ABSTRACT
Escherichia coli senses and responds to trimethylamine-N-oxide (TMAO) in the environment through the TorT-TorS-TorR signal transduction system. The periplasmic protein TorT binds TMAO and stimulates the hybrid kinase TorS to phosphorylate the response regulator TorR through a phosphorelay. Phosphorylated TorR, in turn, activates transcription of the torCAD operon, which encodes the proteins required for anaerobic respiration via reduction of TMAO to trimethylamine. Interestingly, E. coli respires TMAO in both the presence and absence of oxygen, a behavior that is markedly different from the utilization of other alternative electron acceptors by this bacterium. Here we describe an unusual form of regulation by oxygen for this system. While the average level of torCAD transcription is the same for aerobic and anaerobic cultures containing TMAO, the behavior across the population of cells is strikingly different under the two growth conditions. Cellular levels of torCAD transcription in aerobic cultures are highly heterogeneous, in contrast to the relatively homogeneous distribution in anaerobic cultures. Thus, oxygen regulates the variance of the output but not the mean for the Tor system. We further show that this oxygen-dependent variability stems from the phosphorelay.
IMPORTANCE Trimethylamine-N-oxide (TMAO) is utilized by numerous bacteria as an electron acceptor for anaerobic respiration. In E. coli, expression of the proteins required for TMAO respiration is tightly regulated by a signal transduction system that is activated by TMAO. Curiously, although oxygen is the energetically preferred electron acceptor, TMAO is respired even in the presence of oxygen. Here we describe an interesting and unexpected form of regulation for this system in which oxygen produces highly variable expression of the TMAO utilization proteins across a population of cells without affecting the mean expression of these proteins. To our knowledge, this is the first reported example of a stimulus regulating the variance but not the mean output of a signaling system.
INTRODUCTION
Like many bacteria, Escherichia coli has the capacity for respiration in the absence of oxygen by employing alternative compounds as electron acceptors. One example is the reduction of trimethylamine-N-oxide (TMAO) to trimethylamine (TMA) (1). Anaerobic respiration of TMAO or other compounds can provide a significant fitness advantage by enabling more energy-efficient metabolic pathways than are possible with fermentation alone. The primary niches where E. coli encounters TMAO have not been established but are potentially both within and outside animal hosts. TMAO is a naturally abundant osmolyte in algae, fish, and other marine organisms (2). In addition, the compound is secreted in the urine of humans and other animals as a result of TMAO consumption directly via seafood-rich diets (3) and also from the oxidation of TMA that is produced when dietary choline and similar amines are metabolized by gut flora (4–6). TMAO may also have a role in infections of animal hosts. At least one study has demonstrated that the Tor system is activated in urinary tract infections (7). It has also been suggested that TMAO (as well as several other alternative electron acceptors) may be produced from reactive nitrogen and oxygen species at sites of inflammation, potentially providing a competitive advantage for E. coli and related bacteria over strict anaerobes that lack the ability to utilize these compounds (8).
The major pathway for TMAO reduction in E. coli requires expression of the proteins TorC, TorA, and TorD, which are encoded in the torCAD operon (9, 10; for reviews, see references 11 and 12). TorC is a c-type cytochrome and membrane protein that is believed to shuttle electrons from the quinone pool to TorA, a periplasmically localized TMAO reductase. TorD is a chaperone that enables the addition of a molybdenum cofactor to apo-TorA and secretion of mature TorA into the periplasm through the Tat translocon (13–16).
Transcription of torCAD is strictly dependent on TMAO initiating signal transduction through the TorS-TorR phosphorelay (17, 18). TorS is a tripartite hybrid sensor kinase, with histidine kinase, receiver, and histidine-containing phosphotransfer (Hpt) domains, that phosphorylates TorR, a response regulator transcription factor with receiver and DNA binding domains. TMAO sensing is mediated by a periplasmic binding protein, TorT, that binds TorS with high affinity (19). Biochemical and crystallographic studies indicate that TMAO binding to a TorT-TorS heterotetramer triggers conformational changes and likely TorS autophosphorylation to initiate the signaling cascade (19, 20). TorR phosphorylation proceeds by a four-step phosphotransfer from ATP through intermediate states of TorS to TorR-P (21, 22); all four steps are necessary for the activation of torCAD transcription (18). As has been observed for other hybrid kinases, TorS dephosphorylates the phosphorylated form of its partner response regulator, TorR-P, by reverse phosphotransfer upon stimulus removal (23, 24).
In addition to the role of phosphorylated TorR in transcription regulation, TorR represses its own transcription independently of its phosphorylation state (25). The torCAD operon and torR are divergently transcribed, and the small torR-torC intergenic region contains four TorR binding sites with differing binding affinities that are responsible for the regulation of torR and torCAD expression. It has been hypothesized that TorR-P competes with unphosphorylated TorR for occupancy of the high-affinity binding sites and that activation of torCAD transcription is initiated after a critical number of TorR-P molecules is reached (26).
In general, E. coli cells preferentially utilize electron acceptors in a hierarchical order based on their energy yield (27). Since oxygen has the highest standard reduction potential, most alternative respiratory systems are repressed during aerobic growth. However, TMAO is an exception to this rule. Despite the relatively low energy yield of the TMAO-TMA couple, torCAD expression and TMAO reduction occur under both aerobic and anaerobic growth conditions (28). Consistent with this observation, no role for oxygen has been reported to date in the direct control of torCAD transcription or the TorS-TorR phosphorelay. Here we report an unusual form of regulation of torCAD expression by oxygen. We show that although the mean levels of torCAD transcription are the same during aerobic or anaerobic growth in liquid culture, the single-cell behavior within a population is markedly different under the two growth conditions: the cellular level of torCAD transcription is relatively uniform within a population of anaerobically growing E. coli but is highly heterogeneous in a population of aerobically growing cells. Thus, we find that oxygen regulates the variance but not the mean level of torCAD transcription. We further show that this variability is mediated by the TorT-TorS-TorR signaling system.
MATERIALS AND METHODS
Growth media and conditions.
Liquid cultures for fluorescence measurements were grown at 37°C in minimal A medium (29) supplemented with 0.2% glucose and 0.1% Casamino Acids (Difco-BD, Franklin Lakes, NJ). Where indicated, cultures were grown without aeration in screw-cap vials (Wheaton, Millville, NJ) filled to the rim, sealed tightly, and standing at 37°C until the appropriate density was reached. TMAO was purchased from Sigma and used at a final concentration of 10 mM, unless otherwise indicated. TMAO was added when diluting back from overnight cultures. Bacterial cultures to prepare electrocompetent cells were grown in LB (Difco-BD, Franklin Lakes, NJ) or SOB (Amresco, Solon, OH) with the appropriate antibiotic when necessary. To maintain plasmids, antibiotics were added to culture media at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 25 μg/ml; chloramphenicol, 25 or 12 μg/ml for single-copy plasmids (pSMART derivatives). Chromosomal integrants were selected for with chloramphenicol at 12 μg/ml or kanamycin at 20 μg/ml.
Strains and plasmids.
The strains and plasmids used in this study are described in Tables 1 and 2, respectively. The primers used for strain construction are listed in Table 3. Phage P1vir (29) was used for transductions. Transformations of plasmids and of linear DNA for chromosomal integration were performed by electroporation. When necessary, antibiotic resistance cassettes flanked by FLP recombination target (FRT) sites were removed with the plasmid pCP20 as described in reference 30.
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea | Reference or source |
|---|---|---|
| MG1655 | F− λ− ilvG rfb-50 rph-1 | E. coli Genetic Stock Center (CGSC no. 7740) |
| Nissle 1917 | 38 | |
| HS | 39 | |
| EPB47 | MG1655 Φ(ompA+-cfp+) | Goulian lab stock |
| MMR8 | MG1655 attλ::(PtorCAD-yfp) Φ(ompA+-cfp+) | This study |
| MMR60 | MG1655 Δ(xylA xylFG)::PtetA-mCherry | This study |
| MMR72 | MG1655 Δ(lacI lacZYA)::(PtorCAD-cfp) attλ::(PtorCAD-yfp) Δ(xylA xylFG)::PtetA-mCherry | This study |
| MMR84 | MG1655 ΔtorS::(FRT-kan-FRT) Δ(lacI lacZYA)::(PtorCAD-cfp) Δ(xylA xylFG)::PtetA-mCherry | This study |
| MMR125 | MG1655 attλ::(PtorCAD-yfp-FRT-kan-FRT) Φ(ompA+-cfp+) | This study |
| MMR130 | MG1655 attλ::(PtorCAD-yfp-FRT) Φ(ompA+-cfp+) | This study |
| MMR133 | MG1655 ΔtorSTR attλ::(PtorCAD-yfp-FRT-kan-FRT) Φ(ompA+-cfp+) | This study |
| MMR138 | MG1655 ΔtorS ΔtorT torR146::(FRT-cat-FRT) attλ::(PtorCAD-yfp) Φ(ompA+-cfp+) | This study |
| MMR140 | MG1655 ΔtorS ΔtorT torR149::(FRT-cat-FRT) attλ::(PtorCAD-yfp) Φ(ompA+-cfp+) | This study |
| MMR141 | MG1655 Δ(lacI lacZYA)::(PtorC-cfp) Φ(torD+-yfp+) Δ(xylA xylFG)::PtetA-mCherry | This study |
| MMR233 | MG1655 attλ::(PtorCAD-yfp) Φ(ompA+-cfp+) ΔsulA::(FRT-kan-FRT) | This study |
| MMR234 | Nissle1917 attλ::(PtorCAD-yfp) Φ(ompA+-cfp+) ΔsulA::(FRT-kan-FRT) | This study |
| MMR235 | HS attλ::(PtorCAD-yfp) Φ(ompA+-cfp+) ΔsulA::(FRT-kan-FRT) | This study |
| JW5135 | ΔtorS::(FRT-kan-FRT) | 46 |
| JW0941 | ΔsulA::(FRT-kan-FRT) | 46 |
| PIR2 | F− Δlac169 rpoS(Am) robA1 creC510 hsdR514 endA recA1 uidA(ΔMluI)::pir | Invitrogen |
| E. cloni | F− mcrA Δ(mrr-hsdRMS-mcrBC) endA1 recA1 ϕ80lacZΔM15 ΔlacX74 araD139 Δ(ara-leu)7697 galU galK rpsL nupG tonA (attL araC-PBAD-trfA250 bla attR) λ− | Lucigen Corporation |
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
Φ(gene+-cfp+) denotes an operon fusion of a gene and cfp.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant genotype | Reference or source |
|---|---|---|
| pCAH63 | oriRγ cat attPλ Psyn1-uidAf | 31 |
| pCP20 | λcI857(ts) repA101(ts) oriR101 bla cat λpR-FLP | 30 |
| pDSW206 | lacIq Ptrc attenuated promoter | 35 |
| pEB45 | pKD13 yfp-FRT-kan-FRT | 33 |
| pEB48 | pKD13 cfp-FRT-kan-FRT | Goulian lab stock |
| pKD4 | oriγR6K, bla, FRT-kan-FRT | 30 |
| pKD3 | oriγR6K, bla, FRT-cat-FRT | 30 |
| pKD13 | oriγR6K, bla, FRT-kan-FRT | 30 |
| pKD46 | repA101(ts) oriR101 bla ParaB-(gam bet exo) | 30 |
| pSMART | pSMART VC BamHI cat oriV ori2 repE parABC | Lucigen Corporation |
| pMR4 | pCAH63 PtorCAD-yfp | This study |
| pMR14 | pKD13 PtetA-mCherry | This study |
| pMR19 | pKD13 PtorCAD-cfp | This study |
| pMR26 | pDSW206 torT | This study |
| pMR29 | pSMART lacIq-Ptrc-torS | This study |
| pMR32 | pSMART torR | This study |
| pMR46 | pSMART torR146 (TorR K114R R146H) | This study |
| pMR49 | pSMART torR149 (TorR D53A K114R R146H) | This study |
| pTM149 | pBluescript SK PtetA-mCherry | Goulian lab stock |
TABLE 3.
Primers used in this study
| Primer | Sequence | Resulting construct |
|---|---|---|
| torC-L1 | CCGGTACCTGAAGCGATCTTAATGAGCAAATATG | pMR4, torR mutants, pMR32 |
| torC-U2 | CCGGATCCTCAACAATAACAATGTGATGTGGTGTCAGAGG | pMR4 |
| cat-LRed-U1 | ATATCCCAATGGCATCGTAAAGAACATTTTGAGGCATTTCAGTCAGTTGCGCTGGAGCTGCTTCGAA | MMR125, MMR130, MMR133 |
| cat-LRed-L1 | ATGAACCTGAATCGCCAGCGGCATCAGCACCTTGTCGCCTTGCGTATAATATGAATATCCTCCTTAG | MMR125, MMR130, MMR133 |
| Psyn-U1 | CCTGACGGATGGCCTTTTTG | pMR19 |
| gfpNterm-L1 | CATCACCTTCACCCTCTCC | pMR19 |
| Tetafter-MCS-L1 | TATTATCGTGAGGATGCGTC | pMR19 |
| cfpNterm-U1 | CACTGGAGTTGTCCCAATTCTTG | pMR19 |
| Lac-pMR10-U1 | GGTGGCCGGAAGGCGAAGCGGCATGCATTTACGTTGACACCATCGAATGGGCAATCCAGTGCAAAGCTAG | MMR72 |
| Lac-pMR10-L1 | GCCTTCGCACATATCGGTAAATAGCTTGCCTGCTCTTATTCTTTCGGGAGCGATTGTGTAGGCTGGAG | MMR72 |
| torD-yfp-U1 | TAAGCCAGTTATTGCTGGTGTTAGTGGAGTGCGACCACCAAAACAGATAATAGAATTAAAGAGGAGAAAT | MMR141 |
| torD-yfp-L2 | TTAGTGGAGTGCGACCACCAAAACAGATAAAATTAATTCCGGGGATCCGT | MMR141 |
| Xyl-pKD13-U1 | TGCCCGGTATCGCTACCGATAACCGGGCCAACGGACTGCACAGTTAGCCGGCAATCCAGTGCAAAGCTAG | MMR60, MMR72, MMR84, MMR141 |
| Xyl-pKD13-L1 | GTGATTTCAGCCCGGAGAAGCCACCGGATGTCGGTACGGCCAATTTCACTGAGCGATTGTGTAGGCTGGAG | MMR60, MMR72, MMR84, MMR141 |
| torR-SacI-L1 | CCCGAGCTCTTTATCAGCACACATCAGCG | torR mutants, pMR32 |
| torSTRdel-U1 | TACGGCAGAATATGAACAGATATGAACAGAATGAGTAAAACCCTCTGATGATTCCGGGGATCCGTCGACC | MMR133 |
| torSTRdel-L1 | AACTGGCTGATATTGATTCAAATCGCGTTCAGGTCTTTCTTATGCAACCATGTAGGCTGGAGCTGCTTC | MMR133 |
| D53A-FOR | GATTTAATTCTGCTGGCTATCAACTTACCCGA | pMR49, MMR140 |
| D53A-REV | TCGGGTAAGTTGATAGCCAGCAGAATTAAATC | D53A-TorR |
| torR-int-U1 | AGTTTCCGCATAATAACCCCTGTAGAATTATGGTTTAGTGAAGCGATCTTAATGAGCAAA | MMR138, MMR140 |
| torR-cat-rev | TAAGGAGGATATTCATATGGACCATGGGGCAACCAGCCCGGTGAG | MMR138, MMR140 |
| torR-cat-for | CTCACCGGGCTGGTTGCCCCATGGTCCATATGAATATCCTCCTTA | MMR138, MMR140 |
| torR-int-L1 | GCCACTATTCTAGTTGTTCTTAACTGGCTGATATTGATTCAAATCGCGTTCAGGTCTTTCAGATTGCAGCATTACACGTC | MMR138, MMR140 |
| torS-BamHI-U1 | CCACACGGATCCTAATCCGCTTTGCGACTCAACCT | pMR29 |
| torS-HindIII-L1 | CCACACAAGCTTAACTGGCTGATATTGA | pMR29 |
| torT-upper-1 | CCCGGATCCCTAGCATAAAGCCTTATTATTG | pMR26 |
| torT-lower-1 | CCCGTCGACGATTTACTGGTGACGCAACATG | pMR26 |
The PtorCAD-yfp transcriptional fusion fluorescent reporter of strains MMR8, MMR72, MMR233, MMR234, and MMR235 was constructed in a CRIM plasmid and integrated into the chromosome (31). A 140-bp DNA fragment containing the promoter of the torCAD operon, the transcription start site, and all four TorR boxes (26) was amplified by PCR from the chromosome of E. coli K-12 strain MG1655 (32) with primers torC-U2 and torC-L1. The upstream primer was designed to introduce a point mutation that disrupts the torR start codon. Primers added BamHI and KpnI restriction sites for cloning in front of the yfp gene in a derivative of CRIM plasmid pCAH63 (31). The resulting plasmid, pMR4, was integrated in single copy at attλ of MG1655, Nissle 1917, and HS. The cloned torCAD promoter region of pMR4 was verified by sequencing. Following integration into the chromosome, pMR4 was verified to be in single copy by PCR as described in reference 31. For strains MMR8 and MMR72, the fluorescent reporter was introduced by transduction into strain EPB47, which carries an ompA+-cfp+ transcriptional fusion. To generate strains MMR233, MMR234, and MMR235, markerless ompA+-cfp+ was introduced into the PtorCAD-yfp+ MG1655, Nissle 1917, and HS derivatives by linkage with ΔsulA::(FRT-kan-FRT) from JW0941. For strains MMR125 and MMR133, the chloramphenicol resistance cassette of MMR8 was swapped by recombineering with a kanamycin resistance cassette flanked by FRT sites (30). The PCR product for recombination was obtained with primers cat-LRed-U1 and cat-LRed-L1 and template pKD4 (30). Strain MMR130 is identical to MMR125, except that the kanamycin resistance cassette was removed by treatment with pCP20 (30).
The PtorCAD-cfp transcriptional fusion fluorescent reporter of strains MMR72, MMR84, and MMR141 was constructed by recombineering (30). First, plasmid pMR19 was constructed by cloning into pKD13 (30) an overlap extension PCR product that includes the PtorCAD sequence described above and the cfp gene (33). The overlap PCR product was created with template pMR4 and primers psyn-U1 and gfpNterm-L1 and with template pEB48 and primers tet-after-MCS-L1 and cfpNterm-U1. Plasmid pMR19 was used as the template with primers lac-pMR10-U1 and lac-pMR10-L1 to generate a PCR product for integration and substitution of the lac operon. The integration PCR product had flanking regions of homology to the lac operon upstream of the lacI promoter and in the lacA open reading frame (ORF) and includes PtorCAD-cfp with a kanamycin resistance cassette flanked by FRT sites. The integrated transcriptional fusion was verified by sequencing.
The torCAD-yfp operon fusion of strain MMR141 was constructed by recombineering (30) to insert the yfp gene (33), with its own ribosome binding site, immediately downstream of the torD stop codon. The PCR product used for integration was obtained from template pEB45 (33) with primers torD-yfp-U1 and torD-yfp-L1, which have flanking homology to sequences downstream of torD. The constitutively expressed transcriptional fusion PtetA-mCherry of strains MMR60, MMR72, MMR84, and MMR141 was also constructed by recombineering. The filled-in KpnI PtetA-mCherry DNA fragment from plasmid pTM149 was ligated to HincII-digested pKD13 to generate plasmid pMR14. This plasmid was used as the template with primers xyl-pKD13-U1 and xyl-pKD13-L1 to generate a PCR product for integration into the chromosome at the xyl locus, disrupting xylAFG. The insertion was verified by sequencing.
The chromosomal region containing torS, torT, and torR was deleted and replaced with a kanamycin resistance cassette by recombineering to generate strain MMR133. The PCR product for integration was obtained by amplification of the kanamycin resistance cassette and flanking FRT sites of pKD13 with primers torSTRdel-U1 and torSTRdel-L1.
Random mutants of torR were generated by PCR mutagenesis using the GeneMorph II Random Mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Primers torR-SacI-L1 and torC-L1 amplified a region of DNA containing the entire torR sequence and its regulatory region. The PCR product was cloned into the single-copy vector pSMART (Lucigen Corporation, Middleton, WI) to generate a mutant plasmid library. We constructed the control plasmid pMR32 with the nonmutagenized PCR product. The plasmid library was introduced into ΔtorS mutant strain MMR84, which carries the PtorCAD-cfp fluorescent reporter for screening. Transformants carrying torR mutants capable of activating the PtorCAD reporter in the absence of torS and of TMAO were detected as bright fluorescent colonies in a background of dim nonfluorescent colonies. Colony fluorescence was visualized with a home-built fluorescence illuminator described previously (34). The plasmids from selected colonies were purified, and the torR genes were sequenced. Plasmids carrying selected torR alleles were transformed into MMR133, a PtorCAD-cfp reporter strain that has a chromosomal deletion of the entire sequence spanning torS, torT, and torR to verify that PtorCAD transcription activated by the selected torR alleles was truly independent of TorT-TorS-TorR. The torR146 allele, carried by plasmid pMR46, was selected for further study. To disrupt the TorR phosphorylation site in TorR146, we introduced the mutation D53A (18) by overlap extension PCR with mutagenic primers D53A-FOR and D53A-REV. A single nucleotide was substituted, resulting in the loss of an EcoRV site and generating allele torR149 (carried on plasmid pMR49).
Alleles torR146 and torR149 were integrated in single copy at the tor locus into the chromosome of MMR133/pKD46 by recombineering (30) to generate strains MMR138 and MMR140, respectively. The transforming DNA consisted of overlap extension PCR products from primers torR-int-U1 and torR-cat-rev with template pMR46 or pMR49 and from primers torR-int-L1 and torR-cat-for with template pKD3. These PCR products had flanking regions of homology to the torR regulatory region and downstream of the torS ORF. The integrated constructs were verified by sequencing.
Plasmid pMR29 is a derivative of pSMART (Lucigen Corporation) that carries torS driven by an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. The plasmid was constructed by three-way ligation that placed the torS ORF downstream of the NdeI-BamHI fragment from pDSW206 (35), which contains lacIq, lac operators, and a weakened variant of the trc promoter. The NdeI site was filled in and blunt-end ligated to the filled-in EcoRI site of pSMART. The torS gene was amplified by PCR from a genomic MG1655 template with primers torS-BamHI-U1 and torS-HindIII-L1. The upper primer was designed to amplify torS with its native ribosome binding site and to add a BamHI site for ligation of torS downstream of the weakened trc promoter. Additionally, a stop codon was added in frame with the ATG corresponding to the NcoI restriction site to avoid possible translation from the lacZ ribosome binding site sequence in pDSW206. The PCR product was digested with BamHI and HindIII and ligated to the HindIII site of pSMART and the BamHI site of the pDSW206 fragment described above.
Plasmid pMR26 carries torT controlled by the IPTG-inducible trc promoter and was obtained by cloning a torT PCR product into the BamHI and SalI sites of pDSW206 (35). Primers torT-lower-1 and torT-upper-1 were used to amplify torT, including its ribosome binding site, from MG1655 genomic DNA. The upper primer includes a stop codon to avoid translation from the ATG associated with the NcoI site as described above.
Fluorescence microscopy.
Cultures were inoculated from single colonies on LB agar plates, grown in minimal A medium aerobically at 37°C overnight to saturation, diluted 1:1,000 into fresh medium with TMAO as indicated in each experiment, and then grown at 37°C (either aerobically or without oxygenation as described above) to an optical density at 600 nm (OD600) of 0.2 to 0.3. Streptomycin was added to a final concentration of 250 μg/ml to stop protein synthesis. Cultures were then rapidly cooled in an ice-water slurry and kept on ice for at least 1 h. After 1 h on ice, nonoxygenated cultures were aerated in a roller drum at 37°C for 3 h to enable chromophore maturation of the fluorescent reporter proteins. When comparing the absolute fluorescence of oxygenated and nonoxygenated cultures, the oxygenated cultures were aerated in the same fashion as was done for the nonoxygenated cultures to control for possible degradation of fluorescent proteins. Ten microliters of bacterial culture was immobilized between 1% agarose pads and coverslips. Pads were prepared essentially as described previously (36), by placing 100 μl of molten agarose prepared in medium A salts (medium A without glucose or Casamino Acids) between a glass microscope slide and the coverslip. Single-cell fluorescence measurements were made with an inverted microscope (Olympus IX-81) as described previously (37). Image analysis was performed with in-house software by using phase-contrast images to define cell areas.
To grow microcolonies on the microscope, bacterial cultures were first grown aerobically in liquid minimal medium with TMAO as described above but to early exponential phase (OD600 of ∼0.1). Five microliters of bacterial culture was placed in the center of a sterile tissue culture dish (FluoroDish; World Precision Instruments, Inc.), under 5-mm-thick agarose pads containing TMAO that were prewarmed to 37°C. The pads were prepared by dissolving 1% SeaKem GTG agarose (Lonza, Rockland, ME) in minimal medium (with 0.2% glucose and 0.1% Casamino Acids). When the agarose reached a temperature of 55°C, TMAO was added to a final concentration of 1 mM. Culture dishes were placed on the stage of a microscope that was enclosed in a chamber maintained at 37°C by forced airflow (33) and visualized by fluorescence microscopy as described above.
RESULTS
The distribution of torCAD transcription in single cells is heterogeneous under aerobic conditions.
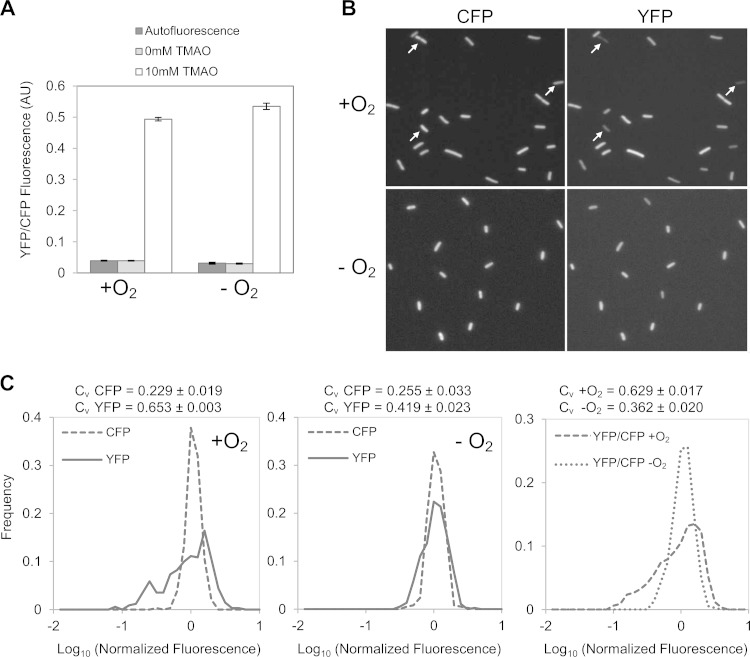
To monitor the output of the TorS-TorR system in single cells, we integrated a copy of the torCAD promoter driving yfp, the gene for yellow fluorescent protein (YFP), at the phage lambda attachment site. The strain also has an operon fusion of cfp, the gene encoding cyan fluorescent protein (CFP), to ompA, which provides an internal fluorescence standard that we have verified is independent of oxygen and TMAO. Cultures were grown to mid-exponential phase in glucose minimal medium in the presence or absence of 10 mM TMAO with or without oxygenation. Cellular fluorescence was recorded by microscopy (see Materials and Methods for details). Fluorescence measurements averaged over the cell population confirm that the torC promoter shows similar levels of activity, on average, for high- and low-oxygen growth conditions (Fig. 1A), as reported previously (28). In both cases, there is a >10-fold increase in fluorescence upon induction with TMAO, with fluorescence in noninduced cells comparable to the background autofluorescence. Interestingly, the YFP fluorescence in the induced (10 mM TMAO) aerobic cultures showed remarkably strong cell-to-cell variability, with fluorescence ranging from close to the background to very bright (Fig. 1B, top right panel). In contrast, CFP fluorescence from the constitutively expressed ompA-cfp fusion in the same cells was much more uniform (Fig. 1B, top left panel). Cells from anaerobic cultures showed a much more homogeneous YFP fluorescence that was similar to the CFP fluorescence from ompA-cfp (Fig. 1B, bottom panels).
FIG 1.
Fluorescence quantification of PtorCAD-yfp reporter expression in the presence or absence of oxygen. Cultures of strain MMR8 [PtorCAD-yfp Φ(ompA+-cfp+)] were grown in minimal medium with or without 10 mM TMAO and with or without aeration, as indicated, and analyzed by fluorescence microscopy as described in Materials and Methods. Autofluorescence in the YFP fluorescence channel was determined from cultures of the parent strain, EPB47 [Φ(ompA+-cfp+)], grown under the same conditions. (A) Average fluorescence from single cells. Values represent the means from two independent experiments; error bars represent the range. More than 300 single cells were analyzed for each condition in each experiment. AU, arbitrary units. (B) Representative fluorescence micrographs from aerobic (upper) and anaerobic (lower) cultures. The CFP [Φ(ompA+-cfp+)] and YFP (PtorCAD-yfp) fluorescence in each field is shown. Arrows highlight cells that are too dim to be easily visualized in the YFP fluorescence image. (C) Distribution of single-cell fluorescence in TMAO-treated cultures. The left and middle panels show the distribution of cellular YFP fluorescence compared to cellular CFP fluorescence in corresponding data sets in aerobic (left) and anaerobic (middle) cultures. The right panel shows a comparison of the distribution of YFP fluorescence normalized by the CFP internal standard in aerobic and anaerobic cultures. Values on the x axis are expressed as log10 fluorescence normalized by the mean value from the corresponding data set. The aerobic and anaerobic data sets consist of measurements of 341 and 379 cells, respectively. The distributions shown in panel C are taken from single experiments and are representative of seven independent experiments. The coefficient of variation (CV = standard deviation/mean) is the mean value from seven independent experiments ± the standard deviation.
The distribution of cellular CFP and YFP fluorescence from induced cultures provides a more quantitative description of cell variability (Fig. 1C). For aerobic cultures, the CFP fluorescence distribution from ompA-cfp is relatively narrow, whereas the distribution of YFP fluorescence from PtorC-yfp in the same population is very broad, with a much higher coefficient of variation (standard deviation/mean) (left panel). In contrast, in cultures grown anaerobically, the YFP fluorescence distribution is much narrower and similar to the distribution of CFP fluorescence (middle panel). Comparison of YFP fluorescence levels normalized by the constitutive CFP fluorescence similarly shows a marked difference between aerobic and anaerobic growth.
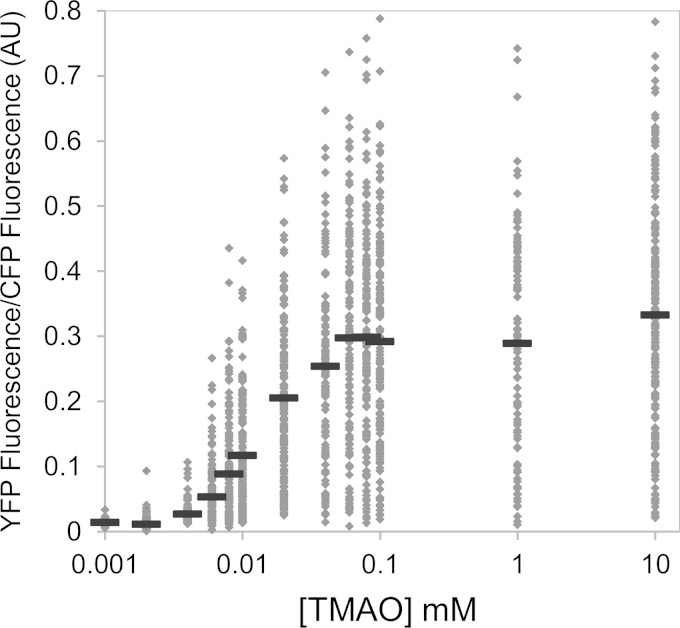
To explore whether the variability seen is dependent on the inducer concentration and whether there were concentrations showing a bimodal distribution, reflecting distinct subpopulations of noninduced and induced cells, we measured the dose response to TMAO of aerobically growing cells (Fig. 2). In the scatter plot shown in Fig. 2, each dot represents the fluorescence value of an individual cell. Mean fluorescence values, represented by horizontal bars, show a sigmoid dependence on TMAO with an approximate Hill coefficient of 2. As is evident from the scatter plot, torCAD transcription is highly heterogeneous across the population of cells throughout the range of inducing levels of TMAO, even at saturating concentrations.
FIG 2.
Scatter plot of the TMAO dose responses of single cells. Cultures of strain MMR8 [PtorCAD-yfp Φ(ompA+-cfp+)] were grown aerobically in minimal medium in the presence of the TMAO concentrations indicated and analyzed by fluorescence microscopy as described in Materials and Methods. Each dot represents the fluorescence value of an individual cell. For each dose, measurements of at least 100 cells were made. Each solid horizontal line represents the mean value for each dose. AU, arbitrary units.
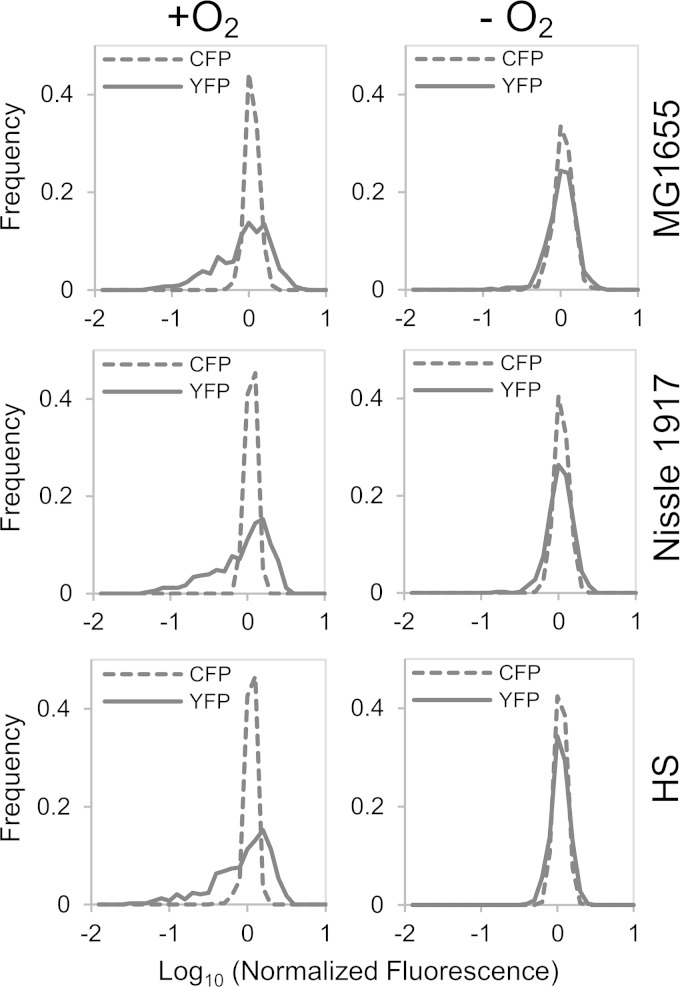
To test whether the heterogeneous transcription of the torC promoter reporter was unique to E. coli MG1655 (the K-12 strain background used in the experiments described above) and thus potentially an artifact of a laboratory strain, we made similar measurements on strains derived from two other E. coli isolates, Nissle 1917 (38) and HS (39). The torCAD promoter sequence of Nissle 1917 is identical to the corresponding MG1655 sequence included in our reporter. The HS sequence has a single nucleotide change 94 bases upstream of the torCAD transcription start site (9), a region not expected to play a role in torCAD transcriptional regulation. Both isolates show induction of the fluorescent reporter upon exposure to 10 mM TMAO and broad YFP fluorescence distributions in single cells that are similar to the behavior of MG1655 (Fig. 3). These results indicate that the aerobic induction and variability of torCAD transcription are not limited to E. coli K-12 strains.
FIG 3.
Fluorescence distributions of the PtorCAD-yfp reporter expression for three different E. coli strains in the presence or absence of oxygen. Cultures of strains MMR234 [Nissle 1917 PtorCAD-yfp Φ(ompA+-cfp+) ΔsulA::(FRT-kan-FRT)], MMR235 [HS PtorCAD-yfp Φ(ompA+-cfp+) ΔsulA::(FRT-kan-FRT)], and MMR233 [MG1655 PtorCAD-yfp Φ(ompA+-cfp+) ΔsulA::(FRT-kan-FRT)] were grown in minimal medium with or without 10 mM TMAO and with or without aeration, as indicated, and analyzed by fluorescence microscopy as described in Materials and Methods. The plots show the distribution of single-cell YFP and CFP fluorescence in TMAO-treated cultures of each strain (top, middle, and bottom panels) with oxygenation (left column) or without oxygenation (right column). Data sets consisted of more than 350 single-cell measurements that were combined from two independent experiments. Values on the x axis are expressed as log10 fluorescence normalized by the mean value of the data set.
The source of variable torCAD transcription acts in trans.
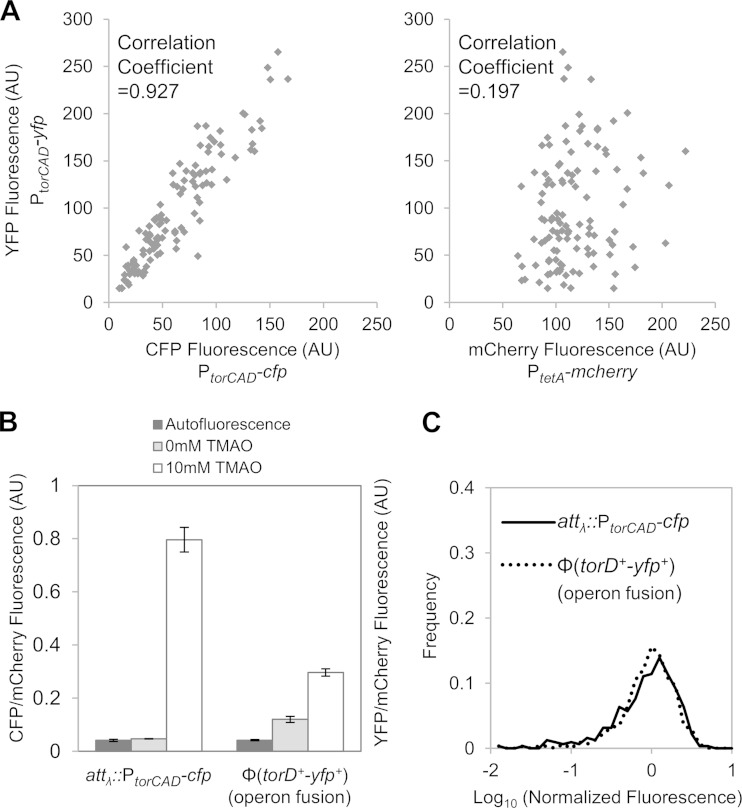
If the cell-to-cell variability in transcription results from factors that act in trans, whether because of deterministic or stochastic effects (extrinsic noise [40]), then two copies of the promoter will show a correlation in their variability across a population of cells. On the other hand, if the source of variability acts in cis, then the cell-to-cell variability between the two promoters will be uncorrelated. We therefore constructed a strain that has two torCAD transcriptional reporters, i.e., a PtorCAD-cfp reporter inserted in place of lacI lacZYA and a PtorCAD-yfp reporter at the lambda phage attachment site. The strain also contains a constitutive tetA promoter driving mcherry integrated at the xylAFG locus. Scatter plots of PtorCAD-yfp versus PtorCAD-cfp single-cell fluorescence show a strong correlation between the two PtorCAD reporters, indicating that the heterogeneous expression of the torCAD operon does not originate from cis-acting factors (Fig. 4A, left panel). The correlation between PtorCAD-yfp and PtetA-mcherry, on the other hand, is much lower (Fig. 4A, right panel). Thus, the results suggest that cell-cell variability in torCAD transcription is due to a trans-acting factor, which could be from the TorT-TorS-TorR signaling system or from other uncharacterized factors acting at the torCAD promoter.
FIG 4.
Comparison of two torCAD reporters in the same cell. (A) Correlation between two promoter fusion reporters (PtorCAD-cfp and PtorCAD-yfp) at different chromosomal loci. Cultures of strain MMR72 (PtorCAD-cfp, PtorCAD-yfp, and PtetA-mcherry) were grown aerobically in minimal medium containing 10 mM TMAO and analyzed by fluorescence microscopy as described in Materials and Methods. Fluorescence values of YFP versus CFP (left panel) and YFP versus mCherry (right panel) are plotted for 113 cells; YFP values are the same in both plots. (B and C) Comparison of promoter and operon fusions in the same cells. Cultures of strain MMR141 [PtorCAD-cfp, Φ(torD+-yfp+), and PtetA-mcherry] were grown aerobically in medium with or without 10 mM TMAO, as indicated. The autofluorescence of cultures of the parent strain, MMR60, grown under the same conditions was determined in the YFP and CFP fluorescence channels. (B) Average fluorescence from the promoter and operon fusions, PtorCAD-cfp and Φ(torD+-yfp+), each normalized by the fluorescence of PtetA-mcherry. Shown are the mean values of two independent experiments, and error bars represent the range. In each experiment, the mean values of >100 cells were determined. (C) Distribution of fluorescence from single cells induced with 10 mM TMAO. For Φ(torD+-yfp+), the mean YFP fluorescence of noninduced cells was subtracted from the fluorescence of induced cells. Values on the x axis are expressed as log10 fluorescence normalized by the mean value of the corresponding data set. The distribution is based on measurements of >300 cells. AU, arbitrary units.
To verify that the heterogeneous expression seen is not an artifact of the transcriptional fusion used in the PtorCAD-yfp reporter, we also constructed an operon fusion in which yfp, with its own ribosome binding site, was inserted downstream of torD in the native torCAD operon [designated Φ(torD+-yfp+)]. This strain also contained the PtorCAD-cfp reporter, as well as PtetA-mcherry as an internal standard. The mean fluorescence from the yfp operon reporter is approximately half of that of PtorCAD-cfp, and the YFP fluorescence is above the background even in the absence of the inducer, which may reflect a second promoter within the operon (Fig. 4B). However, despite these differences, the single-cell fluorescence distributions for the operon and promoter reporters show similar behaviors (Fig. 4C), indicating that the cell-to-cell variability seen is not an artifact of a specific reporter.
The torCAD expression state is not uniform within microcolonies.
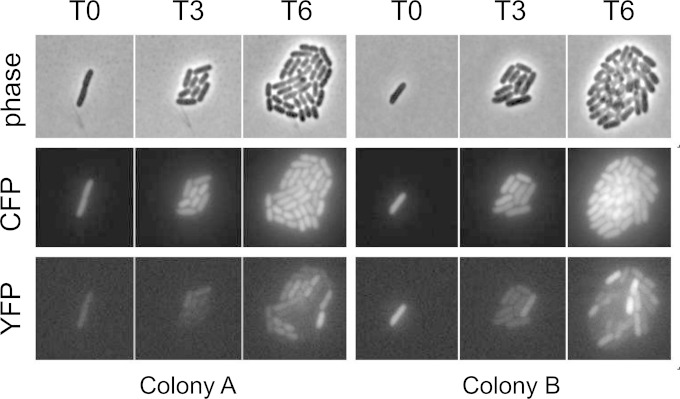
We performed time-lapse microscopy of live cells in order to monitor the level of PtorCAD transcription over several generations. Cells from early-exponential-phase liquid cultures containing TMAO were immobilized on agarose pads composed of the same growth medium with TMAO and maintained at 37°C on the microscope stage so that individual cells would form microcolonies after several rounds of division. Images were acquired soon after immobilization on agarose pads (T0) and then 3 and 6 h after the starting time point (∼4 and ∼8 divisions, respectively [T3 and T6]). If the expression level of the torCAD reporter is inherited for many generations, then we expect to see uniformly bright microcolonies developing from a single bright cell or uniformly dim microcolonies from a dim cell. In contrast, if the expression switches frequently, then we will obtain microcolonies with heterogeneous cellular fluorescence. Figure 5 shows images of two representative microcolonies. In both cases, the CFP fluorescence from the constitutive ompA promoter is homogeneously distributed. In contrast, the YFP fluorescence varies considerably among cells within the colonies, suggesting that the expression state is not strongly inherited in daughter cells and that cells tend to switch the torCAD transcription level over a time scale that is roughly comparable to the division time.
FIG 5.
Time-lapse microscopy of two representative microcolonies. Cultures of strain MMR8 [PtorCAD-yfp Φ(ompA+-cfp+)] were grown aerobically in minimal medium with 10 mM TMAO. Cells were immobilized on 5-mm-thick agarose pads composed of 1% agarose dissolved in the same growth medium with TMAO and maintained on the microscope stage at 37°C with exposure to air. The images shown are of two representative colonies, each derived from a single cell, shortly after immobilization on agarose pads (T0) and then 3 h (T3) and 6 h (T6) after the starting time point (approximately four and eight divisions, respectively). The rows consist of phase-contrast images (top), CFP fluorescence images (middle), and YFP fluorescence images (bottom).
Variability in torCAD transcription is suppressed by a TorR mutant that bypasses TorS.
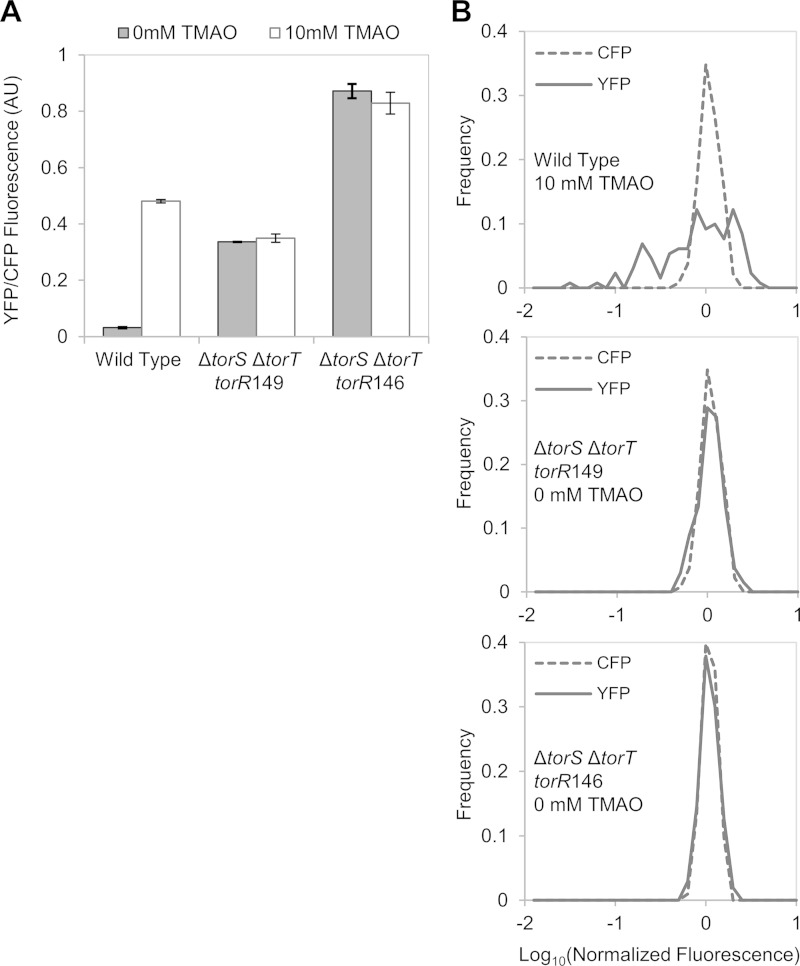
The trans-acting source of variability in torCAD expression could act through TorR-P or through an uncharacterized regulator that functions independently of the TorS-TorR phosphorelay. To distinguish between these possibilities, we isolated constitutively active TorR mutants that bypass the phosphorelay. If the variability in torCAD expression depends on the phosphorelay, then a bypass mutant should show a more uniform distribution of YFP fluorescence among cells. However, if the variability is due to some other regulator, then these mutants should show a high cell-to-cell heterogeneity that is similar to that of wild-type cells. To isolate bypass mutants, we screened a library of mutated torR genes for activation of a PtorCAD-cfp reporter in a torS deletion strain. From this screening, we isolated a torR allele, torR146, encoding a protein with two amino acid substitutions: K114R at the end of the receiver domain and R146H in the DNA binding domain. Since it is possible that TorR146 would still be phosphorylated by alternative phosphodonors, we also constructed a torR146 derivative, torR149, encoding a protein that contains an alanine in place of aspartate at position 53, the site of phosphorylation (18). The corresponding torR alleles were integrated into the chromosome in place of the native torR gene, and the adjacent torT and torS genes were deleted. Both TorR146 and TorR149 activate transcription independently of TorS, TorT, and TMAO, indicating that these TorR mutants can indeed bypass the phosphorelay (Fig. 6A). The lower activity of TorR149 than of TorR146 (∼50%) may indicate that TorR146 can be phosphorylated by a source other than TorS or may simply reflect different intrinsic activities of the two TorR variants. Figure 6B shows the corresponding distributions of cellular PtorCAD-yfp fluorescence; the distributions of the TorR mutant strains are similar and much narrower than the distribution of the wild type (Fig. 6B). These results suggest that the heterogeneous expression of PtorCAD results from variability in TorR-P levels.
FIG 6.
Comparison of PtorCAD-yfp expression in a strain expressing wild-type TorR and strains expressing constitutively active TorR mutants that bypass TorS. Cultures of MMR125 [PtorCAD-yfp Φ(ompA+-cfp+)], MMR138 [ΔtorS ΔtorT torR146 PtorCAD-yfp Φ(ompA+-cfp+)], and MMR140 [ΔtorS ΔtorT torR149 PtorCAD-yfp Φ(ompA+-cfp+)] were grown aerobically in minimal medium with or without 10 mM TMAO and analyzed by fluorescence microscopy as described in Materials and Methods. (A) Average fluorescence from single cells. Shown are the mean values of two independent experiments, and error bars represent the range. Sets of over 100 single cells were analyzed for each experiment. AU, arbitrary units. (B) Distributions of single-cell fluorescence in samples from wild-type (top panel, 131 cells), torR149 (middle panel, 135 cells), and torR146 (bottom panel, 103 cells.) Values on the x axis are expressed as log10 of fluorescence normalized by the mean value of the corresponding data set.
Increased expression of TorS and TorT reduces the cell-to-cell variability of torCAD transcription.
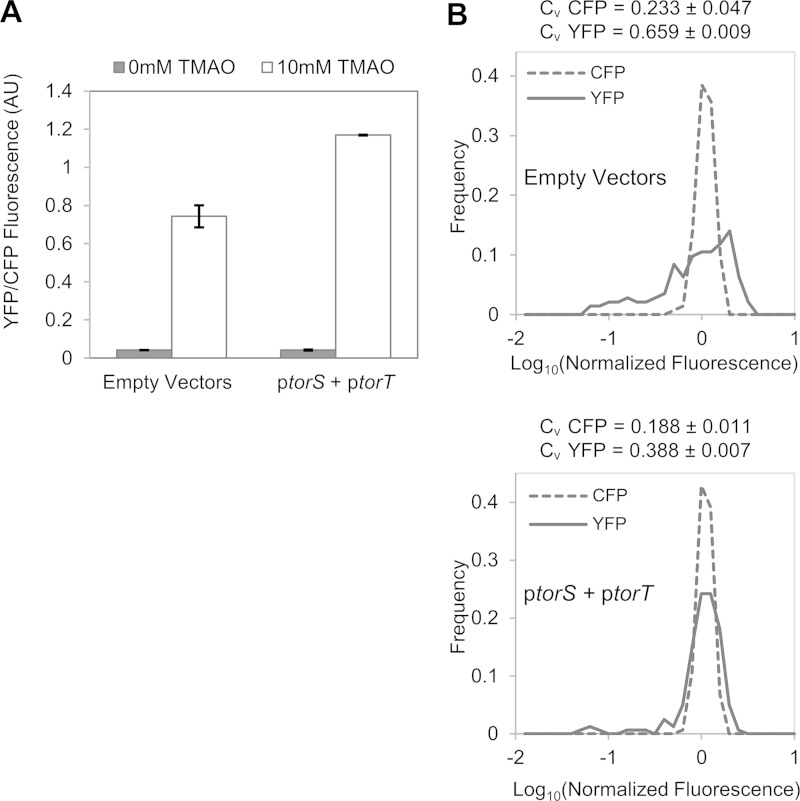
To test whether the variability of torCAD transcription depends on low TorS and TorT expression levels, we added plasmid copies of torS and torT under the control of an IPTG-inducible promoter. Expression of TorS and TorT from plasmids had a relatively modest effect on the PtorC-yfp expression level, increasing the mean by roughly 1.5-fold (Fig. 7A), but produced a marked decrease in cell-to-cell variability (Fig. 7B). These results suggest that fluctuations in the numbers of TorS and TorT proteins could account for the strong cell-to-cell variability in torCAD transcription.
FIG 7.
Effect of extra copies of torS and torT on PtorCAD-yfp reporter expression. Cultures of strains MMR130 [PtorCAD-yfp Φ(ompA+-cfp+)] with plasmids pMR29 and pMR26 (ptorS and ptorT) or with the parent vectors (pSMART and pDSW206) were grown aerobically in minimal medium containing 1 mM IPTG and in the presence or absence of 10 mM TMAO, and analyzed by fluorescence microscopy as described in Materials and Methods. (A) Average fluorescence from single cells. Values are the average fluorescence from two independent experiments, and error bars represent the range of the mean fluorescence from each experiment. Sets of >100 single cells for each condition were analyzed in each experiment. AU, arbitrary units. (B) Distributions of YFP and CFP fluorescence in single cells in samples treated with TMAO. The upper panel represents a strain with both empty vectors (164 cells). The lower panel represents a strain with ptorS and ptorT (164 cells). Values on the x axis are expressed as log10 fluorescence normalized by the mean value of the corresponding data set. The distributions shown in panel B are taken from single experiments and are representative of three independent experiments. Each coefficient of variation (CV = standard deviation/mean) is the mean value of three independent experiments ± the standard deviation.
DISCUSSION
We find that while the average levels of torCAD transcription at maximal induction by TMAO are comparable in E. coli cultures growing aerobically and anaerobically, the single-cell distributions are highly heterogeneous under the former, but not the latter, growth conditions. Thus, oxygen affects the variance but not the mean of the output in this system.
Since two copies of the torCAD promoter in the same cell show strongly correlated transcription (Fig. 4A), the source of variability acts in trans. To determine whether TorR-P mediates the effect, we isolated mutations in torR that decouple torCAD expression from the TorT-TorS-TorR phosphorelay. Strains with these torR alleles show a much more uniform distribution of torCAD transcription in aerobic cultures, suggesting that the source of variability is in the phosphorelay.
It is possible that the variability in gene expression results from fluctuations in low numbers of TorS and TorT molecules. Protein abundance inferred from ribosome profiling data indicates that both TorS and TorT are expressed at exceedingly low levels (~4 molecules/cell for both) (41). Under these conditions, stochastic effects from gene expression and from random partitioning during cell division may produce widely varying cellular abundances of TorS and TorT across the population (40, 42, 43). Cells that lack TorS molecules for extended periods will likely have low levels of TorR-P because of dilution of pre-existing TorR-P from cell growth or spontaneous hydrolysis. On the other hand, if the number of TorS molecules exceeds the number of TorT, then excess TorS that is not bound to TorT will be insensitive to TMAO and should dephosphorylate TorR-P via reverse phosphotransfer (23, 24), again producing lower levels of TorR-P. Indeed, it was previously observed that torS provided in trans in multicopy represses the transcription of the torCAD operon (17, 23). Thus, we expect cells with a TorS/TorT ratio too low (no TorS) or too high (TorS exceeds TorT) to have lower than average TorR-P, making the system sensitive to fluctuations in the levels of both proteins. We note, however, that while stochastic changes in this ratio are likely to contribute to the variability observed, at present, we cannot rule out other mechanisms that might produce cell-to-cell variability in TorS activity.
We do not yet understand the molecular mechanism of the oxygen dependence and the physiological significance of increased variance under aerobic conditions. The fact that diverse E. coli isolates from different phylogenetic groups (MG1655 and HS are in group A, whereas Nissle 1917 is in group B2 [44, 45]) show similar behavior indicates that the phenomenon is not a strain artifact. Of course, it is possible that the variability is an unavoidable consequence of some other aspect of the TorT-TorS-TorR signaling system and does not, on its own, provide E. coli any fitness advantage. It is not understood why E. coli responds to and reduces TMAO under aerobic conditions. One hypothesis proposes that TMA produced by TMAO reduction will increase fitness by counteracting acidification of the environment (28). Interestingly, this mechanism could be effective even if a subset of cells in the population is performing the TMAO reduction. Therefore, depending on the fitness cost of expressing different levels of the torCAD operon, the maximal benefit for the population as a whole could be optimized by a nonuniform level of TMAO reduction across all of the cells in the population. An alternative possibility is that the variability in torCAD expression reflects a bet-hedging strategy whereby the cost of expressing torCAD aerobically is balanced against the benefit of pre-expressed TorCAD should the environment rapidly switch to low oxygen.
ACKNOWLEDGMENTS
This work was supported by NIH award GM080279 to M.G. and by an award from the American Heart Association to M.R.
REFERENCES
- 1.Barrett EL, Kwan HS. 1985. Bacterial reduction of trimethylamine oxide. Annu Rev Microbiol 39:131–149. doi: 10.1146/annurev.mi.39.100185.001023. [DOI] [PubMed] [Google Scholar]
- 2.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. 1982. Living with water stress: evolution of osmolyte systems. Science 217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 3.Zhang AQ, Mitchell SC, Smith RL. 1999. Dietary precursors of trimethylamine in man: a pilot study. Food Chem Toxicol 37:515–520. doi: 10.1016/S0278-6915(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 4.Zeisel SH, daCosta KA, Youssef M, Hensey S. 1989. Conversion of dietary choline to trimethylamine and dimethylamine in rats: dose-response relationship. J Nutr 119:800–804. [DOI] [PubMed] [Google Scholar]
- 5.Treacy EP, Akerman BR, Chow LM, Youil R, Bibeau C, Lin J, Bruce AG, Knight M, Danks DM, Cashman JR, Forrest SM. 1998. Mutations of the flavin-containing monooxygenase gene (FMO3) cause trimethylaminuria, a defect in detoxication. Hum Mol Genet 7:839–845. doi: 10.1093/hmg/7.5.839. [DOI] [PubMed] [Google Scholar]
- 6.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. 2013. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab 17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roos V, Ulett GC, Schembri MA, Klemm P. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect Immun 74:615–624. doi: 10.1128/IAI.74.1.615-624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winter SE, Lopez CA, Baumler AJ. 2013. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Méjean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal MC. 1994. TMAO anaerobic respiration in Escherichia coli: involvement of the Tor operon. Mol Microbiol 11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 10.Gon S, Giudici-Orticoni MT, Méjean V, Iobbi-Nivol C. 2001. Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J Biol Chem 276:11545–11551. doi: 10.1074/jbc.M008875200. [DOI] [PubMed] [Google Scholar]
- 11.McCrindle SL, Kappler U, McEwan AG. 2005. Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv Microb Physiol 50:147–198. doi: 10.1016/S0065-2911(05)50004-3. [DOI] [PubMed] [Google Scholar]
- 12.Cheng V, Weiner J. 2007. S- and N-oxide reductases. EcoSal Plus doi: 10.1128/ecosalplus.3.2.8. [DOI] [PubMed] [Google Scholar]
- 13.Pommier J, Méjean V, Giordano G, Iobbi-Nivol C. 1998. TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J Biol Chem 273:16615–16620. doi: 10.1074/jbc.273.26.16615. [DOI] [PubMed] [Google Scholar]
- 14.Ilbert M, Méjean V, Giudici-Orticoni MT, Samama JP, Iobbi-Nivol C. 2003. Involvement of a mate chaperone (TorD) in the maturation pathway of molybdoenzyme TorA. J Biol Chem 278:28787–28792. doi: 10.1074/jbc.M302730200. [DOI] [PubMed] [Google Scholar]
- 15.Jack RL, Buchanan G, Dubini A, Hatzixanthis K, Palmer T, Sargent F. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J 23:3962–3972. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genest O, Neumann M, Seduk F, Stocklein W, Méjean V, Leimkuhler S, Iobbi-Nivol C. 2008. Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J Biol Chem 283:21433–21440. doi: 10.1074/jbc.M802954200. [DOI] [PubMed] [Google Scholar]
- 17.Jourlin C, Bengrine A, Chippaux M, Méjean V. 1996. An unorthodox sensor protein (TorS) mediates the induction of the tor structural genes in response to trimethylamine N-oxide in Escherichia coli. Mol Microbiol 20:1297–1306. doi: 10.1111/j.1365-2958.1996.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 18.Jourlin C, Ansaldi M, Méjean V. 1997. Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J Mol Biol 267:770–777. doi: 10.1006/jmbi.1997.0919. [DOI] [PubMed] [Google Scholar]
- 19.Baraquet C, Théraulaz L, Guiral M, Lafitte D, Méjean V, Jourlin-Castelli C. 2006. TorT, a member of a new periplasmic binding protein family, triggers induction of the Tor respiratory system upon trimethylamine N-oxide electron-acceptor binding in Escherichia coli. J Biol Chem 281:38189–38199. doi: 10.1074/jbc.M604321200. [DOI] [PubMed] [Google Scholar]
- 20.Moore JO, Hendrickson WA. 2012. An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure 20:729–741. doi: 10.1016/j.str.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 22.Perraud AL, Weiss V, Gross R. 1999. Signalling pathways in two-component phosphorelay systems. Trends Microbiol 7:115–120. doi: 10.1016/S0966-842X(99)01458-4. [DOI] [PubMed] [Google Scholar]
- 23.Ansaldi M, Jourlin-Castelli C, Lepelletier M, Théraulaz L, Mejéan V. 2001. Rapid dephosphorylation of the TorR response regulator by the TorS unorthodox sensor in Escherichia coli. J Bacteriol 183:2691–2695. doi: 10.1128/JB.183.8.2691-2695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peña-Sandoval GR, Kwon O, Georgellis D. 2005. Requirement of the receiver and phosphotransfer domains of ArcB for efficient dephosphorylation of phosphorylated ArcA in vivo. J Bacteriol 187:3267–3272. doi: 10.1128/JB.187.9.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansaldi M, Simon G, Lepelletier M, Méjean V. 2000. The TorR high-affinity binding site plays a key role in both torR autoregulation and torCAD operon expression in Escherichia coli. J Bacteriol 182:961–966. doi: 10.1128/JB.182.4.961-966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon G, Jourlin C, Ansaldi M, Pascal MC, Chippaux M, Méjean V. 1995. Binding of the TorR regulator to cis-acting direct repeats activates tor operon expression. Mol Microbiol 17:971–980. doi: 10.1111/j.1365-2958.1995.mmi_17050971.x. [DOI] [PubMed] [Google Scholar]
- 27.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320:217–234. doi: 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 28.Ansaldi M, Théraulaz L, Baraquet C, Panis G, Méjean V. 2007. Aerobic TMAO respiration in Escherichia coli. Mol Microbiol 66:484–494. doi: 10.1111/j.1365-2958.2007.05936.x. [DOI] [PubMed] [Google Scholar]
- 29.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 30.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 33.Batchelor E, Goulian M. 2006. Imaging OmpR localization in Escherichia coli. Mol Microbiol 59:1767–1778. doi: 10.1111/j.1365-2958.2006.05048.x. [DOI] [PubMed] [Google Scholar]
- 34.Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol 70:494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyashiro T, Goulian M. 2007. Single-cell analysis of gene expression by fluorescence microscopy. Methods Enzymol 423:458–475. doi: 10.1016/S0076-6879(07)23022-8. [DOI] [PubMed] [Google Scholar]
- 37.Miyashiro T, Goulian M. 2007. Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc Natl Acad Sci U S A 104:16305–16310. doi: 10.1073/pnas.0700025104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 41.Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. 2002. Regulation of noise in the expression of a single gene. Nat Genet 31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 43.Huh D, Paulsson J. 2011. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet 43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lasaro M, Liu Z, Bishar R, Kelly K, Chattopadhyay S, Paul S, Sokurenko E, Zhu J, Goulian M. 2014. Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J Bacteriol 196:1723–1732. doi: 10.1128/JB.01296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 46.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]