147, p < 0.001; Fig. 2B,C), similar to previous studies profiling gene expression in these MSNs (Lobo et al., 2006; Heiman et al., 2008; Ena et al., 2013; Rothwell et al., 2014). We then examined Egr3 gene expression in each MSN subtype in NAc after repeated cocaine exposure (7 d, 20 mg/kg), followed by 24 h withdrawal, in the D1-Cre-RT and D2-Cre-RT mice. Repeated cocaine induced Egr3 in D1-MSNs while reducing it in D2-MSNs (Student's t test, **p < 0.01, ***p < 0.001; D1-MSN: n = 4–6 per group, t(8) = 4.15, p < 0.01; D2-MSN: n = 4–5 per group, t(7) = 5.85, p < 0.001; Fig. 2D). We found no detectable change in Egr3 mRNA levels in D1-MSNs versus D2-MSNs at the basal level (data not shown).
Figure 1.
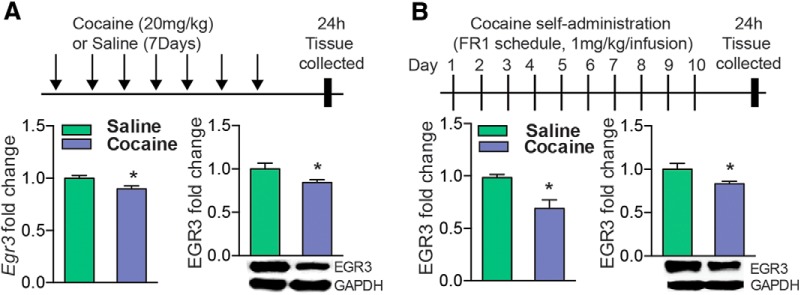
Egr3 expression after repeated cocaine exposure. A, qRT-PCR and Western blot analysis of Egr3 demonstrate that mRNA and protein levels are decreased in NAc after repeated cocaine exposure (7 d, 20 mg/kg) followed by 24 h of withdrawal in mice. B, Rats that self-administer cocaine display a similar reduction of Egr3 mRNA and protein. All values were normalized to GAPDH. Error bars indicate SEM.
Figure 2.
D1-Cre-RT and D2-Cre-RT validation and Egr3 ribosome-associated mRNA levels in MSN subtypes after cocaine exposure. A, Illustration of HA-tagged Rpl22 in D1-MSNs or D2-MSNs by crossing RT mice to D1-Cre or D2-Cre mice. Polyribosomes are immunoprecipitated from D1-MSNs or D2-MSNs with HA-tagged magnetic beads and then cell-specific ribosome-associated mRNA is isolated. B, C, MSN subtypes display enrichment of ribosome-associated mRNA for known MSN subtype genes. D, qRT-PCR analysis of Egr3 ribosome-associated mRNA in D1-Cre-RT and D2-Cre-RT NAc tissue after repeated cocaine (7 d, 20 mg/kg) or saline exposure. Repeated cocaine exposure induces Egr3 ribosome-associated mRNA in D1-MSNs while decreasing Egr3 in D2-MSNs. Error bars indicate SEM.
Egr3 overexpression in MSN subtypes oppositely mediates behavioral responses to cocaine
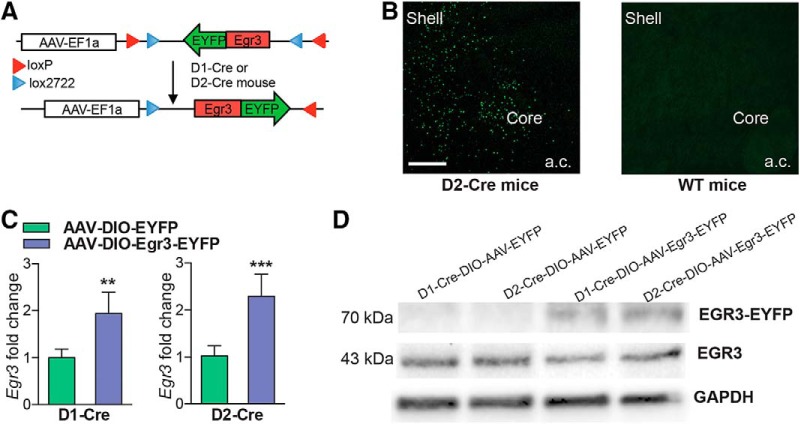
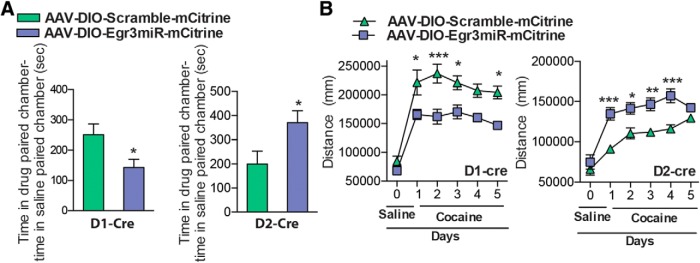
To understand the behavioral significance of the opposite induction of Egr3 in NAc MSN subtypes, we generated Cre-inducible AAVs to overexpress or knock-down Egr3 in each NAc MSN. We investigated two behaviors, CPP, which provides an indirect measure of drug reward, and cocaine-induced locomotor activity, the latter over a 5 d period. We first overexpressed Egr3 in each MSN subtype by injecting the double-floxed inverted open reading frame (DIO) Cre-dependent AAV vector, expressing Egr3 fused to enhanced yellow fluorescent protein (EYFP), into the NAc of D1-Cre or D2-Cre mice (Fig. 3A). Cre specificity of the virus was confirmed by immunostaining images, which demonstrated expression of Egr3-EYFP in NAc of Cre lines but not wild-type mice (Fig. 3B, data not shown). The efficiency of Egr3 overexpression was confirmed by qRT-PCR and Western blot. We observed an increase of Egr3 mRNA and expression of the Egr3-EYFP protein in D1-Cre and D2-Cre NAc receiving the AAV-Egr3-EYFP compared with control virus (Student's t test, **p < 0.01, ***p < 0.001; D1-Cre: n = 3–5 per group, t(6) = 4.27, p < 0.01, D2-Cre: n = 5–6 per group, t(9) = 5.89, p < 0.001; Fig. 3C,D). Next, behavioral studies demonstrated that Egr3 overexpression in D1-MSNs versus D2-MSNs oppositely mediates cocaine CPP and cocaine-induced locomotor responses. Egr3 overexpression in D1-MSNs enhances time spent in the cocaine-paired chamber in CPP and increased cocaine-induced locomotion on days 2–5. In contrast, Egr3 overexpression in D2-MSNs blunts these behaviors (CPP: Student's t test, *p < 0.05; D1-Cre: n = 9–13 per group, t(20) = 2.13, p < 0.05, D2-Cre: n = 8–13 per group, t(19) = 2.11, p < 0.05: locomotion: repeated measures two-way ANOVA, D1-Cre: interaction F(5,105) = 4.39; p<0.01, Bonferroni posttest: *p < 0.05, **p < 0.01. D2-Cre: Interaction F(5,105) = 4.73; p < 0.001 Bonferroni posttest: *p < 0.05, ***p < 0.001; n = 9–14; Fig. 4A,B).
Figure 3.
Cre-inducible AAV overexpression of Egr3 in D1-MSN and D2-MSN subtypes. A, Schematic of the double-floxed, inverted, open reading frame Cre-dependent AAV vector expressing Egr3-EYFP. B, Image of a D2-Cre NAc demonstrating expression of Egr3-EYFP in NAc but no expression is visible in wild-type (WT) mice. Scale bar, 100 μm. C, D, AAV-DIO-Egr3-EYP overexpression D1-Cre and D2-Cre NAc results in the upregulation of Egr3 mRNA (C) and expression of 70 kDa Egr3-EYFP protein in NAc (D) compared with mice receiving the AAV-DIO-EYFP control virus. Error bars indicate SEM.
Egr3 knock-down in MSN subtypes oppositely mediates behavioral responses to cocaine
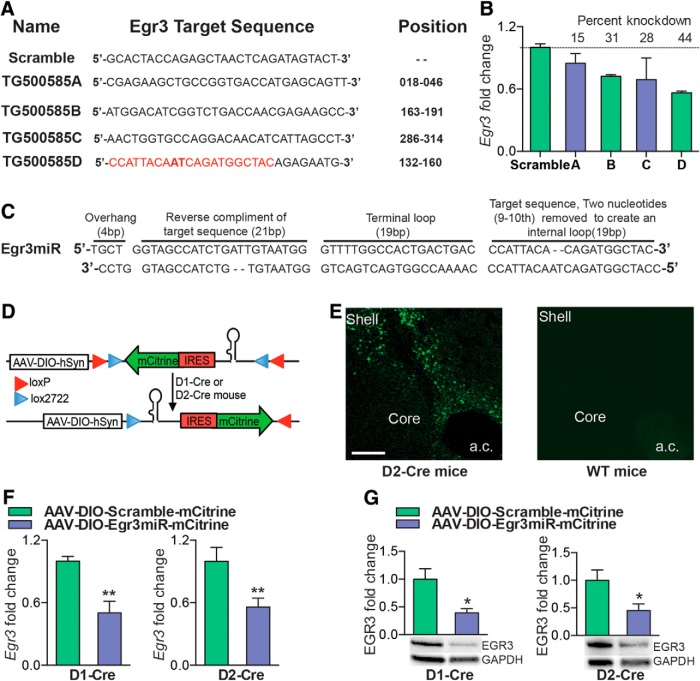
We next investigated whether miRNA-mediated knock-down of Egr3 in each MSN causes opposite behaviors to those observed with overexpression. We chose an miRNA approach as opposed to an shRNA approach, which can be toxic in vivo and requires the RNA polymerase II promoter, which could limit expression level (McBride et al., 2008; Ehlert et al., 2010). To address this issue, we developed an artificial miRNA that contained a portion of the shRNA target sequence. This approach allows the expression of miRNA, along with a reporter gene mCitrine, under the human synapsin (hSyn) promoter in Cre-expressing neurons. We chose the most efficient shRNA sequence, which displayed a 44% knock-down in Neuro2a cells, for miRNA engineering (Fig. 5A,B). After generation of the Egr3miR sequence, we cloned it into the Cre-inducible vector AAV-DIO-IRES-mCitrine (Fig. 5C,D). To evaluate the in vivo efficiency of the Egr3miR knock-down, we injected the virus into NAc of D1-Cre and D2-Cre mice. Two weeks later, we observed expression of mCitrine in NAc of Cre lines, but not wild-type mice (Fig. 5E, data not shown). The Egr3 knock-down efficiency was confirmed by qRT-PCR and protein levels by Western blot, which demonstrated a 50% and 44% knock-down of Egr3 mRNA and 60% and 55% knock-down of Egr3 protein in NAc of D1-Cre and D2-Cre mice, respectively (Student's t test, *p < 0.05, **p < 0.01; mRNA: D1-Cre: n = 4 per group, t(6) = 4.15, p < 0.01; D2-Cre: n = 4–5 per group, t(7) = 2.94, p < 0.01; protein: D1-Cre: n = 5 per group, t(8) = 3.11, p < 0.05; D2-Cre: n = 5 per group, t(8) = 2.48, p < 0.05; Fig. 5F,G). We next investigated whether the MSN subtype knock-down of Egr3 can also modulate cocaine CPP and cocaine-induced locomotion behaviors. Egr3 knock-down in D1-MSNs reduced the time spent in the drug-paired chamber in CPP and reduced the locomotor response to cocaine, whereas Egr3 knock-down in D2-MSNs enhanced these behaviors (CPP: Student's t test, *p < 0.05; D1-Cre: n = 8–12 per group, t(18) = 1.90, p < 0.05; D2-Cre: n = 5 per group, t(8) = 2.36, p < 0.05; locomotion: repeated-measures two-way ANOVA, D1-Cre: interaction F(5,70) = 2.410; p < 0.05, Bonferroni posttest: *p < 0.05, ***p < 0.0001. D2-Cre: Interaction F(5,35) = 3.691; p < 0.01, Bonferroni posttest: *p < 0.05, **p < 0.01, ***p < 0.001; n = 5–9; Fig. 6A,B).
Figure 5.
Cre-inducible AAV miRNA knock-down of Egr3 in D1-MSN and D2-MSN subtypes. A, Mouse Egr3 shRNA sequences and their targeting of the Egr3 gene. The position is relative to the ATG start site of Egr3 cDNA. B, qRT-PCR analysis of Egr3 demonstrates Egr3shRNA knock-down in Neuro2a cells. C, Illustration of the miRNA engineering sequence. The first 21 bp of the Egr3 target sequence (red) of shRNA is used for miRNA engineering. D, Schematic of the double-floxed, inverted, open reading frame Cre-dependent AAV vector expressing Egr3miR and mCitrine. E, Image of a D2-Cre NAc demonstrating expression of mCitrine in NAc, but no visible expression is observed in wild-type (WT) NAc. Scale bar, 100 μm. F, G, D1-Cre and D2-Cre mice expressing AAV-DIO-Egr3miR-mCitrine in NAc display reduction of Egr3 mRNA (F) and protein (G) compared with AAV-DIO-Scramble mCitrine controls. Error bars indicate SEM.
Figure 6.
Egr3miR knock-down in MSN subtypes alters cocaine CPP and cocaine-induced locomotor activity. A, Cocaine (7.5 mg/kg) CPP is decreased in D1-Cre mice that received AAV-DIO-Egr3miR-mCitrine into NAc, whereas it is increased in D2-Cre mice that received AAV-DIO-Egr3miR-mCitrine compared with AAV-DIO-scramble-mCitrine controls. Error bars indicate SEM. B, Cocaine (10 mg/kg)-induced locomotor activity is decreased in D1-Cre mice (days 1, 2, 3, and 5), whereas it is increased in D2-Cre mice expressing AAV-DIO-Egr3miR-mCitrine in NAc compared with controls expressing AAV-DIO-Scramble.
Egr3 regulates cocaine-associated genes transcriptionally
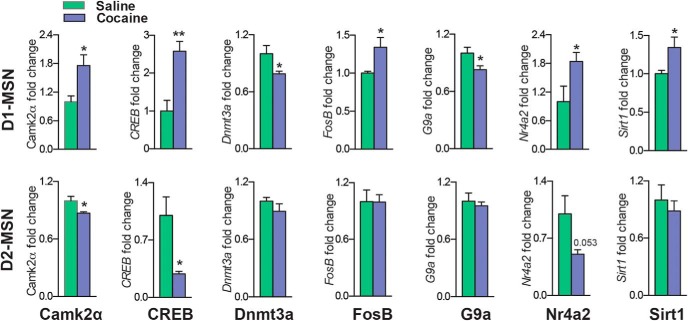
Finally, to address the role of Egr3 as a transcription factor, we investigated Egr3 binding to promoters of genes critical for cocaine-induced cellular and behavioral plasticity (Carlezon et al., 1998; Xi et al., 2002; Heusner and Palmiter, 2005; Anderson et al., 2008; Moussawi et al., 2009; Renthal et al., 2009; LaPlant et al., 2010; Maze et al., 2010; Kourrich et al., 2012; Vialou et al., 2012; Ferguson et al., 2013; Grueter et al., 2013; Lobo et al., 2013; Robison et al., 2013; Rogge et al., 2013; Bilbao et al., 2014). We used promoter profiling of Egr3-binding sites to determine which genes to examine (www.gene-regulation.com). These genes include the signaling molecule Camk2α; the transcription factors CREB, FosB, and Nr4a2; the histone- or DNA-modifying enzymes Dnmt3a, G9a, and Sirt1; and the glutamate receptors NMDAR1 and mGluR2. We performed ChIP using an Egr3 antibody on NAc tissue from mice that received repeated cocaine (7 d, 20 mg/kg) followed by 24 h withdrawal. Egr3 binding was increased on the promoter regions of Camk2α, CREB, FosB, Nr4a2, and Sirt1 and decreased on the promoter regions of the histone and DNA methylation enzymes G9a and Dnmt3a in the cocaine group (Student's t test, *p < 0.05; n = 6 per group; Camk2α t(10) = 2.78, p < 0.05, CREB t(10) = 2.47, p < 0.05, Dnmt3a t(10) = 2.56, p < 0.05, FosB t(10) = 2.99, p < 0.05, G9a t(10) = 1.55, p < 0.05, Nr4a2 t(10) = 3.07, p < 0.05, Sirt1 t(10) = 2.06, p < 0.05; Fig. 7). However, we found no change in the binding affinity of Egr3 on the promoter regions of glutamate receptors with the Egr3-binding sites mGluR2 and NMDAR1 in the cocaine group compared with saline controls. We next examined the cell-type-specific regulation of target genes after repeated cocaine administration (7 d, 20 mg/kg) followed by 24 h withdrawal. We observed an increase of CAMK2α and CREB ribosome-associated mRNA in D1-MSNs and a decrease of these genes in D2-MSNs in the cocaine group [Student's t test *p < 0.05, **p < 0.01; n = 6 per group; Camk2α: D1-MSN t(8) = 2.88, p < 0.05, D2-MSN t(8) = 2.75, p < 0.05; CREB: D1-MSN t(6) = 4.12, p < 0.01 (n = 4–6 per group), D2-MSN t(8) = 3.12, p < 0.05; Fig. 8]. FosB, Nr4a2, and Sirt1 were increased only in D1-MSNs, with no change in D2-MSNs in the cocaine group (FosB: n = 4–6 per group; t(8) = 2.13, p < 0.05; Nr4a2: n = 4–5, t(7) = 2.37, p < 0.05; Sirt1: n = 5 per group, t(8) = 2.33, p < 0.05). In contrast G9a, and Dnmt3a ribosome-associated mRNA was reduced in D1-MSNs with no change observed in D2-MSNs in the cocaine group compared with saline controls (G9a: n = 5–6 per group, t(9) = 2.42, p < 0.05; Dnmt3a: n = 5 per group, t(8) = 2.35, p < 0.05; Fig. 8).
Figure 7.
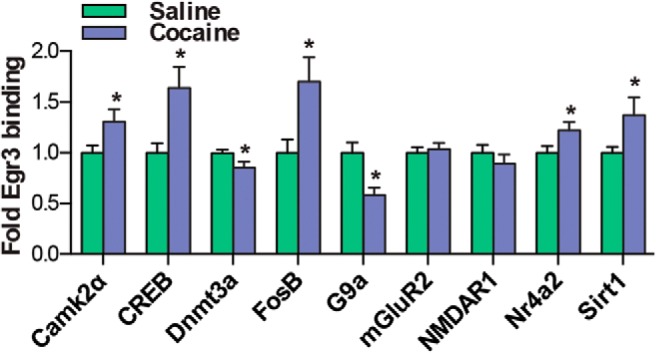
Egr3 transcriptional regulation of cocaine-associated genes after repeated cocaine exposure. ChIP using an Egr3 antibody to immunoprecipitate Egr3 bound to promoters of cocaine-associated genes after 7 d of cocaine (20 mg/kg) or saline exposure followed by 24 h withdrawal. Egr3 binding to the Camk2α, CREB, FosB, Nr4a2, and Sirt1 promoters is increased, whereas binding to the Dnmt3a and G9a promoters is decreased in the cocaine group. Error bars indicate SEM.
Figure 8.
Egr3-regulated genes are differentially expressed in MSN subtypes after repeated cocaine exposure. Cell-type-specific ribosome-associated mRNA analysis from D1-Cre-RT and D2-Cre-RT NAc mice after repeated cocaine (7 d, 20 mg/kg) exposure followed by 24 h withdrawal demonstrates altered induction of Egr3-regulated genes. Ribosome-associated mRNA for Camk2α and CREB is increased in D1-MSNs, whereas it is decreased in D2-MSNs in the cocaine group. FosB, Nr4a2, and Sirt1 are increased in D1-MSNs, with no change in D2-MSNs in the cocaine group. Dnmt3a and G9a are decreased in D1-MSNs, with no change in D2-MSNs in the cocaine group. Error bars indicate SEM.
Discussion
Our study demonstrates a role for the transcription factor Egr3 in NAc D1-MSNs versus D2-MSNs in the actions of cocaine. We first establish that repeated cocaine exposure causes a bidirectional induction of Egr3 in NAc MSN subtypes. Egr3 is increased in D1-MSNs and decreased in D2-MSNs in these conditions. The decrease of Egr3 in D2-MSNs corresponds to the decrease observed after contingent and noncontingent cocaine in total NAc tissue, which includes all MSNs, interneurons, and glia. The decrease that we observed in the total NAc in these conditions is consistent with other studies demonstrating a reduction or return to control levels of immediate early genes such as c-Fos, Egr1 (Zif268), and FosB in striatum after repeated psychostimulant exposure compared with acute exposure (Hope et, al., 1992; Ennulat et al., 1994; Renthal et al., 2008). However, a previous study demonstrated an increase in FosB in D1-MSNs after repeated cocaine administration using a similar ribosomal epitope-tagged approach (Heiman et al., 2008). Consistent with that study, here, we observed an increase in Egr3 in the D1-MSN population by repeated cocaine administration, which was masked in our total NAc tissue. Our findings emphasize the importance of cell-subtype-selective profiling because critical molecular adaptations can be missed when whole tissue is examined.
Consistent with ribosome-associated mRNA profiling, which demonstrated an increase of Egr3 in D1-MSNs after cocaine exposure, we found that enhancing levels of Egr3 in D1-MSNs potentiates cocaine-mediated reward using the CPP paradigm and increases cocaine-induced locomotion. Similarly, reducing Egr3 levels using an miRNA knock-down strategy to mimic the reduction of Egr3 observed after repeated cocaine administration in D2-MSNs also enhanced these behaviors. When we performed the reverse genetic manipulations of Egr3 in each MSN, knock-down in D1-MSNs and overexpression in D2-MSNs, we were able to blunt place preference and blunt locomotor responses to cocaine. In addition, our studies indicate the feasibility of bidirectionally altering levels of genes of interest, in this case Egr3, using Cre-inducible AAVs. A number of studies have used Cre-inducible viruses to overexpress genes of interest or to perform shRNA knock-down in MSN populations (Ena et al., 2013; Grueter et al., 2013; Dias et al., 2014; Rothwell et al., 2014); however, we used a cell-type-selective miRNA approach to express the Egr3miR under the hSyn promoter and effectively knock down Egr3 levels in each MSN subtype.
Most studies implicate a predominate role for D1-MSNs in mediating the positive rewarding and sensitizing effects of psychostimulants (Hikida et al., 2010; Lobo et al., 2010; Ferguson et al., 2011; Lobo and Nestler, 2011; Bock et al., 2013; Chandra et al., 2013; Lenz and Lobo, 2013), with many cocaine-induced molecular adaptations occurring in the D1-MSNs (Lee et al., 2006a; Lobo and Nestler, 2011; Grueter et al., 2013; Lobo et al., 2013; Rogge et al., 2013; Maze et al., 2014). Our results demonstrating an enhancement of Egr3 in D1-MSNs and showing that AAV-mediated increase of Egr3 in this MSN subtype promotes positive cocaine-induced behavioral output is consistent with these studies. However, the D2-MSNs can also promote positive cocaine-induced responses. We demonstrate that a reduction of Egr3 in the D2-MSNs is critical for the rewarding and locomotor responses to cocaine. Further, we demonstrated previously that the enhanced BDNF signaling that occurs through the VTA-NAc circuit has a predominate role in the D2-MSNs (Lobo et al., 2010). Because other studies have demonstrated that Egr3 is regulated through BDNF signaling (Roberts et al., 2006; Kim et al., 2012), BDNF may regulate the reduction of Egr3 in D2-MSNs, possibly through a negative feedback mechanism in which Egr3 blunts its own transcription through enhancing levels of its corepressor NGF1-A binding protein 2 (Nab2). In contrast, the Egr3 increase in D1-MSNs may occur through its positive feedback loop by which Egr3 binds to its own promoter to enhance Egr3 transcription (Kumbrink et al., 2010). Another potential mechanism for the bidirectional induction of Egr3 in MSN subtypes could be through D1 receptor signaling in D1-MSNs by enhancing PKA (Surmeier et al., 2007) and subsequently CREB activity (Carlezon et al., 2005), which can induce Egr3 transcription (Suehiro et al., 2010) and D2 receptor signaling, in D2-MSNs. This blunts PKA (Surmeier et al., 2007), potentially preventing CREB from inducing Egr3 levels and causing reduced Egr3 levels from basal conditions. Future studies can determine the precise signaling and transcriptional mechanisms that mediate this bidirectional induction of Egr3 in each MSN subtype.
Finally, we examined the role of Egr3 as a transcription factor to regulate expression of genes critical for psychostimulant-induced cellular and behavioral plasticity (Carlezon et al., 1998; Xi et al., 2002; Heusner and Palmiter, 2005; Anderson et al., 2008; Moussawi et al., 2009; Renthal et al., 2009; LaPlant et al., 2010; Maze et al., 2010; Kourrich et al., 2012; Vialou et al., 2012; Ferguson et al., 2013; Grueter et al., 2013; Lobo et al., 2013; Robison et al., 2013; Bilbao et al., 2014). Repeated cocaine exposure altered binding of Egr3 to promoters for the signaling molecule:Camk2α; the transcription factors CREB, FosB, and Nr4a2; and the histone- and DNA-modifying enzymes G9a, Dnmt3a, and Sirt1. Ribosome-associated mRNA, for the majority of the genes that displayed increased Egr3 binding to their promoters (Camk2α, CREB, FosB, Nr4a2, and Sirt1), was increased in D1-MSNs with repeated cocaine exposure. Two of these genes, Camk2α and CREB, also displayed a reduction in D2-MSNs under these conditions. The increase in FosB in NAc D1-MSNs is consistent with a previous study showing increased FosB in these MSNs in striatum with repeated cocaine (Heiman et al., 2008). Potentially, this increase in FosB is representative of the long-lasting isoform of FosB, ΔFosB, which we and others have demonstrated to be induced in D1-MSNs under repeated contingent and noncontingent cocaine conditions (Nye et al., 1995; Moratalla et al., 1996; Lee et al., 2006a; Lobo et al., 2013; Robison et al., 2013). Consistent with these findings, enhanced ΔFosB in D1-MSNs is important for the positive behavioral and synaptic plasticity outcomes mediated by cocaine (Kelz et al., 1999; Grueter et al., 2013). In addition, a previous study demonstrated that ΔFosB is critical for the induction of Camk2α in D1-MSNs and that Camk2α can mediate positive behavioral outcomes to cocaine (Robison et al., 2013). Our results demonstrating increased binding of Egr3 to the Camk2α promoter implicates a role for Egr3 in Camk2α-mediated cocaine actions.
We also observed increased Egr3 binding on the CREB promoter and CREB was induced in D1-MSNs, whereas it was reduced in D2-MSNs with cocaine exposure. Previous studies have indicated that enhanced CREB levels in the entire NAc are responsible for blunting cocaine reward (Carlezon et al., 1998), whereas increased forms of a dominant-negative CREB in the NAc or exclusively in striatal D1-MSNs, as well as a knock-out of CREB in NAc, enhances the rewarding and/or locomotor responses to cocaine (Carlezon et al., 1998; Vialou et al., 2012; Bilbao et al., 2014). Our results suggest that the Egr3 regulation of CREB activity in D1-MSNs can act as negative modulator of the actions of cocaine.
Another gene that is enhanced in the NAc and positively mediates behavioral and cellular plasticity changes by cocaine is the class III histone deacetylase Sirt1 (Renthal et al., 2009; Ferguson et al., 2013). Our studies demonstrate that Egr3 regulates this positive induction of Sirt1 and that the rewarding effects of Sirt1 likely occur through D1-MSNs because ribosome-associated mRNA for this gene is increased in D1-MSNs after repeated cocaine exposure. In contrast, other epigenetic-related enzymes, the repressive histone methylation enzyme G9a and the DNA methylation enzyme Dnmt3a, display reduced Egr3 binding to their promoters and reduced ribosome-associated mRNA in D1-MSNs with no change in D2-MSNs. Reduced levels of G9a or Dnmt3a potentiate cocaine reward, whereas enhancing the levels of these genes attenuates this behavior (LaPlant et al., 2010; Maze et al., 2010). Our results suggest that these behavioral effects likely occur through D1-MSNs. However, a previous study demonstrated that G9a levels are reduced in D1-MSNs and D2-MSNs in total striatum after repeated cocaine exposure (Maze et al., 2014). Our NAc-selective profiling suggests that the reduction of G9a in D2-MSNs with cocaine may be occurring mainly in dorsal striatum. In addition, knock-out of G9a in striatal D2-MSNs shifts these MSNs to a D1-MSN phenotype and enhances behavioral responses to cocaine (Maze et al., 2014), further demonstrating a role for G9a in D1-MSNs.
Collectively, our studies demonstrate a novel and dynamic role for Egr3 in NAc cell subtypes in mediating behavioral plasticity to cocaine. We show that bidirectional induction of Egr3 in MSN subtypes by repeated cocaine is critical for mediating cocaine reward and locomotor responses. We further demonstrate that Egr3 can regulate transcriptionally signaling molecules, transcription factors, and histone- or DNA-modifying enzymes that are critical for psychostimulant-induced cellular and behavioral plasticity. Our ChIP studies and our profiling of these genes in MSN subtypes after repeated cocaine exposure establishes a molecular signature for these molecules and their regulation by Egr3 in MSN subtypes. This detailed understanding of the transcriptional underpinnings occurring in each MSN subtype has implications for therapeutic targeting of molecules selective to MSN subtypes in cocaine abuse.
Footnotes
This work was supported by the National Institutes of Health (NIH Grant RO1DA038613). P.K. is supported by the NIH (Grants R01DK090918 and R01DK039308) and the National Kidney Foundation. D.M.D. is supported by NIH Grant R01DA037257. A.M.G is supported by NIH Grant T-32-AA007583. We thank B.L. Roth (University of North Carolina) and K. Deisseroth (Stanford University) for the AAV backbone vectors used in this study and A. Nectow (Rockefeller University) for advice on the RT procedure.
The authors declare no competing financial interests.
References
- Anderson SM, Famous KR, Sadri-Vakili G, Kumaresan V, Schmidt HD, Bass CE, Terwilliger EF, Cha JH, Pierce RC. CaMKII: a biochemical bridge linking accumbens dopamine and glutamate systems in cocaine seeking. Nat Neurosci. 2008;11:344–353. doi: 10.1038/nn2054. [DOI] [PubMed] [Google Scholar]
- Arango-Lievano M, Schwarz JT, Vernov M, Wilkinson MB, Bradbury K, Feliz A, Marongiu R, Gelfand Y, Warner-Schmidt J, Nestler EJ, Greengard P, Russo SJ, Kaplitt MG. Cell-type specific expression of p11 controls cocaine reward. Biol Psychiatry. 2014;76:794–801. doi: 10.1016/j.biopsych.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Santini E, Shen W, Birnbaum S, Valjent E, Surmeier DJ, Fisone G, Nestler EJ, Greengard P. Distinct subclasses of medium spiny neurons differentially regulate striatal motor behaviors. Proc Natl Acad Sci U S A. 2010;107:14845–14850. doi: 10.1073/pnas.1009874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbao A, Rieker C, Cannella N, Parlato R, Golda S, Piechota M, Korostynski M, Engblom D, Przewlocki R, Schütz G, Spanagel R, Parkitna JR. CREB activity in dopamine D1 receptor expressing neurons regulates cocaine-induced behavioral effects. Front Behav Neurosci. 2014;8:212. doi: 10.3389/fnbeh.2014.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Shin JH, Kaplan AR, Dobi A, Markey E, Kramer PF, Gremel CM, Christensen CH, Adrover MF, Alvarez VA. Strengthening the accumbal indirect pathway promotes resilience to compulsive cocaine use. Nat Neurosci. 2013;16:632–638. doi: 10.1038/nn.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, Duman RS, Neve RL, Nestler EJ. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- Chandra R, Lenz JD, Gancarz AM, Chaudhury D, Schroeder GL, Han MH, Cheer JF, Dietz DM, Lobo MK. Optogenetic inhibition of D1R containing nucleus accumbens neurons alters cocaine-mediated regulation of Tiam1. Front Mol Neurosci. 2013;6:13. doi: 10.3389/fnmol.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, Dietz DM, Lobo MK, Ghose S, Mouzon E, Neve RL, Tamminga CA, Nestler EJ. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;4:656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, Scobie K, Bagot R, LaBonté B, Ribeiro E, Liu X, Kennedy P, Vialou V, Ferguson D, Peña C, Calipari ES, Koo JW, Mouzon E, Ghose S, Tamminga C, Neve R, Shen L, Nestler EJ. Beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. 2014;516:51–55. doi: 10.1038/nature13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert EM, Eggers R, Niclou SP, Verhaagen J. Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC Neurosci. 2010;11:20. doi: 10.1186/1471-2202-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ena SL, De Backer JF, Schiffmann SN, de Kerchove d'Exaerde A. FACS array profiling identifies Ecto-5′ nucleotidase as a striatopallidal neuron-specific gene involved in striatal-dependent learning. J Neurosci. 2013;33:8794–8809. doi: 10.1523/JNEUROSCI.2989-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennulat DJ, Babb Sm, Cohen BM. Persistent reduction of immediate early gene mRNA in rat forebrain following single or multiple doses of cocaine. Brain Res Mol Brain Res. 1994;26:106–112. doi: 10.1016/0169-328X(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C, Laitman B, Stockman V, LaPlant Q, Cahill ME, Nestler EJ, Shen L. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D, Koo JW, Feng J, Heller E, Rabkin J, Heshmati M, Renthal W, Neve R, Liu X, Shao N, Sartorelli V, Shen L, Nestler EJ. Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. J Neurosci. 2013;33:16088–16098. doi: 10.1523/JNEUROSCI.1284-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Eskenazi D, Ishikawa M, Wanat MJ, Phillips PE, Dong Y, Roth BL, Neumaier JF. Transient neuronal inhibition reveals opposing roles of indirect and direct pathways in sensitization. Nat Neurosci. 2011;14:22–24. doi: 10.1038/nn.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ, Jr, Sibley DR. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter BA, Robison AJ, Neve RL, Nestler EJ, Malenka RC. FosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc Natl Acad Sci U S A. 2013;110:1923–1928. doi: 10.1073/pnas.1221742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusner CL, Palmiter RD. Expression of mutant NMDA receptors in dopamine D1 receptor-containing cells prevents cocaine sensitization and decreases cocaine preference. J Neurosci. 2005;28:6651–6657. doi: 10.1523/JNEUROSCI.1474-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida T, Kimura K, Wada N, Funabiki K, Nakanishi S. Distinct roles of synaptic transmission in direct and indirect striatal pathways to reward and aversive behavior. Neuron. 2010;66:896–907. doi: 10.1016/j.neuron.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Hope B, Kosofsky B, Hyman SE, Nestler EJ. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci U S A. 1992;89:5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvert P, Dietrich JB, Aunis D, Zwiller J. Differential rat brain expression of EGR proteins and of the transcriptional corepressor NAB in response to acute or chronic cocaine administration. Neuromolecular Med. 2002;1:137–151. doi: 10.1385/NMM:1:2:137. [DOI] [PubMed] [Google Scholar]
- Kelz MB, Chen J, Carlezon WA, Jr, Whisler K, Gilden L, Beckmann AM, Steffen C, Zhang YJ, Marotti L, Self DW, Tkatch T, Baranauskas G, Surmeier DJ, Neve RL, Duman RS, Picciotto MR, Nestler EJ. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- Kim JH, Roberts DS, Hu Y, Lau GC, Brooks-Kayal AR, Farb DH, Russek SJ. Brain-derived neurotrophic factor uses CREB and Egr3 to regulate NMDA receptor levels in cortical neurons. J Neurochem. 2012;120:210–219. doi: 10.1111/j.1471-4159.2011.07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Park BH, Lee JH, Park SK, Kim JH. Cell type-specific alterations in the nucleus accumbens by repeated exposures to cocaine. Biol Psychiatry. 2011;69:1026–1034. doi: 10.1016/j.biopsych.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Kourrich S, Klug JR, Mayford M, Thomas MJ. AMPAR-independent effect of striatal αCaMKII promotes the sensitization of cocaine reward. J Neurosci. 2012;32:6578–6586. doi: 10.1523/JNEUROSCI.6391-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbrink J, Kirsch KH, Johnson JP. EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. J Cell Biochem. 2010;111:207–217. doi: 10.1002/jcb.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Kim Y, Kim AM, Helmin K, Nairn AC, Greengard P. Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proc Natl Acad Sci U S A. 2006a;103:3399–3404. doi: 10.1073/pnas.0511244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006b;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JD, Lobo MK. Optogenetic insights into striatal function and behavior. Behav Brain Res. 2013;255:44–54. doi: 10.1016/j.bbr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Nestler EJ. The striatal balancing act in drug addiction: distinct roles of direct and indirect pathway medium spiny neurons. Front Neuroanat. 2011;5:41. doi: 10.3389/fnana.2011.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Covington HE, 3rd, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, Dietz DM, Zaman S, Koo JW, Kennedy PJ, Mouzon E, Mogri M, Neve RL, Deisseroth K, Han MH, Nestler EJ. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Zaman S, Damez-Werno DM, Koo JW, Bagot RC, DiNieri JA, Nugent A, Finkel E, Chaudhury D, Chandra R, Riberio E, Rabkin J, Mouzon E, Cachope R, Cheer JF, Han MH, Dietz DM, Self DW, Hurd YL, Vialou V, Nestler EJ. DeltaFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J Neurosci. 2013;33:18381–18395. doi: 10.1523/JNEUROSCI.1875-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Chaudhury D, Dietz DM, Von Schimmelmann M, Kennedy PJ, Lobo MK, Sillivan SE, Miller ML, Bagot RC, Sun H, Turecki G, Neve RL, Hurd YL, Shen L, Han MH, Schaefer A, Nestler EJ. G9a influences neuronal subtype specification in striatum. Nat Neurosci. 2014;17:533–539. doi: 10.1038/nn.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty JF, Whitfield TW, Jr, Berglind WJ. Brain-derived neurotrophic factor and cocaine addiction. Brain Res. 2010;1314:183–193. doi: 10.1016/j.brainres.2009.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratalla R, Vallejo M, Elibol B, Graybiel AM. D1-class dopamine receptors influence cocaine-induced persistent expression of Fos-related proteins in striatum. Neuroreport. 1996;8:1–5. doi: 10.1097/00001756-199612200-00001. [DOI] [PubMed] [Google Scholar]
- Moussawi K, Pacchioni A, Moran M, Olive MF, Gass JT, Lavin A, Kalivas PW. N-Acetylcysteine reverses cocaine-induced metaplasticity. Nat Neurosci. 2009;12:182–189. doi: 10.1038/nn.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye HE, Hope BT, Kelz MB, Iadarola M, Nestler EJ. Pharmacological studies of the regulation of chronic FOS-related antigen induction by cocaine in the striatum and nucleus accumbens. J Pharmacol Exp Ther. 1995;275:1671–1680. [PubMed] [Google Scholar]
- O'Donovan KJ, Baraban JM. Major Egr3 isoforms are generated via alternate translation start sites and differ in their abilities to activate transcription. Mol Cell Biol. 1999;19:4711–4718. doi: 10.1128/mcb.19.7.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan S, Gashler A, Siegel MG, Chang LC, Joseph LJ, Shows TB, Le Beau MM, Sukhatme VP. EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene. 1991;6:917–928. [PubMed] [Google Scholar]
- Prasad KM, Smith RS, Xu Y, French BA. A single direct injection into the left ventricular wall of an adeno-associated virus 9 (AAV9) vector expressing extracellular superoxide dismutase from the cardiac troponin-T promoter protects mice against myocardial infarction. J Gene Med. 2011;6:333–341. doi: 10.1002/jgm.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, 3rd, Truong HT, Alibhai I, Kumar A, Montgomery RL, Olson EN, Nestler EJ. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28:7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, 3rd, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ. Genome wide analysis of chromatin regulation by cocaine reveals a novel role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried MU, Girod A, Leike K, Büning H, Hallek M. Adeno-associated virus capsids displaying immunoglobulin-binding domains permit antibody-mediated vector retargeting to specific cell surface receptors. J Virol. 2002;76:4559–4566. doi: 10.1128/JVI.76.9.4559-4566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DS, Hu Y, Lund IV, Brooks-Kayal AR, Russek SJ. Brain-derived neurotrophic factor (BDNF)-induced synthesis of early growth response factor 3 (Egr3) controls the levels of type A GABA receptor alpha 4 subunits in hippocampal neurons. J Biol Chem. 2006;281:29431–29435. doi: 10.1074/jbc.C600167200. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Vialou V, Mazei-Robison M, Feng J, Kourrich S, Collins M, Wee S, Koob G, Turecki G, Neve R, Thomas M, Nestler EJ. Behavioral and structural responses to chronic cocaine require a feedforward loop involving DeltaFosB and calcium/calmodulin-dependent protein kinase II in the nucleus accumbens shell. J Neurosci. 2013;33:4295–4307. doi: 10.1523/JNEUROSCI.5192-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Singh H, Dang R, Wood MA. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci. 2013;33:6623–6632. doi: 10.1523/JNEUROSCI.4472-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, Südhof TC. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Wilkinson MB, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V, Renthal W, Graham A, Birnbaum SG, Green TA, Robison B, Lesselyong A, Perrotti LI, Bolaños CA, Kumar A, Clark MS, Neumaier JF, Neve RL, Bhakar AL, Barker PA, Nestler EJ. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29:3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, Nestler EJ. The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010;33:267–276. doi: 10.1016/j.tins.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Evanoff R, Quintana A, Evans E, Miller JA, Ko C, Amieux PS, Griswold MD, McKnight GS. RiboTag analysis of actively translated mRNAs in Sertoli and Leydig cells in vivo. PLoS One. 2013;8:e66179. doi: 10.1371/journal.pone.0066179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self DW. Regulation of drug-taking and -seeking behaviors by neuroadaptations in the mesolimbic dopamine system. Neuropharmacology. 2004;47:242–255. doi: 10.1016/j.neuropharm.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Lobo MK, Spencer S, Kalivas PW. Cocaine-induced adaptations in D1 and D2 accumbens projection neurons (a dichotomy not necessarily synonymous with direct and indirect pathways) Curr Opin Neurobiol. 2013;23:546–552. doi: 10.1016/j.conb.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suehiro J, Hamakubo T, Kodama T, Aird WC, Minami T. Vascular endothelial growth factor activation of endothelial cells is mediated by early growth response-3. Blood. 2010;115:2520–2532. doi: 10.1182/blood-2009-07-233478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Ding J, Day M, Wang Z, Shen W. D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci. 2007;30:228–235. doi: 10.1016/j.tins.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Vialou V, Feng J, Robison AJ, Ku SM, Ferguson D, Scobie KN, Mazei-Robison MS, Mouzon E, Nestler EJ. Serum response factor and cAMP response element binding protein are both required for cocaine induction of DeltaFosB. J Neurosci. 2012;32:7577–7584. doi: 10.1523/JNEUROSCI.1381-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang GJ, Baler R, Telang F. Imaging dopamine's role in drug abuse and addiction. Neuropharmacology. 2009;56:3–8. doi: 10.1016/j.neuropharm.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi ZX, Ramamoorthy S, Baker DA, Shen H, Samuvel DJ, Kalivas PW. Modulation of group II metabotropic glutamate receptor signaling by chronic cocaine. J Pharmacol Exp Ther. 2002;303:608–615. doi: 10.1124/jpet.102.039735. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Kaufmann WE, Lanahan A, Papapavlou M, Barnes CA, Andreasson KI, Worley PF. Egr3/Pilot, a zinc finger transcription factor, is rapidly regulated by activity in brain neurons and colocalizes with Egr1/zif268. Learn Mem. 1994;1:140–152. [PubMed] [Google Scholar]